ABSTRACT
ArgO and LysE are members of the LysE family of exporter proteins and ordinarily mediate the export of l-arginine (Arg) in Escherichia coli and l-lysine (Lys) and Arg in Corynebacterium glutamicum, respectively. Under certain conditions, ArgO also mediates Lys export. To delineate the arrangement of ArgO in the cytoplasmic membrane of E. coli, we have employed a combination of cysteine accessibility in situ, alkaline phosphatase fusion reporters, and protein modeling to arrive at a topological model of ArgO. Our studies indicate that ArgO assumes an Nin-Cout configuration, potentially forming a five-transmembrane helix bundle flanked by a cytoplasmic N-terminal domain (NTD) comprising roughly its first 38 to 43 amino acyl residues and a short periplasmic C-terminal region (CTR). Mutagenesis studies indicate that the CTR, but not the NTD, is dispensable for ArgO function in vivo and that a pair of conserved aspartate residues, located near the opposing edges of the cytoplasmic membrane, may play a pivotal role in facilitating transmembrane Arg flux. Additional studies on amino acid substitutions that impair ArgO function in vivo and their derivatives bearing compensatory amino acid alterations indicate a role for intramolecular interactions in the Arg export mechanism, and some interactions are corroborated by normal-mode analyses. Lastly, our studies suggest that ArgO may exist as a monomer in vivo, thus highlighting the requirement for intramolecular interactions in ArgO, as opposed to interactions across multiple ArgO monomers, in the formation of an Arg-translocating conduit.
IMPORTANCE The orthologous proteins LysE of C. glutamicum and ArgO of E. coli function as exporters of the basic amino acids l-arginine and l-lysine and the basic amino acid l-arginine, respectively, and LysE can functionally substitute for ArgO when expressed in E. coli. Notwithstanding this functional equivalence, studies reported here show that ArgO possesses a membrane topology that is distinct from that reported for LysE, with substantial variation in the topological arrangement of the proximal one-third portions of the two exporters. Additional genetic and in silico studies reveal the importance of (i) the cytoplasmic N-terminal domain, (ii) a pair of conserved aspartate residues, and (iii) potential intramolecular interactions in ArgO function and indicate that an Arg-translocating conduit is formed by a monomer of ArgO.
INTRODUCTION
In growing bacteria, the steady-state intracellular level of an amino acid is determined by the balance of its biosynthesis, transmembrane (TM) transport, utilization for anabolic processes such as protein synthesis, and conversion to other metabolites. Whereas amino acid transport is usually associated with its uptake from the medium, the phenomenon of amino acid export has only recently been recognized (reviewed in references 1 and 2). The physiological basis of the existence of exporters of amino acids that are essential cellular metabolites is enigmatic. It is thought that exporters may function as outward valves to mitigate toxicity arising from elevated levels of their substrates in the cytoplasm, particularly under conditions where the cytoplasm possesses a limited capacity for their catabolism (1, 3). In the case of sugar exporters, it is believed that their function may serve to mitigate cellular toxicity arising under conditions of excessive accumulation of phosphorylated sugar intermediates (4).
LysE from Corynebacterium glutamicum was the first amino acid exporter to be functionally characterized (5). LysE exports the basic amino acids l-arginine (Arg) and l-lysine (Lys) (6, 7), with export being energized by the proton motive force (PMF), correlating with the ΔΨ component of the PMF (8). The ortholog of LysE in Escherichia coli, ArgO ordinarily mediates the export only of Arg (9) and another membrane protein in E. coli, LysO, mediates Lys export (10, 11, 12). However, under conditions where expression of argO is dissociated from the repressive effect of Lys on its expression, which occurs via the ArgP transcription factor (9), the Lys export potential of ArgO is rendered apparent (10). Thus, expression of argO from the Ptrc promoter complements the thialysine-sensitive phenotype of a lysO deletion mutant (10), indicating that ordinarily the Lys export potential of ArgO in E. coli is rendered cryptic. The Lys export capacity of ArgO is easy to come to terms with, given that the two proteins are 35% identical and 50% similar. Moreover, the absence of ArgO from E. coli renders it highly sensitive to the presence of the Arg antimetabolite canavanine (Can) in the medium (9) and we have noted that Ptrc-driven expression of LysE bearing an abutted N-terminal hemagglutinin (HA) tag can complement the Can-supersensitive (Canss) phenotype of the argO mutant (data not shown). Other workers have also reported observations similar to ours regarding the functionality of LysE in E. coli (13).
The LysE protein superfamily originally comprised three protein families, namely, LysE, RhtB, and CadD (13). However, a recent study included additional protein families in this superfamily (14). Members of the LysE family are widely distributed and are found in many bacterial genomes, with some genomes, such as that of Mycobacterium tuberculosis H37Rv, bearing the potential to encode multiple paralogs of LysE. These proteins contain on an average 200 to 220 amino acids and are largely thought to mediate amino acid export, and predictions of their topology are supportive of a six-TM helical arrangement (14). While LysE and ArgO remain the functionally best-characterized members of the LysE family, the absence of structural information limits structure-function analysis of this protein family. As an initial step toward determination of the molecular basis of Arg export by ArgO, we have undertaken a detailed analysis of its topology in E. coli. Our results indicate that ArgO appears to adopt a topology that is distinct from that predicted in silico, with an indispensable cytoplasmic N-terminal domain (NTD), a dispensable short periplasmic C-terminal region (CTR), and a TM domain present between the NTD and the CTR. Protein modeling predicts the TM domain to adopt a five-helical-bundle configuration. Additional genetic studies with argO mutants and their second-site intragenic suppressors led us to postulate the molecular determinants participating in the Arg export mechanism. Finally, the topological model proposed in this study of ArgO differs from the one proposed for LysE (13) with respect to the topology of the proximal one-third portions of the two exporters in the cytoplasmic membrane.
MATERIALS AND METHODS
Growth media and bacterial strains.
LB medium and glucose minimal A (MA) medium were used as rich and defined synthetic media, respectively (15). The antibiotics ampicillin (Amp), kanamycin (Kan), and tetracycline (Tet) and the Ptrc promoter inducer isopropyl-β-d-thiogalactopyranoside (IPTG) were used at appropriate concentrations. Bacterial cultures were cultivated at 37°C. Antibiotic selection was maintained during all cultivations involving strains bearing plasmids. The bacterial strains used in this study are derivatives of E. coli K-12, and some were constructed by P1 transduction (15). GJ4823 is an argO205::Tn10dTet derivative of MC4100 and bears a Tn10dTet insertion in argO eliminating ArgO function and has been described previously (9). UTL2 is a galU derivative of strain UT5600 [F− araC14 leuB6 secA206 lacY1 proC14 tsx-67 Δ(ompT-fepC)266 entA403 glnX44 trpE38 rfbC1 rpsL109 xylA5 mtl-1 thiE1] (16, 17). GJ9099.1 is an argO205::Tn10dTet hnsFL::Kan derivative of UTL2. The chromosomal hnsFL allele encodes the histone-like protein H-NS bearing a C-terminal 3× FLAG epitope (H-NSFL). The argO205::Tn10dTet and hnsFL::Kan alleles were introduced into UTL2 by P1 transduction and were obtained from GJ4823 and an hnsFL::Kan derivative of MG1655 (18), respectively. The genotype of GJ9006 is GJ4823 araD+ ΔphoA::Kan. The ΔphoA::Kan mutation was obtained from strain JW0374 of the Keio collection (19) and introduced via P1 transduction into GJ4823.
Plasmids and PCR primers.
The plasmids used in this study are derivatives of pTrc99A (20), pOK12 (21), and pCL1920 (22) and are described in Table S1 in the supplemental material, whereas primers for PCR and site-specific mutagenesis are listed in Table S2. Plasmid pHYD3025 is a derivative of pTrc99A and was described previously (10). Standard molecular biology procedures were employed for plasmid construction, and site-directed mutagenesis was performed by overlap extension PCR (23). The site-directed mutagenesis procedure employed for the construction of translational fusions of argO to phoA was the QuikChange method. In all overlap extension PCRs, a pair of primers, JGLPTRFP (5′-CGACATCATAACGGTTCTGG-3′) and JGLPTRRP (5′-TGGGACCACCGCGCTA-3′), flanking the cloned gene were used in conjunction with the primer pair that contained the nucleotide substitutions responsible for the argO alteration introduced. The integrity of all plasmids was verified by DNA sequencing. The ArgO amino acid residue or argO codon numbering described in this study refers to the naturally occurring amino acid or codon numbers. The codons or amino acids of the abutted epitope tags are not considered.
In vivo test of ArgO function.
The functionality of proteins encoded by the various argO open reading frames (ORFs) was gauged by spotting 5 μl of 10-fold serially diluted (for Fig. 1) or 10,000-fold-diluted (for other relevant figures) stationary-phase cultures of the argO mutant GJ4823 bearing plasmid-encoded versions of ArgO on MA agar containing 2 μg/ml Can with the required antibiotic selection. A strain lacking ArgO is rendered hypersensitive to Can, a toxic analogue of Arg (9). The ability of plasmid encoded versions of ArgO to complement the Canss phenotype of GJ4823 was assessed.
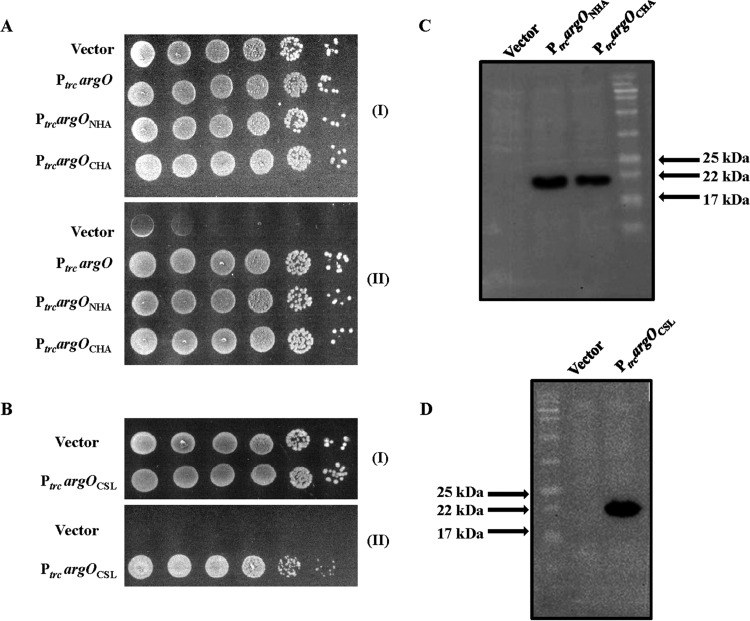
FIG 1.
Functionality of HA epitope-tagged ArgO and its Cys-less derivative in vivo. Tenfold serial dilutions of GJ4823 (MC4100 argO205::Tn10dTet) bearing the vector pHYD3025 and plasmids encoding ArgO (PtrcargO; pHYD2869A), ArgO with an N-terminal (PtrcargONHA; pHYD2869) or a C-terminal (PtrcargOCHA; pHYD2835) HA tag appended (A), and Cys-less ArgO bearing an appended N-terminal HA tag (PtrcargOCSL; pHYD2870) (B) were spotted onto the surface of an MA agar plate (I) and an MA agar plate containing 2 μg/ml Can (II). Immunodetection of proteins encoded by argOCHA, argONHA (C), and argOCSL (D) was done with anti-HA antibody.
Isolation of argO mutants and their second-site intragenic suppressors.
To identify amino acids critical for ArgO function, the argO ORF (argOCHA) encoding ArgO with an appended C-terminal HA tag present within the NdeI and XhoI sites of plasmid pHYD2805 (see Table S1 in the supplemental material) was used as a template. Error-prone PCR was performed with the primer pair T7FP (5′-TAATACGACTCACTATAGGG-3′) and T7RP (5′-GCTAGTTATTGCTCAGCGGT-3′), whose DNA synthesis priming sites flank argOCHA, with pHYD2805 as the template. Error-prone PCR was performed by altering the PCR conditions (24–27). The pool of PCR products was digested with NdeI and XhoI and ligated to NdeI- and SalI-cut pHYD3025, bringing the cloned argO ORFs under the transcriptional control of the Ptrc promoter. The ligation mixture was introduced into DH5α, and transformants were isolated on LB agar plates containing 0.2% glucose and Amp. Around 5,000 transformants were pooled, plasmid DNA isolated from this pool was introduced into GJ4823 by transformation, and 500 individual transformants were screened for the inability to complement the Canss phenotype of GJ4823. Retransformation experiments demonstrated that the inability to complement GJ4823 was plasmid borne. From a collection of 50 transformants with the aforementioned phenotype, 6 were chosen that yielded plasmids expressing a variant ArgO to a level equivalent to the level of the nonmutated ArgO protein encoded by parental plasmid pHYD2835. In this way, a set of argOCHA ORFs encoding ArgO with impaired activity was identified. The plasmids bearing these ORFs were designated pHYD2835.1, pHYD2835.2, pHYD2835.3, pHYD2835.4, pHYD2835.5, and pHYD2835.6 (see Table S1 in the supplemental material).
In order to identify amino acid substitutions compensating for the defect in the ArgO variants encoded by the above-mentioned plasmids, pools of PCR products obtained after error-prone PCR on the argOCHA templates present in these plasmids were cloned separately into the NdeI and HindIII sites of plasmid pHYD3025. JGLPTRFP and JGLPTRRP served as the PCR primer pair for this purpose, and transformants of DH5α were recovered following the separate cloning of six pools of PCR products in pHYD3025. Six pooled plasmid DNA preparations were introduced into GJ4823, and Can- and Amp-resistant (Canr and Ampr, respectively) transformants were selected on MA glucose agar containing 2 μg/ml Can and an appropriate concentration of Amp. Plasmid DNA isolated from individual transformants was introduced into GJ4823 to show that the Canr phenotype was plasmid borne, following which the argOCHA insert present in the plasmid was sequenced. In some instances, the Canr phenotype resulted from reversion of the original (primary) missense mutation present in argOCHA, accounting for the Canss-to-Canr conversion imparted by that argOCHA-expressing plasmid. In other cases, the primary missense mutation was present along with another missense mutation, indicating that the Canss-to-Canr conversion resulted from the presence of an intragenic (secondary) suppressor mutation. Plasmids belonging to this category were saved for further study. For each instance of isolation of a compensatory amino acid substitution, the phenotype imparted by that amino acid substitution alone was also tested by engineering argOCHA ORFs bearing the cognate codon alteration and argOCHA ORFs bearing combinations of primary and secondary missense mutations were also engineered.
Construction of argO-phoA translational fusions.
Translational fusions of portions of argO extending from its translational initiation codon to phoA encoding alkaline phosphatase lacking its signal sequence, namely, codons for the first 26 amino acids (28), were generated in a two-step procedure. The translational fusions were assembled on plasmid pOK12 (see Table S1 in the supplemental material) and subsequently transferred to plasmid pHYD3025, bringing the expression of the fusion-encoding ORF under the control of the Ptrc promoter. The procedure for in-frame fusion of the 8th codon of argO to the 27th codon of phoA is described. Identical procedures were used for fusions of other argO codons to the 27th codon of phoA. We constructed plasmid pHYD2869A, which is a derivative of pOK12 in which an argO ORF (argONHA) encoding ArgO with an appended N-terminal HA tag but lacking a promoter was contained within the NdeI and XhoI sites. After the 8th codon of argO, encoding glycine, a SalI restriction site was introduced by the QuikChange method with the primer pair JGLARGOG8FP and JGLARGOG8RP (see Table S2 in the supplemental material) and the resultant plasmid was designated pHYD2869B8. An XhoI fragment containing phoA lacking its first 26 codons but bearing its natural translational termination codon was released from plasmid pHYD2865 by digestion with XhoI and ligated to SalI-cut pHYD2869B8, and plasmids bearing properly oriented argONHA-phoA translational fusions were identified by restriction digestion and DNA sequencing. One such plasmid, designated pHYD2869C8, was chosen, and the argONHA-phoA translational fusion from this plasmid was isolated by digestion with NdeI and XhoI and cloned into the NdeI and SalI sites of plasmid pHYD3025, generating plasmid pHYD2869D8, wherein the expression of the argONHA-phoA translational fusion was placed under the control of the Ptrc promoter of pHYD3025. Identical procedures were used for the construction of fusions of codons 36, 66, 110, 146, 181, and 201 of argO to codon 27 of phoA, and in each case, the hybrid protein produced contained two amino acids, valine and glutamate, preceding the PhoA portion of the hybrid. The ArgO-PhoA fusions constructed in this study are of the type where PhoA is appended to the C-terminal end of ArgO protein fragments of differing lengths, all starting from its N terminus.
PhoA activities of the ArgO-PhoA hybrids following their expression in vivo were determined as described in reference 29, with modifications. Overnight cultures of GJ9006 bearing plasmids encoding the ArgO-PhoA hybrids were subcultured and cultivated to mid-log phase in LB broth containing Amp and IPTG (10 μM) at 37°C. Iodoacetamide at 1 mM was added to these cultures to eliminate probable spontaneous oxidation of cytoplasmically located PhoA of a given ArgO-PhoA hybrid (30), and the cultures were kept on ice for 10 min. The cultures were centrifuged at 6,000 rpm for 5 min at room temperature and washed twice with 2 volumes of a buffer comprising 10 mM Tris (pH 8.0), 10 mM MgSO4, and 1 mM iodoacetamide. The cells were resuspended in a buffer containing 1 M Tris (pH 8.0), 0.1 mM ZnCl2, and 0.002% SDS; 20 μl of chloroform was used to permeabilize the cells; and the assay of PhoA was initiated at room temperature by the addition of 200 μl of 4-mg/ml p-nitrophenylphosphate prepared in 1 M Tris (pH 8.0). The assay was terminated by the addition of 120 μl of a buffer comprising 165 mM K2HPO4 and 80 mM EDTA. Units of PhoA activity were calculated as described in reference 29 and are presented as means ± standard errors of values obtained from two independent assays performed in duplicate. In vivo levels of ArgO-PhoA hybrids were detected by immunoblotting with anti-HA antibody.
Cysteine accessibility studies with protein PEGylation.
Cultures of GJ9099.1 containing plasmids encoding single-cysteine-substituted derivatives of cysteine (Cys)-less ArgO with an appended N-terminal HA tag (see Table S1 in the supplemental material) were grown overnight under mild agitation in MA medium containing 40 μg/ml each l-leucine, l-proline, and l-tryptophan with appropriate antibiotic selection and subcultured in the same medium supplemented with 1 mM IPTG. In the case of the plasmid encoding ArgO bearing the S135C substitution or the one encoding ArgO with the V142C substitution, the subculturing medium contained 0.1 mM IPTG owing to toxicity associated with elevated expression of the encoded proteins. Cultures were grown to mid-log phase and normalized to an A600 of 2. The cells were centrifuged and washed with 50 mM HEPES buffer (pH 6.5) and resuspended in 1 ml of the same buffer. All centrifugation steps during cell processing were performed at 3,000 rpm at room temperature for 5 min. For the culture of a given single-cysteine derivative, 3 cell aliquots corresponding to an A600 of 0.2 were transferred to three Eppendorf tubes marked as untreated (U), treated (T), and sonicated (S). Sample S was sonicated with the Diagenode Bioruptor UCD-200 as described in reference 10. MAL-PEG (methoxypolyethylene glycol maleimide) prepared in dimethyl sulfoxide (DMSO) was added to the T and S samples to a final concentration of 5 mM. An equal volume of DMSO was added to sample U. The MAL-PEG cross-linking reaction was quenched by adding freshly prepared dithiothreitol (DTT) to a final concentration of 100 mM after 1 h of incubation at room temperature. Samples were solubilized in 2% of SDS with the DTT concentration maintained at 100 mM and sonicated briefly (final sample volume 170 μl), and then the total cell protein in each sample was precipitated by the methanol-chloroform extraction method (31). The precipitated protein was solubilized in 35 μl of an SDS-DTT buffer (2% SDS, 100 mM DTT), and an appropriate volume of 6× SDS-PAGE gel loading dye was added. Equal volumes of whole-cell proteins in the SDS-PAGE gel loading buffer were loaded onto two 15% SDS-PAGE gels and, after electrophoresis and transfer to polyvinylidene difluoride (PVDF) membranes, probed with anti-HA and anti-FLAG antibodies to detect a shift in the mobility of the HA-tagged ArgO species and the resident 3× FLAG-tagged H-NS protein caused by cross-linking with MAL-PEG. Single-cysteine-substituted ArgO variants that did not exhibit a cross-linked species with MAL-PEG following sonication were thought to possess a Cys residue residing in the cytoplasmic membrane. Cells expressing such variants were cultivated until mid-log phase, normalized to an A600 of 2, washed, and processed as described above. Cells corresponding to an A600 of 0.2 were solubilized in the presence of 2% SDS and briefly sonicated, and whole-cell protein extracted by methanol-chloroform extraction was solubilized in 2% SDS. The solubilized protein was treated with MAL-PEG at 5 mM, and reaction quenching and further sample processing were done as described above.
For detection of ArgO–MAL-PEG adducts in the cytoplasmic membrane, mid-log-phase cultures of GJ9099.1 containing the plasmid expressing Cys-less ArgO and a set of derivative plasmids encoding single Cys substitutions in Cys-less ArgO were cultivated as described above and normalized to an A600 2 in 50 mM HEPES buffer (pH 6.5). A culture volume corresponding to an A600 of 0.8 was sonicated to clarity with a bath sonicator. MAL-PEG (at 5 mM) was added to the sonicated samples. Following 1 h of incubation at room temperature, the cross-linking reaction was quenched by the addition of DTT (at 100 mM). The sonicated samples were centrifuged at 350,000 × g for 10 min in a Beckman ultracentrifuge with a 100 Ti fixed-angle rotor at 4°C. The supernatant was decanted, and the translucent (crude membrane) pellet was solubilized in 50 mM HEPES buffer containing 2% SDS and 1% Triton X-100. The Thermo Scientific bicinchoninic acid (BCA) assay kit was used to quantify the total membrane protein, and 20 μg of the protein was loaded onto a 15% SDS-PAGE gel. Following electrophoresis and transfer to a PVDF membrane, an anti-HA antibody was used to probe the transferred blot.
Cross-linking with DSS.
Overnight cultures of GJ4823 bearing plasmids encoding C-terminally HA-tagged versions of ArgO and MscL were subcultured in MA medium with 10 μM IPTG. Cultures were grown to mid-log phase and normalized to an A600 of 2, centrifuged at 6,000 rpm for 5 min at room temperature, and washed twice with 1 ml of phosphate-buffered saline. Cells at an A600 of 0.2 were transferred to two Eppendorf tubes, and disuccinimidyl suberate (DSS) freshly prepared in DMSO was added at a final concentration of 1 mM to one Eppendorf tube and an equivalent volume of DMSO was added to the other. The reaction was quenched by the addition of Tris (pH 8.0) to a final concentration of 100 mM after incubation at room temperature for 30 min. The Eppendorf tubes were centrifuged at 6,000 rpm for 5 min at room temperature, and the cell pellet was resuspended in 30 μl of 1× SDS-gel loading dye, sonicated briefly, and loaded onto a 15% SDS-PAGE gel. Cross-linked products were visualized by immunoblotting with anti-HA antibody.
Immunoblotting.
Mid-log-phase cultures of appropriate strains were normalized to an A600 of 2, and cells at an A600 of 2 were solubilized in 30 μl of 1× SDS-PAGE loading dye. Samples were incubated at 37°C for 10 min before being loaded onto a 15% SDS-PAGE gel. The wet transfer method (transfer buffer comprising SDS running buffer plus 20% methanol) was employed to transfer the proteins to a PVDF membrane. Transferred proteins were stained with amido black to ascertain that the same amount of protein was loaded into each lane, following which the membrane was destained with a solution containing 45% methanol, 45% water, and 10% glacial acetic acid. The membrane was washed three times with 1× phosphate-buffered saline (pH 7.5)–0.1% Tween 20; this was followed by a blocking procedure with 5% fat-free milk powder for half an hour. Following further washing, the membrane was probed with the primary antibody, either an anti-HA or an anti-FLAG M2 monoclonal antibody (Sigma), at a dilution of 1:20,000 for 3 h and with a horseradish peroxidase-conjugated secondary antibody (1:20,000) for 1 h. Western signals were detected with the enhanced chemiluminescence solution kit, and signal images were captured by a ProteinSimple ChemiDoc. For the immunoblot assay pertaining to the detection of ArgO-PhoA hybrids, the samples were processed as follows. Three-milliliter mid-log-phase cultures of GJ9006 containing plasmids encoding ArgO-PhoA hybrids were centrifuged. Cells were solubilized in radioimmunoprecipitation assay buffer (Abcam) and sonicated briefly, the whole-cell protein was quantitated by the Thermo Scientific Pierce BCA Protein Assay kit, and an amount corresponding to 20 μg of whole-cell protein was loaded onto a 15% SDS-PAGE gel. Following electrophoresis and transfer to a PVDF membrane, the hybrids were detected with anti-HA antibody. Representative images of Western blot assays performed twice independently are shown. Expression of the cloned argO genes was induced with inclusion of IPTG in the growth medium prior to the immunoblotting experiments. For Fig. 1C, 5, 6, 7, and 8 (see also Fig. S4 in the supplemental material), plasmid-bearing strains were cultivated in MA medium containing 10 μM IPTG, whereas for Fig. 3, plasmid-bearing strains were cultivated in LB medium containing 10 μM IPTG. Expression of Cys-less argO (Fig. 1D) was induced by the inclusion of IPTG at 1 mM in the MA medium. The IPTG concentrations and the growth medium used for the immunoblot assays described in Fig. 2 (see also Fig. S1 and S2 in the supplemental material) are described above in the section on cysteine accessibility studies using protein PEGylation.
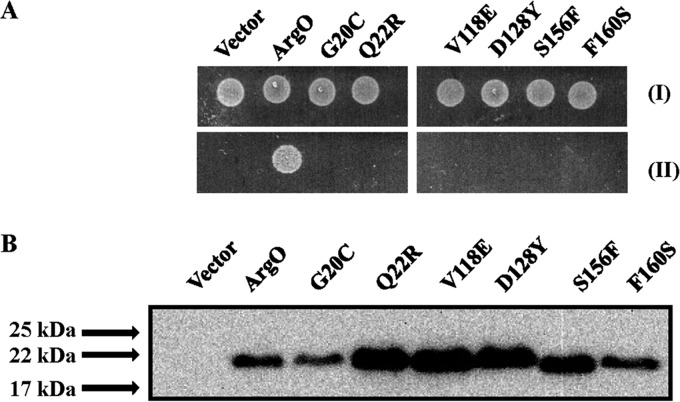
FIG 5.
Compromised ArgO function effected by plasmids encoding ArgO bearing amino acid substitutions. (A) Samples of 5 μl of 10,000-fold-diluted stationary-phase cultures of GJ4823 bearing the vector or plasmid pHYD2835 encoding ArgO bearing an appended C-terminal HA tag and its derivatives bearing the amino acid substitutions indicated were spotted onto the surface of an MA agar plate (I) and an MA agar plate containing 2 μg/ml Can (II). (B) Levels of the corresponding ArgO proteins detected by immunoblotting with anti-HA antibody. A600-normalized sample loading was performed, and the ArgO-encoding plasmids indicated are described in Table S1 in the supplemental material.
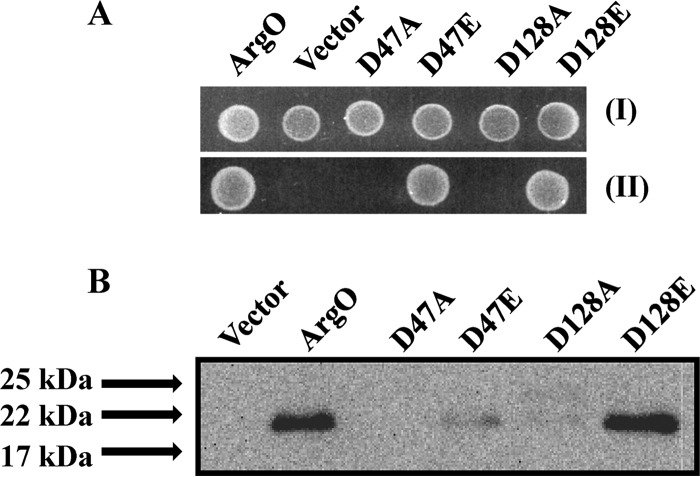
FIG 6.
Effect of replacements of a pair of conserved aspartate residues on ArgO function. (A) Samples of 5 μl of 10,000-fold-diluted stationary-phase cultures of GJ4823 bearing the vector or plasmid pHYD2835 encoding C-terminally HA-tagged ArgO and its plasmid derivatives encoding ArgO with the amino acid substitutions indicated were spotted onto the surface of an MA agar plate (I) and an MA agar plate containing 2 μg/ml Can (II). (B) Levels of substitution-bearing ArgO proteins detected by immunoblotting with anti-HA antibody. Sample loading was done as described in the legend to Fig. 5, and the ArgO-encoding plasmids indicated are described in Table S1 in the supplemental material.
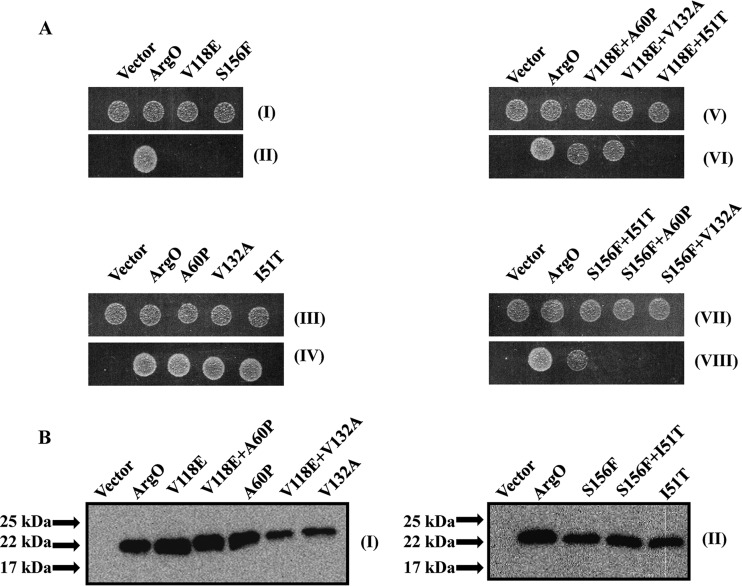
FIG 7.
Restoration of compromised ArgO function by compensatory amino acid substitutions. (A) Samples of 5 μl of 10,000-fold-diluted stationary-phase cultures of GJ4823 bearing the vector or plasmid pHYD2835 encoding C-terminally HA-tagged ArgO and its plasmid derivatives encoding ArgO bearing the single and double amino acid substitutions indicated were spotted onto the surface of an MA agar plate (I, III, V, and VII) and an MA agar plate containing 2 μg/ml Can (II, IV, VI, and VIII). (B) Levels of the single- and double-substitution-bearing ArgO proteins indicated were detected by immunoblotting with anti-HA antibody. Sample loading was done as described in the legend to Fig. 5, and the ArgO-encoding plasmids indicated are described in Table S1 in the supplemental material.
FIG 8.
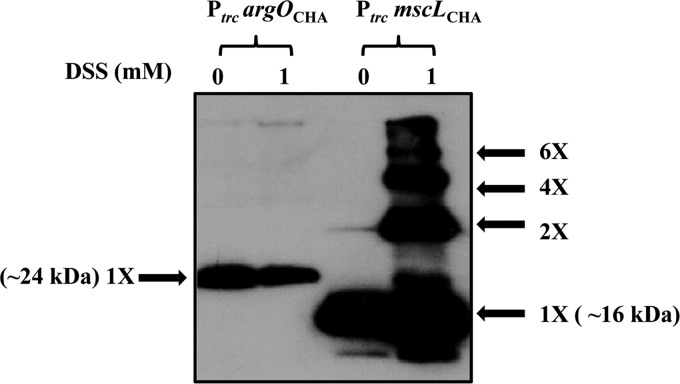
Electrophoretic mobility of HA epitope-tagged ArgO and MscL on SDS-PAGE gels following treatment with DSS. Following their growth, cultures of GJ4823 bearing plasmids encoding HA epitope-tagged ArgO (PtrcargOCHA; pHYD2835) and MscL (PtrcmscLCHA; pHYD2868) were either treated with 1 mM DSS (lanes 1) or not treated with DSS (lanes 0). Following reaction quenching, the samples were processed for SDS-PAGE and Western blotting with anti-HA antibody. The approximate molecular masses of the monomeric species of ArgO and MscL are indicated, and the other oligomeric species of MscL are marked.
FIG 3.
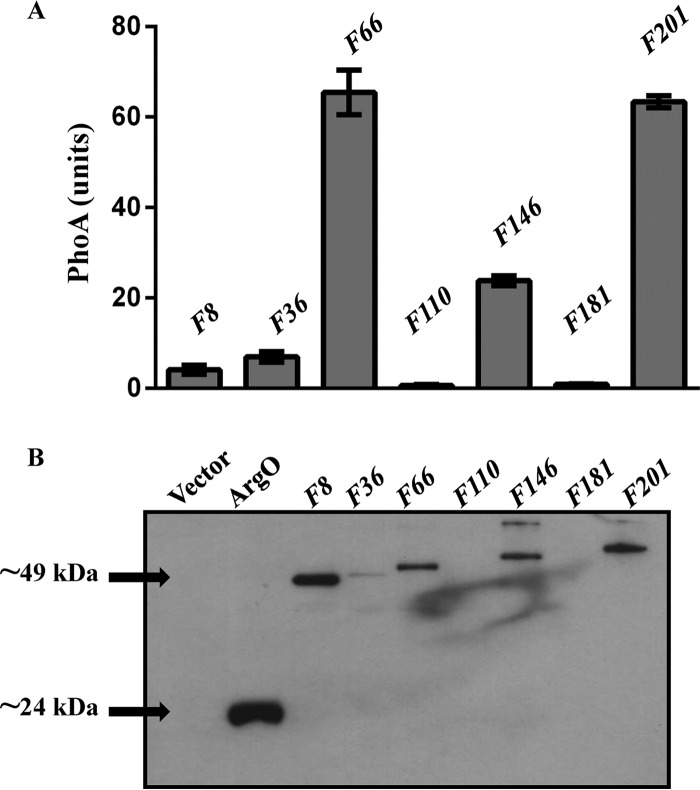
(A) PhoA activities of plasmid-encoded N-terminal fusions (F) of ArgO to PhoA in strain GJ9006. The fusion numbering indicates the length of the amino acid segment of ArgO with PhoA appended at its C terminus. For example, translational fusion F8 encodes a hybrid protein comprising the N-terminal eight amino acids of ArgO fused to PhoA lacking its signal sequence. An HA tag is present at the N terminus of each ArgO-PhoA hybrid. (B) Levels of the ArgO-PhoA hybrids detected by immunoblotting with anti-HA antibody in exponential-phase cultures of GJ9006 bearing plasmids expressing the hybrids indicated. A 20-μg sample of whole-cell protein was loaded into each well. The lane marked ArgO displays the Western blot assay signal of GJ9006 bearing plasmid pHYD2869 expressing ArgO abutted with an N-terminal HA tag. Arrowheads indicate the approximate molecular masses of ArgO (∼24 kDa) and the F8 hybrid (∼49 kDa), and plasmids encoding the ArgO-PhoA hybrids indicated are described in Table S1 in the supplemental material.
FIG 2.
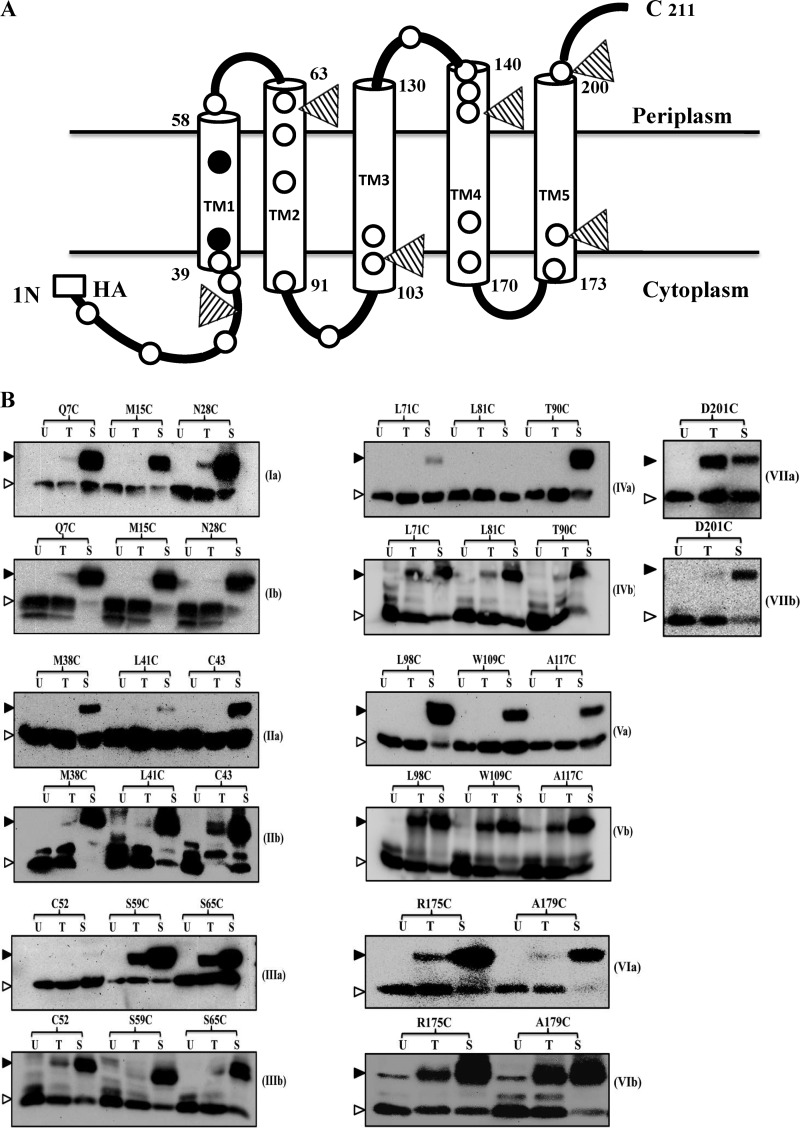
Membrane topology of ArgO. (A) Two-dimensional topological model of ArgO. The lengths of the TM segments were set to those obtained for the in silico model of ArgO described in Fig. 9. The open and closed circles represent, respectively, the inferred positions of the Cys substitutions engineered into Cys-less ArgO and two of the three naturally occurring Cys residues, Cys43 and Cys52, in ArgO. The positions of PhoA fusions to ArgO are marked with triangles, and the HA tag is shown. (B) Cys accessibility depicting immunodetection of the ArgO–MAL-PEG adducts. Cultures of GJ9099.1 containing plasmids encoding the single-Cys-substituted ArgO proteins indicated were processed, and the ArgO–MAL-PEG (Ia-VIIa) and H-NS–MAL-PEG (Ib-VIIb) adducts were detected by Western blotting with anti-HA and anti-FLAG antibodies, respectively. U, T, and S stand for samples that were not treated with MAL-PEG, treated with MAL-PEG, and treated with MAL-PEG following sonication, respectively. The ArgO–MAL-PEG and H-NS–MAL-PEG adducts and free ArgO and H-NS species are marked with filled and open triangles, respectively.
Molecular modeling of ArgO.
ArgO showed poor sequence similarity to other structurally characterized membrane transporters. TM helix prediction based on sequence features suggested a topology with six TM helices. Putative TM and/or α-helical segments were identified with TMHMM v.2.0 (32) and TMpred (33). The sequence and TM helix predictions were used to evaluate potential models of ArgO structure. The online server i-TASSER (34), which uses a hierarchical approach for protein structure prediction, was used to obtain a molecular model of ArgO. While structural templates are first identified by the multiple threading approach from the Protein Data Bank with LOMETS (35) in this procedure, full-length models are constructed by iterative template fragment assembly simulations (36).The first 38 residues of ArgO were deleted from the model, as this segment was demonstrated to be cytoplasmic while hierarchical models co-opted this polypeptide stretch as a part of the TM segment. The final model has a TM component comprising residues 39 to 211 with an Nin-Cout topology. The quality of the model was improved by iterative rounds of structure idealization by implementation of the refinement program Refmac5 in Coot (37). This step also served to reconcile the outliers in the Ramachandran plot (38). Low-frequency normal modes were studied for the collective motion of atoms in the ArgO model with the online server ElNémo (39). Scaled and observed B factors of residues obtained after these simulations were examined alongside the structural models.
RESULTS AND DISCUSSION
HA epitope-tagged versions of ArgO are functional in vivo.
For the studies described here, we constructed versions of argO encoding ArgO proteins with the HA epitope attached to the N- or C-terminal end, and the corresponding genes were designated argONHA and argOCHA, respectively. A strain lacking ArgO is rendered highly sensitive to the Arg antimetabolite Can (9), and basal-level expression of both argOCHA and argONHA from the Ptrc promoter of plasmids pHYD2835 (10) and pHYD2869, respectively, complemented the Canss phenotype of GJ4823, a strain that lacks ArgO (9) (Fig. 1A), indicating that attachment of the HA epitope to the two termini of ArgO does not perturb ArgO function. The presence of the Ptrc inducer IPTG at concentrations of >10 μM caused significant growth retardation of GJ4823 bearing plasmids pHYD2835 and pHYD2869 in MA agar (data not shown), indicating that elevated expression of these genes led to cellular toxicity. As expected, expression of the native argO gene under the control of the Ptrc promoter also complemented the Canss phenotype of GJ4823 (Fig. 1A) and its overexpression also impaired cell growth (data not shown). In addition, we constructed a variant of argONHA in which codons encoding the three naturally occurring cysteines of ArgO were replaced with those encoding alanine. The resulting gene (argOCSL) encoded a cysteine (Cys)-less ArgO protein whose basal-level expression from the Ptrc promoter in plasmid pHYD2870 in GJ4823 also complemented its Canss phenotype (Fig. 1B). Overexpression of argOCSL induced by the inclusion of 1 mM IPTG in the medium did not yield any toxic phenotype (data not shown), which was in contrast to the case observed for argOCHA, argONHA, and native argO expression. The corresponding HA-tagged proteins were immunodetected with anti-HA antibody (Fig. 1C and D). Although the predicted molecular masses of both C- and N-terminally HA-tagged ArgO (and also of Cys-less ArgO) are approximately 24.5 kDa, we consistently noted that epitope-tagged ArgO displayed some anomalous mobility, migrating as an approximately 22-kDa species (Fig. 1C and D). It may be noted that, in the strictest sense, complementation of the argO mutant by the expression of a given ArgO variant need not qualify that variant to possess an Arg export property identical to that of wild-type ArgO. It is possible that the variant possesses an attenuated Arg export property, which nonetheless is sufficient for complementation.
The membrane topology of ArgO.
To delineate the topology of ArgO in situ, two approaches were employed, namely, cysteine (Cys) accessibility and analysis of ArgO-PhoA fusion protein reporters. In the first approach, Cys-less ArgO proteins bearing a unique engineered Cys residue were assessed for Cys accessibility by Mal-PEG cross-linking. MAL-PEG reacts with the thiol group of Cys to form a PEG-maleimide-thiol adduct. Adduct formation in a protein increases its molecular mass by 5,000 Da, which can be detected as a shift in the mobility of the protein in SDS-PAGE after Western blotting with a protein-specific antiserum. The retardation of the mobility of the PEG-maleimide-protein thiol adduct can also be detected if the protein in question is labeled with an epitope tag naturally devoid of Cys residues like the HA tag, in which case antisera against the epitope serve the purpose. By employing site-specific mutagenesis, we generated 24 plasmids that are derivatives of argOCSL-expressing plasmid pHYD2870, each encoding ArgO bearing a single cysteine substitution at a specific site in ArgO. Visible expression and an ability to complement the argO mutant GJ4823 were the two criteria for selection of the Cys substitutions. The coverage of Cys substitutions is thus restricted by these requirements. All encoded ArgO variants were functional, as their expression in strain GJ9099.1 complemented the Canss phenotype associated with the presence of the argO205::Tn10dTet mutation (data not shown).
MAL-PEG is not able to permeate membranes, and despite the limited permeability of the outer membrane (OM) of E. coli, MAL-PEG cannot traverse it, owing to the 500-Da size exclusion cutoff imposed by the OM porins. Hence, to allow MAL-PEG to diffuse into the periplasm, we employed strain GJ9099.1 for this study. GJ9099.1 is a derivative of strain UTL2 (16) whose OM is leaky, permitting passive diffusion of bulky compounds into the periplasm. A mutation in galU is thought to lead to the leaky OM of UTL2 (17). The other two relevant features of GJ9099.1 are that it lacks ArgO and encodes a chromosomally encoded version of the nucleoid protein H-NS bearing a 3× FLAG tag abutting its C terminus. The relevance of the second feature is described below.
An assessment of the three possible locations of an amino acid residue in ArgO, proxied by a Cys substitution, namely, periplasmic, TM, and cytoplasmic, can be made depending upon its reactivity with MAL-PEG. A Cys residue located in the periplasm would react with MAL-PEG in intact (unbroken) cells, whereas a Cys at a cytoplasmic location would react upon sonication of the cell suspension. A Cys residue buried in the TM region would remain nonreactive, regardless of the physical state of the cell, but would react only when the corresponding membrane protein is extracted from its native location and solubilized in SDS. For assessment of the location of a particular Cys residue, a culture of strain GJ9099.1 expressing single-Cys-substituted ArgO was divided into three portions. MAL-PEG was not added to the portion marked U. The portion marked T was treated with MAL-PEG, and the portion marked S was sonicated and then treated with MAL-PEG. MAL-PEG adduct formation in the T sample indicated that the interrogated Cys residue lay in the periplasm, whereas adduct formation in the S but not the T sample suggested a cytoplasmic location of the Cys residue. Absence of adduct in the T and S samples was suggestive of a probable intramembrane location of the Cys residue. Adduct formation was detected by Western blotting of the portions of samples marked U, T, and S with anti-HA antiserum. As expected, Cys-less ArgO did not display any adduct with MAL-PEG when its host strain was subjected to the above-described conditions for detection of the ArgO–MAL-PEG adduct (see Fig. S1 and S2 in the supplemental material).
To assess the integrity of the cells during sample processing, the reactivity with MAL-PEG of a single Cys residue of a chromosomally encoded cytoplasmic protein, H-NS, labeled with a C-terminal 3× FLAG epitope (H-NSFL) by the host cell GJ9099.1 was simultaneously determined. The H-NSFL–MAL-PEG adduct was detected by Western blotting of an equivalent portion of the U, T, and S samples with an anti-FLAG antibody. The FLAG epitope also lacks Cys residues. We noted that in all of the instances examined, the MAL-PEG adducts with ArgO increased its molecular mass by 5,000 Da in our Western blot assays, which accorded with expectations; however, for poorly understood reasons, the molecular mass increment for H-NS with the MAL-PEG adduct was approximately 10 kDa (data not shown). Two additional features of our Western blot assays are to be noted. First, we observed the presence of doublet anti-FLAG immunoreactive species in some instances pertaining to the detection of the H-NS–MAL-PEG adducts (Fig. 2B; see Fig. S1 and S2 in the supplemental material). Second, in many instances, we observed enhanced detection of the HA-tagged ArgO species following MAL-PEG cross-linking (Fig. 2B; see Fig. S1 and S2). While a reason for the former is not known, the latter observation may be related to more efficient transfer and/or binding of PEGylated ArgO to the PVDF membrane during electroblotting. The aforementioned explanation has been previously provided by others (40) to account for enhanced immunodetection of a PEGylated membrane protein. By using this protein PEGylation approach, we arrived at a topological model of ArgO with an overall topology compatible with an Nin-Cout configuration (Fig. 2A). The features of this model are described below.
ArgO possesses an NTD that is located in the cytoplasm.
The Cys accessibility of ArgO variants bearing single Cys substitutions of amino acid residues 7, 15, 28, 38, and 41 and the ArgO variant bearing the single naturally occurring Cys 43 residue indicates that the region of ArgO comprising roughly up to residues 38 to 43 is located in the cytoplasm. This region is referred to as the NTD (Fig. 2B, Ia and IIa). The weakened ArgO–MAL-PEG adduct seen with ArgO bearing the L41C substitution (Fig. 2, IIa) may result from diminished reactivity of the thiol group.
TM, periplasmic, and cytoplasmic loop regions of ArgO.
The topological model of ArgO contains a TM domain comprising five segments, TM1 to TM5 (Fig. 2A). The lengths of the TM segments in Fig. 2A have been set to those obtained for the in silico model of ArgO (see Fig. 9). Of these, the first two N-terminal segments, TM1 and TM2, are better interrogated than the others, which therefore remain putative TM segments in the ArgO model. This conclusion is based on the Cys accessibilities of the ArgO variant bearing the naturally occurring Cys as the 52nd amino acid of ArgO and two other variants bearing the L71C and L81C substitutions. These Cys residues are clearly located in an intramembrane environment because they were either not accessible to MAL-PEG whether their host cell envelopes were left intact or disrupted by sonication (Fig. 2B, IIIa and IVa). However, they were readily modified by MAL-PEG when the corresponding proteins were removed from the native membrane environment by whole-cell protein precipitation and SDS solubilization of the precipitated proteins (see Materials and Methods; also see Fig. S1I in the supplemental material).
FIG 9.
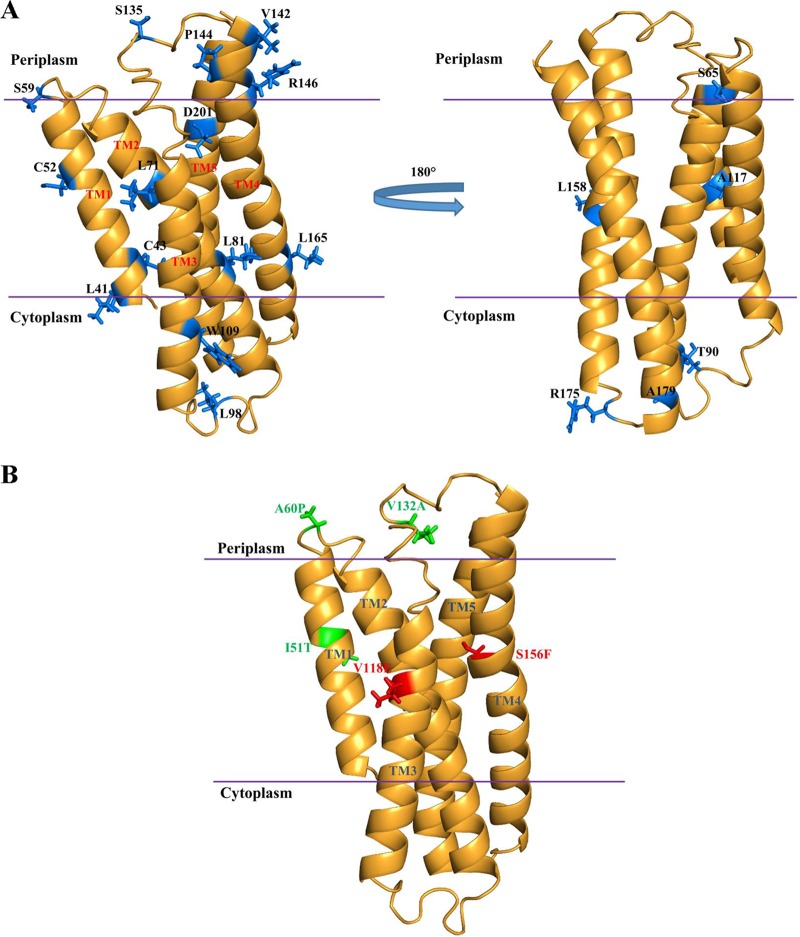
(A) Molecular model of ArgO. The first 38 residues comprising the cytoplasmic NTD are not included in this model. The locations of the cysteine substitutions that formed the basis for the topological assignment are indicated. On the basis of experimental observations, a monomer model of ArgO was designed. (B) The positions of two amino acid substitutions that disable ArgO function (red) and their compensatory amino acid substitutions (green) in the ArgO model are shown.
Cys accessibility studies allowed the delineation of the periplasmic and cytoplasmic loop regions of ArgO. The ready modification of Cys substitutions at positions 59 and 65 in intact cells of GJ9099.1 (Fig. 2B, IIIa) supports a periplasmic location of these Cys residues in ArgO, revealing the existence of a short periplasmic loop between TM1 and TM2. Similarly, Cys substitutions at positions 135, 142, 144, and 146 are also inferred to be periplasmic (see Fig. S1IIa and IIIa in the supplemental material) because they exhibited significant modification by MAL-PEG in intact cells, thus outlining the second short periplasmic loop in ArgO connecting putative TM3 and TM4. The MAL-PEG accessibility of the Cys substitutions at positions 90, 98, 109, and 117 in ArgO is indicative of cytoplasmic positioning of these Cys residues (Fig. 2B, IVa and Va) and aids in the delineation of a cytoplasmic loop between TM2 and putative TM3. A second cytoplasmic loop in ArgO between putative TM4 and TM5 can be inferred on the basis of accessibilities of Cys substitutions at positions 175 and 179 (Fig. 2B, VIa) and the Cys substitution at position 165 in ArgO (see Fig. S1IIIa in the supplemental material). Finally, Cys accessibility studies indicated the existence of a short periplasmic CTR in ArgO as gauged by the accessibility of the Cys substitution at position 201 to MAL-PEG in intact cells (Fig. 2B, VIIa). This observation, in addition, allows one to infer the existence of the fifth TM segment in ArgO, namely, TM5 (Fig. 2A).
Following sonication and treatment with MAL-PEG, we isolated cell membranes from strains expressing a set of Cys-substituted ArgO variants bearing the Q7C, N28C, L98C, L109C, L165C, and R175C substitutions, in which the Cys residues were inferred to be in the cytoplasm on the basis of the aforementioned studies. Immunoblotting with anti-HA antibody showed the presence of the ArgO–MAL-PEG adducts for all of the ArgO variants chosen (see Fig. S2 in the supplemental material), indicating that the ArgO–MAL-PEG adducts resulted from ArgO variants integrated into the cytoplasmic membrane. Overall, these studies indicated that the topology of ArgO is consistent with an Nin-Cout configuration and that the TM domain of ArgO appears to comprise five TM segments flanked at its N and C termini by a cytoplasmic NTD and a short CTR.
Assessment of ArgO topology by using ArgO-PhoA hybrids.
Fusions of PhoA (alkaline phosphatase) to N-terminal segments of a polytopic inner membrane protein are also employed as probes of its topology (41). PhoA displays compartment-specific folding in E. coli, being enzymatically active in the periplasm but not in the cytoplasm. The level of PhoA activity of a hybrid protein comprising the N-terminal segment of a membrane protein fused to PhoA usually serves as a reliable indicator of the location of the hybrid fusion joint. We constructed a set of argO-phoA translational fusions and placed their expression under the control of the Ptrc promoter of plasmid pHYD3025 (Fig. 2A). In addition, an N-terminal HA epitope was appended to the encoded ArgO-PhoA hybrids. The fusion joints were chosen such that the PhoA portion in the encoded hybrid protein was present at the C-terminal ends of hydrophilic portions (loops) of ArgO, inferred from in silico predictions of the TMHMM suite. Positioning the reporter fusion in this manner is thought to minimize fusion joint-dependent perturbations of topogenic signals in polytopic membrane proteins (42), particularly those conforming to the positive-inside rule (43). Expression in vivo of fusions of PhoA to the N-terminal 66, 146, and 201 amino acyl segments of ArgO yielded significantly high PhoA activity, and the hybrids could be immunodetected (Fig. 3). This observation indicated a potential periplasmic location of the fusion joints of these hybrids. On the other hand, expression of ArgO-PhoA hybrids bearing fusions of the N-terminal 8, 36, 110, and 181 amino acyl segments to PhoA produced negligible PhoA activity, and the corresponding proteins, barring the case of hybrid F8 (Fig. 3), were not detected by Western blotting (Fig. 3). In this regard, an earlier study has shown that PhoA retained in the cytoplasm is proteolytically unstable and is rapidly degraded (44), and thus, we would like to place the locations of the fusion joints in these hybrids tentatively as cytoplasmic. However, on the basis of the results obtained with the F8 hybrid, one can assign the location of the N-terminal 8 amino acids of ArgO as cytoplasmic with a certain degree of confidence (Fig. 3). Reasons for the apparent stability of the F8 hybrid are ill understood. Among the ArgO-PhoA hybrids, the only hybrid retaining ArgO function was the one containing the N-terminal 201 amino acids of ArgO fused to PhoA, as its expression in argO mutant strain GJ4823 complemented its Canss phenotype (data not shown).
Comparisons of topological assignments of amino acyl residues of ArgO from Cys accessibility and PhoA reporter fusion studies.
Overall, a high degree of concordance was obtained for the topological assignments of the interrogated amino acids of ArgO between the two approaches used for assessing the topology of ArgO. Thus, the periplasmic assignments of Cys-substituted residues 65, 144/146, and 201 (Fig. 2B; see Fig. S1 in the supplemental material) correlated with the high PhoA activity yielding ArgO-PhoA hybrids F66, F146, and F201, respectively (Fig. 3). On the other hand, the cytoplasmic assignments of Cys-substituted residues 7, 38, 109, and 175/179 (Fig. 2B) correlated with the ArgO-PhoA hybrids with low PhoA activity, namely, F8, F36, F110, and F181, respectively (Fig. 3).
Similarities and differences between topological models of ArgO and LysE.
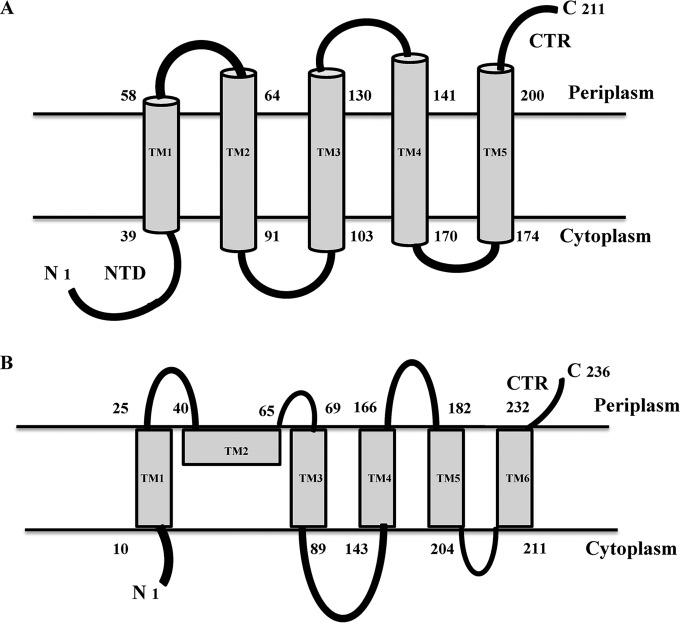
A comparison of the two-dimensional topological models of LysE (13) and ArgO proposed in this study reveals that the major difference between the two models lies in the arrangement of the first two N-terminal putative helical segments (Fig. 4). The cytoplasmic NTD and TM1 proposed for ArgO, in the context of the LysE model, are placed such that the segment homologous to the ArgO NTD in LysE spans the membrane and the segment corresponding to ArgO TM1 in LysE is positioned on the periplasmic side lying parallel to the plane of the membrane (13). Both models propose an overall Nin-Cout configuration with a TM domain comprising five TM segments and a short CTR, and the arrangements of the putative four TM helices in the distal two-thirds of the two exporters are similar. Given our unpublished observation that expression of epitope-tagged LysE can substitute for ArgO function in E. coli, the marked difference in the topology of LysE and ArgO in the N-terminal region is somewhat enigmatic. It seems likely that the difference may arise from the two approaches used to determine their topologies. In contrast to Vrljic et al. (13), who have exclusively used PhoA-LacZ fusions to determine LysE topology, our analysis of ArgO topology, performed with biologically active ArgO variants, rests mainly on the scanning of cysteine accessibility, which is the least invasive (in terms of perturbing protein structure) means of determining membrane protein topology, as opposed to the PhoA-LacZ fusion approach. If our topological model of ArgO is valid, then one would, in principle, expect LysE to possess a topology similar to that of ArgO. Perhaps abutment of certain N-terminal segments of LysE to PhoA in the analysis performed by Vrljic et al. (13) led to an altered localization of PhoA.
FIG 4.
Comparison of the experimentally derived two-dimensional topological schemes for ArgO (A, from this study) and LysE (B). The scheme for LysE and the designations of its TM segments are from reference 13. The cytoplasmic NTD of ArgO and the CTR of ArgO and LysE are indicated.
Amino acid substitutions in positions conserved in the LysE family impair ArgO activity.
We employed a PCR mutagenesis approach (see Materials and Methods) to screen and isolate versions of argOCHA that encoded proteins with impaired ArgO activity. Among a collection of 50 such plasmids carrying mutant argO genes that failed to complement the Canss phenotype of strain GJ4823, 6 plasmids were identified that expressed a functionally defective ArgO protein (Fig. 5A) bearing amino acid substitutions to a level equivalent to or, in some instances, slightly greater than that of wild-type ArgO (Fig. 5B). The G20C, Q22R, V118E, D128Y, S156F, and F160S substitutions in ArgO were isolated in this screening. Barring the V118E substitution, all amino acid replacements abolishing ArgO function were substitutions of amino acids that are conserved in orthologs of the LysE family (see Fig. S3 in the supplemental material). The first two lie in the proposed cytoplasmic NTD of ArgO, and the rest are located in the TM domain of ArgO. In the alignment shown in Fig. S3 in the supplemental material, valine at 118 in ArgO is conserved in five out of seven orthologs, with the position generally occupied by a hydrophobic amino acid.
In addition we tested the effects of specific deletions engineered in argOCHA and argONHA on the ability of the truncated proteins to complement the Canss phenotype of GJ4823. Expression of an ArgO variant lacking the last 10 amino acids could complement the Canss phenotype of the argO mutant, though somewhat weakly, whereas expression of ArgO variants with deletions of the N-terminal 28 or C-terminal 40 amino acids could not complement the Canss phenotype (see Fig. S4 in the supplemental material). None of the deletion-bearing ArgO variants displayed detectable levels of protein when expressed in GJ4823 (see Fig. S4 in the supplemental material). Given this, meaningful inferences about the essentiality of the N-terminal 28 and C-terminal 40 amino acids of ArgO for its function cannot be drawn from deletion studies. However, given that expression of an ArgO variant lacking the last 10 amino acids could complement the Canss phenotype of the argO mutant permits the conclusion that the last 10 amino acids of ArgO, constituting the CTR, are not absolutely necessary for ArgO function. Furthermore, the identification of multiple amino acid substitutions in the NTD, namely, the G20C and Q22R substitutions, that disable ArgO function without impairing the level of the corresponding ArgO protein, provide a more reliable indication of the essentiality of the NTD for ArgO function.
Conservative substitutions of a pair of conserved aspartate residues do not perturb ArgO function in vivo.
Perusal of the amino acid alignment of orthologs of ArgO revealed the conservation of a pair of aspartates, D47 and D128 (see Fig. S3 in the supplemental material). Of the two, the importance of D128 was independently inferred by the isolation of the D128Y ArgO-disabling substitution described earlier (Fig. 5). Conversions of D47 and D128 individually to alanine abolished ArgO function (Fig. 6A). However, ArgO bearing single glutamate substitutions of the two aspartate residues, namely, the D47E and D128E substitutions, retained activity and could complement the Canss phenotype of argO mutant strain GJ4823 (Fig. 6A). The protein corresponding to ArgOD128E was produced at a level comparable to that of wild-type ArgO, whereas ArgOD47E was produced at a level much below that of wild-type ArgO (Fig. 6B). Proteins corresponding to ArgOD47A and ArgOD128A could not be detected (Fig. 6B). With respect to the two-dimensional topological representation of ArgO (Fig. 2A), D47 and D128 are positioned at the base of TM1, toward the cytoplasmic face and near the apex of TM3 at the periplasmic face, respectively, of the cytoplasmic membrane. This off (membrane)-centered positioning, which is positioning at the membrane periphery of the two aspartate residues, is compatible with earlier studies of the natural distributions of amino acids in TM domains, which have noted that charged residues in TM helices generally tend to occupy positions near the aqueous interfaces on the two sides of the membrane (45; reviewed in reference 46). Negatively charged amino acids, glutamates, positioned in an off-centered fashion are thought to be critical in Tet efflux mediated by the TetA(P) protein of Clostridium perfringens (47). Overall, these observations reveal the importance of aspartates in the TM domain of ArgO.
Second-site suppressor studies indicate the importance of intramolecular interactions in ArgO function.
Perturbations of ArgO function caused by amino acid substitutions may occur if some substitutions lead to loss of intramolecular interactions such as inter- or intrahelical interactions (reviewed in references 48 and 49), thereby disabling ArgO activity. In order to identify such interactions, amino acid replacements compensating for the original (primary) substitution in ArgO were sought. For this purpose, an error-prone PCR mutagenesis strategy was employed (see Materials and Methods) to isolate second-site suppressor mutations in argO. The six plasmids encoding ArgO bearing the aforementioned primary substitutions were used as templates in an error-prone PCR. Following cloning of the PCR products, the plasmid pool was introduced into strain GJ4823 and transformants exhibiting a Canr phenotype indicative of restoration of ArgO function were selected on Can-supplemented MA agar plates. Compensatory substitutions were obtained for the ArgO variants with the V118E and the S156F primary substitutions. The defect caused by the V118E substitution was compensated for by either the A60P or the V132A amino acid substitution, whereas the one caused by the S156F replacement was restored by the I51T substitution (Fig. 7A). ArgO bearing the I51T, A60P, and V132A substitutions alone retained ArgO function (Fig. 7A). Furthermore, the effect of the compensatory substitution was specific to a given primary substitution. Whereas ArgOV118E A60P or ArgOV118E V132A retained ArgO function, ArgOV118E I51T was inactive. Similar results were obtained with the S156F I51T compensatory pair in that the defect caused by the S156F substitution was compensated for only by the I51T substitution and not by the A60P or the V132A substitution (Fig. 7A). In addition, proteins corresponding to the various substitutions bearing argO ORFs were, in most cases, produced at levels comparable to those of wild-type ArgO (Fig. 7B). The molecular model of ArgO described later provides a probable rationale for functional compensation in ArgO.
Evidence of a monomeric state of ArgO in situ.
In order to probe the oligomeric state of ArgO in vivo, cross-linking was performed with the membrane-permeating amine cross-linker DSS. DSS provides a spacer arm of 11.4 Å, and six primary amines in ArgO, including the N-terminal one, are available as substrates for cross-linking. The appended HA tag is devoid of a primary amine. Treatment of a culture of GJ4823 expressing HA epitope-tagged ArgO with DSS at 1 mM, followed by SDS-PAGE and Western blotting, led to the detection of an ArgO signal corresponding only to the monomer species (Fig. 8). On the other hand, treatment of GJ4823 expressing an HA epitope-tagged version of the mechanosensitive channel MscL known to form multiple oligomers with DSS treatment (50) yielded signals corresponding to the monomeric, dimeric, tetrameric, and hexameric species of MscL following Western blotting (Fig. 8). The pattern of mobilities of various species of MscL detected by Western blotting following DSS treatment is consistent with an earlier report (50). In a related approach, we tested the effects of overproduction of the six functionally defective ArgO polypeptides (Fig. 5) on the function of chromosomally encoded ArgO of the wild-type strain. If the functional state of ArgO in vivo is an oligomer (at the least a dimer), then overproduction of functionally compromised ArgO would interfere with wild-type ArgO and cause the wild-type strain to exhibit sensitivity to Can. This approach is analogous to one used with the multidrug exporter EmrE wherein the overproduction of functionally compromised EmrE variants impaired the function of wild-type EmrE in vivo, permitting the inference that the functional state of EmrE in vivo was an oligomer (51). However, none of the overproduced defective ArgO variants led to any discernible sensitivity to Can in the wild-type strain (see Fig. S5 in the supplemental material). A 10-fold higher concentration of Can (from 2 to 20 μg/ml) was used to detect marginal growth inhibition of the wild type, which was not observed. The aforementioned evidence, albeit indirect, is supportive of a monomeric functional state of ArgO but is not conclusive.
Molecular model of ArgO.
The experimental data on the topology of ArgO formed the basis of a molecular model of ArgO under the premise that the monomeric state of ArgO is valid. ArgO has 211 residues, with residues 1 to 38 forming the cytoplasmic component of ArgO. The lack of a suitable structural template for this polypeptide segment precluded efforts to construct a three-dimensional model of the cytoplasmic domain. Residues 39 to 211 incorporate five TM helices (along with the CTR) and were evaluated for their consistency with experimental data (Fig. 9; see Fig. S3 in the supplemental material). The approximate lengths of TM segments TM1 to TM5 in the ArgO model are 20, 28, 27, 30, and 38 amino acids, respectively. In the model of ArgO (Fig. 9), the length of TM5 is inclusive of the CTR. The average length of TM segments has been thought to vary from 20 to 40 residues (52). Furthermore, a recent analysis has catalogued the lengths of α-helical TM segments in a high-resolution data set to note that the median TM segment length can vary from 16 to 30 amino acids (53). The assigned lengths of the TM segments in ArgO are compatible overall with the aforementioned findings on TM lengths in integral membrane proteins It is worth noting, in this context, that while PhoA reporter fusion analysis of C. glutamicum LysE suggested a five-TM α-helical spanner topology (13), members of this family are believed to adopt a six-TM topology (14). Indeed, sequence-based predictions for both ArgO and LysE suggest six TM helices (see Fig. S3 in the supplemental material). Given this inconsistency, the experimental data on the TM domain and the boundaries of the TM helices were used to guide the molecular modeling studies. The locations of the Cys substitutions inferred from Cys accessibility studies (Fig. 2) thus served as markers for iterative rounds of model building (Fig. 9). The dynamic rearrangement of TM helices in ArgO was examined by an analysis of the low-frequency normal modes. Normal-mode analysis is particularly suited for this analysis, as the rigid body motions (in this case, the TM helices) involve larger amplitudes (1 to 10 Å) and a time scale of 10−9 to 1 s (39). The underlying hypothesis is that the vibrational normal modes with the lowest frequencies describe the largest movements in a protein. A comparison of the initial model and observed fluctuations was used to evaluate the functional data derived from genetic analysis and mutant phenotypes.
The evidence of a monomeric state of ArgO in vivo suggested that functionally relevant interactions would be largely confined to the TM helical segments of the ArgO monomer. The ArgO model was subsequently examined in the context of dynamic fluctuations predicted by the low-frequency normal modes, a snapshot of which is depicted in Fig. S6 in the supplemental material, to obtain a probable rationale for the functional compensation of two ArgO-disabling amino acid substitutions, namely, V118E and S156F. Normal-mode analysis of the ArgO model suggested a degree of conformational flexibility in the TM helices of ArgO. While one or two low-frequency normal modes are sufficient to provide a fair description of the conformational change (see Fig. S6 in the supplemental material), B factors computed from the mean squared displacement of the first 100 lowest-frequency normal modes alongside linear scaling to observed B factors in the reference structure provide a measure of the flexibility of the macromolecule (see Fig. S7 in the supplemental material). That the V118E mutant is inactive is not surprising in a structural context, given that an additional charge is introduced into the TM helix (TM3) that in the ArgO model stacks against the TM2 helix. However, its compensation by the A60P substitution, with an inferred periplasmic location suggests that the basis for its functional compensation of the V118E substitution probably involves a compensatory reorientation of one or more TM segments of ArgO. Normal-mode-derived B factors of the ArgO model (see Fig. S7 in the supplemental material) indicate that V132, which is predicted to lie in a loop connecting TM3 with TM4, displays considerable dynamic fluctuation, which may account for the ability of the V132A substitution to compensate for the defect in the V118E mutant. The compensation in this case may be restricted to TM3. While the substitution of a phenylalanine instead of a serine as the 156th amino acid inactivates ArgO, the ability of the I51T mutant to compensate for the S156F mutation remains to be rationalized. The locations of the aforementioned ArgO-disabling substitutions and their compensatory amino acid replacements in the ArgO model are depicted in Fig. 9B. The fact that all of the compensatory amino acid substitutions identified in this study occur at positions that are not absolutely conserved in LysE family orthologs (see Fig. S3 in the supplemental material) may account for their tolerance when present as individual substitutions in ArgO.
In conclusion, a model of ArgO is proposed that is based on experimentation involving cysteine accessibility, PhoA reporter fusions, and protein modeling. While our data are consistent with the periplasmic location of the C terminus of ArgO proposed earlier (54), we note that the cytoplasmic NTD is a distinctive feature of ArgO. A mechanistic role for the NTD and the pair of conserved aspartates in ArgO-mediated Arg export remains to be identified. Furthermore, the experimentally guided molecular model of ArgO provides a probable rationale for the effects of compensatory amino acid substitutions in ArgO function. Lastly, in the model of ArgO proposed in this study, a skewed distribution of the positively charged amino acids Arg and Lys is apparent, with six Arg and four Lys residues of ArgO located in the cytoplasm and two Arg residues and one Lys residue displaying a periplasmic location. This feature suggests that the overall topology of ArgO conforms to the positive-inside rule (43, 55).
Supplementary Material
ACKNOWLEDGMENTS
We thank Ethan Bibi, J. Gowrishankar, T. Mori, and Aswin Seshasayee for strains used in this study; Sudipta Mukherjee and Tulika Sharma for assistance in the construction of cysteine substitutions in ArgO and ArgO-PhoA hybrids, respectively; and Kapil Goutam for advice.
A.P. was supported by fellowships from the Council of Scientific and Industrial Research, Government of India, and the Centre for DNA Fingerprinting and Diagnostics. Additional fellowship support to A.P. from the Centre of Excellence in Microbial Biology and DST-ANR proposal no. ID-06 through the Department of Biotechnology, Government of India, and the Department of Science and Technology, Government of India, respectively, is also acknowledged. A.K.G. and S.D. were supported by fellowships from the Indian Institute of Science, Bangalore, and the Department of Biotechnology, Government of India, respectively. This work was supported by a Centre of Excellence in Microbial Biology grant from the Department of Biotechnology, Government of India, to A.A.S.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00423-16.
REFERENCES
- 1.Burkovski A, Krämer R. 2002. Bacterial amino acid transport proteins: occurrence, functions, and significance for biotechnological applications. Appl Microbiol Biotechnol 58:265–274. doi: 10.1007/s00253-001-0869-4. [DOI] [PubMed] [Google Scholar]
- 2.Marin K, Krämer R. 2007. Amino acid transport systems in biotechnologically relevant bacteria. p 289–325. In Wendisch V. F. (ed), Amino acid biosynthesis: pathways, regulation and metabolic engineering. Microbiology monographs, vol. 5 Springer, Berlin, Germany. [Google Scholar]
- 3.Erdmann A, Weil B, Krämer R. 1993. Lysine secretion by wild-type Corynebacterium glutamicum triggered by dipeptide uptake. J Gen Microbiol 139:3115–3122. doi: 10.1099/00221287-139-12-3115. [DOI] [Google Scholar]
- 4.Koita K, Rao CV. 2012. Identification and analysis of the putative pentose sugar efflux transporters in Escherichia coli. PLoS One 7:e43700. doi: 10.1371/journal.pone.0043700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bröer S, Krämer R. 1991. Lysine excretion by Corynebacterium glutamicum. 1 Identification of a specific secretion carrier system. Eur J Biochem 202:131–135. [DOI] [PubMed] [Google Scholar]
- 6.Vrljic M, Sahm H, Eggeling L. 1996. A new type of transporter with a new type of cellular function: l-lysine export from Corynebacterium glutamicum. Mol Microbiol 22:815–826. doi: 10.1046/j.1365-2958.1996.01527.x. [DOI] [PubMed] [Google Scholar]
- 7.Bellmann A, Vrljic M, Patek M, Sahm H, Krämer R, Eggeling L. 2001. Expression control and specificity of the basic amino acid exporter LysE of Corynebacterium glutamicum. Microbiology 147:1765–1774. doi: 10.1099/00221287-147-7-1765. [DOI] [PubMed] [Google Scholar]
- 8.Bröer S, Krämer R. 1991. Lysine excretion by Corynebacterium glutamicum. 2. Energetics and mechanism of the transport system. Eur J Biochem 202:137–143. [DOI] [PubMed] [Google Scholar]
- 9.Nandineni MR, Gowrishankar J. 2004. Evidence for an arginine exporter encoded by yggA (argO) that is regulated by the LysR-type transcriptional regulator ArgP in Escherichia coli. J Bacteriol 186:3539–3546. doi: 10.1128/JB.186.11.3539-3546.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pathania A, Sardesai AA. 2015. Distinct paths for basic amino acid export in Escherichia coli: YbjE (LysO) mediates export of l-lysine. J Bacteriol 197:2036–2047. doi: 10.1128/JB.02505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueda T, Nakai Y, Gunji Y, Takikawa R, Joe Y. September 2009. l-Amino acid-producing microorganism and method for producing l-amino acid. US patent EP 1,664,318 B1.
- 12.Ueda T, Nakai Y, Gunji Y, Takikawa R, Joe Y. December 2009. l-Amino acid-producing microorganism and method for producing l-amino acid. US patent US 7,629,142 B2.
- 13.Vrljic M, Garg J, Bellmann A, Wachi S, Freudl R, Malecki MJ, Sahm H, Kozina VJ, Eggeling L, Saier MH Jr. 1999. The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradyme [sic] for a novel superfamily of transmembrane solute translocators. J Mol Microbiol Biotechnol 1:327–336. [PubMed] [Google Scholar]
- 14.Tsu BV, Saier MH Jr. 2015. The LysE superfamily of transport proteins involved in cell physiology and pathogenesis. PLoS One 10:e0137184. doi: 10.1371/journal.pone.0137184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 16.Beja O, Bibi E, Kaback HR. 1996. Functional expression of mouse Mdrl in an outer membrane permeability mutant of Escherichia coli. Proc Natl Acad Sci U S A 93:5969–5974. doi: 10.1073/pnas.93.12.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harel YM, Bailone A, Bibi E. 1999. Resistance to bacitracin as modulated by an Escherichia coli homologue of the bacitracin ABC transporter BcrC subunit from Bacillus licheniformis. J Bacteriol 181:6176–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandraprakash D, Seshasayee AS. 2014. Inhibition of factor-dependent transcription termination in Escherichia coli might relieve xenogene silencing by abrogating H-NS-DNA interactions in vivo. J Biosci 39:53–61. doi: 10.1007/s12038-014-9413-4. [DOI] [PubMed] [Google Scholar]
- 19.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006–0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amann E, Ochs B, Abel KJ. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 21.Vieira J, Messing J. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189–194. doi: 10.1016/0378-1119(91)90365-I. [DOI] [PubMed] [Google Scholar]
- 22.Lerner CG, Inouye M. 1990. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res 18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 24.Zhou Y, Zhang X, Ebright RH. 1991. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res 19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eckert KA, Kunkel TA. 1990. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res 18:3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cadwell RC, Joyce GF. 1994. Mutagenic PCR. PCR Methods Appl 3:S136–140. doi: 10.1101/gr.3.6.S136. [DOI] [PubMed] [Google Scholar]
- 27.Chen JC, Minev M, Beckwith J. 2002. Analysis of ftsQ mutant alleles in Escherichia coli: complementation, septal localization, and recruitment of downstream cell division proteins. J Bacteriol 184:695–705. doi: 10.1128/JB.184.3.695-705.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manoil C, Beckwith J. 1985. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A 82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brickman E, Beckwith J. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol 96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 30.Derman AI, Beckwith J. 1995. Escherichia coli alkaline phosphatase localized to the cytoplasm slowly acquires enzymatic activity in cells whose growth has been suspended: a caution for gene fusion studies. J Bacteriol 177:3764–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wessel D, Flügge UI. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 32.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann K, Stoffel W. 1993. TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler 347:166. [Google Scholar]
- 34.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu S, Zhang Y. 2007. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramachandran GN, Ramakrishnan C, Sasisekharan V. 1963. Stereochemistry of polypeptide chain configurations. J Mol Biol 7:95–99. doi: 10.1016/S0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 39.Suhre K, Sanejouand Y-H. 2004. ElNémo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res 32:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch S, Fritsch MJ, Buchanan G, Palmer T. 2012. Escherichia coli TatA and TatB proteins have N-out, C-in topology in intact cells. J Biol Chem 287:14420–14431. doi: 10.1074/jbc.M112.354555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manoil C, Beckwith J. 1986. A genetic approach to analyzing membrane protein topology. Science 233:1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- 42.Boyd D, Traxler B, Beckwith J. 1993. Analysis of the topology of a membrane protein by using a minimum number of alkaline phosphatase fusions. J Bacteriol 175:553–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Heijne G. 1992. Membrane protein structure prediction, hydrophobicity analysis and the positive inside rule. J Mol Biol 225:487–494. doi: 10.1016/0022-2836(92)90934-C. [DOI] [PubMed] [Google Scholar]
- 44.Derman AI, Puziss JW, Bassford PJ Jr, Beckwith J. 1993. A signal sequence is not required for protein export in prlA mutants of Escherichia coli. EMBO J 12:879–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ulmschneider MB, Sansom MS. 2001. Amino acid distributions in integral membrane protein structures. Biochim Biophys Acta 1512:1–14. doi: 10.1016/S0005-2736(01)00299-1. [DOI] [PubMed] [Google Scholar]
- 46.von Heijne G. 2006. Membrane-protein topology. Nat Rev Mol Cell Biol 7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 47.Kennan RM, McMurry LM, Levy SB, Rood JI. 1997. Glutamate residues located within putative transmembrane helices are essential for TetA(P)-mediated tetracycline efflux. J Bacteriol 179:7011–7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Curran AR, Engelman DM. 2003. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol 13:412–417. doi: 10.1016/S0959-440X(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 49.Senes A, Engel DE, DeGrado WF. 2004. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol 14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Blount P, Sukharev SI, Moe PC, Schroeder MJ, Guy HR, Kung C. 1996. Membrane topology and multimeric structure of a mechanosensitive channel protein of Escherichia coli. EMBO J 15:4798–4805. [PMC free article] [PubMed] [Google Scholar]
- 51.Yerushalmi H, Lebendiker M, Schuldiner S. 1996. Negative dominance studies demonstrate the oligomeric structure of EmrE, a multidrug antiporter from Escherichia coli. J Biol Chem 271:31044–31048. doi: 10.1074/jbc.271.49.31044. [DOI] [PubMed] [Google Scholar]
- 52.Papaloukas C, Granseth E, Viklund H, Elofsson A. 2008. Estimating the length of transmembrane helices using Z-coordinate predictions. Protein Sci 17:271–278. doi: 10.1110/ps.073036108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shelar A, Bansal M. 2014. Sequence and conformational preferences at termini of α-helices in membrane proteins: role of the helix environment. Proteins 82:3420–3436. doi: 10.1002/prot.24696. [DOI] [PubMed] [Google Scholar]
- 54.Daley DO, Rapp M, Granseth E, Melen K, Drew D, von Heijne G. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321–1323. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 55.Heijne G. 1986. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the transmembrane topology. EMBO J 5:3021–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.