Nucleosomes present a strong, sequence-dependent barrier for moving replisomes and can survive during DNA replication in vitro.
Keywords: Replication, chromatin, nucleosome, mechanism
Abstract
Efficient overcoming and accurate maintenance of chromatin structure and associated histone marks during DNA replication are essential for normal functioning of the daughter cells. However, the molecular mechanisms of replication through chromatin are unknown. We have studied traversal of uniquely positioned mononucleosomes by T7 replisome in vitro. Nucleosomes present a strong, sequence-dependent barrier for replication, with particularly strong pausing of DNA polymerase at the +(31–40) and +(41–65) regions of the nucleosomal DNA. The exonuclease activity of T7 DNA polymerase increases the overall rate of progression of the replisome through a nucleosome, likely by resolving nonproductive complexes. The presence of nucleosome-free DNA upstream of the replication fork facilitates the progression of DNA polymerase through the nucleosome. After replication, at least 50% of the nucleosomes assume an alternative conformation, maintaining their original positions on the DNA. Our data suggest a previously unpublished mechanism for nucleosome maintenance during replication, likely involving transient formation of an intranucleosomal DNA loop.
INTRODUCTION
The position and integrity of nucleosomes in the genome are important for normal cellular functions, cell differentiation, aging, and cancer development (1). The processive enzymes involved in DNA replication and transcription often alter the nucleosome positioning and chromatin structure (2–7), raising a question about the mechanisms that guarantee the maintenance of genomic patterns of histone modifications during these processes. It has been observed that nucleosomes are assembled immediately behind the replication fork (8). Parental nucleosomes are preserved and randomly distributed to both daughter DNA helices after replication of the SV40 viral chromatin (9, 10). A recent study in budding yeast showed that most of the parental histones after replication are localized within 400 base pairs (bp) from their original locations (11). Parental histones are detected in both daughter cells and randomly segregated to both arms of the fork in bulk genome (12–14). The majority of parental (H3-H4)2 tetramers are segregated together to the same daughter DNA, whereas a fraction of tetramers containing histone variants split during replication (15, 16). Thus, most of the parental histones are distributed to both daughter cells and remain near their original positions on the DNA. However, studying the detailed mechanism of histone survival on DNA during replication is extremely difficult because of the absence of a highly purified and efficient experimental system that faithfully recapitulates chromatin replication in vitro.
In the only study using a highly purified bacteriophage T4 replisome system and a plasmid template that has randomly positioned nucleosomes, it has been shown that nucleosomes slow down the replisome, survive replication, and most often segregate to the leading strand (17). However, the lack of precise nucleosome positioning and the presence of an excess of histone-free DNA during replication allowed only a limited analysis of the mechanism of chromatin replication.
Here, we have established a highly purified, tractable in vitro system to study the detailed mechanism of replication through a single positioned nucleosome by the T7 replisome (18). This system recapitulates the ability of nucleosomes to survive DNA replication and remain bound near the original DNA regions observed in vivo. Analysis of the strong nucleosomal barrier to DNA replication using quantitative, time-resolved approaches revealed a previously unpublished mechanism of replication through chromatin that involves nucleosome survival and a new role for the exonuclease activity in overcoming the nucleosomal barrier.
RESULTS
Development of an experimental approach for analysis of replication through chromatin in vitro
The goal of this work is to investigate the detailed mechanism of T7 polymerase–helicase traversal through a nucleosome. The efficiency of an available purified eukaryotic replication system is not sufficient for direct analysis of the effect of replication on a chromatin structure [only ~10% of the templates are replicated by the most efficient purified eukaryotic (yeast) replisome in vitro] (19). Therefore, one of the best-characterized prokaryotic replication systems (T7 replisome) progressing through a single, precisely positioned nucleosome in vitro was used. There is substantial conservation of the replisome components, including helicase, primase, and polymerase, between prokaryotes and eukaryotes. One important difference between the eukaryotic and prokaryotic replisome is in the helicase. Replicative eukaryotic helicases (MCM2-7 ring) encircle the leading strand, and replicative prokaryotic helicases encircle the lagging strand. However, it was suggested that active CMG (cdc45, Mcm2-7, and GINS) may also encircle the lagging strand (20, 21). Thus, the forefront of the replication fork structure appears to be conserved (22, 23). On the other hand, because our system is prokaryotic and completely defined, it reveals chromatin properties that are inherent to nucleosomes themselves.
It is relatively easy to set up a highly active T7 replisome system from purified proteins in comparison to other replisomes (24), and it has many advantages: (i) an intact, functional replisome is efficiently formed using a minimal set of proteins; (ii) the proteins are well characterized biochemically, and the high-resolution structures are available (25–27); (iii) the T7 replisome is fast, processive, faithful, and stable, thus recapitulating many properties of eukaryotic replication; and (iv) detailed ensemble and single-molecule studies of DNA replication were conducted in the past (18, 24–28).
The T7 replisome is composed of a DNA polymerase T7 gp5 (DNAP), a processivity factor [Escherichia coli thioredoxin (trx)], and a helicase/primase (T7 gp4). T7 gp5 is an 80-kDa protein containing a 5′-3′ polymerization activity and a 3′-5′ proofreading exonuclease activity, similar to eukaryotic polymerases. T7 gp5 (DNAP) interacts with E. coli trx, which increases the processivity of DNA synthesis more than 100-fold (18). The exonuclease activity efficiently removes misincorporated bases and increases the fidelity of DNA synthesis during replication. Disruption of the exonuclease activity of T7 DNAP (T7 DNAP/exo−) does not affect its polymerization activity on DNA and the stability of the replication complex (29). The helicase T7 gp4 contains primase and helicase activities. It forms a hexamer complex and moves in a 5′-3′ direction on the lagging DNA strand, helping to melt the double-stranded DNA (dsDNA) template in a 3′-deoxythymidine 5′-triphosphate hydrolysis–dependent reaction (30, 31). Helicase T7 gp4 hexamer can form a stable and active complex with T7 DNAP that is involved in the initiation and elongation of replication (32).
Precisely positioned mononucleosomes have been extensively used to study the mechanisms of various processes involving processive enzymes, such as mechanisms of transcription through chromatin (33–35). Nucleosomes formed on strong positioning and natural sequences behave similarly in different transcriptional assays (36, 37). This system has several advantages: (i) the nucleosome is located at a unique, known position of the DNA template and is structurally homogeneous; (ii) the high-resolution structure of a positioned nucleosome was solved (38); (iii) many methods are available for the analysis of the process and outcome of replication through the nucleosome, including time-resolved, structural, and single-molecule techniques (24, 32); and (iv) the nucleosome composed of modified histones, different histone variants, or subnucleosomes can be easily assembled and studied.
To obtain a DNA template that supports mononucleosome assembly and replication by the T7 replisome, we ligated a synthetic preformed DNA replication fork to a long duplex DNA template or to a positioned nucleosome (Fig. 1A). Next, a 5′-end, 32P–labeled 24-mer DNA primer is annealed to the fork. The replisome is then assembled by the addition of T7 DNAP and T7 helicase in the replication buffer containing deoxynucleotide triphosphates (dNTPs). The reaction is activated by the addition of magnesium ions and then quenched by chelating EDTA after different time intervals of DNA synthesis. The 5′-end–labeled DNA products synthesized from the leading strand replication are analyzed by denaturing or native polyacrylamide gel electrophoresis (PAGE).
Fig. 1. Nucleosomes cause strong pausing of the T7 replisome.
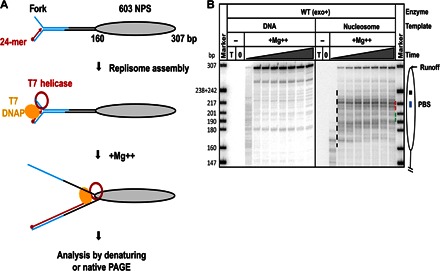
(A) Experimental approach for analysis of replication through chromatin. Each template contains the fork DNA structure, linker DNA, and the strong 603 NPS. The fork DNA structure (in blue) is ligated to the nucleosomal template (gray oval). After ligation, the P32-end–labeled 24-mer DNA primer (red arrow) is annealed to the template DNA strand. Next, DNAP, which was preassembled with E. coli trx and T7 helicase (red ring), is added to the reaction and forms a replisome in the presence of all dNTPs. The reaction is then activated by the addition of Mg++. Replication products are analyzed by denaturing or native PAGE. (B) Analysis of labeled products after replication of 603 DNA (histone-free or organized in a nucleosome) for different time intervals (0, 2, 5, 10, 30, 60, 120, 240, and 480 s) by denaturing PAGE. The locations of the nucleosome (oval), the nucleosome dyad (square), the PBS (blue line), and the runoff transcript and nucleosome-specific pausing (black dashed line) are shown. T, DNA template only; 0, reaction before addition of Mg++. Note that the nucleosomal pausing regions +(31–40) and +(41–65) are indicated by green and red dashed lines, respectively. Markers are pBR322 DNA–Msp I digest marker (New England Biolabs). The sizes of marker DNA fragments are indicated on the left side.
A replisome encounters a strong, DNA sequence–dependent nucleosomal barrier
The replisome efficiently and processively replicated the histone-free 307-bp replication fork–containing DNA: the reaction was completed on 65 to 75% of starting templates within 5 s (Fig. 1B). The fraction of templates completed was calculated as a fraction of the total signal present in the lane and corresponding to the replication of nucleosomal DNA. This high efficiency of replication can be achieved only by the intact replisome complex (RC) because DNAP without helicase failed to reach the nucleosome positioning sequence (NPS) region of the DNA template with or without the nucleosome (fig. S1). The nucleosome assembled on the 603 NPS forms a strong barrier to the replisome: the reaction was nearly completed after 4 min and only on 22% of the templates (Fig. 1B). Strong nucleosome-specific pausing was detected at multiple positions in the nucleosome, especially at the +(41–65) DNA region (41 to 65 bp from the nucleosome boundary; Fig. 1B), where the replisome was arrested on ~30% of the templates. Only minimal replisome pausing was detected after the +59 region, indicating that histones were either displaced from or translocated along the DNA after the replisome proceeded past this region.
A similar nucleosomal pausing has been observed during transcription through the nucleosome by various RNA polymerases, where two mechanisms of progression through chromatin have been described [Pol II– and Pol III–type, respectively (6, 39)]. The Pol II–type mechanism of transcription through chromatin is characterized by a high nucleosomal barrier to transcription and by displacement of a single histone H2A/H2B dimer (36, 37). The remaining subnucleosome (DNA-bound histone hexamer) remains at the original position on the DNA. A considerably different, Pol III–type mechanism involves transfer of a complete histone octamer from in front of the transcribing enzyme to behind it (40, 41). Because of the difference in the mechanisms of progression through chromatin, nucleosomes formed on different NPSs, such as 601 and 603 NPSs with different locations (Fig. 1B and fig. S2) of the polar barrier sequences (PBS), which dictate the high affinity of nucleosomal DNA to core histones (33), are characterized by different pausing patterns, characteristic of the Pol II– and Pol III–type mechanisms, respectively (35, 36). In particular, 601 nucleosomes with the PBS sequence in promoter-distal location present a high barrier only to Pol II (35, 36).
To determine the type of the nucleosome pausing mechanism used during DNA replication, we replicated the nucleosomes formed on the 601 NPS (fig. S2). The replisome progressed on the 601 histone-free DNA as efficiently as on the 603 template (fig. S2). However, the +59 nucleosomal pausing was considerably stronger on the 601 nucleosome than on the 603 nucleosome, resulting in only less than 5% of the replisomes overcoming the 601 nucleosomal barrier (fig. S2). A similar discrimination between the 601 and 603 nucleosomes was observed in the case of the Pol II–type but not the Pol III–type mechanism of transcription (36). Thus, the data suggest that a mechanism similar to the Pol II–type mechanism of transcription through chromatin is used during DNA replication.
In summary, nucleosomes present a strong, DNA sequence–dependent barrier for moving the replisome. The sequence dependence of the nucleosomal barrier indicates that the replisome and RNA polymerase II use similar mechanisms of progression through chromatin. Because the Pol II–type mechanism is characterized by nucleosome survival at the original position on the DNA, a similar mechanism likely operates during progression of the replisome.
Exonuclease activity of DNAP facilitates the overcoming of the nucleosomal barrier to replication
In eukaryotic cells, both the leading strand and lagging strand replicative DNA polymerases (Pol δ and Pol ε) have exonuclease activities that affect replication fidelity; the corresponding loss-of-function mutations are involved in carcinogenesis (42). T7 DNAP also contains the 3′-5′ exonuclease domain localized at the N terminus of the protein (25, 29). Mutant polymerase, exo− T7 DNAP does not have exonuclease activity because of D5A and E7A mutations (29). The mutant (exo−) and wild-type (exo+) DNA polymerases replicate DNA at similar rates and have similar activities in the replisome assembly and initiation of replication (29). How this exonuclease activity affects DNA replication through chromatin is unknown.
As expected, the rates and processivities of 603 DNA replication by the exo+ and exo− T7 replisome are comparable (compare the data in Figs. 1B and 2A). However, exonuclease activity significantly affects the rate and efficiency of replication through the nucleosome (Fig. 2A). The yields of full-length products were 12 and 22% after replication by the exo− and exo+ T7 replisome, respectively, suggesting that exonuclease activity facilitates replication through chromatin.
Fig. 2. Exonuclease activity increases the efficiency of replication through the nucleosome.
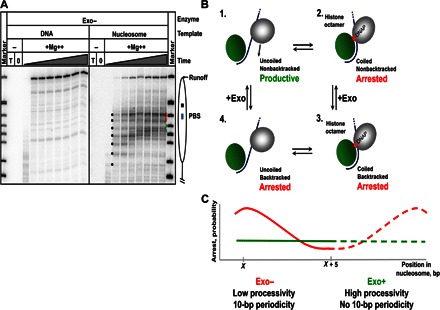
(A) Analysis of labeled products after replication of 603 DNA or nucleosomal templates by the exo− T7 replisome for different time intervals (0, 2, 5, 10, 30, 60, 120, 240, and 480 s) by denaturing PAGE. Note the ~10-bp periodic pausing pattern (indicated by black dots; also see fig. S3) detected after replication for short time periods that is not present after replication by the exo+ T7 replisome (Fig. 1B). (B) Proposed mechanism explaining the 10-bp periodic nucleosomal pausing patterns. It is proposed that discrete 10-bp DNA regions of nucleosomal DNA are uncoiled from the octamer stepwise, after the T7 replisome encounters DNA-histone interactions. As the T7 replisome proceeds along uncoiled DNA (complex 1), it arrests after encountering DNA-histone interactions (complex 2) and possible backtracking (complex 3). When the replisome is backtracked, the DNA may also recoil to bind back to the octamer, which leads to peeling of the primer end from the template, resulting in a more stable arrest. To proceed further, DNA has to be uncoiled from the octamer (complex 4) and the replisome has to recover from the backtracked state. The recovery is likely facilitated by the exonuclease activity that can excise the peeled primer end and regenerate the functionally active, fully annealed primer template. The exo− enzyme must wait for DNA uncoiling to move forward, resulting in a lower efficiency and 10-bp periodicity of replication through chromatin. Nucleosomal DNA and histone octamer are shown in blue and green. DNA polymerase is in gray. The arrow indicates the direction of replisome progression. (C) The expected efficiency of arrest of exo− and exo+ replisomes (red and green lines, respectively) in a nucleosome.
The nucleosomal pausing pattern also strongly depends on the presence of exonuclease activity of T7 DNAP (compare Figs. 1B and 2A). After replication for a short time (less than 30 s), the pausing pattern has 10-bp periodicity during replication by the exo−, but not the exo+, replisome. At the later time points, the pausing patterns are also different (see Fig. 3A and fig. S3 for the side-by-side comparison). Because spontaneous DNA uncoiling likely occurs in 10-bp intervals (Fig. 2B), 10-bp periodicity is expected to be more pronounced for the polymerases that are more easily stalled or arrested. Such stalled or arrested states can be produced from nucleosome recoiling and from relocating the newly synthesized primer end from the template, which would prevent the next nucleotide binding and forward motion (Fig. 2B). We propose the idea that the 10-bp periodic pattern is related to the production of these states, which also decreases the yield of runoff products (see Discussion and Fig. 2C). The pausing patterns characteristic of later time points during replication through the nucleosome are more similar between the exo+ and exo− replisomes, suggesting that here, the enzymes encounter very strong nucleosomal barriers where exonuclease activity is less helpful. In summary, the exonuclease activity of T7 DNAP suppresses the formation of the 10-bp periodic pattern, facilitating progression of the replisome through chromatin.
Fig. 3. Kinetic analysis of replication through a nucleosome by exo+ and exo− replisomes.
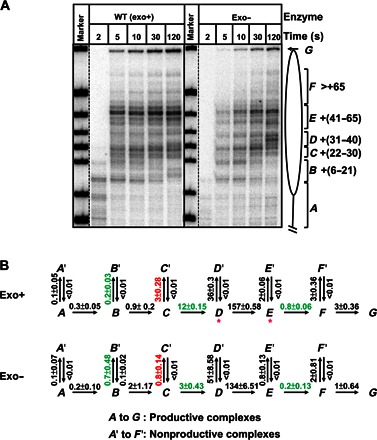
(A) 603 nucleosomes were replicated by exo+ and exo− replisomes for indicated time intervals. End-labeled DNA was analyzed by denaturing PAGE. The intranucleosomal pauses and runoff (from A to G) were quantified using a Phosphorimager and the OptiQuant software. (B) The quantified data were analyzed using an elongation model that produces a good fit of the experimental data to the calculated curves (fig. S4). The fitting curves (fig. S4) and kinetic parameters were obtained using the KinTek Explorer software. All rate constants were averages from three independent experiments. Rate constants that are more than threefold different between the two forms of DNAPs are marked by green and red colors (for positive and negative effects on processivity of the exo+ replisome, respectively). Note that the exo+ replisome has a higher overall rate of replication through the nucleosome than exo−; the rates of replication through the +(31–40) and +(41–65) regions have the largest differences.
To evaluate the effect of exonuclease activity on different steps during chromatin replication, we analyzed the time courses of replication through the nucleosome by exo+ and exo− replisomes (Fig. 3A). The paused DNA intermediates were quantified, grouped into species from A to F, plotted against time, and fitted to a sequential multistep model (Fig. 3B), which was previously developed for transcribed chromatin (43), where A is the starting species and G is the runoff species, using the KinTek Explorer software (44). A good fit of the entire set to the sequential model (fig. S4) was obtained only when we included nonproductive complexes from each intermediate (A′ to F′; Fig. 3B) and reversible steps for the formation of nonproductive complexes at each step during elongation. Kinetic parameters for the major steps during nucleosome replication by the two different enzymes were obtained and compared (Fig. 3B). For both exo− and exo+ RCs, the main parts of the nucleosomal barrier to replication [pausing at the +(31–40) and +(41–65) regions] are dictated by the rates of transition from the productive D complex to the nonproductive complex D′ and by the rate of transition between the E and F complexes, respectively.
Consistent with the overall higher efficiency of replication, the exo+ enzyme has a higher forward rate than the exo− replisome during several steps of replication through the nucleosome (Fig. 3B). This forward rate is a net rate that includes several nucleotide incorporation steps and all corresponding pauses. In particular, the data suggest that the exonuclease activity is critical for replication through the +(31–40) and +(41–65) regions of the nucleosome. First, the net forward rate of the C-to-D transition is about fourfold higher in the case of the exo+ replisome. The lower rate of D-to-D′ and the higher rate of D-to-E transitions for the exo+ enzyme are also observed in this region, consistent with less efficient pausing of the exo+ replisome in the +(31–40) region (region D). Second, the net rate of the D-to-E transition is about four orders of magnitude higher than the rate of the E-to-F step in both reactions, suggesting that the critical step is replication through the +(41–65) region (region E) of the nucleosome. The exo+ enzyme progresses through the +(41–65) region with a net rate about fourfold faster than that of the exo− replisome. Third, the rate of the B-to-B′ transition is about threefold lower in the exo+ reaction than in the exo− reaction, indicating that the inactive complexes at this region are formed less efficiently by the exo+ replisome.
In summary, the analysis of the kinetics (Fig. 3B) is consistent with our hypothesis (Fig. 2B), stating that the main effect of exonuclease activity is to promote efficient bypass through certain positions in the nucleosome. In the absence of this activity, the replisome is more likely to be arrested in the nucleosome. The critical intermediates (nonproductive and productive complexes) that dictate the +(31–40) and +(41–65) nucleosomal barriers to replication through a nucleosome by the T7 replisome have been identified.
Nucleosomes survive at the original positions on the DNA after replication
To determine the nucleosome fate after replication, we compared the expected and experimentally observed products of the reaction (Fig. 4). Most of the histone-free DNA templates were replicated to completion after 5 s (Fig. 1B), resulting in the accumulation of end-labeled full-length, double-stranded 603 DNA. Three main complexes were obtained after replication through the nucleosome: arrested RCs with low, heterogeneous mobility in the gel, nucleosomes, and dsDNA (Fig. 4, A and B). Nucleosomes and dsDNA were identified by comparing their mobility in the gel with the mobility of the corresponding purified species. The identity of nucleosomes was further verified by a restriction enzyme sensitivity assay (see below). Nucleosomes corresponded to ~45 and ~65% of the replication products in the case of the exo+ and exo− T7 replisome, respectively (Fig. 4B). This ratio is close to 50%, suggesting that ~50% of the histones segregated to the second DNA strand (unlabeled and invisible in the assay) or dissociated into solution. Mobility of nucleosomes in the native gel depends on their positioning within a given DNA fragment (45, 46). Because mobility of nucleosomes after replication and of end-positioned nucleosomes assembled on the dsDNA is similar (Fig. 4B), nucleosomes either remain at the original locations or are translocated to the other end of the DNA fragment. To discriminate between these possibilities, we incubated the nucleosomes in the presence of different restriction enzymes that have single sites at different positions on the DNA fragment (Fig. 5A) either before or after gel purification (from the Nu band in Fig. 4B). Replicated nucleosomes and dsDNA were completely digested by Bss SI or Msl I enzymes, which have single sites beyond the 603 NPS. In contrast, nucleosomal templates were not sensitive to Cac8 I or Cla I, although histone-free DNA was readily digested by the enzymes (Fig. 5B). The data suggest that the Cac8 I or Cla I site (localized within the 603 NPS) is protected by the positioned nucleosomes, and this nucleosome positioning is minimally affected by DNA replication. Similar results were obtained after replication with exo− T7 replisome enzymes without gel purification of nucleosomes (fig. S5). However, nucleosomes formed after replication with the exo+ replisome were sensitive to all restriction enzymes, suggesting that they have an alternative conformation, which relaxes into the canonical structure after gel purification. Because mobility of nucleosomes after replication in the native gel is somewhat heterogeneous, a possibility of nucleosome translocation over a short distance (up to 30 bp) cannot be excluded.
Fig. 4. Nucleosomes survive after replication by the T7 replisome.
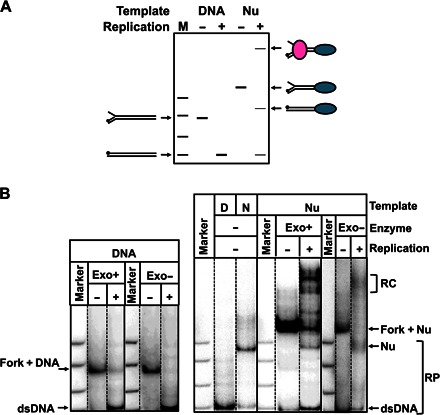
(A) The diagram shows mobility of the substrates and the products of replication in native gel. The nucleosome is shown as blue, and DNAP are shown as pink ovals. The labeled DNA end is indicated by a black circle. M, pBR322–Msp I digest. (B) Analysis of labeled templates after replication by the exo+/− T7 replisome of the 603 DNA or nucleosome for 240 s by native PAGE. Some RCs are stalled in the nucleosome. The nucleosomes (N) assembled on the 307-bp dsDNA (D) are the expected replication products (RP). Marker is pBR322–Msp I digest. Nucleosomes correspond to ~45 and ~65% of the replication product (average of three experiments) in the case of the exo+ and exo− T7 replisome, respectively.
Fig. 5. Nucleosomes remain at the original position on the DNA after replication by the exo+ T7 replisome.
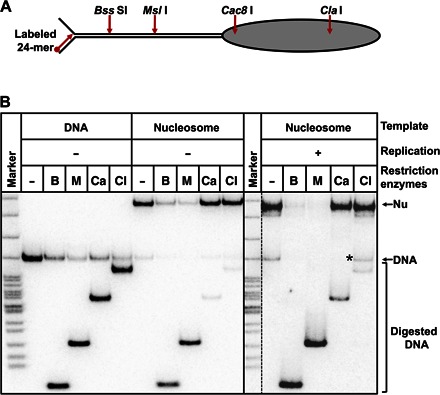
(A) Positions of sites for restriction enzymes on the nucleosomal template. (B) Analysis of nucleosome fate using restriction enzyme sensitivity assay (B, Bss SI; M, Msl I; Ca, Cac8 I; Cl: Cla I). The PAGE-purified nucleosomes after replication, 307-bp dsDNA, and the nucleosomes assembled on the 307-bp dsDNA were incubated in the presence of an excess of indicated restriction enzymes and analyzed by native PAGE. DNA fragment resistant to digestion by Cla I (likely due to dissociation of nucleosomal DNA, resistant to the enzyme, during the electrophoresis) is indicated by asterisks.
To evaluate a possibility that histones are transiently displaced and rebound at the original positions on DNA after replication, we added a mixture of histone H2A/H2B dimers and H3/H4 tetramers to the DNA before replication. Analysis of the replication products by native PAGE revealed that no nucleosomes were formed after DNA replication (fig. S6). This result suggests that histones cannot be completely displaced from the DNA during replication under our experimental conditions.
In summary, the data suggest that at least 50% of the histone octamer complexes segregate as single units to the dsDNA formed after replication, and a considerable fraction of the remaining histones is associated with the single-stranded DNA. Histones never completely leave DNA during replication, further supporting the proposal about the formation of an intermediate containing an intranucleosomal DNA loop during replication (see Discussion). Nucleosomes that survive replication remain at nearly original positions on the DNA, although they assume an alternative, less stable conformation after replication by the exo+ replisome.
The length of DNA upstream of the nucleosome modulates the efficiency of replication
A 600 to 1000–bp, partially nucleosome-depleted DNA region upstream of the replisome was observed in vivo (10). To investigate the possible role of the nucleosome-free DNA in the replication through chromatin, we performed replication using templates that have linker DNA between the fork and the 603 nucleosome of different lengths—136, 279, or 479 bp (Fig. 6A). The efficiency of overcoming the nucleosomal barrier is increased nearly twofold as the length of the linker DNA is increased to 479 bp, reaching ~50% of overall efficiency of DNA replication (Fig. 6B). A comparable (~2-fold) increase in the efficiency of replication through the nucleosome was detected using the exo− enzyme (Fig. 6C).
Fig. 6. The length of the spacer DNA dictates the efficiency of replication through the nucleosome.
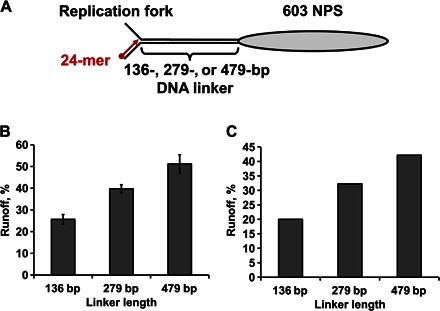
(A) Design of the templates containing linker DNA between the replication fork and the nucleosome of different lengths. (B and C) Efficiency of replication of nucleosomal templates is directly proportional to the length of the linker DNA. The templates of different lengths were replicated by the exo+ (B) or exo− (C) T7 replisome for 8 min, as described in Fig. 1. Runoff products of replication by the T7 replisome of the nucleosome templates were quantified after separation by denaturing PAGE and normalized to the amount of runoff products detected after replication of the DNA templates. The fraction of all RCs capable of overcoming the nucleosomal barrier is shown. Average values from three (B) or two experiments (C) with SDs are shown.
How can the length of linker DNA affect progression of the replisome through chromatin? The presence of DNA could induce either displacement or translocation of nucleosomes. In a mechanistically similar experimental system (with mononucleosomes transcribed by Pol II), the presence of upstream DNA longer than 150 bp induced backward nucleosome translocation (47). The histone octamer surface that becomes partially exposed during replication through the nucleosome could be captured by DNA that is present in close proximity. The presence of longer, more flexible DNA behind the replisome resulted in an increase of the local DNA concentration in the vicinity of the exposed octamer surface, resulting in a more efficient nucleosome translocation. Thus, longer linker DNA functions as a more efficient competitor for the histone octamer during replication, increasing the efficiency of nucleosome translocation. The lower nucleosome barrier on longer templates was likely observed because of more efficient nucleosome translocation during the replication, clearing the path for the moving replisome.
In summary, the data indicate that the presence of extensive regions of histone-free DNA upstream of replicated chromatin results in more efficient overcoming of the nucleosomal barrier by the replisome, most likely due to more efficient nucleosome translocation during the replication.
DISCUSSION
We have established a new experimental system assembled from highly purified proteins and DNA-protein complexes, including structurally defined templates for analysis of the mechanism of DNA replication in chromatin (Fig. 1A). This experimental system recapitulates important properties of replicated chromatin observed in vivo—nucleosome survival and histone survival after the replication. With this system, it has been shown that nucleosomes present a strong, DNA sequence–dependent barrier for moving replisomes (Figs. 1 to 3). The sequence dependence of the nucleosomal barrier indicates that replisomes and RNA polymerase II use similar mechanisms of progression through chromatin, likely involving formation of a transient intranucleosomal DNA loop (36). Nucleosomes survive during replication at the original position on the dsDNA (Figs. 4 and 5). Exonuclease activity of T7 DNAP increases processivity and the overall rate of progression of the replisome through chromatin, most likely by resolving nonproductive DNA-protein states to functionally active states of the replisome (Figs. 2 and 3). The presence of histone-free DNA upstream of the replication fork facilitates replication through chromatin (Fig. 6), likely due to the more efficient nucleosome translocation during replication.
Comparison of the mechanism of replication through chromatin with other related, well-studied processes (for example, transcription through chromatin by Pol II– and Pol III–type mechanisms) suggests that chromatin replication is a unique process. The overall height and sequence dependence of the nucleosomal barrier to replication (Fig. 1B and fig. S2) as well as the fate of nucleosomes after replication (Figs. 4 and 5) are more similar to the Pol II–type mechanism (33, 37), suggesting that the general features of the Pol II–type mechanism are likely applicable in the case of chromatin replication. Transcription through chromatin using the Pol II–type mechanism involves the formation of an extremely small intranucleosomal DNA loop (Ø-loop) that contains transcribing Pol II, which mediates nucleosome survival (33). A similar intermediate likely mediates nucleosome survival during DNA replication. However, the size of the intranucleosomal DNA loop is likely to be larger by 10 to 30 bp to accommodate a different structure of the replisome. As a result, the original nucleosome positioning is likely changed during replication by 10 to 30 bp (see Results).
At the same time, Pol II transcription through a nucleosome shows strongly diminished pausing after position +50 (33, 36), which contrasts the pausing characteristics of the T7 replisome that shows extended pausing up to the +65 region of the nucleosomal DNA (Fig. 1B and fig. S2). The earlier release of Pol II–specific pausing likely occurs because of the displacement of the promoter-distal H2A/H2B dimer (6, 33), and the absence of this early release of pausing during replication could mean that the dimer remains associated with the nucleosome after replication, as was observed during transcription by SP6 and T7 RNA polymerases (34, 36, 41). In agreement with this proposal, no discrete bands corresponding to hexasomes [nucleosomes missing the H2A/H2B dimer (37)] were observed after replication (Fig. 4B). However, because of the heterogeneity of nucleosome mobility after replication, we cannot entirely exclude the possibility that some of the complexes are subnucleosomes missing one or both H2A/H2B dimer(s).
The different nucleosomal pausing patterns (Fig. 3A) and slightly different efficiencies of nucleosome survival (Fig. 4B) characteristic of exo+ or exo− RCs (45 and 65% of the templates, respectively) indicate that exonuclease activity could play an important role during replication through chromatin, helping to resolve the backtracked intermediates and supporting more efficient replication (Fig. 3B). Exo+ polymerases are more processive than exo− polymerases (48, 49). Together, the data suggest the following model for the mechanism of DNA replication in chromatin in vitro (fig. S7). When the replisome enters into the nucleosome, DNA is partially uncoiled from the octamer (fig. S7, complex 1). The T7 replisome pauses and backtracks at several positions within the nucleosome (fig. S7, complexes 2 and 2′). DNAP containing exonuclease activity can cut the peeled primer end and convert the backtracked state into a productive state (fig. S7, complex 2). The different efficiencies of progression of exo+ and exo− enzymes through the nucleosome result in the formation of slightly different intranucleosomal DNA loops (smaller or larger, complexes 3 and 3′, respectively; fig. S7) and likely dictate either nucleosome survival or backward nucleosome translocation after replication. Longer upstream DNA adjacent to the replisome dictates an increased probability of formation of the intermediate with a larger DNA loop (fig. S7, complex 3′) and therefore results in a more efficient nucleosome translocation after replication. Similarly, it has been shown that the size of the intranucleosomal DNA loop dictates the nucleosome fate during transcription by different RNA polymerases (34).
In agreement with the previous study using a highly purified T4 replisome (17), we also observed strong nucleosome barriers to the replisome and nucleosome survival during replication. Our defined system also allowed mapping and analysis of the nucleosomal pausing patterns and identification of the critical intermediates, allowing us to propose a detailed mechanism of T7 replisome traversal of a nucleosome (see below). In the previously described purified system, it was technically difficult to track positions of individual nucleosomes after replication (17). Our results suggest that in the absence of an intramolecular competitor DNA, a large fraction of nucleosomes (at least 50%) survives after replication at the original positions on DNA.
Overall, our data identify three pathways for the reassembly of chromatin during/after replication in vivo (Fig. 7), where nucleosome density behind the replication fork is transiently decreased over 600 to 1000 bp (10). Some nucleosomes will likely be translocated as a complete unit to the histone-free DNA, and some nucleosomes will survive intact at nearly original positions. As a result, nucleosomes and perhaps some subnucleosomes will be equally distributed between the two arms of the replication fork (Fig. 7, pathways 1 and 2), and at least some of them will remain at the locations that are close to the original nucleosome positions (Fig. 7, pathway 1). Histones are also assembled on the DNA de novo after replication (Fig. 7, pathway 3). Consistently, it has been shown that two types of nucleosome-like structures, possibly octamers and tetramers, are detected on the daughter DNA strand (10). In yeast, the parental nucleosomes after replication are localized close (within 400 bp) to their original positions (11). Moreover, parental (H3-H4)2 tetramers mostly equally segregate to the genomes of the daughter cells as single units (16). Parental histones are associated with nascent chromatin immediately after replication in the absence of protein synthesis in human cells (50). The histone chaperone FACT (facilitates chromatin transcription) complex is recruited to the replisome complex during replication and has a possible function in histone segregation (51, 52).
Fig. 7. Proposed model of chromatin reassembly during or after replication.
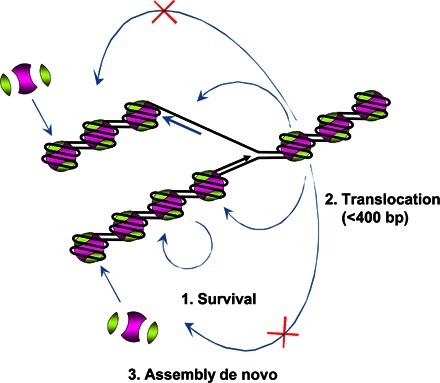
The original histones are nearly quantitatively recovered on leading strand (after survival and translocation) and lagging strand (after translocation). Three pathways of nucleosome reformation after replication are proposed: (i) original histone octamers survive at the original positions on DNA, (ii) original histone octamers are transferred within the 400-bp region upstream of the replisome, or (iii) de novo histone assembly after replication.
The spreading model has been proposed in the past to explain the inheritance of epigenetic markers located on histones (5, 53). Parental histones H3-H4 that carry most of the epigenetic and regulatory marks are semiconservatively distributed on the daughter DNA strands and serve as templates for enzymes (writers) to modify newly synthesized histones on the adjacent chromatin regions (54). For example, the PRC2 (polycomb repressive complex 2) could bind H3K27 trimethylated nucleosomes and catalyze H3K27me3 at adjacent nucleosomes. These modifications can be symmetrically or asymmetrically distributed in one nucleosome after replication (55, 56). Most of the canonical (H3-H4)2 tetramers segregate to daughter genomes as one unit (16). Our data are consistent with these observations and suggest that (H3-H4)2 tetramer survival occurs through the pathways 1 and 2 (Fig. 7). According to our model, nucleosomes having parental histones are assembled next to nucleosomes containing new histones; then, the “old” histones could serve as templates for writers of histone modifications that, in turn, will spread the histone modifications in cis after replication. Thus, our model is consistent with the spreading model for epigenetic inheritance and suggests a particular mechanism that mediates the inheritance of histone-associated epigenetic markers.
MATERIALS AND METHODS
Protein purification
Exo– T7 gp5 (D5A and E7A) and T7 helicase (T7 gp4A′) were purified as described (24, 29, 57). Trx was obtained from Sigma. T7 gp5 was purchased from New England Biolabs. Purification of -H1 chicken erythrocyte chromatin was conducted as previously described (45).
DNA templates, replication fork construction, and nucleosome reconstitution
Primers to assemble the fork structure template were modified from the study of Pandey et al. (24). The fork was obtained by annealing of two long overlapping oligonucleotides (90 and 100 bp) that contain a nonpairing fork structure. 603 and 601 NPS templates were prepared as described before (36, 58). In summary, NPS templates were amplified by polymerase chain reaction (PCR) and purified by gel electrophoresis using a gel extraction kit (Omega BioTek). Nucleosomes were reconstituted on the DNA templates as previously described (59). In summary, NPS templates were mixed with purified chicken erythrocyte H2A/H2B dimers and H3/H4 tetramers at a 1:1.8:1.2 molar ratio in the presence of salmon testes DNA (present in threefold weight excess over NPS templates) in the following buffer: 2 M NaCl, 10 mM tris-HCl (pH 7.4), 0.1% NP-40, and 0.2 mM EDTA (pH 8). The DNA/histone mixtures were then dialyzed against buffers containing 10 mM tris-HCl (pH 7.4), 0.1% NP-40, and 0.2 mM EDTA (pH 8), and progressively decreasing (2, 1.5, 1, 0.75, 0.5 M, and 10 mM) NaCl at 4°C, at each step for 2 hours.
The fork DNA and NPS templates (DNA or mononucleosomes) were ligated at the Tsp RI (New England Biolabs) restriction site by T4 DNA Ligase (Promega). After ligation, the 24-mer DNA primer was annealed to the 3′ terminus of the leading strand. The control 307-bp dsDNA fragment for restriction enzyme mapping and DNase I footprinting was PCR-amplified using the template (the fork ligated to 603 NPS) (58) and primers (24-mer DNA primer and reverse primer of 603 NPS). All DNA sequences and templates are described in table S1.
Replication assay and nucleosome fate
T7 DNAP was first incubated with trx (Sigma) at a 1:5 molar ratio to form a 1:1 complex in T7 replication buffer [50 mM tris-HCl (pH 7.5), 40 mM KCl, and 5 mM dithiothreitol (DTT)]. Trx-activated T7 DNAP (twofold molar excess to total DNA) and helicase (sixfold molar excess to DNAP as a hexamer) were sequentially incubated with templates (DNA or nucleosomes containing fork structure) in T7 replication buffer with 2 mM EDTA, 1 mM DTT, bovine serum albumin (BSA; 0.1 mg/ml), and 0.2 mM dNTPs at 4°C for 30 min. Replication reaction was started by the addition of 10 mM MgCl2 at 20°C and then stopped after different time periods (2 to 480 s) by the addition of EDTA to the final concentration of 300 mM. After the replication reaction, the DNA products were purified and then analyzed using either denaturing or native PAGE. For analysis of nucleosome fate, replication was conducted for 240 s. The data were quantified using the ImageQuant software.
PAGE purification of nucleosomes
The in vitro replication was performed as described above. After 4 min of replication, different complexes were separated using native PAGE (5.4%, 39:1 acrylamide/bis-PAGE, 0.5× tris-borate EDTA buffer) at 4°C. Gel regions containing nucleosomes were cut, fragmented, and incubated in equal volume of the buffer containing 10 mM Hepes-NaOH (pH 8.0), 0.2 mM EDTA-NaOH (pH 8.0), and BSA (0.2 mg/ml) for ~16 hours at 4°C. The samples were centrifuged at 3000g, 4°C for 3 min, and the supernatant was collected and concentrated on an Amicon Ultra-10K.
Restriction enzyme mapping
Nucleosome products were PAGE-purified after replication as described above. PAGE-purified replicated nucleosomes, control dsDNA, and end-positioned nucleosomes were incubated in the presence of an excess of Bss SI, Msl I, Cac8 I, or Cla I restriction enzyme (New England Biolabs) for 30 min at 20°C before or after gel purification. Labeled DNA templates were then analyzed by native PAGE.
Kinetic analysis and rate constants determination
Data from time-course experiments were computationally fitted to the kinetic model using the KinTek Explorer software (24, 43). All parameters were obtained after the simulation. Average rate constants and SDs were obtained from three experiments.
Supplementary Material
Acknowledgments
Funding: This work was supported by NIH RO1 grants to V.M.S. (GM58650) and to S.S.P. (GM55310) and by a Russian Science Foundation grant (RSF 14-24-00031). Author contributions: H.-W.C., O.I.K., S.S.P., and V.M.S. designed the experiments, and H.-W.C., M.P., and V.M.S. performed the experiments. H.-W.C., S.S.P., and V.M.S. wrote the manuscript. All authors discussed the results and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/11/e1601865/DC1
fig. S1. Helicase activity is essential for processive replication by the T7 replisome.
fig. S2. Strong nucleosomal barrier affects the processivity of the T7 replisome.
fig. S3. Analysis of the nucleosomal pausing patterns formed during replication by exo+ and exo− replisomes.
fig. S4. Analysis of time courses of replication through chromatin by the exo+ (A) or exo− replisome (B) using the KinTek Explorer software.
fig. S5. Mapping of nucleosome positions after replication by the T7 replisome using restriction enzyme sensitivity assay.
fig. S6. Nucleosomes are not formed de novo during or after T7 replication.
fig. S7. Proposed role for exonuclease activity during replication through a nucleosome.
table S1. Sequences of oligonucleotides and DNA templates.
REFERENCES AND NOTES
- 1.Struhl K., Segal E., Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 20, 267–273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annunziato A. T., The fork in the road: Histone partitioning during DNA replication. Genes 6, 353–371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das C., Tyler J. K., Histone exchange and histone modifications during transcription and aging. Biochim. Biophys. Acta 1819, 332–342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramachandran S., Henikoff S., Replicating nucleosomes. Sci. Adv. 1, e1500587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitehouse I., Smith D. J., Chromatin dynamics at the replication fork: There’s more to life than histones. Curr. Opin. Genet. Dev. 23, 140–146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulaeva O. I., Hsieh F.-K., Chang H.-W., Luse D. S., Studitsky V. M., Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim. Biophys. Acta 1829, 76–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smolle M., Workman J. L., Venkatesh S., reSETting chromatin during transcription elongation. Epigenetics 8, 10–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith D. J., Whitehouse I., Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 483, 434–438 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugasawa K., Ishimi Y., Eki T., Hurwitz J., Kikuchi A., Hanaoka F., Nonconservative segregation of parental nucleosomes during simian virus 40 chromosome replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 89, 1055–1059 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasser R., Koller T., Sogo J. M., The stability of nucleosomes at the replication fork. J. Mol. Biol. 258, 224–239 (1996). [DOI] [PubMed] [Google Scholar]
- 11.Radman-Livaja M., Verzijlbergen K. F., Weiner A., van Welsem T., Friedman N., Rando O. J., van Leeuwen F., Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLOS Biol. 9, e1001075 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Annunziato A. T., Split decision: What happens to nucleosomes during DNA replication? J. Biol. Chem. 280, 12065–12068 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Prior C. P., Cantor C. R., Johnson E. M., Allfrey V. G., Incorporation of exogenous pyrene-labeled histone into Physarum chromatin: A system for studying changes in nucleosomes assembled in vivo. Cell 20, 597–608 (1980). [DOI] [PubMed] [Google Scholar]
- 14.Jackson V., Chalkley R., Histone segregation on replicating chromatin. Biochemistry 24, 6930–6938 (1985). [DOI] [PubMed] [Google Scholar]
- 15.Jackson V., Deposition of newly synthesized histones: New histones H2A and H2B do not deposit in the same nucleosome with new histones H3 and H4. Biochemistry 26, 2315–2325 (1987). [DOI] [PubMed] [Google Scholar]
- 16.Xu M., Long C., Chen X., Huang C., Chen S., Zhu B., Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science 328, 94–98 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Bonne-Andrea C., Wong M. L., Alberts B. M., In vitro replication through nucleosomes without histone displacement. Nature 343, 719–726 (1990). [DOI] [PubMed] [Google Scholar]
- 18.Hamdan S. M., Richardson C. C., Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 78, 205–243 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Yeeles J. T. P., Deegan T. D., Janska A., Early A., Diffley J. F. X., Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519, 431–435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa A., Ilves I., Tamberg N., Petojevic T., Nogales E., Botchan M. R., Berger J. M., The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 18, 471–477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ilves I., Petojevic T., Pesavento J. J., Botchan M. R., Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 37, 247–258 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly T. J., SV40 DNA replication. J. Biol. Chem. 263, 17889–17892 (1988). [PubMed] [Google Scholar]
- 23.Kurth I., O’Donnell M., New insights into replisome fluidity during chromosome replication. Trends Biochem. Sci. 38, 195–203 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandey M., Syed S., Donmez I., Patel G., Ha T., Patel S. S., Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature 462, 940–943 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doublié S., Tabor S., Long A. M., Richardson C. C., Ellenberger T., Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391, 251–258 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Hollis T., Stattel J. M., Walther D. S., Richardson C. C., Ellenberger T., Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl. Acad. Sci. U.S.A. 98, 9557–9562 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singleton M. R., Sawaya M. R., Ellenberger T., Wigley D. B., Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell 101, 589–600 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Akabayov B., Akabayov S. R., Lee S.-J., Tabor S., Kulczyk A. W., Richardson C. C., Conformational dynamics of bacteriophage T7 DNA polymerase and its processivity factor, Escherichia coli thioredoxin. Proc. Natl. Acad. Sci. U.S.A. 107, 15033–15038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel S. S., Wong I., Johnson K. A., Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry 30, 511–525 (1991). [DOI] [PubMed] [Google Scholar]
- 30.Nandakumar D., Pandey M., Patel S. S., Cooperative base pair melting by helicase and polymerase positioned one nucleotide from each other. eLife 4, e06562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandey M., Patel S. S., Helicase and polymerase move together close to the fork junction and copy DNA in one-nucleotide steps. Cell Rep. 6, 1129–1138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel S. S., Pandey M., Nandakumar D., Dynamic coupling between the motors of DNA replication: Hexameric helicase, DNA polymerase, and primase. Curr. Opin. Chem. Biol. 15, 595–605 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulaeva O. I., Gaykalova D. A., Pestov N. A., Golovastov V. V., Vassylyev D. G., Artsimovitch I., Studitsky V. M., Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 16, 1272–1278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang H.-W., Shaytan A. K., Hsieh F.-K., Kulaeva O. I., Kirpichnikov M. P., Studitsky V. M., Structural analysis of the key intermediate formed during transcription through a nucleosome. Trends Cell Mol. Biol. 8, 13–23 (2013). [PMC free article] [PubMed] [Google Scholar]
- 35.Chang H.-W., Kulaeva O. I., Shaytan A. K., Kibanov M., Kuznedelov K., Severinov K. V., Kirpichnikov M. P., Clark D. J., Studitsky V. M., Analysis of the mechanism of nucleosome survival during transcription. Nucleic Acids Res. 42, 1619–1627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bondarenko V. A., Steele L. M., Újvári A., Gaykalova D. A., Kulaeva O. I., Polikanov Y. S., Luse D. S., Studitsky V. M., Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24, 469–479 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Kireeva M. L., Walter W., Tchernajenko V., Bondarenko V., Kashlev M., Studitsky V. M., Nucleosome remodeling induced by RNA polymerase II: Loss of the H2A/H2B dimer during transcription. Mol. Cell 9, 541–552 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Vasudevan D., Chua E. Y. D., Davey C. A., Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J. Mol. Biol. 403, 1–10 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Studitsky V. M., Walter W., Kireeva M., Kashlev M., Felsenfeld G., Chromatin remodeling by RNA polymerases. Trends Biochem. Sci. 29, 127–135 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Studitsky V. M., Kassavetis G. A., Geiduschek E. P., Felsenfeld G., Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science 278, 1960–1963 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Studitsky V. M., Clark D. J., Felsenfeld G., A histone octamer can step around a transcribing polymerase without leaving the template. Cell 76, 371–382 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Heitzer E., Tomlinson I., Replicative DNA polymerase mutations in cancer. Curr. Opin. Genet. Dev. 24, 107–113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsieh F.-K., Kulaeva O. I., Patel S. S., Dyer P. N., Luger K., Reinberg D., Studitsky V. M., Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 110, 7654–7659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson K. A., Simpson Z. B., Blom T., FitSpace explorer: An algorithm to evaluate multidimensional parameter space in fitting kinetic data. Anal. Biochem. 387, 30–41 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Walter W., Kireeva M. L., Tchernajenko V., Kashlev M., Studitsky V. M., Assay of the fate of the nucleosome during transcription by RNA polymerase II. Methods Enzymol. 371, 564–577 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Walter W., Studitsky V. M., Construction, analysis, and transcription of model nucleosomal templates. Methods 33, 18–24 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Kulaeva O. I., Studitsky V. M., Mechanism of histone survival during transcription by RNA polymerase II. Transcription 1, 85–88 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foury F., Vanderstraeten S., Yeast mitochondrial DNA mutators with deficient proofreading exonucleolytic activity. EMBO J. 11, 2717–2726 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Studwell P. S., O’Donnell M., Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J. Biol. Chem. 265, 1171–1178 (1990). [PubMed] [Google Scholar]
- 50.Alabert C., Bukowski-Wills J.-C., Lee S.-B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A., Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16, 281–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K., GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8, 358–366 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Foltman M., Evrin C., De Piccoli G., Jones R. C., Edmondson R. D., Katou Y., Nakato R., Shirahige K., Labib K., Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 3, 892–904 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Probst A. V., Dunleavy E., Almouzni G., Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10, 192–206 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Campos E. I., Stafford J. M., Reinberg D., Epigenetic inheritance: Histone bookmarks across generations. Trends Cell Biol. 24, 664–674 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Margueron R., Justin N., Ohno K., Sharpe M. L., Son J., Drury W. J. III, Voigt P., Martin S. R., Taylor W. R., De Marco V., Pirrotta V., Reinberg D., Gamblin S. J., Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Voigt P., LeRoy G., Drury W. J. III, Zee B. M., Son J., Beck D. B., Young N. L., Garcia B. A., Reinberg D., Asymmetrically modified nucleosomes. Cell 151, 181–193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Patel S. S., Rosenberg A. H., Studier F. W., Johnson K. A., Large scale purification and biochemical characterization of T7 primase/helicase proteins. Evidence for homodimer and heterodimer formation. J. Biol. Chem. 267, 15013–15021 (1992). [PubMed] [Google Scholar]
- 58.Lowary P. T., Widom J., New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 59.Gaykalova D. A., Kulaeva O. I., Bondarenko V. A., Studitsky V. M., Preparation and analysis of uniquely positioned mononucleosomes. Methods Mol. Biol. 523, 109–123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/11/e1601865/DC1
fig. S1. Helicase activity is essential for processive replication by the T7 replisome.
fig. S2. Strong nucleosomal barrier affects the processivity of the T7 replisome.
fig. S3. Analysis of the nucleosomal pausing patterns formed during replication by exo+ and exo− replisomes.
fig. S4. Analysis of time courses of replication through chromatin by the exo+ (A) or exo− replisome (B) using the KinTek Explorer software.
fig. S5. Mapping of nucleosome positions after replication by the T7 replisome using restriction enzyme sensitivity assay.
fig. S6. Nucleosomes are not formed de novo during or after T7 replication.
fig. S7. Proposed role for exonuclease activity during replication through a nucleosome.
table S1. Sequences of oligonucleotides and DNA templates.


