Abstract
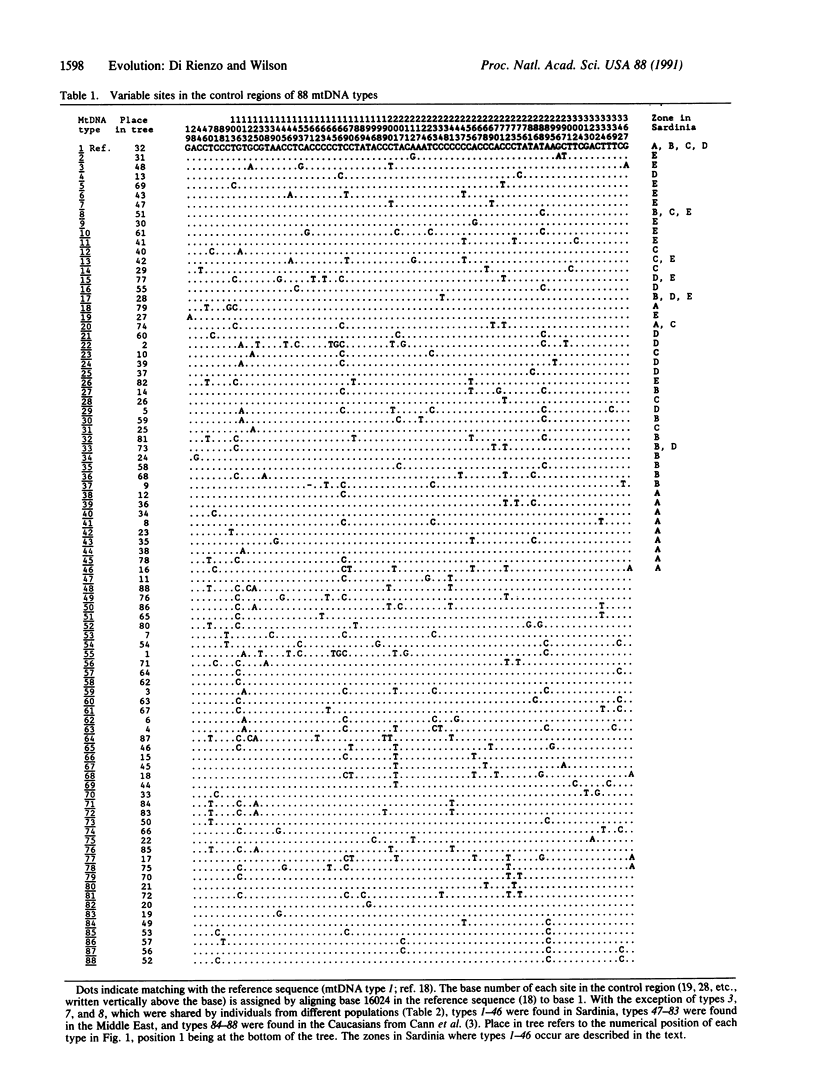
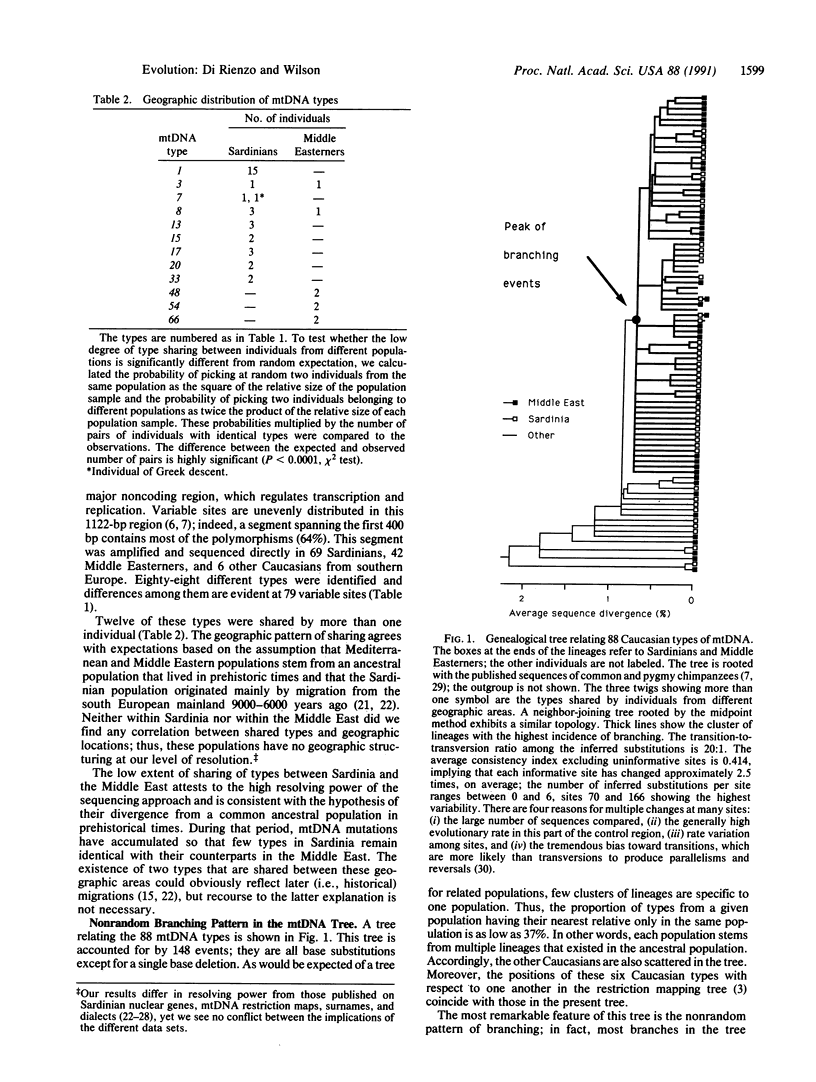
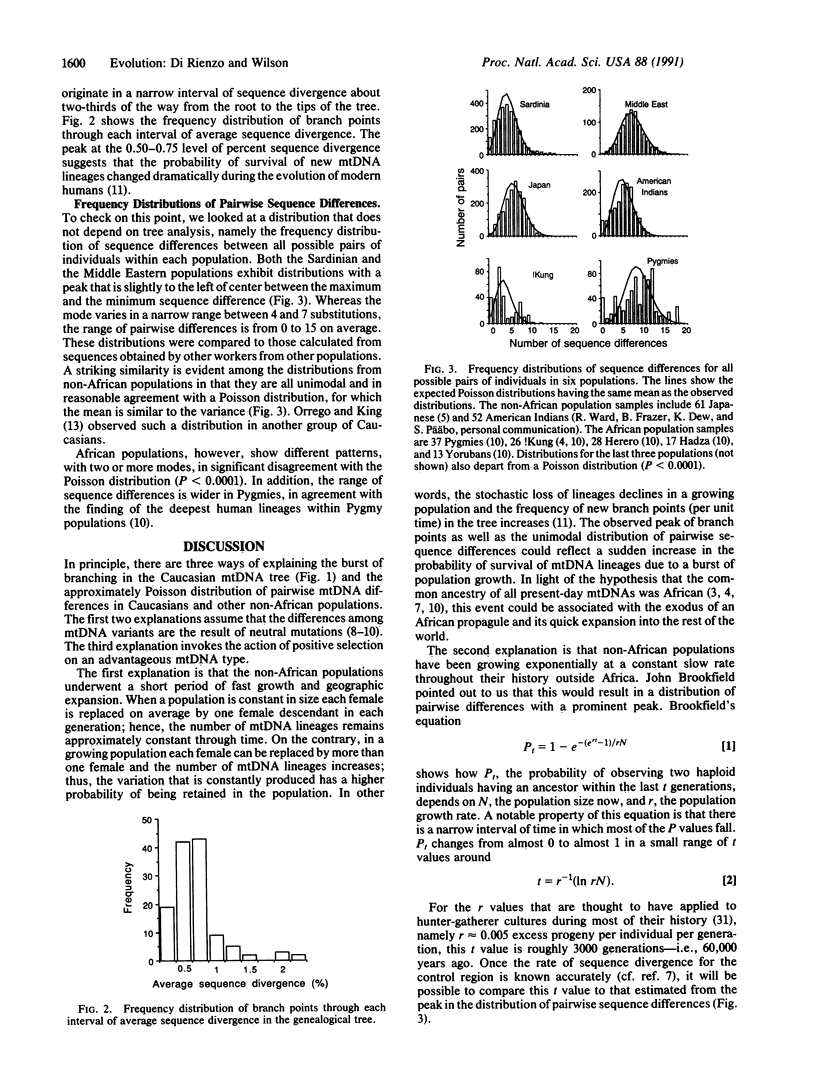
Eighty-eight types of mitochondrial (mt) DNA were found by sequencing the most variable part of the control region from 117 Caucasians. In the tree relating those types, most of the branching events occur about two-thirds of the way from the root of the tree to the tips of the branches. Moreover, the distribution of sequence differences between all possible pairs of individuals is approximately Poisson. Other non-African populations show a similar pattern. Assuming a neutral model, these findings imply that the probability of survival of new lineages has undergone dramatic changes, probably due to population expansion. Conversely, African populations show multimodal distributions fitting with a model of constant population size.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Avise J. C., Ball R. M., Arnold J. Current versus historical population sizes in vertebrate species with high gene flow: a comparison based on mitochondrial DNA lineages and inbreeding theory for neutral mutations. Mol Biol Evol. 1988 Jul;5(4):331–344. doi: 10.1093/oxfordjournals.molbev.a040504. [DOI] [PubMed] [Google Scholar]
- Avise J. C., Neigel J. E., Arnold J. Demographic influences on mitochondrial DNA lineage survivorship in animal populations. J Mol Evol. 1984;20(2):99–105. doi: 10.1007/BF02257369. [DOI] [PubMed] [Google Scholar]
- Brega A., Scozzari R., Maccioni L., Iodice C., Wallace D. C., Bianco I., Cao A., Santachiara Benerecetti A. S. Mitochondrial DNA polymorphisms in Italy. I. Population data from Sardinia and Rome. Ann Hum Genet. 1986 Oct;50(Pt 4):327–338. doi: 10.1111/j.1469-1809.1986.tb01754.x. [DOI] [PubMed] [Google Scholar]
- Brown W. M., Prager E. M., Wang A., Wilson A. C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Cann R. L., Stoneking M., Wilson A. C. Mitochondrial DNA and human evolution. Nature. 1987 Jan 1;325(6099):31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- DEEVEY E. S., Jr The human population. Sci Am. 1960 Sep;203:195–204. [PubMed] [Google Scholar]
- Foran D. R., Hixson J. E., Brown W. M. Comparisons of ape and human sequences that regulate mitochondrial DNA transcription and D-loop DNA synthesis. Nucleic Acids Res. 1988 Jul 11;16(13):5841–5861. doi: 10.1093/nar/16.13.5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg B. D., Newbold J. E., Sugino A. Intraspecific nucleotide sequence variability surrounding the origin of replication in human mitochondrial DNA. Gene. 1983 Jan-Feb;21(1-2):33–49. doi: 10.1016/0378-1119(83)90145-2. [DOI] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai S., Hayasaka K. Intraspecific nucleotide sequence differences in the major noncoding region of human mitochondrial DNA. Am J Hum Genet. 1990 Apr;46(4):828–842. [PMC free article] [PubMed] [Google Scholar]
- Piazza A., van Loghem E., de Lange G., Curtoni E. S., Ulizzi L., Terrenato L. Immunoglobulin allotypes in Sardinia. Am J Hum Genet. 1976 Jan;28(1):77–86. [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Santachiara Benerecetti A. S., Scozzari R., Semino O., Torroni A., Brega A., Wallace D. C. Mitochondrial DNA polymorphisms in Italy. II. Molecular analysis of new and rare morphs from Sardinia and Rome. Ann Hum Genet. 1988 Jan;52(Pt 1):39–56. doi: 10.1111/j.1469-1809.1988.tb01076.x. [DOI] [PubMed] [Google Scholar]
- Sartoris S., Varetto O., Migone N., Cappello N., Piazza A., Ferrara G. B., Ceppellini R. Mitochondrial DNA polymorphism in four Sardinian villages. Ann Hum Genet. 1988 Oct;52(Pt 4):327–340. doi: 10.1111/j.1469-1809.1988.tb01112.x. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989 Nov;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. H., Schaffner W., Wilson A. C., Päbo S. DNA phylogeny of the extinct marsupial wolf. Nature. 1989 Aug 10;340(6233):465–467. doi: 10.1038/340465a0. [DOI] [PubMed] [Google Scholar]
- Vigilant L., Pennington R., Harpending H., Kocher T. D., Wilson A. C. Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Clark A. G., Stoneking M., Cann R. L., Wilson A. C. Allelic variation in human mitochondrial genes based on patterns of restriction site polymorphism. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9611–9615. doi: 10.1073/pnas.83.24.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman E., Zei G., Moroni A., Cavalli-Sforza L. L. Surnames in Sardinia. II. Computation of migration matrices from surname distributions in different periods. Ann Hum Genet. 1984 Jan;48(Pt 1):65–78. doi: 10.1111/j.1469-1809.1984.tb00836.x. [DOI] [PubMed] [Google Scholar]
- Zei G., Guglielmino Matessi R., Siri E., Moroni A., Cavalli-Sforza L. Surnames in Sardinia. I. Fit of frequency distributions for neutral alleles and genetic population structure. Ann Hum Genet. 1983 Oct;47(Pt 4):329–352. doi: 10.1111/j.1469-1809.1983.tb01003.x. [DOI] [PubMed] [Google Scholar]



