Abstract
The sirtuins (SIRT1–7) are a family of nicotinamide adenine dinucleotide (NAD+)-dependent deacylases with remarkable abilities to prevent diseases and even reverse aspects of ageing. Mice engineered to express additional copies of SIRT1 or SIRT6, or treated with sirtuin-activating compounds (STACs) such as resveratrol and SRT2104 or with NAD+ precursors, have improved organ function, physical endurance, disease resistance and longevity. Trials in non-human primates and in humans have indicated that STACs may be safe and effective in treating inflammatory and metabolic disorders, among others. These advances have demonstrated that it is possible to rationally design molecules that can alleviate multiple diseases and possibly extend lifespan in humans.
The past 100 years have witnessed unprecedented advances in our ability to prevent and treat disease. Unfortunately, most medicines are designed to only treat one specific condition while ignoring other comorbidities. Thus, although the inhabitants of most nations are living longer, healthspan is not increasing. In the United Kingdom, for example, the percentage of men’s lifespan spent in poor health has risen from 20% to 40% between 1995 and 2006 (REF. 1). Recent progress in longevity research, however, may soon enable doctors to treat diseases that affect multiple organs with one medicine (or just a few in combination), to significantly increase healthspan and compress the morbid period of our lives.
“To eat when you are sick is to feed your sickness” wrote Hippocrates, the father of Western medicine, almost 2,400 years ago. Today, his views seem remarkably prescient. Calorie restriction without malnutrition is considered the gold standard in biogerontology as the most robust way to delay ageing and age-related diseases. Since the discovery almost 80 years ago that calorie restriction can extend the lifespan of rats2, great strides have been made in understanding why reducing calorie intake results in profound health benefits. Early theories, which included a delay in development and reduced metabolic rate, were subsequently ruled out owing to inconsistent experimental observations3.
Numerous studies of simple and complex model organisms over the past 20 years have lent credence to the idea that a set of evolutionarily conserved longevity pathways are responsible for the effects of calorie restriction on lifespan4 (FIG. 1). Dozens of genes and pathways have now been uncovered that compress the period of morbidity and extend the lifespan of model organisms, from yeast to rodents. Major signalling targets include insulin/insulin-like growth factor 1 (IGF1) signalling, target of rapamycin (TOR), adenosine monophosphate-activated protein kinase (AMPK) and the nicotinamide adenine dinucleotide (NAD+)-dependent sirtuin deacylases5,6. It is believed that these pathways evolved to sense and respond to the nutritional environment and to promote cellular defence mechanisms in the face of extrinsic adversity (FIG. 1).
Figure 1. Nutrient-responsive signalling pathways that maintain health and extend lifespan.
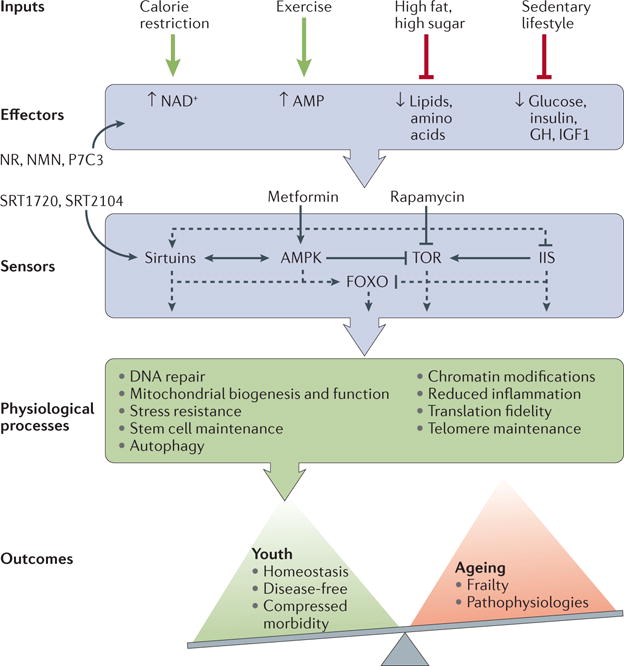
Calorie or dietary restriction increases the concentrations of metabolic effectors such as nicotinamide adenine dinucleotide (NAD+) and AMP while reducing the concentrations of glucose, amino acids and lipids. Exogenous administration of nicotinamide riboside (NR), nicotinamide mononucleotide (NMN) or the nicotinamide phosphoribosyltransferase (NAMPT) activator P7C3 can increase NAD+ levels. Calorie restriction also reduces the concentrations of the hormonal effectors insulin, insulin-like growth factor 1 (IGF1) and growth hormone (GH). These effectors stimulate or inhibit the activity of metabolic sensors such as the sirtuins (SIRTs), AMP kinase (AMPK), target of rapamycin (TOR), insulin–IGF1 signalling (IIS) and forkhead box O (FOXO) transcription factors. Sirtuin-activating compounds (STACs) such as SRT1720 and SRT2104 can directly activate SIRT1, whereas rapamycin is a direct inhibitor of TOR. Metformin indirectly activates AMPK. These metabolic sensors regulate downstream activities such as DNA repair, mitochondrial biogenesis and function, stress resistance, stem cell and telomere maintenance, autophagy, chromatin modifications, reduced inflammation, and translation fidelity. The net effect is to tip the scale in favour of homeostasis and compressed morbidity, resulting in a disease-free, more youthful-like state.
Interestingly, several naturally occurring molecules can activate these survival pathways and extend lifespan in rodents7,8. Rapamycin, a drug first discovered in a bacterium from Easter Island, reduces organ transplant rejection by inhibiting mechanistic TOR (mTOR). Rapamycin is thought to extend lifespan by mimicking diets that have low levels of essential amino acids such as methionine or tryptophan9,10. Another potential longevity drug is metformin, an AMPK-activating molecule from the Hellebore buttercup plant that is a frontline therapy for type 2 diabetes11. Clinical trials testing the ability of these molecules to slow aspects of ageing are underway or in the planning stages12. As discussed below, a promising approach to discovering longevity drugs is to design compounds de novo based on a molecular understanding of longevity. By designing increasingly potent compounds that activate sirtuins, it has been possible to pharmacologically mimic some of the physiological effects of calorie restriction both in animals and humans, with the ultimate goal of creating medicines that treat multiple age-related diseases and prevent many others.
The yeast silent information regulator 2 (SIR2) gene, which gave sirtuins their name, was first shown to extend lifespan in yeast almost 20 years ago, making sirtuins one of the first families of longevity genes to be discovered13–16. Since then, multiple lines of evidence, from yeast to humans, have implicated sirtuins in mediating the health benefits of dietary restriction and exercise. Although sirtuins have attracted considerable attention, they have also attracted controversy, including finally settled debates about how sirtuins activators work and even whether sirtuins have a role in ageing17,18. In this Review, we discuss the trials and tribulations of sirtuin research, which have constructively led to a clearer understanding of sirtuin structure, enzymatic activity and biological roles. We describe the ongoing quest to discover and develop safe and potent sirtuin-activating compounds (STACs) as means to combat multiple age-associated diseases. The use of such compounds in clinical trials is discussed in the context of the challenges that lie ahead if they are to reach their full potential in medicine.
Sirtuins in metabolism and longevity
Sirtuins were initially discovered in the budding yeast Saccharomyces cerevisiae by virtue of their essential function in gene silencing19 and subsequently found to dictate ‘replicative lifespan’, which is defined by the number of daughter cells a mother cell can produce15. Sir2 silences transcription at three loci: the HM mating-type loci, telomeres and the ribosomal DNA (rDNA). As cells age, the Sir2–Sir3–Sir4 complex relocalizes away from telomeres and the mating-type locus to accumulate at the rDNA locus, ostensibly to slow the formation and toxic accumulation of extrachromosomal rDNA circles (ERCs) that cause replicative ageing in yeast20,21. Inserting an extra copy of SIR2 (but not SIR3 or SIR4) into the yeast genome suppresses ERC formation and extends its lifespan22. This finding was corroborated by an unbiased genome-wide association study that identified the SIR2 locus as the most significant regulator of yeast replicative lifespan23. In 2000, Sir2 was reported to have histone deacetylase activity, which results in chromatin compaction and requires NAD+, a cofactor also required for redox reactions and whose levels change in response to nutrients and stress24,25. This striking discovery raised the intriguing possibility that yeast Sir2 and its homologues might act as nutrient and metabolic sensors that relay nuclear changes in the NAD+/NADH ratio to alter both transcription and genome stability24.
Sirtuins in other species were rapidly identified by virtue of their homology to yeast Sir2, specifically to the conserved central catalytic core26. In the nematode Caenorhabditis elegans27 and the fruitfly Drosophila melanogaster28, sirtuins were shown to control stress resistance and longevity, although the experimental approaches and magnitude of the effects have been debated18 (Supplementary information S1 (box)). By now, numerous research groups have shown that sirtuin overexpression in the nematode29–31 and the fruitfly32,33 results in a reproducible increase in longevity.
On the heels of the work performed in these organisms, there has been considerable effort to determine the role of the seven mammalian sirtuins (SIRT1–7) and to find small molecules to modulate their activity for pharmaceutical purposes. There are many points at which to intervene. Sirtuins are regulated at the level of transcription, translation, protein stability and oxidation. Sirtuins are also regulated by protein–protein interactions, natural inhibitors such as nicotinamide, microRNAs, localization within the cell and within organelles, as well as by substrate and co-substrate availability34–36. Similar to the yeast sirtuins, the mammalian sirtuins SIRT1, SIRT6 and SIRT7 act as transcription regulators37,38. These sirtuins also serve many additional roles, including the control of energy metabolism, cell survival, DNA repair, tissue regeneration, inflammation, neuronal signalling and even circadian rhythms (reviewed in REFS 39,40). SIRT1, which is predominately a nuclear protein, deacetylates histones H3, H4 and H1 (REFS 37,38) but also modifies more than 50 non-histone proteins41, including transcription factors and DNA repair proteins. Examples of transcription factors regulated by SIRT1 include p53 (REFS 42,43), nuclear factor-κB (NF-κB)44, peroxisome proliferator-activated receptor-γ co-activator 1α (PGC1α)45 and sterol regulatory element-binding protein (SREBP)46. Examples of SIRT1-regulated DNA repair proteins include Ku70 (REFS 47,48), poly(ADP-ribose) polymerase 1 (PARP1)49 and Werner syndrome ATP-dependent helicase (WRN)50,51. Other mammalian sirtuins localize to various subcellular compartments where they target non-histone proteins41: SIRT2 is cytosolic; SIRT3, SIRT4 and SIRT5 are mitochondrial; and SIRT6 and SIRT7 are nuclear (FIG. 2). Adding to the complexity, there is a growing list of chemical reactions that the mammalian sirtuins catalyse, including demalonylation, desuccinylation, decrotonylation, depropionylation, delipoamidation, other long-chain fatty acid deacylations and mono-ADP-ribosylation52–56, which are generally referred to as deacylation reactions, not to be confused with deacetylation.
Figure 2. Localization, enzymatic activity and modulation of sirtuins by small molecules.
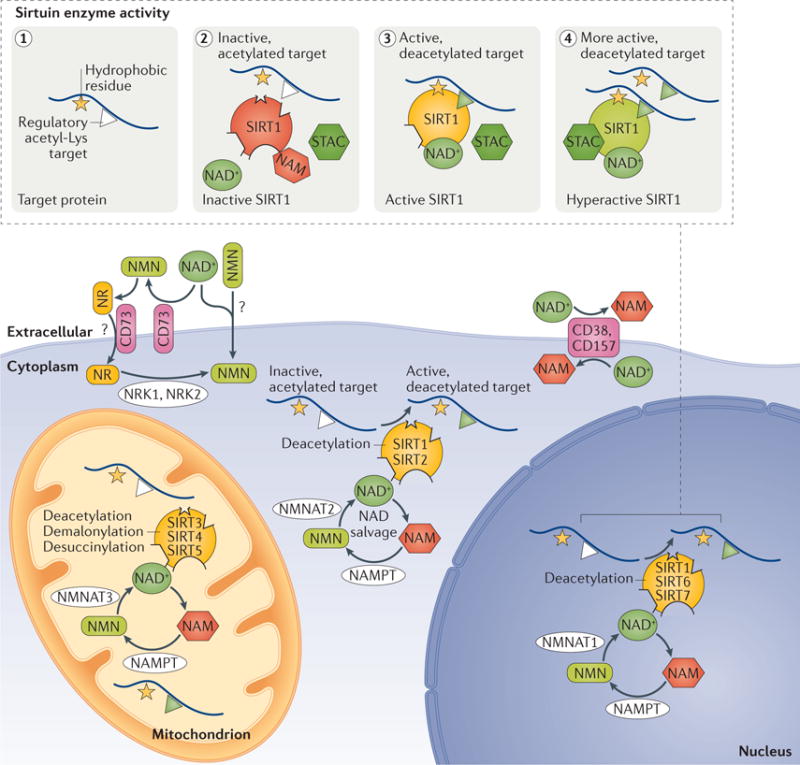
The mammalian sirtuins (SIRT1−7) are a family deacylases that are localized to various cellular compartments. SIRT1, SIRT6 and SIRT7 are localized primarily to the nucleus, whereas SIRT3, SIRT4 and SIRT5 are present in the mitochondria. SIRT1 (in some cell types) and SIRT2 are localized to the cytosol. Their activities are regulated by nicotinamide dinucleotide (NAD+), which is synthesized de novo from Trp, Asp or niacin (not shown). NAD+ is also recycled by the NAD salvage pathway from nicotinamide (NAM) by nicotinamide phosphoribosyltransferase (NAMPT), which converts NAM to nicotinamide mononucleotide (NMN). NMN is converted to NAD+ by nicotinamide mononucleotide adenylyltransferase1 (NMNAT1), NMNAT2 or NMNAT3, which are also capable of converting nicotinic acid mononucleotide (NaMN) to NAD+. Additional NAD+ precursors used in the NAD salvage pathway include nicotinamide riboside (NR) and nicotinic acid riboside, which are converted to NMN and NaMN, respectively (not shown) by nicotinamide riboside kinase 1 (NRK1) and NRK2. The cell membrane-bound glycohydrolases CD38 and CD157 degrade NAD+ to NAM135,142. CD73 converts NAD+ to NMN and NMN to NR; it is a transmembrane protein that may also act as a NR transporter177,178. There is evidence to indicate that NAD+ and NMN can be directly transported across the cell membrane in some cell types, although the mechanism is not yet known. The inset shows that SIRT1 activation is stimulated by the presence of a hydrophobic amino acid adjacently to the target Lys (panel 1). When NAM levels are sufficiently high, it binds into the carboxy-terminal pocket of SIRT1 and inhibits it58 (panel 2). SIRT1 utilizes NAD+ as a co-substrate to promote the binding and deacetylation of specific protein substrates (panel 3). Sirtuin-activating compounds (STACs) activate SIRT1 by binding to the STAC-binding domain and primarily lowering the Km for the peptide substrate, thereby increasing its catalytic activity (panel 4).
During sirtuin-mediated deacetylation of target Lys residues, NAD+ is converted to nicotinamide (NAM) and O-acetyl-ADP-ribose57. NAM then acts as a sirtuin inhibitor by binding to the C-pocket of sirtuins, which lies adjacent to the NAD+-binding pocket58,59. NAM remains one of the most effective and widely used sirtuin inhibitors. Recent studies have also revealed a link between NAM and its methylated form 1-methylnicotinamide (MNA) and health. In the nematode, both NAM and MNA increase longevity, and so does overexpression of the nematode gene required for formation of MNA, anmt-1, which is a homologue of the mammalian NAM-N-methyltransferase (NNMT), via a pathway seemingly independent of sirtuin activity30. In mammals, knockdown of NNMT protects against diet-induced obesity60, although a more recent study found that liver NNMT is increased by calorie restriction and seemed to improve glucose and cholesterol metabolism61. Additional studies are needed to understand the potential benefits of sirtuin-mediated by-products of NAD metabolism in mammals.
Although the results from experiments in which sirtuins were upregulated genetically or pharmacologically are generally promising (TABLE 1), the large number of pathways in which sirtuins are involved is a potential double-edged sword. Sirtuins interact with all the major conserved longevity pathways, for example AMPK62 and insulin–IGF1 signalling63–68, including targets such as protein kinase A (PKA)63, mTOR64–66, forkhead box O (FOXO)67 and IGF1 (REF. 68) (FIG. 1). Whether the multiple pathways in which sirtuins are involved will prove to be advantageous or deleterious for therapy in humans is unclear, but in the past 10 years, numerous sirtuin-overexpressing mouse strains have been generated, and at least two of them live longer. Notably, however, a transgenic mouse with high levels of Sirt1 overexpression recapitulates many of the benefits of calorie restriction, including increased metabolic activity, reduced blood lipid levels and improved glucose metabolism, but not a longer lifespan69. Another transgenic mouse with moderate overexpression of Sirt1 is protected from inflammation, liver cancer, diabetes and hepatic steatosis, but it too does not live longer70,71. A brain-specific Sirt1-overexpressing mouse strain (BRASTO) has a 9–16% longer mean lifespan, depending on the sex of the mice, and a significant increase in maximal longevity72. Male mice ubiquitously overexpressing Sirt6 live ~15% longer than wild-type mice, although this effect is not observed in females73. Sirt6-overexpressing mice are also resistant to hypoxia and to the detriments of a high-fat diet74. In humans, loss of SIRT6 activity has been directly implicated in the progression of ductal pancreatic adenocarcinoma, a highly fatal form of pancreatic cancer75. These findings indicate that molecules that activate SIRT1 and/or SIRT6 in humans may provide both broad health benefits and potent antitumour activities76. Achieving this therapeutic potential, however, would require a detailed molecular understanding of how sirtuins can be activated.
Table 1.
Sirtuins and sirtuin-activating compounds in stress resistance, calorie restriction, exercise and ageing
| Species | Sirtuin knockout shortens longevity |
Sirtuin overexpression extends longevity |
STACs extend longevity |
Sirtuins mediate the effects of CR on longevity |
Sirtuin expression or activity altered by CR or fasting |
Sirtuin implicated in ageing- stress pathways |
Sirtuin expression or activity altered by exercise |
|---|---|---|---|---|---|---|---|
| Yeast (Saccharomyces cerevisiae) | Yes (20–50%)15,22 | Yes (30–50%)15,22 | • Resveratrol (30–58%)77 • Resveratrol failed86 |
• Sir2 is essential132,180 • Hst2-dependent181 • Sir2-independent182,183 |
• Yes132,180 • NAD+ levels unchanged184,185 |
>100 citations | Not tested |
| Worm (Caenorhabditis elegans) | Yes (~5–10%)157,186 | • * High expression (2.1 line 50%)27 • Low gene copy number (10–25%)29,30,157,187,188 • Eleven independent low copy overexpressing lines (~5–15%)188 |
• Resveratrol (~10–65%)157,159,189,190 • SRT1720 failed158 |
• Essential31,186 • Non-essential191,192 |
Not tested | >40 citations | Not tested |
| Fruit fly (Drosophila melanogaster) | • Yes (~50%)193 • No change194,195 |
• * Yes (~18–29%)28,161 • Fat-body specific dSir2 (REFS 32,196) • Dose-dependent197 |
• Resveratrol (8–29%)157,197 • Resveratrol failed190 |
• * Essential28 • Fat-body specific dSir2 (REFS 32,196) |
Yes195,197 | >24 citations | Not tested |
| Honey bee (Apis mellifera) | Not tested | Not tested | Resveratrol (33–38%)166 | Not tested | Not tested | Not tested | Not tested |
| Fish (Nothobranchius furzeri and Nothobranchius guentheri) | Not tested | Not tested | Resveratrol (27–59%)163–165 | Not tested | Not tested | Not tested | Not tested |
| Mouse (Mus musculus) | • Elevated embryonic and postnatal lethality198 • Yes (~20–50%)68,199,200 |
• Brain Sirt1 (9–16%)72 • Sirt6 in males (9–17%)73 |
• Resveratrol in animals fed with high-fat diets167 • SRT1720 (~8–22%)124 • SRT2104 in males (~10%)125 |
Essential199,200 | >100 citations | >800 citations | >100 citations |
Multiple studies from the same laboratory were removed to limit citations. CR, calorie restriction; Hst2, homologous to Sir2; NAD+, nicotinamide adenine dinucleotide; Sir2, silent information regulator 2; SIRTs, sirtuins; STACs, sirtuin-activating compounds.
Disputed by REF. 18.
Sirtuin-activating compounds
The search for molecules that activate sirtuins began more than a decade ago. In the past 3 years, many of the challenges facing the development of STACs as medicines have been overcome, including resolving technical issues (see below), increasing the bioavailability of STACs and determining which diseases to prioritize in clinical trials.
How do sirtuin activators work?
The first STACs were discovered for SIRT1 in 2003, the most potent of which was resveratrol. This initial discovery was important because it proved that allosteric activation of sirtuins was possible77. High-throughput screening and medicinal chemical efforts have since identified more than 14,000 STACs from a dozen chemical classes, including stilbenes (for example, resveratrol), chalcones (for example, butein) and flavones (for example, quercetin) from plants77. Synthetic STACs include imidazothiazoles (for example, SRT1720)78, thiazolopyridines (for example, STAC-2), benzimidazoles (for example, STAC-5) and bridged ureas (for example, STAC-9)79,80 (BOX 1). All of these chemical classes activate SIRT1 by lowering the Km value of the substrate through a K-type allosteric activation mechanism. Interestingly, recent work demonstrated that SIRT6 deacetylase activity can be activated by endogenous fatty acids at the amino terminus of the enzyme56, hinting that SIRT6 may also be amenable to activation in vivo by synthetic molecules. This finding presents an exciting possibility given the ability of SIRT6 to enhance DNA repair, to modulate the metabolism of cancer cells to prevent malignancy, and to extend mouse lifespan73,75,81–84.
Box 1. The hunt for sirtuin activators.
As soon as yeast cells were shown to live longer with an extra copy of the sirtuin-encoding gene silent information regulator 2 (SIR2), a molecule that can activate sirtuins was quickly sought after. Generally, although activators are rare and much harder to study than inhibitors, they have distinct advantages: they are less prone to be rendered ineffective by residual enzymatic activity, they often have greater target specificity within an enzyme family and they elicit fewer side effects90. To date, the sirtuin that has received the most attention as a drug target is SIRT1. The first generation of sirtuin-activating compounds (STACs) were a group of related plant polyphenols that included, among others, quercetin, butein and resveratrol, which is a molecule found in red wine77. These molecules lower the binding affinity of SIRT1 for the substrate to increase enzymatic activity by tenfold77. Resveratrol rapidly became the molecule of choice to test SIRT1 activation because it was potent, nontoxic, naturally available and inexpensive. Many studies have since shown that resveratrol can extend maximal lifespan in yeast154–156, worms157,158, flies159–161, fish162–165 and even honeybees166 (TABLE 1). In mice, resveratrol protects against many of the deleterious effects of a high-calorie diet, delays age-related diseases, increases exercise endurance, and can mimic many of the transcriptional changes caused by calorie restriction167–171. These findings have led to the prevailing model in which SIRT1 imparts many of the benefits of a calorie restriction diet69–71,136. Additional screens for more potent, soluble and efficacious STACs have been successful172,173.
In mice, STACs mimic calorie restriction, increase mitochondrial function, protect from fatty liver and muscle wasting, and have strong antidiabetic, cardioprotective and anti-inflammatory effects in disease models78. STACs can also extend a mouse’s lifespan by up to 15%, even when initially given at 1 year of age123–125. In dozens of studies, the effects of STACs were shown to be either partially or completely SIRT1-dependent (reviewed in REF. 174). For example, in mice, the ability of resveratrol to prevent skin tumours is blunted in the Sirt1 germline knockout175, and mitochondrial function in skeletal muscles is not induced in a Sirt1-knockout strain105. Furthermore, synthetic STACs that are much more specific such as SRT1720 and SRT2104 in mice increase mitochondrial mass in muscle and liver124,125. Mechanistically, STACs trigger the deacetylation of peroxisome proliferator-activated receptor-γ co-activator 1α (PGC1α) to increase mitochondrial biogenesis; these effects were also shown to be SIRT1-dependent176.
The original fluorescence-based screens used to discover STACs were criticized because STACs only seemed to activate SIRT1 when bulky and hydrophobic fluorescent moieties were attached to the peptide substrates85,86. The most critical paper concluded that resveratrol and the chemically distinct STACs SRT1720 and SRT2104 were simply binding to the hydrophobic fluorophores17. This challenge, which spanned a few years, led to the discovery that SIRT1 has several structural and positional requirements of amino acids that are adjacent to the acetylated Lys residue for enabling substrate recognition87 and SIRT1 activation79,85,86. A major clue came from the discovery that the activation of SIRT1 is favoured when a large hydrophobic residue is present on the carboxy- terminal side of the acetylated Lys88. A study performed on natural peptide substrates revealed that bulky hydrophobic amino acids (Trp, Tyr or Phe) at the +1 and +6 positions relative to the acetyl-Lys could substitute for the fluorescent groups in the original assays89. These findings indicated that SIRT1 prefers specific hydrophobic amino acids in locations adjacent to the target Lys for substrate recognition. The same study also identified a mutation in SIRT1 (substitution of Glu230 with Lys) that prevented its activation by resveratrol89 and, strikingly, also by more than 100 synthetic STACs from multiple chemical scaffolds89, indicating that both natural and synthetic STACs activate SIRT1 by a common mechanism.
According to Zorn and Wells90, “one of the simplest ways to activate an enzyme with a small molecule is to bind to an allosteric site within the catalytic domain of that enzyme and induce a conformational change which changes the affinity of that enzyme for its native substrate.” Evidence from crystal structures and enzymological and biophysical studies has indicated that STACs function not by binding within the catalytic domain but by binding to a relatively rigid helix-turn-helix N terminus called the STAC-binding domain (SBD)91. Deletion studies of human SIRT1 have indicated that the N terminus is a key mediator of allosteric activation. The N terminus is necessary for activation by resveratrol and by synthetic STACs, which physically bind to this region89, and itself can activate SIRT1 in trans92. These data indicate that the N-terminal domain may function to activate SIRT1 in trans when SIRT1 is a dimer, or in cis to enhance substrate binding to the SIRT1 catalytic core92.
Recent crystallographic and amino acid substitution studies of human SIRT1 have identified the residues with which STACs come in contact and suggest an activation mechanism that is consistent with a ‘bend-at-the-elbow’ model91. STACs bind to the α-helical SBD in the N terminus, allowing the domain to flip over the site of interaction between the substrate and catalytic core91 (FIG. 3a). Next to the SBD are the hinge residues Arg234 and Glu230, located within a polybasic linker (KRKKRK), which allow the STAC-bound SBD to interact with the catalytic domain through a salt bridge formed between the guanidinium group of the SBD and the carboxylate group of Asp475 and hydrogen bonds to His473 and Val459 (FIG. 3b). In this model, the negatively charged Glu230 makes contacts with the positively charged Arg446, which explains why the SIRT1 substitution of Glu230 to Lys (a negative to positive charge) blocks SIRT1 activation and why subsequently replacing Arg446 with Glu (a positive to negative charge) restores the ability of STACs to activate SIRT1.
Figure 3. The allosteric activation mechanism of SIRT1.
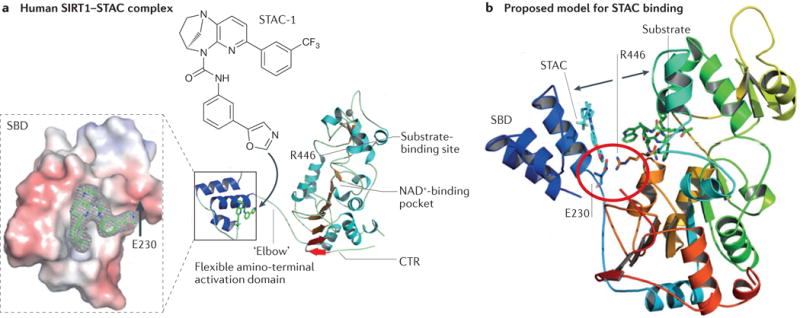
a| The structure of a truncated human sirtuin 1 (SIRT1) in complex with a synthetic sirtuin-activating compound (STAC) showing the binding site of STAC-1 at the amino-terminal STAC-binding domain (SBD). STAC binding promotes the interaction of the SBD with the central catalytic domain to lower the Km for the substrate, thereby increasing SIRT1 activity. The carboxy-terminal regulatory segment (CTR) stabilizes the deacetylase activity. Substitution of Glu230 (E230) with a Lys residue impairs enzymatic activation by reducing the electrostatic interaction between Glu230 on the SBD and Arg446 on the substrate-binding site. b | Model of how STACs activate SIRT1. Structural analysis and mutagenesis indicated that the amino acids Glu230 and Arg446 form a salt bridge (red circle) to facilitate binding of the amino terminus of SIRT1 with its catalytic core (movement indicated by arrows). The protein ribbon is rainbow-coloured from blue at the amino terminus to red at the carboxyl terminus. A STAC1 and an acetylated p53 peptide substrate are shown in cyan and green, respectively. Adapted from REF. 91, Nature Publishing Group.
Additional clues to the mechanism of the allosteric activation of sirtuins have emerged from studies of the yeast Sir2 protein, which is the catalytic component of a Sir2–Sir3–Sir4 complex. Molecular dynamic simulations have indicated that the substrate-binding channel toggles between an open and closed conformation, with Sir4 maintaining the N-terminal helix in an active conformation89. Interestingly, Glu230 of mammalian SIRT1 structurally aligns with a similar negatively charged residue in yeast Sir2 (Asp223), which is required for gene silencing93.
Specificity of SIRT1 activators in vivo
Resveratrol is a nonspecific compound, meaning it interacts with numerous proteins within the cell94. In addition to SIRT1, reported targets of resveratrol include AMPK95,96, phoshodiesterases97, F1-ATPase98, complex III of the mitochondrial electron transport chain99, PARP1 and Tyr-tRNA synthetase100. Elucidating which effects of resveratrol are mediated by SIRT1 and which by other effectors has been a considerable task; for example, whether resveratrol acts on SIRT1 or AMPK. Metformin, a pro-longevity drug that activates AMPK and produces similar physiological effects to resveratrol, increases NAD+ levels by activating the NAD salvage pathway enzyme nicotinamide phosphoribosyltransferase (NAMPT) (FIG. 2) and increases the NAD+/NADH ratio, both of which increase SIRT1 activity96,101. Other studies have shown that resveratrol acts on SIRT1 to activate AMPK by deacetylating and activating the AMPK kinase LKB1 (also known as STK11)102–105. Thus, SIRT1 can activate AMPK and AMPK can activate SIRT1. Recent evidence suggests that resveratrol can activate both AMPK and SIRT1 in vivo; however, the predominant target is highly dose-dependent105.
The identification of a SIRT1 mutation that blocked STAC activation in vitro89 has provided the perfect means to test whether STACs alter physiology by activating SIRT1 directly. In human HeLa cells and in mouse primary myocytes engineered to express the E230K SIRT1 mutation, resveratrol and synthetic STACs (SRT1720 and SRT2014) failed to induce the expected changes in mitochondrial mass, gene expression and cellular ATP concentrations. These results provided strong evidence that these STACs work by directly activating SIRT1 (REF. 89). To address the question of how STACs function in vivo, it would be interesting to repeat these experiments in a mouse carrying a SIRT1 E230K knock-in mutation.
Human and non-human primate studies
Studies using rodents have demonstrated that STACs promote health during ageing or metabolic stress, including protection from cancer, neurodegeneration, cardiovascular disease and diabetes, and even lifespan extension. Because regulatory agencies currently do not consider ageing a disease, the first sirtuin activator is most likely to be used clinically to treat an age-related disease such as diabetes, non-alcoholic fatty liver disease or an inflammatory disease (FIG. 4).
Figure 4. Sirtuin activation and its disease relevance.
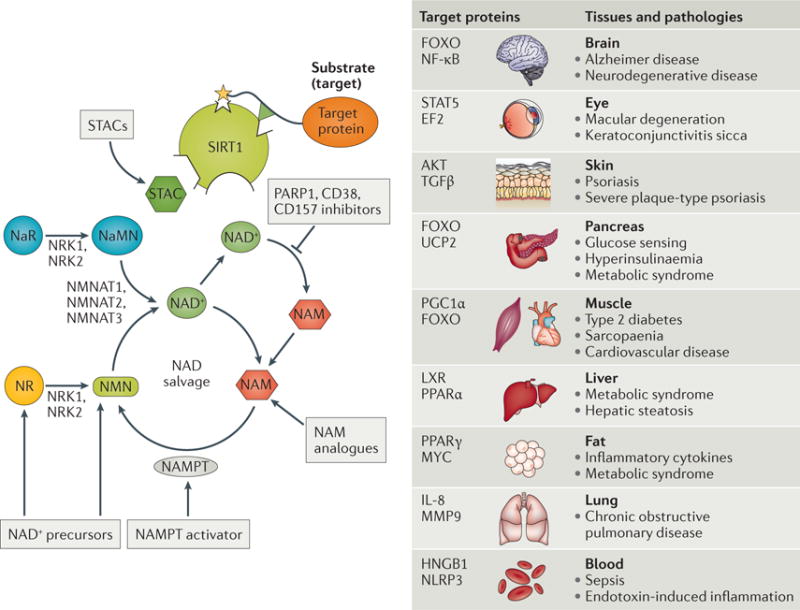
Sirtuin activity can be stimulated through various mechanisms: allosterically by sirtuin-activating compounds (STACs), such as resveratrol, SRT1720 and SRT2104, which lower the Km for the target proteins; increasing nicotinamide adenine dinucleotide (NAD+) levels by providing its precursors nicotinamide riboside (NR) or nicotinamide mononucleotide (NMN); inhibiting the glycohydrolases CD38 or CD157 (with apigenin, quercetin, GSK 897-78c)142,143; inhibiting poly(ADP-ribose) polymerases (PARPs)34; activating nicotinamide phosphoribosyltransferase (NAMPT) with P7C3 (REF. 152); or by providing nicotinamide (NAM) analogues, such as methyl-NAM. Nicotinamide riboside kinase 1 (NRK1) and NRK2 convert NR and nicotinic acid riboside (NaR) is to NMN and nicotinic acid mononucleotide (NaMN), respectively. NaMN and NMN are converted to NAD+ by nicotinamide mononucleotide adenylyltransferase 1 (NMNAT1), NMNAT2 and NMNAT3. Sirtuins directly or indirectly target more than 100 signalling proteins with relevance to human disease in a variety of tissues ranging from brain to blood (reviewed in REF. 41). Although STACs have therapeutic potential, certain diseases, such as cancer in certain contexts and Parkinson disease, may benefit from sirtuin inhibition (reviewed in REF. 179). EF2, elongation factor 2; FOXO, forkhead box O; HNGB1, high-mobility group box 1; IL, interleukin; LXR, liver X receptor; MMP9, matrix metalloproteinase 9; NF-κB, nuclear factor-κB; NLRP3, NOD-, LRR- and pyrin domain-containing 3; PGC1α, peroxisome proliferator-activated receptor-γ co-activator 1α; PPAR, peroxisome proliferator-activated receptor; STAT5, signal transducer and activator of transcription 5; TGFβ, transforming growth factor-β; UCP2, mitochondrial uncoupling protein 2.
The big question is whether sirtuin activators will work in humans106. Resveratrol as a proof-of-concept molecule is providing valuable clues. Even before it was shown to activate SIRT1, epidemiological studies in the 1980s suggested that resveratrol may explain aspects of the ‘French paradox’, the phenomenon whereby the French have a low incidence of cardiovascular disease even though they consume high amounts of saturated fats107. However, the validity of the French paradox has since been challenged108. A conglomerate of laboratories at the National Institute on Aging in the United States evaluated the benefits of resveratrol in controlled studies in non-human primates109–111. Following a 2-year high-fat and high-sugar feeding regimen, they determined that a 2-year resveratrol treatment prevented high-fat and high-sugar-induced arterial wall inflammation and increased arterial pulse wave velocity110, which are a major cause of cardiovascular morbidity and mortality in Western societies112. Additional studies from the National Institute on Aging using the same experimental approach found that resveratrol led to improvements in insulin sensitivity of visceral adipose tissue compared with control monkeys109. Moreover, preservation of pancreatic β-cell morphology and maintenance, and expression of essential β-cell transcription factors were observed111.
At least six clinical studies have now evaluated the safety and effects of resveratrol in humans (TABLE 2). The majority of studies observed improvements in glucose metabolism and in multiple indicators of protection from cardiovascular disease113–118. In one randomized double-blind crossover study, a group of healthy obese men were administered resveratrol (150 mg per day) or placebo for 30 days. The men receiving resveratrol exhibited a significantly reduced resting metabolic rate, reduced systolic blood pressure and improved HOMA index118. Muscle samples from those patients showed increased SIRT1 and PGC1α protein expression, and increased AMPK activity, mitochondrial respiration and fatty acid oxidation118. Another study in non-obese men, however, found that resveratrol failed to provide any measurable physiological improvements117. More recently, a phase II study evaluating resveratrol in patients with Alzheimer disease showed that resveratrol can delay cognitive decline in the ability to perform daily tasks119, a finding consistent with studies of resveratrol and SIRT1 in mouse models of Alzheimer disease120. A comprehensive summary and meta-analysis of clinical data has been recently published, the conclusion of which is that the human data are consistent with rodent studies, although more studies are warranted121.
Table 2.
Clinical trials of sirtuin-activating compounds (2000–2016)
| Sirtuin-activating compound | Phase I (safety) | Phase II (safety and efficacy) | Phase III–IV (pivotal and post-marketing) |
|---|---|---|---|
| Resveratrol or trans-resveratrol |
|
|
|
| SRT2104 |
|
|
None |
| Nicotinamide riboside |
|
|
None |
Clinical trials of sirtuin-activating compounds are listed in the table with their unique ClinicalTrials.gov identifier number. At the time of this publication, we excluded studies with unknown status.
The reason for variability in the efficacy of resveratrol in clinical trials is not yet known106. One explanation is the choice of human subjects. Studies that have carefully prescreened patients for advanced age and/or for insulin resistance have seen the greatest benefit of resveratrol, suggesting that SIRT1 activation restores homeostasis and is most effective in the context of dysfunctional physiology114. In mice, for example, the strongest effects of resveratrol are on obese mice. Another confounding issue is that resveratrol and many of the early STACs are hydrophobic, with low solubility and bioavailability. Thus, the efficacy of STACs probably depends on the formulation, the size of the chemical particles and whether the compounds are taken with food106. Nevertheless, for resveratrol to have metabolic benefits it might not need to be available to all tissues: a recent study using rats found that resveratrol activates SIRT1 in the gut endothelium to stimulate the secretion of the metabolic hormone glucagon- like peptide 1, thereby reducing glucose levels without even having to enter the circulation122.
More than 14,000 synthetic STACs have been synthesized to date, and dozens of these have been tested in animal models of type 2 diabetes, colitis, neurodegeneration, fatty liver and atherosclerosis, among others78,89,123–127. Currently, STACs are in their fifth generation78–80 and have more than 1,000-fold greater potency in vitro than resveratrol, with an EC50 in the low nanomolar range. The STAC named SRT2104, which mimics aspects of calorie restriction and extends male mouse lifespan125, has advanced through multiple phase I trials with few, if any, side effects128–130 to phase II trials (TABLE 2). Two SRT2104 clinical trials in elderly volunteers and otherwise healthy smokers showed a slight reduction in body weight, a 15–30% improvement in the cholesterol ratio and a 19% decrease in triglyceride levels129,130. A separate study of patients with the inflammatory condition plaque-type psoriasis showed a significant reduction in disease manifestation following 84 days of oral administration of 500 or 1,000 mg per kg SRT2104 (REF. 131). Pharmaceutical companies are continuing to develop STACs that have improved pharmaceutical properties and to conduct additional clinical trials (J. Ellis, personal communication).
NAD+ boosters as sirtuin activators
It has been known since 2003 that upregulation of the NAD salvage pathway, which recycles NAD+ from NAM, can extend lifespan and mimic calorie restriction in yeast132,133. The gene responsible for the rate-limiting step in the NAD salvage pathway in yeast, PNC1 (which encodes nicotinamidase), is activated by diverse stresses and calorie restriction, resulting in greater flux through the pathway and increased Sir2 activity. This early observation helped to explain how diverse stresses such as calorie restriction, amino acid restriction, heat stress and osmotic stress extend yeast lifespan.
In mammals, the homologue of PNC1 is NAMPT, which also catalyses the rate-limiting step in NAD salvage. NAMPT catalyses the formation of nicotinamide mononucleotide (NMN) from NAM, which is then converted to NAD by nicotinamide mononucleotide adenylyltransferase 1 (NMNAT1), NMNAT2 and NMNAT3 (FIG. 4). Nicotinamide riboside, a naturally occurring precursor of NAD+, enters the salvage pathway after being converted to NMN by the nicotinamide riboside kinase (NRK) enzymes. CD38 and its homologue CD157 are glycohydrolases that degrade NAD+ (REF. 134). As humans and mice age, levels of NAD+ decline, possibly because the consumption of NAD+ by CD38 outweighs its synthesis by the kynurenine pathway135.
NAD-boosting molecules constitute a newer class of STACs gaining attention as a way to restore NAD+ levels in elderly individuals and potentially activate all seven sirtuins with a single compound. Examples of NAD-boosting molecules include NMN, nicotinamide riboside67,136–141 and inhibitors of CD38 such as apigenin142, quercetin142 and GSK 897-78c143. In rodents, these molecules have been administered via various routes, including intraperitoneal, gavage and in drinking water, at doses ranging from 100 to 1,000 mg per kg per day67,136–141 for more than 6 months without any apparent adverse side effects140. The effects of NAD-boosting STACs on physiology are surprisingly broad, with improvements in glucose metabolism and mitochondrial function, and the recovery from injury of the heart, ears and eyes67,144–147 (FIG. 4). Nicotinamide riboside supplementation in mice starting at 24 months of age (a very advanced aged for mice), resulted in a modest (~5%) yet significant increase in longevity141. Nicotinamide riboside also prevents high-fat diet-induced glucose dysregulation145 and protects mice from DNA damage148,149, noise-induced hearing loss150, cardiac injury151 and stem cell-niche depletion141. Treatment with the related molecule NMN also protected 2-year old mice against a high-fat diet147 and restored youthful levels of mitochondrial function, ATP production and insulin sensitivity in muscle, ostensibly through SIRT1 activation137,147. Other indicators of inflammation and muscle wasting were also reduced137,147. Other NAD+ precursors such as nicotinic acid adenine dinucleotide or nicotinic acid riboside have yet to be tested but may be equally or even more efficacious. Non-naturally occurring derivatives of these compounds are also worth exploring given that the natural compounds have relatively low stability and short half-lives145.
Targeting the enzymes that regulate NAD+ levels, such as CD38, CD157 and NAMPT, may also be worth exploring for their therapeutic potential (FIG. 4). For example, a recent study indicated that a class of neuroprotective compounds (for example, P7C3) work by allosterically activating NAMPT152. This finding is consistent with a study showing that using NMN to increase NAD+ can protect neurons from the degeneration caused by SARM1, a protein that triggers a precipitous decline in NAD+ levels101. It will be interesting to test the effects of P7C3 and related molecules and whether they might have benefits similar to or better than the currently available STACs.
Future perspectives
The sirtuins have generated a considerable amount of excitement and scrutiny over the past 20 years. There is now consensus that sirtuins underlie aspects of calorie restriction and that they are key regulators of ageing and age-related diseases. Moreover, STACs can prevent and treat a variety of diseases in model organisms and in humans by directly binding to and activating SIRT1.
The path forwards now seems clear. For many years, synthetic STACs have been sufficiently potent to justify their use in the clinic but proved to be limiting in their bioavailability. Now with more soluble and specific compounds available89,91,123,125,127, STACs are re-entering clinical trials. Currently, given the promising results in patients with psoriasis treated with SRT2014 (REF. 131), further clinical trials for this disease seems to be a good path to follow. Compounds that raise NAD+ levels, such as nicotinamide riboside and NMN, also show great promise as calorie restriction mimetics to treat numerous age-related conditions, and possibly extend lifespan. Other possible trials include those to treat rare diseases such as a the DNA damage syndromes trichothiodystrophy or Cockayne syndrome (which are both alleviated by increased NAD+ levels in mice), an inflammatory disorder such as psoriasis, or metabolic diseases such as type 2 diabetes or non-alcoholic fatty liver disease.
Although many questions have been resolved in recent years, many still remain. For example, the physiological roles of the newly described activities of the sirtuins — namely, demalonylation, desuccinylation, decrotonylation, depropionylation and delipoamidation — remain unclear55. The pharmacokinetics and pharmacodynamics of NAD precursors and how they are affected by the route of delivery need clarification. In addition, identifying the NAD+ pools responsible for eliciting their benefits, both within the body and within subcellular compartments, is important. Moreover, are other sirtuins in addition to SIRT1 and SIRT6 amenable to allosteric activation? And what are the transport mechanisms for NAD+ precursors across cell membranes into the bloodstream and into cells153? This last question is important given new data indicating that there is an intracellular form of NAMPT (iNAMPT) and an extracellular form (eNAMPT), which is secreted from fat tissue and controls both systemic and hypothalamic NAD+ bioavailability153. Perhaps the most practical question is whether STACs will ever be approved as a drug to treat ageing or age-related diseases in humans. With the elderly population increasing worldwide and health-care costs threatening the global economy, the answer to that question cannot come soon enough.
Supplementary Material
Acknowledgments
The authors thank M. B. Schultz for suggestions and edits and are grateful for financial support from the National Institute on Aging, the National Institutes of Health, the Paul F. Glenn Foundation for Medical Research, Edward Schulak, and Ovaxon.
Glossary
- Replicative ageing
In yeast, the number of daughter cells produced by a mother cell before senescence
- Redox reactions
Oxidation–reduction (redox) reactions involving the transfer of electrons between two chemical species
- Hepatic steatosis
Also known as fatty liver, is a term used to describe the accumulation of fat in the liver cells
- Allosteric activation
Activation of an enzyme by binding of a ligand, which enhances the binding of substrates at other binding sites
- Km
Michaelis constant, which reflects the affinity of an enzyme for its substrate. The Km is measured as the substrate concentration at which the reaction rate is half of its maximum rate
- K-type allosteric activation
Refers to the major type of allosteric activation, in which the main feature that is altered is the Michaelis constant (Km)
- HOMA index
The homeostatic model assessment (HOMA) index is a clinical measure used to predict the function of pancreatic β-cells and insulin resistance
- Bioavailability
The degree and rate at which a substance is absorbed and is made available at the site of physiological activity
- EC50
The concentration of substrate that elicits a half-maximal enzymatic response
- Plaque-type psoriasis
The most common form of the disease, which is manifested as raised, red patches covered with a silvery white build-up of dead skin cells or scale
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
DATABASES:ClinicalTrials.gov: https://clinicaltrials.gov
SUPPLEMENTARY INFORMATION: See online article: S1 (box)
References
- 1.Robine JM, et al. The joint action on healthy life years (JA: EHLEIS) Arch Public Health. 2013;71:2. doi: 10.1186/0778-7367-71-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nutrition. 1989;5:155–171. A pioneering study reporting that reduced calorie intake and reduced body size leads to extended longevity. [PubMed] [Google Scholar]
- 3.Anderson RM, Weindruch R. The caloric restriction paradigm: implications for healthy human aging. Am J Hum Biol. 2012;24:101–106. doi: 10.1002/ajhb.22243. An important review outlining what lessons have been learnt from calorie restriction studies and how they can be applied to human ageing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 5.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller RA, et al. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell. 2014;13:468–477. doi: 10.1111/acel.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Montalvo A, et al. Metformin improves healthspan and lifespan in mice. Nat Commun. 2013;4:2192. doi: 10.1038/ncomms3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercken EM, Carboneau BA, Krzysik-Walker SM, de Cabo R. Of mice and men: the benefits of caloric restriction, exercise, and mimetics. Ageing Res Rev. 2012;11:390–398. doi: 10.1016/j.arr.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spindler SR. Caloric restriction: from soup to nuts. Ageing Res Rev. 2010;9:324–353. doi: 10.1016/j.arr.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Phung OJ, Sobieraj DM, Engel SS, Rajpathak SN. Early combination therapy for the treatment of type 2 diabetes mellitus: systematic review and meta-analysis. Diabetes Obes Metab. 2014;16:410–417. doi: 10.1111/dom.12233. [DOI] [PubMed] [Google Scholar]
- 12.Check-Hayden E. Anti-ageing pill pushed as bona fide drug. Nature. 2015;522:265–266. doi: 10.1038/522265a. [DOI] [PubMed] [Google Scholar]
- 13.Friedman DB, Johnson TE. Three mutants that extend both mean and maximum life span of the nematode, Caenorhabditis elegans, define the age-1 gene. J Gerontol. 1988;43:B102–B109. doi: 10.1093/geronj/43.4.b102. Arguably the first evidence to indicate that genes may control longevity in worms. [DOI] [PubMed] [Google Scholar]
- 14.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. The first study to show that sirtuins are involved in controlling yeast longevity. [DOI] [PubMed] [Google Scholar]
- 16.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang RA. C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. These findings provided undisputed evidence that a single gene mutation can robustly extend longevity in the worm C. elegans. [DOI] [PubMed] [Google Scholar]
- 17.Pacholec M, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burnett C, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy BK, et al. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 21.Sinclair DA, Guarente L. Extrachromosomal rDNA circles — a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. The first study to show that replicative lifespan is mediated by the accumulation of ERCs. [DOI] [PubMed] [Google Scholar]
- 22.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stumpferl SW, et al. Natural genetic variation in yeast longevity. Genome Res. 2012;22:1963–1973. doi: 10.1101/gr.136549.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. The first study to determine the mechanism for Sir2 was a NAD-dependent histone deacetylase. [DOI] [PubMed] [Google Scholar]
- 25.Landry J, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 27.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 28.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizki G, et al. The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet. 2011;7:e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmeisser K, et al. Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat Chem Biol. 2013;9:693–700. doi: 10.1038/nchembio.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moroz N, et al. Dietary restriction involves NAD+-dependent mechanisms and a shift toward oxidative metabolism. Aging Cell. 2014;13:1075–1085. doi: 10.1111/acel.12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee KK, et al. dSir2 in the adult fat body, but not in muscles, regulates life span in a diet-dependent manner. Cell Rep. 2012;2:1485–1491. doi: 10.1016/j.celrep.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 33.Whitaker R, et al. Increased expression of Drosophila Sir2 extends life span in a dose-dependent manner. Aging (Albany, NY) 2013;5:682–691. doi: 10.18632/aging.100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oberdoerffer P, et al. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinclair DA, Mills K, Guarente L. Molecular mechanisms of yeast aging. Trends Biochem Sci. 1998;23:131–134. doi: 10.1016/s0968-0004(98)01188-8. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toiber D, Sebastian C, Mostoslavsky R. Characterization of nuclear sirtuins: molecular mechanisms and physiological relevance. Handb Exp Pharmacol. 2011;206:189–224. doi: 10.1007/978-3-642-21631-2_9. [DOI] [PubMed] [Google Scholar]
- 39.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. A thorough review of the various biological mechanisms of sirtuin function in mammalian systems and physiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa T, Guarente L. SnapShot: sirtuins, NAD, and aging. Cell Metab. 2014;20:192. doi: 10.1016/j.cmet.2014.06.001. Lists the many sirtuin signalling protein targets. [DOI] [PubMed] [Google Scholar]
- 42.Luo J, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 43.Vaziri H, et al. hSIR2SIRT1 functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 44.Yeung F, et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodgers JT, et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 46.Walker AK, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 48.Cohen HY, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 49.Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2α deacetylase activity. J Biol Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 50.Vaitiekunaite R, et al. Expression and localization of Werner syndrome protein is modulated by SIRT1 and PML. Mech Ageing Dev. 2007;128:650–661. doi: 10.1016/j.mad.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 51.Li K, et al. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. J Biol Chem. 2008;283:7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- 52.Peng C, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10:M111.012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du J, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 55.Feldman JL, et al. Kinetic and structural basis for acyl-group selectivity and NAD dependence in sirtuin-catalyzed deacylation. Biochemistry. 2015;54:3037–3050. doi: 10.1021/acs.biochem.5b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 59.Landry J, Slama JT, Sternglanz R. Role of NAD+ in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- 60.Kraus D, et al. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature. 2014;508:258–262. doi: 10.1038/nature13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong S, et al. Nicotinamide N-methyltransferase regulates hepatic nutrient metabolism through Sirt1 protein stabilization. Nat Med. 2015;21:887–894. doi: 10.1038/nm.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Liang Y, Vanhoutte PM. SIRT1 and AMPK in regulating mammalian senescence: a critical review and a working model. FEBS Lett. 2011;585:986–994. doi: 10.1016/j.febslet.2010.11.047. [DOI] [PubMed] [Google Scholar]
- 63.Gerhart-Hines Z, et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD+ Mol Cell. 2011;44:851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Armour SM, et al. Inhibition of mammalian S6 kinase by resveratrol suppresses autophagy. Aging (Albany, NY) 2009;1:515–528. doi: 10.18632/aging.100056. These findings provided an interesting link between mammalian sirtuins and mTOR signalling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghosh HS, McBurney M, Robbins PD. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS ONE. 2010;5:e9199. doi: 10.1371/journal.pone.0009199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu M, et al. Resveratrol inhibits mTOR signaling by promoting the interaction between mTOR and DEPTOR. J Biol Chem. 2010;285:36387–36394. doi: 10.1074/jbc.M110.169284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mouchiroud L, et al. The NAD+/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Longo VD. Linking sirtuins, IGF-I signaling, and starvation. Exp Gerontol. 2009;44:70–74. doi: 10.1016/j.exger.2008.06.005. This paper provides a strong link between calorie restriction, IGF1 signalling and sirtuins. [DOI] [PubMed] [Google Scholar]
- 69.Bordone L, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 70.Banks AS, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Satoh A, et al. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 74.Kanfi Y, et al. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 75.Kugel S, et al. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell. 2016;165:1401–1415. doi: 10.1016/j.cell.2016.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chalkiadaki A, Guarente L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer. 2015;15:608–624. doi: 10.1038/nrc3985. [DOI] [PubMed] [Google Scholar]
- 77.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 78.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dai H, et al. SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J Biol Chem. 2010;285:32695–32703. doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hubbard BP, Sinclair DA. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol Sci. 2014;35:146–154. doi: 10.1016/j.tips.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mao Z, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van MM, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu Z, et al. SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle. 2015;14:269–276. doi: 10.4161/15384101.2014.980641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sebastian C, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Borra MT, Langer MR, Slama JT, Denu JM. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry. 2004;43:9877–9887. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- 86.Kaeberlein M, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 87.Chen Y, et al. Quantitative acetylome analysis reveals the roles of SIRT1 in regulating diverse substrates and cellular pathways. Mol Cell Proteomics. 2012;11:1048–1062. doi: 10.1074/mcp.M112.019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lakshminarasimhan M, Rauh D, Schutkowski M, Steegborn C. Sirt1 activation by resveratrol is substrate sequence-selective. Aging (Albany, NY) 2013;5:151–154. doi: 10.18632/aging.100542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hubbard BP, et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. A report of a sirtuin amino acid peptide screen that revealed the essential amino acid required for STAC binding to SIRT1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zorn JA, Wells JA. Turning enzymes ON with small molecules. Nat Chem Biol. 2010;6:179–188. doi: 10.1038/nchembio.318. [DOI] [PubMed] [Google Scholar]
- 91.Dai H, et al. Crystallographic structure of a small molecule SIRT1 activator–enzyme complex. Nat Commun. 2015;6:7645. doi: 10.1038/ncomms8645. The determination of the crystal structure for a truncated SIRT1 bound to the activator STAC-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ghisays F, et al. The N-terminal domain of SIRT1 is a positive regulator of endogenous SIRT1-dependent deacetylation and transcriptional outputs. Cell Rep. 2015;10:1665–1673. doi: 10.1016/j.celrep.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cuperus G, Shafaatian R, Shore D. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 2000;19:2641–2651. doi: 10.1093/emboj/19.11.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Howitz KT, Sinclair DA. Xenohormesis: sensing the chemical cues of other species. Cell. 2008;133:387–391. doi: 10.1016/j.cell.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fulco M, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Canto C, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park SJ, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gledhill JR, Montgomery MG, Leslie AG, Walker JE. Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc Natl Acad Sci USA. 2007;104:13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zini R, Morin C, Bertelli A, Bertelli AA, Tillement JP. Effects of resveratrol on the rat brain respiratory chain. Drugs Exp Clin Res. 1999;25:87–97. [PubMed] [Google Scholar]
- 100.Sajish M, Schimmel P. A human tRNA synthetase is a potent PARP1-activating effector target for resveratrol. Nature. 2015;519:370–373. doi: 10.1038/nature14028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, Milbrandt J. Neurobiology. SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science. 2015;348:453–457. doi: 10.1126/science.1258366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hou X, et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008;283:20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ivanov VN, et al. Resveratrol sensitizes melanomas to TRAIL through modulation of antiapoptotic gene expression. Exp Cell Res. 2008;314:1163–1176. doi: 10.1016/j.yexcr.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J Biol Chem. 2008;283:27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Price NL, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tome-Carneiro J, et al. Resveratrol and clinical trials: the crossroad from in vitro studies to human evidence. Curr Pharm Des. 2013;19:6064–6093. doi: 10.2174/13816128113199990407. A meta-analysis of the effects of resveratrol in clinical trials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Walsh GP. Does diet or alcohol explain the French paradox. Lancet. 1995;345:528. [PubMed] [Google Scholar]
- 108.Semba RD, et al. Resveratrol levels and all-cause mortality in older community-dwelling adults. JAMA Intern Med. 2014;174:1077–1084. doi: 10.1001/jamainternmed.2014.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jimenez-Gomez Y, et al. Resveratrol improves adipose insulin signaling and reduces the inflammatory response in adipose tissue of rhesus monkeys on high-fat, high-sugar diet. Cell Metab. 2013;18:533–545. doi: 10.1016/j.cmet.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mattison JA, et al. Resveratrol prevents high fat/sucrose diet-induced central arterial wall inflammation and stiffening in nonhuman primates. Cell Metab. 2014;20:183–190. doi: 10.1016/j.cmet.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fiori JL, et al. Resveratrol prevents β-cell dedifferentiation in nonhuman primates given a high-fat/high-sugar diet. Diabetes. 2013;62:3500–3513. doi: 10.2337/db13-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.AlGhatrif M, et al. Longitudinal trajectories of arterial stiffness and the role of blood pressure: the Baltimore Longitudinal Study of Aging. Hypertension. 2013;62:934–941. doi: 10.1161/HYPERTENSIONAHA.113.01445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bhatt JK, Thomas S, Nanjan MJ. Resveratrol supplementation improves glycemic control in type 2 diabetes mellitus. Nutr Res. 2012;32:537–541. doi: 10.1016/j.nutres.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 114.Crandall JP, et al. Pilot study of resveratrol in older adults with impaired glucose tolerance. J Gerontol A Biol Sci Med Sci. 2012;67:1307–1312. doi: 10.1093/gerona/glr235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wong RH, et al. Acute resveratrol supplementation improves flow-mediated dilatation in overweight/obese individuals with mildly elevated blood pressure. Nutr Metab Cardiovasc Dis. 2011;21:851–856. doi: 10.1016/j.numecd.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 116.Magyar K, et al. Cardioprotection by resveratrol: a human clinical trial in patients with stable coronary artery disease. Clin Hemorheol Microcirc. 2012;50:179–187. doi: 10.3233/CH-2011-1424. [DOI] [PubMed] [Google Scholar]
- 117.Poulsen MM, et al. High-dose resveratrol supplementation in obese men: an investigator-initiated, randomized, placebo-controlled clinical trial of substrate metabolism, insulin sensitivity, and body composition. Diabetes. 2013;62:1186–1195. doi: 10.2337/db12-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Timmers S, et al. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011;14:612–622. doi: 10.1016/j.cmet.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Turner RS, et al. A randomized, double-blind, placebo-controlled trial of resveratrol for Alzheimer disease. Neurology. 2015;85:1383–1391. doi: 10.1212/WNL.0000000000002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim D, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hausenblas HA, Schoulda JA, Smoliga JM. Resveratrol treatment as an adjunct to pharmacological management in type 2 diabetes mellitus — systematic review and meta-analysis. Mol Nutr Food Res. 2015;59:147–159. doi: 10.1002/mnfr.201400173. [DOI] [PubMed] [Google Scholar]
- 122.Cote CD, et al. Resveratrol activates duodenal Sirt1 to reverse insulin resistance in rats through a neuronal network. Nat Med. 2015;21:498–505. doi: 10.1038/nm.3821. [DOI] [PubMed] [Google Scholar]
- 123.Minor RK, et al. SRT1720 improves survival and healthspan of obese mice. Sci Rep. 2011;1:70. doi: 10.1038/srep00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mitchell SJ, et al. The SIRT1 activator SRT1720 extends lifespan and improves health of mice fed a standard diet. Cell Rep. 2014;6:836–843. doi: 10.1016/j.celrep.2014.01.031. Describes the effects of feeding the STAC SRT1720 on healthspan and lifespan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mercken EM, et al. SRT2104 extends survival of male mice on a standard diet and preserves bone and muscle mass. Aging Cell. 2014;13:787–796. doi: 10.1111/acel.12220. Long-term administration of STAC SRT2104 extends healthspan and longevity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Graff J, et al. A dietary regimen of caloric restriction or pharmacological activation of SIRT1 to delay the onset of neurodegeneration. J Neurosci. 2013;33:8951–8960. doi: 10.1523/JNEUROSCI.5657-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miranda MX, et al. The Sirt1 activator SRT3025 provides atheroprotection in Apoe−/− mice by reducing hepatic Pcsk9 secretion and enhancing Ldlr expression. Eur Heart J. 2015;36:51–59. doi: 10.1093/eurheartj/ehu095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hoffmann E, et al. Pharmacokinetics and tolerability of SRT2104, a first-in-class small molecule activator of SIRT1, after single and repeated oral administration in man. Br J Clin Pharmacol. 2013;75:186–196. doi: 10.1111/j.1365-2125.2012.04340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Libri V, et al. A pilot randomized, placebo controlled, double blind phase I trial of the novel SIRT1 activator SRT2104 in elderly volunteers. PLoS ONE. 2012;7:e51395. doi: 10.1371/journal.pone.0051395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Venkatasubramanian S, et al. Cardiovascular effects of a novel SIRT1 activator, SRT2104, in otherwise healthy cigarette smokers. J Am Heart Assoc. 2013;2:e000042. doi: 10.1161/JAHA.113.000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Krueger JG, et al. A randomized, placebo-controlled study of SRT2104, a SIRT1 activator, in patients with moderate to severe psoriasis. PLoS ONE. 2015;10:e0142081. doi: 10.1371/journal.pone.0142081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Anderson RM, et al. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 134.Malavasi F, et al. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev. 2008;88:841–886. doi: 10.1152/physrev.00035.2007. [DOI] [PubMed] [Google Scholar]
- 135.Camacho-Pereira J, et al. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 2016;23:1127–1139. doi: 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Braidy N, et al. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in Wistar rats. PLoS ONE. 2011;6:e19194. doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gomes AP, et al. Declining NAD+ induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ramsey KM, Mills KF, Satoh A, Imai S. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in β cell-specific Sirt1-overexpressing (BESTO) mice. Aging Cell. 2008;7:78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153:1448–1460. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gong B, et al. Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-gamma coactivator 1α regulated beta-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiol Aging. 2013;34:1581–1588. doi: 10.1016/j.neurobiolaging.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang H, et al. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352:1436–1443. doi: 10.1126/science.aaf2693. [DOI] [PubMed] [Google Scholar]
- 142.Escande C, et al. Flavonoid apigenin is an inhibitor of the NAD+ ase CD38: implications for cellular NAD+ metabolism, protein acetylation, and treatment of metabolic syndrome. Diabetes. 2013;62:1084–1093. doi: 10.2337/db12-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Haffner CD, et al. Discovery, synthesis, and biological evaluation of thiazoloquin(az)olin(on)es as potent CD38 inhibitors. J Med Chem. 2015;58:3548–3571. doi: 10.1021/jm502009h. [DOI] [PubMed] [Google Scholar]
- 144.Mouchiroud L, Houtkooper RH, Auwerx J. NAD+ metabolism: a therapeutic target for age-related metabolic disease. Crit Rev Biochem Mol Biol. 2013;48:397–408. doi: 10.3109/10409238.2013.789479. A thorough review outlining the potential mechanisms and roles of NAD in metabolism and disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Canto C, et al. The NAD+ precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Khan NA, et al. Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol Med. 2014;6:721–731. doi: 10.1002/emmm.201403943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tummala KS, et al. Inhibition of de novo NAD+ synthesis by oncogenic URI causes liver tumorigenesis through DNA damage. Cancer Cell. 2014;26:826–839. doi: 10.1016/j.ccell.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 149.Scheibye-Knudsen M, et al. A high-fat diet and NAD+ activate Sirt1 to rescue premature aging in cockayne syndrome. Cell Metab. 2014;20:840–855. doi: 10.1016/j.cmet.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brown KD, et al. Activation of SIRT3 by the NAD+ precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 2014;20:1059–1068. doi: 10.1016/j.cmet.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xu W, et al. Lethal cardiomyopathy in mice lacking transferrin receptor in the heart. Cell Rep. 2015;13:533–545. doi: 10.1016/j.celrep.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang G, et al. P7C3 neuroprotective chemicals function by activating the rate-limiting enzyme in NAD salvage. Cell. 2014;158:1324–1334. doi: 10.1016/j.cell.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yoon MJ, et al. SIRT1-mediated eNAMPT secretion from adipose tissue regulates hypothalamic NAD+ and function in mice. Cell Metab. 2015;21:706–717. doi: 10.1016/j.cmet.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jarolim S, et al. A novel assay for replicative lifespan in Saccharomyces cerevisiae. FEMS Yeast Res. 2004;5:169–177. doi: 10.1016/j.femsyr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 155.Yang H, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Morselli E, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging (Albany, NY) 2009;1:961–970. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 158.Zarse K, et al. Differential effects of resveratrol and SRT1720 on lifespan of adult Caenorhabditis elegans. Horm Metab Res. 2010;42:837–839. doi: 10.1055/s-0030-1265225. [DOI] [PubMed] [Google Scholar]
- 159.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 160.Bauer JH, Goupil S, Garber GB, Helfand SL. An accelerated assay for the identification of lifespan-extending interventions in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:12980–12985. doi: 10.1073/pnas.0403493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bauer JH, et al. dSir2 and Dmp53 interact to mediate aspects of CR-dependent lifespan extension in D. melanogaster. Aging (Albany, NY) 2009;1:38–48. doi: 10.18632/aging.100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Genade T, Lang DM. Resveratrol extends lifespan and preserves glia but not neurons of the Nothobranchius guentheri optic tectum. Exp Gerontol. 2013;48:202–212. doi: 10.1016/j.exger.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 163.Liu T, et al. Resveratrol attenuates oxidative stress and extends lifespan in the annual fish Nothobranchius guentheri. Rejuven Res. 2015;18:225–233. doi: 10.1089/rej.2014.1618. [DOI] [PubMed] [Google Scholar]
- 164.Valenzano DR, Cellerino A. Resveratrol and the pharmacology of aging: a new vertebrate model to validate an old molecule. Cell Cycle. 2006;5:1027–1032. doi: 10.4161/cc.5.10.2739. [DOI] [PubMed] [Google Scholar]
- 165.Yu X, Li G. Effects of resveratrol on longevity, cognitive ability and aging-related histological markers in the annual fish Nothobranchius guentheri. Exp Gerontol. 2012;47:940–949. doi: 10.1016/j.exger.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 166.Rascon B, Hubbard BP, Sinclair DA, Amdam GV. The lifespan extension effects of resveratrol are conserved in the honey bee and may be driven by a mechanism related to caloric restriction. Aging (Albany, NY) 2012;4:499–508. doi: 10.18632/aging.100474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. The first evidence to indicate that resveratrol could reverse many of the detriments of feeding a high-fat diet in mice and extend longevity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Barger JL, Kayo T, Pugh TD, Prolla TA, Weindruch R. Short-term consumption of a resveratrol-containing nutraceutical mixture mimics gene expression of long-term caloric restriction in mouse heart. Exp Gerontol. 2008;43:859–866. doi: 10.1016/j.exger.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 169.Barger JL, et al. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS ONE. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Park SK, et al. Gene expression profiling of aging in multiple mouse strains: identification of aging biomarkers and impact of dietary antioxidants. Aging Cell. 2009;8:484–495. doi: 10.1111/j.1474-9726.2009.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 172.Nakata A, et al. Potent SIRT1 enzyme-stimulating and anti-glycation activities of polymethoxyflavonoids from Kaempferia parviflora. Nat Prod Commun. 2014;9:1291–1294. [PubMed] [Google Scholar]
- 173.Nayagam VM, et al. SIRT1 modulating compounds from high-throughput screening as anti-inflammatory and insulin-sensitizing agents. J Biomol Screen. 2006;11:959–967. doi: 10.1177/1087057106294710. [DOI] [PubMed] [Google Scholar]
- 174.Lamming DW, Sabatini DM, Baur JA. Pharmacologic means of extending lifespan. J Clin Exp Pathol. 2012;(Suppl 4):7327. doi: 10.4172/2161-0681.S4-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Boily G, He XH, Pearce B, Jardine K, McBurney MW. Sirt1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene. 2009;28:2882–2893. doi: 10.1038/onc.2009.147. [DOI] [PubMed] [Google Scholar]
- 176.Feige JN, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 177.Garavaglia S, et al. The high-resolution crystal structure of periplasmic Haemophilus influenzae NAD nucleotidase reveals a novel enzymatic function of human CD73 related to NAD metabolism. Biochem J. 2012;441:131–141. doi: 10.1042/BJ20111263. [DOI] [PubMed] [Google Scholar]
- 178.Grozio A, et al. CD73 protein as a source of extracellular precursors for sustained NAD+ biosynthesis in FK866-treated tumor cells. J Biol Chem. 2013;288:25938–25949. doi: 10.1074/jbc.M113.470435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Carafa V, et al. Sirtuin functions and modulation: from chemistry to the clinic. Clin Epigenetics. 2016;8:61. doi: 10.1186/s13148-016-0224-3. An in-depth review discussing sirtuin inhibition as a potential therapeutic target. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 181.Lamming DW, et al. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 182.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tsuchiya M, et al. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell. 2006;5:505–514. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- 184.Mei SC, Brenner C. Calorie restriction-mediated replicative lifespan extension in yeast is non-cell autonomous. PLoS Biol. 2015;13:e1002048. doi: 10.1371/journal.pbio.1002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Evans C, et al. NAD+ metabolite levels as a function of vitamins and calorie restriction: evidence for different mechanisms of longevity. BMC Chem Biol. 2010;10:2. doi: 10.1186/1472-6769-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 187.van der Horst A, Schavemaker JM, Pellis-van BW, Burgering BM. The Caenorhabditis elegans nicotinamidase PNC-1 enhances survival. Mech Ageing Dev. 2007;128:346–349. doi: 10.1016/j.mad.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 188.Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–E2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- 189.Gruber J, Tang SY, Halliwell B. Evidence for a trade-off between survival and fitness caused by resveratrol treatment of Caenorhabditis elegans. Ann NY Acad Sci. 2007;1100:530–542. doi: 10.1196/annals.1395.059. [DOI] [PubMed] [Google Scholar]
- 190.Bass TM, Weinkove D, Houthoofd K, Gems D, Partridge L. Effects of resveratrol on lifespan in Drosophila melanogaster and Caenorhabditis elegans. Mech Ageing Dev. 2007;128:546–552. doi: 10.1016/j.mad.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 191.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mair W, Panowski SH, Shaw RJ, Dillin A. Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS ONE. 2009;4:e4535. doi: 10.1371/journal.pone.0004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Astrom SU, Cline TW, Rine J. The Drosophila melanogaster sir2+ gene is nonessential and has only minor effects on position-effect variegation. Genetics. 2003;163:931–937. doi: 10.1093/genetics/163.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Newman BL, Lundblad JR, Chen Y, Smolik SMA. Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span. Genetics. 2002;162:1675–1685. doi: 10.1093/genetics/162.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pallos J, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington’s disease. Hum Mol Genet. 2008;17:3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hoffmann J, Romey R, Fink C, Yong L, Roeder T. Overexpression of Sir2 in the adult fat body is sufficient to extend lifespan of male and female Drosophila. Aging (Albany, NY) 2013;5:315–327. doi: 10.18632/aging.100553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Parashar V, Rogina B. dSir2 mediates the increased spontaneous physical activity in flies on calorie restriction. Aging (Albany, NY) 2009;1:529–541. doi: 10.18632/aging.100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.McBurney MW, et al. The mammalian SIR2α protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Li Y, Xu W, McBurney MW, Longo VD. Sirt1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008;8:38–48. doi: 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


