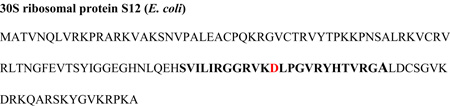Abstract
The bacterial ribosomal protein S12 contains a universally conserved D88 residue on a loop region thought to be critically involved in translation due to its proximal location to the A site of the 30S subunit. While D88 mutants are lethal this residue has been found to be post-translationally modified to β-methylthioaspartic acid, a post-translational modification (PTM) identified in S12 orthologs from several phylogenetically distinct bacteria. In a previous report focused on characterizing this PTM, our results provided evidence that this conserved loop region might be involved in forming proteins-protein interactions (1). To follow-up on this study, the D88 containing loop was probed to identify candidate binders employing a two-step complementary affinity purification strategy. The first involved an endogenously expressed S12 protein containing a C-terminal tag for capturing S12 binding partners. The second strategy utilized a synthetic biotinylated peptide representing the D88 conserved loop region for capturing S12 loop interaction partners. Captured proteins from both approaches were detected by utilizing SDS-PAGE and one-dimensional liquid chromatography tandem mass spectrometry. The results presented in this report revealed proteins that form direct interactions with the 30S subunit and elucidated which are likely to interact with S12. In addition, we provide evidence that two proteins involved in regulating ribosome and/or mRNA transcript levels under stress conditions, RNase R and Hfq, form direct interactions with the S12 conserved loop suggesting that it is likely part of a protein binding interface.
Keywords: Affinity pull-down protein purification; Peptide affinity pull-downs; PTM: post-translational modification; SPA: Sequence Peptide Affinity; SPA-S12: SPA tagged ibosomal protein S12; 1D-LC/MS/MS: One-dimensional liquid chromatography tandem mass spectrometry; FDR: False positive discovery rate, Metabolic labeling, relative quantification
Introduction
Bacterial ribosomes are composed of a small subunit (30S) containing about 20 proteins and a single rRNA (16S), and a large subunit (50S) consisting of over 30 proteins and two rRNAs (5S and 23 S). Many studies have indicated that rRNAs and several ribosomal proteins contain post-transcriptional modifications as well as post-translational modifications (PTMs) respectively. While a large amount of biochemical and genetic information has been obtained about the structure and functional relationships concerning rRNA modifications (2, 3), very little has been learned about the biological significance of ribosomal protein PTMs. In particular, the small subunit (30S) Escherichia coli (E. coif) ribosomal protein S12 contains a novel PTM, β-methylthiolation, located on the universally conserved Asp 88 (4) (see Figure 1). Structural data indicate that D88 is located in a conserved loop that is projected toward a site on the 30S subunit (5) involved in translation and tRNA decoding. Site-directed mutagenesis reveals that substitutions at the D88 position are lethal. Nearby substitutions are however tolerated (6). β-methylthiolation of Asp88 has been identified in S12 orthologs from phylogenetically distant bacteria and appears to be unique to prokaryotes (4, 7–10). A recent genetic knockout study (in E. coli) revealed a completely viable strain in the absence of the PTM thereby indicating that β-methylthiolation is not essential for the conserved loop function but is likely to be important under specific yet to be defined conditions (11).
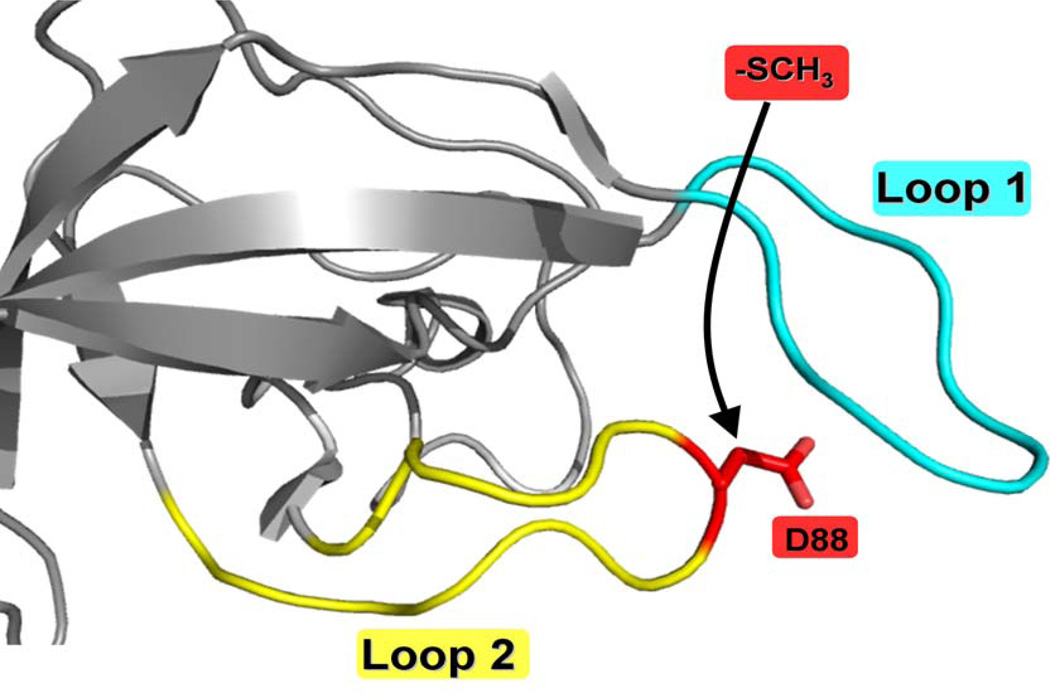
Figure 1.
The region surrounding D88 in S12, as determined in the structure from Thermus thermophilus 30S subunit (47) (Protein Data Back accession no. 1FJG). This region consists of 2 highly conserved loops (Loopl and Loop2) that are directed toward a site on the 30S subunit involved in translation. β-methylthio-D88 (highlighted in red) is located on the second conserved Loop 2 (highlighted in yellow) that connects two anti-parallel β strands This figure was created using the PyMol Molecular Graphics System (DeLano Scientific, Palo Alto, California, US).
To date, several reports have indicated that RimO is the methylthiotransferase involved in β-methylthiolation of D88 (11)-(12) (13). The rimO gene is homologous to the complete gene sequence of MiaB, a bi-functional enzyme characterized in E. coli that methylthiolates transfer RNA (14–17). There currently remains inconclusive data at the molecular level about how D88 is modified by RimO, or the nature of the PTM’s biochemical function (11–13). We recently reported a study suggesting that RimO modifies the ribosome-assembled form of S12 with the aid of a previously uncharacterized protein, YcaO (1). Our quantitative mass spectrometry data using genetic knockouts (representing rimO and ycaO genes) correlated with previous reports regarding RimO and also indicated that YcaO had a substantial impact on the modification status of S12. Specifically we found that S12 was predominantly in the modified form in the wildtype strain, was abundantly found in both forms in the ΔycaO knockout and completely abolished in the ΔrimO knockout. Additionally, expression microarray performed on these genetic knockouts resulted in overlapping transcriptional changes suggesting a link between β-methylthiolation and the transcription of anaerobic genes. This transcriptomic phenotype suggests that the PTM and conserved loop perform a specific function involving additional yet unidentified proteins; however, the molecular details are currently unknown.
In order to expand upon our recent characterization of this PTM we coordinated a complimentary proteomics approach that included the previously described endogenously expressed C-terminally tagged S12 (SPA-S12) and a peptide affinity approach to identify candidate S12 interacting proteins. In this report we compare SPA-S12 associated complexes from various experimental conditions and present a complimentary peptide affinity approach that elucidates which candidate proteins most likely form direct interactions with the 30S subunit and/or S12.
Experimental Procedure
Construction of Recombinant E. coli SPA-S12
The SPA (Sequence Peptide Affinity) purification system used in this work adds a C-terminal tag consisting of three modified FLAG sequences (3X FLAG) and a calmodulin binding peptide (CBP) to the chromosomal gene to produce an endogenously expressed bait protein for capturing native protein complexes from E. coli (18). The resulting protein complexes captured by interactors would therefore be biologically relevant. To generate the SPA-S12 containing strain (NCMS1) we obtained an SPA cassette from Jack Greenblatt's laboratory (University of Toronto, Canada). The lambda recombineering experiments used to insert an SPA tag to the COOH end of the rspL chromosomal gene for constructing SPA-S12 have been described previously. SPA-S12 protein expression was confirmed by western blot analysis and the resulting E. coli strain displayed a growth rate similar to wild-type, indicating the C-terminal tag had minimal effect on SPA-S12 structure and function.
Culturing of E. coli Strains used for Recombinant SPA-S12 and Peptide Pull-downs
W3110 E.coli (negative control strain lacking the SPA tag) and W3110 strains in which a bait protein had been SPA tagged were cultured in Terrific Broth (TB) media (Quality Biological, Inc. Gaithersburg, MD) at 37 °C to mid-log phase or early stationary phase. Cells were harvested and pellets were frozen at −80 °C until purification. For peptide pull-downs, W3110 E. coli was grown at 37 °C to early stationary phase using M9 minimal synthetic media supplemented either with labeled [15N]-ammonium chloride (99%) (Cambridge Isotope Laboratories, Inc. Andover, MA) or the 14N isotope as previously described (19). Cells were harvested and pellets frozen at −80 °C until used for peptide pull-down experiments. To ensure complete 15N incorporation serial inoculations into 8 consecutive 15N minimal media overnights were performed. Mass spectrometric analysis was used to confirm complete 15N incorporation.
Isolation of SPA-S12 Complexes from Polysome Depleted Supernatant
Lysates and/or polysome depleted supernatants were prepared by lysing E. coli cells representing NCMS1 or W3110 (~2 gram) cell pellets. Briefly, recombinant SPA-S12 pull-downs were performed under 3 different experimental conditions (total of 9 replicates; 3 per experimental condition) in parallel with a negative control strain lacking an SPA tag sequence. The negative control was used to compare and subtract non-specific contaminants identified in the experimental pull-downs. A single Anti-FLAG affinity purification step was used for each pull-down to preserve transient non-stoichiometric interactions. The first SPA-S12 experimental (crude) condition was performed by combining 100 µg washed Anti-FLAG agarose beads (Sigma Chemical Company, St. Louis, MO) with ~28 mg/ml whole cell lysates and incubating for 2 hours at 4 °C using an orbital rotator. The second and third experimental conditions included using polysome depleted extracts from either mid-log phase or stationary phase cells. Polysomes were depleted from samples prior to performing pull-downs by centrifuging the resulting supernatant at 100,000 × g for 4 hours as previously described (1). The resulting polysome depleted supernatants (~8 mg/ml) were incubated overnight at 4 °C using an orbital rotator with 100 µg washed Anti-FLAG agarose beads added to ~8 mg/ml supernatant. For all pull-downs, SPA-S12 complexes were washed extensively with TBS buffer, eluted using 100 µg/ml 3X FLAG peptides and then concentrated using an Ultracel YM-3 Centrifugal device (Millipore, Billerica, MA). Concentrated SPA-S12 and negative control eluates were quantified using the BCA assay with BSA as a standard (Thermo, Rockford, IL). The eluted proteins were denatured in SDS sample buffer by heating at 70 °C for 7 minutes and separated by SDS-PAGE (10%). The gel was then stained with SimplyBlue Safestain (Invitrogen, Carlsbad, CA). After each SDS-PAGE analysis the eluted proteins were subjected to in-gel digestions followed by mass spectrometry as described below.
Peptide Synthesis and Pull-downs
Bait peptides consisted of biotinylated tagged (at amino terminus) 25 mers with a molecular mass of 2620 daltons (New England Peptides LLC. Gardner, MA). The purity of the peptides was confirmed by the manufacturer using mass spectrometric analysis. The peptides used are as follows (see table 4 for respective amino acid sequences): the experimental S12 second conserved loop region representing residues 77–100 containing D88 (S12 peptide) and a decoy control peptide with the same amino acid content but with residues in random order. The S12 peptide sequence represents the entire loop and flanking regions of connecting anti-parallel β-strands; β-strand backbone hydrogen bonding interactions may potentially stabilize the naturally occurring loop structure. The random decoy peptide sequence was chosen because it shares no homology with any sequences in genome space. Circular dichroism spectroscopy analysis indicates that both synthetic peptides contain similar secondary structure that resembles a β-hairpin loop (data not shown). To immobilize peptide baits and saturate streptavidin binding sites 1 mg (100 µl) of M280 streptavidin-coated Dynabeads (Dynal, Carlsbad, CA) were pre-loaded with 2 µg (~ 800 picomoles) of biotinlylated peptide prior to incubation with cell lysates. Peptide pull-down experiments were performed in parallel such that immobilized control biotinlylated peptides (scrambled peptide) were incubated in the lysate from W3110 cells grown in 15N containing minimal media and immobilized experimental biotinlylated peptides (S12 peptide) were separately incubated in the lysate from cells grown in 14N containing minimal media. To further reduce the variability associated with background protein binding, cross experiments were performed in which the S12 peptide was incubated in the lysate from 15N labeled cells in one pull-down (the scrambled peptide would be incubated in 14N labeled lysate) and 14N labeled lysate was in the next pull-down. Each pull-down was performed overnight at 4 °C in ~2mg/ml ribosome-free lysate using constant rotation. W3110 cells grown in either 15N or 14N media were lysed and treated to remove ribosomes using the same procedure as described above for SPA-S12 pull-downs. Lysate supernatant protein concentration was determined using the BCA assay. After extensive washes using TBS buffer, bound proteins were eluted from the immobilized peptides by boiling in SDS sample buffer. Eluate proteins from control and experimental bait peptides were combined and separated by SDS-PAGE (4–12%) and then analyzed using in-gel digestions (a gel lane represents combined eluates) and mass spectrometry as described below.
Table 4.
| S12 biotinylated peptide | Bio-SVILIRGGRVKDLPGVRYHTVRGA-amide |
|---|---|
| Scrambled biotinylated peptide | Bio-VHGRDLKRPVLAGTGIRVGVYIRS-amide |
ID LC-MS/MS analysis
Gel lanes from recombinant SPA-S12 and peptide pull-downs were manually excised top to bottom into 20 ~ 2-mm bands. In-gel tryptic digestion, peptide extraction and one-dimensional liquid chromatography tandem mass spectrometry (1D-LC-MS/MS) analysis was performed using a Nano LC ID Proteomics HPLC system (Eksigent, Dublin, Ca) coupled online to an (LTQ)-Orbitrap mass spectrometer (Thermo Fisher, San Jose, CA) equipped with a Nanomate nanoelectrospray ionization source (Advion, Ithaca, NY) as previously described (1). ID LC/MS/MS experiments for SPAS12 pull-downs were operated such that spectra were acquired for 60 minutes in the data dependent mode with dynamic exclusion enabled. The top 5 peaks in the 400–2,000 m/z range of every MS survey scan were fragmented. Survey spectra were acquired with 60,000 resolution in the Orbitrap mass analyzer and fragmented in the LTQ ion trap. ID LC/MS/MS experiments for peptide pull-downs were operated in a similar manner except the top 3 peaks in the 400–2,000 m/z range of every MS survey scan were fragmented and survey spectra were acquired with 30,000 resolution in the Orbitrap mass analyzer.
Analysis of SPAS12 pull-downs
All Thermo LTQ-Orbitrap Xcalibur raw files were processed by the automation client Mascot Daemon (version 2.3), which allows batch processing and analysis of data by Mascot. Mascot Daemon utilizes the Thermo utility extract_msn (version 5.0) to convert Xcalibur data into DTA format peak lists and submits these to a Mascot Server for sequence searches (all DTA files for a given raw file are merged into an MGF file). Fragmentation spectra were searched using the Mascot version 2.3 search engine (Matrix Sciences, London, UK) against an E. coli database (created from the Uniprot Knowledgebase Release 2010_08; date July 13th 2010) containing the porcine trypsin and human keratin sequences. Alternative isoform sequences, where relevant, were retrieved from the varsplic annotation file in UniProtKB. This E. coli database contains all ECOLI records and is supplemented with ECOLX for completeness (contains 5,103 sequences; 1,603,703 residues). All recombinant data were searched against a decoy database composed of reversed translated sequences in order to determine false positive discovery rates. Search parameters were as follows: trypsin specificity, 2 missed cleavages, carboxamidomethylation of cysteine as a static modification, methionine oxidation (+16 daltons) and aspartic acid methylthiolation (+46 daltons) as variable modifications, and +1 through +4 charge states. Subsequent database searches were conducted to consider a variety of common chemical modifications (e.g., pyroGlu from N-terminal Gin, and pyroCys from N-terminal carboxamidomethylCys) and PTMs of interest (e.g., N-acetylation of proteins). The precursor ion mass tolerance was ± 50 ppm and the fragment ion mass tolerance was ± 0.6 Da. A total of 9 experimental pull-downs and 5 negative controls were performed where 14 LTQ datasets were acquired (mid-log phase available publicly via NCBI peptide data resource accession# PSE145; all other data have been submitted to the PeptideAtlas database repository and can be accessed at http://www.peptideatlas.org/repository/). The mid-log phase data is from a previous report (1). The experimental and negative control data were analyzed separately and the resultant output files were concatenated using the in-house software MassSieve (20). MassSieve reassembles peptide identifications into protein identifications and facilitates parsimonious comparisons of the concatenated experimental data with negative control data (resultant peptide and protein identifications are presented in supplementary tables). Proteins were considered true interactors if identified in at least 2 out of 3 experimental runs and were either absent in the negative control or substantially enriched in the SPA-S12 pull-down. Peptide identifications were the primary metric for inclusion of a protein; a minimum of 2 peptides was required for a protein to be considered sufficiently identified. Spectra matched to unique peptides were used as a secondary metric to estimate abundance of those proteins that were captured in the SPA-S12 pull-downs relative to the control (at least 4-fold higher number of either peptide identifications or spectra matched to peptides). A peptide could still be present at concentration levels below the detection limit required to trigger MS/MS. MassSieve filters were adjusted to include only peptide identifications with Mascot Ion Scores equal to or exceeding their identity scores (corresponds to ≥ 95% confidence). This resulted in a calculated false discovery (FDR) rate of 2.62% for all peptide identifications representing negative control and SPA-S12 pull-downs. FDR = 2 Nd/(Nd + Nt); where Nd and Nt are the number of matched decoy (resulting from the reverse translated sequence) and target peptides passing the above cutoff, respectively.
Analysis of peptide pull-downs
Fragmentation spectra from peptide pull-downs and corresponding crossover experiments were searched using the SEQUEST search engine (version 27) with the same search parameters and E. coli database described above. SEQUEST was used for these data because the quantification software ProRata 1.1 requests SEQUEST output files and is incompatible with Mascot. However, we have found that SEQUEST searches (using the same criteria for Mascot) with recombinant SPA-S12 data are qualitatively consistent to the Mascot results with regard to protein identifications that passed the criteria used in this work (data not shown). A minimum of 2 peptides was required for a protein to be considered sufficiently identified for quantification. For these data the calculated FDR was 2.2%. Samples representing all gel lanes (combined eluates) involved two iterative SEQUEST searches. In the first iteration, unlabeled amino acids were used. In the second iteration, 15N for amino acid molecular masses were used (0.98 Da added to glycine, alanine, serine, proline, valine, threonine, cysteine, leucine, isoleucine, aspartic acid, glutamic acid, methionine, phenylalanine and tyrosine; 1.96 Da added to asparagine, glutamine, lysine and tryptophan; 2.94 Da added to histidine; and 3.92 Da added to arginine). Results from these 14N and 15N searches were merged using the “Merge Directories” tool of ProRata. Peptide identifications were filtered, organized and assembled using DTASelect 2.06 (21). DTASelect filters based on +1, +2 and +3 charged peptides with Xcorr scores ≥ 1.8, ≥ 2.5, ≥ 3.5, respectively and a ΔCN score ≥ 0.08. Multiple MS-MS scans for identified peptide sequences were retained by specifying DTASelect locus filter −t 0. Peptide Quantification was performed using ProRata 1.1 as previously described (22, 23). Briefly, the ProRata program extracted selected ion chromatograms, estimated peptide abundance ratios and assembled peptide quantifications into protein quantification lists (using default settings) based on proteins identified by SEQUEST from the DTASelect output text file. For each direct comparison, identified peptides were quantified and filtered with a minimum profile signal-to noise ratio cutoff of 2. Proteins consisting of at least two quantified peptides were evaluated for quantification. Confidence intervals were calculated for every protein as an error bar of their abundance ratio estimate. Protein abundance was considered significant if the log ratio (log2 abundance ratio) of the protein is equal to or greater than two (at least a 4 fold difference) and if the confidence interval width does not exceed three. A small CI width corresponds to a more accurate estimation of the ratio difference. The MS raw data and the data analysis configuration files will be available with the other files.
Results
SPA-S12 is effective for affinity capture of ribosomal subunits and associated proteins
We recently constructed an endogenously expressed recombinant S12 protein (SPA-S12) with a C-terminal tag to qualitatively identify candidate S12 interacting proteins. All SPA-S12 and negative control pull-downs were performed with a single affinity chromatographic step and the resulting proteins were separated by SDS-PAGE and subjected to in-gel digestion as previously described (Figure 2). The single affinity chromatographic step was utilized to capture transient interactions. After 1D-LC-MS/MS analysis the Mascot algorithm identified 4251 unique peptides representing proteins from any of nine biological replicates representing three different experimental conditions (see Supplemental Protein Tables S1A, SIB and SIC). These experimental conditions included: whole cell lysate, polysome depleted mid-log phase supernatant and polysome depleted stationary phase supernatant. For each experimental condition we considered protein identifications based on two or more unique peptides and detected in at least two of three biological replicates. Protein identifications were considered SPA-S12 specific interactors if they were unique to (or estimated to be more abundant compared to control) the SPA-S12 pull-downs. A total of 170 SPA-S12 specific proteins were identified this way (see Supplemental Table S2, S3 and S4). All identified proteins from this work were categorized into three major groups using gene annotations which include: 1) ribosomal proteins, 2) ribosomal associated proteins and 3) non-specific proteins (non-ribosomal). The second category included those proteins that were previously reported in the literature to interact with ribosomes, conserved uncharacterized proteins and proteins that may be indirectly associated via polysomes. A majority of the proteins from this work belong in the first two categories; however, a third category consists of proteins with no apparent functional relationship to the ribosome and/or polysome. To better understand the nature of the protein identifications from our work we additionally utilized the EMBL-EBI IntAct molecular interaction database (http://www.ebi.ac.uk/intact/main.xhtml) to compare proteins identified from all three experimental conditions with other published interaction data. When proteins from this work are listed as bait (tagged species) our pull-down results match 72% of the captured protein identifications listed in the IntAct database; the number is considerably smaller (45%) when proteins are listed as prey (captured proteins) (see supplemental table S5). A key observation from this analysis is that many of the apparent non-specific identifications in our supplemental section are among those consistently identified in the IntAct database by other groups.
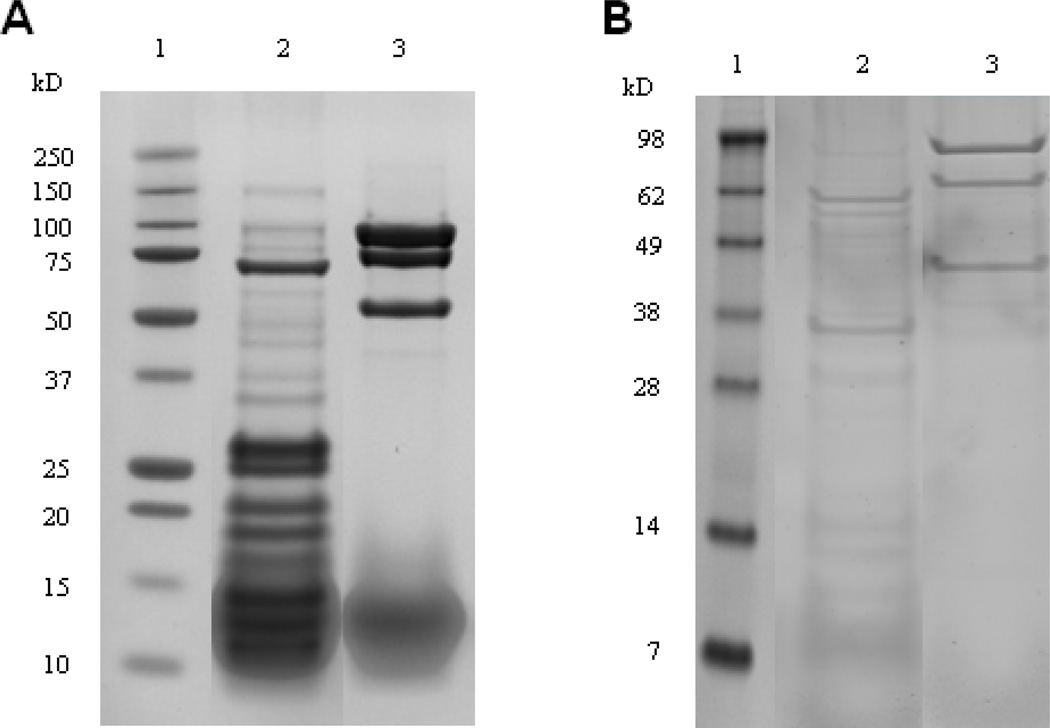
Figure 2.
SPA tagged S12 (SPA-S12) pull-downs in whole cell lysates enriched for proteins representing a heterogeneous population of isolated 30S subunits, 70S ribosomes and/or translationally active polysomes of ribosomal small subunit associated complexes. Pull-downs performed in polysome depleted extract resulted in the enrichment of ribosomal small subunit associated complexes (A) Polypeptides from eluates representing SPA-S12 purified from whole cell lysates (lane 2) and negative control (lane 3) (B) Polypeptides from eluates representing SPA-S12 purified from polysome depleted extract (lane 2) and negative control (lane 3). Pull-downs were separated by SDS-PAGE (10% polyacrylamide gels) prior to in-gel tryptic digestion and ID LC tandem mass spectrometry. Molecular weight standards are represented in lane 1. The image representing (B) have been displayed in a previous report (18).
SPA-S12 pull-down of polysomes from whole cell lysate
The first set of pull-downs performed in whole cell lysate resulted in the capture of proteins (from categories one and two) most likely associated with a heterogeneous population of isolated 30S subunits, 70S ribosomes and/or translationally active polysomes (clusters of ribosomes joined by a common mRNA template). The number of 30S protein identifications and spectra (spectral hits) matched to small subunit peptides suggests the 30S subunit to be the most abundant (See table 1). All 21 small subunit proteins were identified compared to only 11 large subunit proteins (out of 34 possible 50S proteins). Furthermore, the average sequence coverage per small subunit protein was approximately 77% and the average number of spectra matched to 30S proteins was 892 per protein. These values were much smaller for large subunit proteins (38% average sequence coverage and an average of 43 spectra matched per protein). The capture of ribosome associated proteins listed in table 2A and 2B suggests that a sub-population of polysomes were also isolated. These proteins are involved in several co-linear processes (including translation and transcription) that occur on bacterial polysomes (24)–(25) (26–34). Many ribosomal associated proteins identified from this pull-down strategy were among those previously found to be associated with sucrose gradient purified E. coli ribosomes (and subunits) from separate iTRAQ studies (listed in table 2A and 2B) (35)-(36). In those studies the authors verified the ribosome association of select proteins and in several cases localized the association to a specific subunit. The sample complexity represented by our whole cell lysate pull-downs prevented the definitive assignment to a ribosomal subunit. Since our goal was to identify S12 interacting proteins we pursued experimental conditions aimed at substantially reducing this sample complexity.
Table 1.
Small and large ribosomal subunit proteins from SPA-S12 pull-downs performed in whole cell lysates
| Matched | ||||
|---|---|---|---|---|
| Accession | Protein Name | Unique Peptides | Spectra | % Sequence Coverage |
| P0AG67 | S1 | 68 | 4548 | 98 |
| P0A7V0 | S2 | 32 | 2312 | 100 |
| P0A7V3 | S3 | 21 | 1580 | 82.8 |
| P0A7V8 | S4 | 24 | 1016 | 72.3 |
| P0A7W1 | S5 | 20 | 1179 | 95.2 |
| P02358 | S6 | 12 | 577 | 85.9 |
| P02359 | S7 | 26 | 1504 | 100 |
| P0A7W7 | S8 | 20 | 836 | 100 |
| P0A7X3 | S9 | 13 | 471 | 72.3 |
| P0A7R5 | S10 | 13 | 602 | 97 |
| P0A7R9 | S11 | 8 | 408 | 61.2 |
| P0A7S3 | S12 | 8 | 303 | 49.2 |
| P0A7S9 | S13 | 19 | 748 | 100 |
| P0AG59 | S14 | 7 | 146 | 43.5 |
| P0ADZ4 | S15 | 10 | 213 | 61.7 |
| P0A7T3 | S16 | 11 | 720 | 78 |
| P0AG63 | S17 | 10 | 274 | 65.1 |
| P0A7T7 | S18 | 10 | 520 | 84 |
| P0A7U3 | S19 | 10 | 339 | 70.6 |
| P0A7U7 | S20 | 8 | 225 | 60.9 |
| P68679 | S21 | 7 | 231 | 34.7 |
| P60422 | L2 | 12 | 111 | 43.3 |
| P60438 | L3 | 3 | 24 | 22.9 |
| P60723 | L4 | 5 | 55 | 32.8 |
| P62399 | L5 | 10 | 68 | 58.1 |
| P0AG55 | L6 | 10 | 52 | 49.7 |
| P0A7K2 | L7/L12 | 5 | 27 | 64.5 |
| P0ADY3 | L14 | 2 | 12 | 30.1 |
| P02413 | L15 | 5 | 37 | 43.1 |
| P0ADY7 | L16 | 4 | 71 | 33.8 |
| P61175 | L22 | 6 | 17 | 44.5 |
Table 2.
| A. Ribosomal maintenance/translational/folding proteins from SPA-S12 pull-downs performed in whole cell lysates | |||||
|---|---|---|---|---|---|
| Acession | Unique | Matched | % Seq. | Comment | |
| Interacting Protein | Peptides | Spectra | Cov. | ||
| P06992 | RsmA | 10 | 116 | 58.6 | Ribo. RNA 30S methyltransferase A |
| P36929 | RsmB | 18 | 141 | 59.7 | Ribo. RNA 30S methyltransferase B |
| P39406 | RsmC | 21 | 171 | 77.5 | Ribo. RNA 30S methyltransferase C |
| P0ADX9 | RsmD | 4 | 19 | 27.7 | Ribo. RNA 30S methyltransferase D |
| P0AGL7 | RsmE | 9 | 65 | 66.2 | Ribo. RNA 30S methyltransferase E |
| P60390 | RsmH | 14 | 102 | 52.4 | Ribo. RNA 30S methyltransferase H |
| P75876 | Rlml | 15 | 85 | 47 | Ribo. RNA 50S methyltransferase I |
| P0AA39 | RluC* | 6 | 20 | 21.9 | Ribo. 50S pseudouridine synthase C |
| P55135 | RumA | 5 | 12 | 15.5 | 23S rRNA (uracil-5-)-methyltransferase |
| P0AGJ5 | yfiF* | 14 | 174 | 46.4 | Unchar. tRNA/rRNA methyltransferas |
| P76550 | yffS | 16 | 143 | 56.5 | Uncharacterized protein |
| P0A8A8 | RimP* | 5 | 58 | 28.6 | Ribosome maturation factor |
| P0A7X6 | RimM* | 9 | 37 | 58.2 | Ribosome maturation factor |
| P43329 | HrpA | 43 | 312 | 41.4 | ATP-dependent RNA helicase |
| P69222 | IF-1 | 4 | 27 | 94.4 | Translation initiation factor |
| P0A705 | IF-2* | 44 | 475 | 59.8 | Translation initiation factor |
| P0A707 | IF-3* | 14 | 568 | 77.2 | Translation initiation factor |
| P0A6M8 | Elongation factor G | 22 | 111 | 49.4 | Elongation factor G |
| P21499 | RNase R* | 52 | 625 | 71.2 | Ribonuclease R |
| P21338 | RNase I* | 11 | 86 | 66 | Ribonuclease I |
| P30850 | Exoribonuclease 2 | 11 | 29 | 21.1 | Exoribonuclease 2 |
| P0A9J0 | RNase G | 13 | 43 | 37 | Ribonuclease G |
| P0A6X3 | Protein hfq* | 6 | 35 | 64.7 | Protein hfq |
| P05055 | PNP transferase * | 15 | 58 | 29.8 | Polyribonucleotide nucleotidyltransferase |
| P10408 | SecA | 22 | 64 | 35.8 | Protein translocase subunit secA |
| P0A6Z3 | HtpG | 4 | 11 | 8.9 | Chaperone protein htpG |
| P08622 | DnaJ | 6 | 22 | 23.7 | Chaperone protein dnaJ |
| P0A850 | Trigger factor* | 16 | 99 | 47.9 | Trigger factor |
| P63284 | ClpB* | 21 | 78 | 38.5 | Chaperone protein clpB |
| B. Transcription proteins from SPA-S12 pull-downs performed in whole cell lysates | |||||
|---|---|---|---|---|---|
| Acession | Protein | Unique | Matched | % Seq | Comments |
| Name | Peptides | Spectra | Cov. | ||
| P0A7Z4 | RpoA* | 13 | 37 | 52.3 | DNA-directed RNA polymerase subunit alpha |
| P0A8V2 | RpoB* | 37 | 142 | 36.9 | DNA-directed RNA polymerase subunit beta |
| P0A8T7 | RpoC* | 47 | 290 | 41.5 | DNA-directed RNA polymerase subunit beta' |
| P0A8W0 | NanR | 4 | 11 | 23.9 | Transcriptional regulator |
| P0ACN4 | AllR | 8 | 61 | 35 | HTH-type transcriptional repressor |
| P03030 | LysR | 5 | 11 | 27.6 | Transcriptional activator protein |
| P37671 | YiaJ | 9 | 24 | 50.4 | HTH-type transcriptional regulator |
| P0ACP5 | GntR | 10 | 35 | 43.8 | HTH-type transcriptional regulator |
| P0AG30 | Rho | 6 | 20 | 17.9 | Transcription termination factor |
| P39356 | yjhU | 3 | 6 | 12.2 | Uncharacterized transcriptional regulator |
| P30864 | YafC | 8 | 35 | 28.3 | Uncharacterized HTH-type transcriptional regulator |
| P77300 | YagI | 6 | 24 | 30.9 | Uncharacterized HTH-tvpe transcriptional regulator |
Polysome depletion results in capture of small subunit ribonucleoprotein particles (RNPs) and associated proteins
We attempted to completely remove ribosomes and subunits by subjecting whole cell lysates to 100K centrifugation (see materials and methods). Affinity pull-downs were then performed in the resulting supernatant to potentially capture the unassembled form of SPA-S12 and directly associating partners under mid-log and stationary phase conditions. The subsequent affinity captures were free of polysomes (and 50S large subunit proteins) but surprisingly resulted in the capture of SPA-S12 in a complex with small ribonucleoprotein particles (RNPs) possibly representing partially assembled small subunit RNPs (see table 3A). We therefore referred to these samples as polysomal depleted extract (PDE) samples. Nearly identical small subunit proteins were found under both growth conditions which included only primary binding proteins (S4, S7, S8, S15 and S20) or secondary binding proteins (S5, S6, S9, S11, S12, S13 and S18); both are categorized by the stage or order at which they assemble during 30S assembly (37)-(38). We previously demonstrated that these were co-isolated with 16S ribosomal RNA (rRNA) thereby confirming the capture of small subunit RNPs.
Table 3.
| A. Ribosomal proteins from SPA-S12 pull-downs performed in polysome-free (PDE) mid-log and stationary phase lysates | |||||||
|---|---|---|---|---|---|---|---|
| Midlog | Stationary | ||||||
| Acession | Protein | Unique | Matched | % Seq. | Unique | Matched | % Seq. |
| Name | Peptides | Spectra | Cov. | Peptides | Spectra | Cov. | |
| P0AG67 | S1 | 26 | 176 | 50.8 | 42 | 417 | 93.7 |
| P0A7V8 | S4 | 16 | 142 | 58.7 | 15 | 190 | 68.9 |
| P0A7V3 | S3 | ** | 11 | 143 | 56.2 | ||
| P0A7W1 | S5 | 12 | 114 | 82.6 | 12 | 323 | 41.7 |
| P02358 | S6 | 8 | 80 | 62.9 | 6 | 24 | 61.5 |
| P02359 | S7 | 15 | 147 | 69.8 | 14 | 240 | 60.8 |
| P0A7W7 | S8 | 8 | 82 | 67.7 | 10 | 170 | 79 |
| P0A7X3 | S9 | 6 | 58 | 42.3 | 8 | 133 | 48.4 |
| P0A7R5 | S10 | 5 | 31 | 43.7 | 9 | 114 | 53.4 |
| P0A7R9 | S11 | 7 | 29 | 79.8 | 5 | 56 | 50 |
| P0A7S3 | S12 | 6 | 71 | 24.2 | 8 | 96 | 55.6 |
| P0A7S9 | S13 | 9 | 60 | 61 | 9 | 92 | 67.8 |
| P0ADZ4 | S15 | 8 | 90 | 68.5 | 8 | 36 | 60 |
| P0A7T7 | S18 | 5 | 63 | 50.6 | 5 | 99 | 50.6 |
| P0A7U7 | S20 | 4 | 22 | 55.2 | ** | ||
| B. Candidate S12 binders from polysome-free (PDE) mid-log and stationary | |||||||
|---|---|---|---|---|---|---|---|
| Midlog | Sationary | ||||||
| Acession | Protein Name | Unique | Matched | % Seq. | Unique | Matched | % Seq. |
| Peptides | Spectra | Cov. | Peptides | Spectra | Cov. | ||
| P0AEI4 | MiaB like Methyltransferase RimO | 9 | 34 | 24.3 | ** | ||
| P75838 | uncharacterized ycaO | 21 | 237 | 44.3 | 13 | 103 | 24.5 |
| P76116 | uncharacterized yncE | 10 | 44 | 38.8 | 4 | 11 | 18.4 |
| P33355 | uncharacterized yehS | 7 | 22 | 50 | ** | ||
| P0AF93 | uncharacterized yjgf | 7 | 52 | 77.4 | 8 | 32 | 69.5 |
| P0CE47 | Elongation factor Tu | 34 | 1370 | 84.5 | ** | ||
| P0A6P1 | Elongation factor Ts | 16 | 52 | 55.8 | ** | ||
| P0A6M8 | Elongation factor G | 43 | 482 | 73 | 25 | 131 | 53.8 |
| P0A6N4 | Elongation factor P | 5 | 17 | 26 | ** | ||
| P21499 | RNaseR | 22 | 67 | 41.2 | 31 | 145 | 50.5 |
| P0A6X3 | Hfq-1 | 7 | 25 | 54 | 5 | 20 | 44 |
| P0A805 | Ribosome Recycling Factor | 6 | 21 | 47 | 5 | 12 | 35.7 |
| P0A8A8 | Ribosome maturation factor rimP | 6 | 40 | 33 | 6 | 80 | 35.3 |
Protein was either not detected at all or was below the baseline level.
Several ribosome associated proteins (see table 3B) identified in the PDE datasets were designated as candidate S12 interactors, including those implicated in previous reports to interact with the 30S subunit (or S12), to be linked to β-methylthiolation of S12 and conserved proteins that do not have a defined function (39, 40) (33), (117), (13), (1).
Peptide affinity pull-downs capture two S12 specific proteins
Our interest in S12 β-methylthiolation prompted the design of a strategy aimed at capturing proteins that recognize and interact with the D88 containing conserved loop. Toward this goal, mass spectrometry was used to compare proteins that bind to a biotinylated peptide that mimics the S12 D88 conserved loop (S12 peptide) and/or a biotinylated negative decoy control peptide (see table 4). The decoy control is an isomer with a scrambled amino acid sequence that is not homologous to any known sequence in genomic databases; proteins binding to the negative control will therefore be nonspecific. Proteins that are significantly enriched and/or bind specifically to the S12 peptide are therefore likely to form interactions with S12. A similar peptide bait strategy has been employed to identify specific regions where proteins bind and/or to elucidate the effect a PTM may have on binding (41).
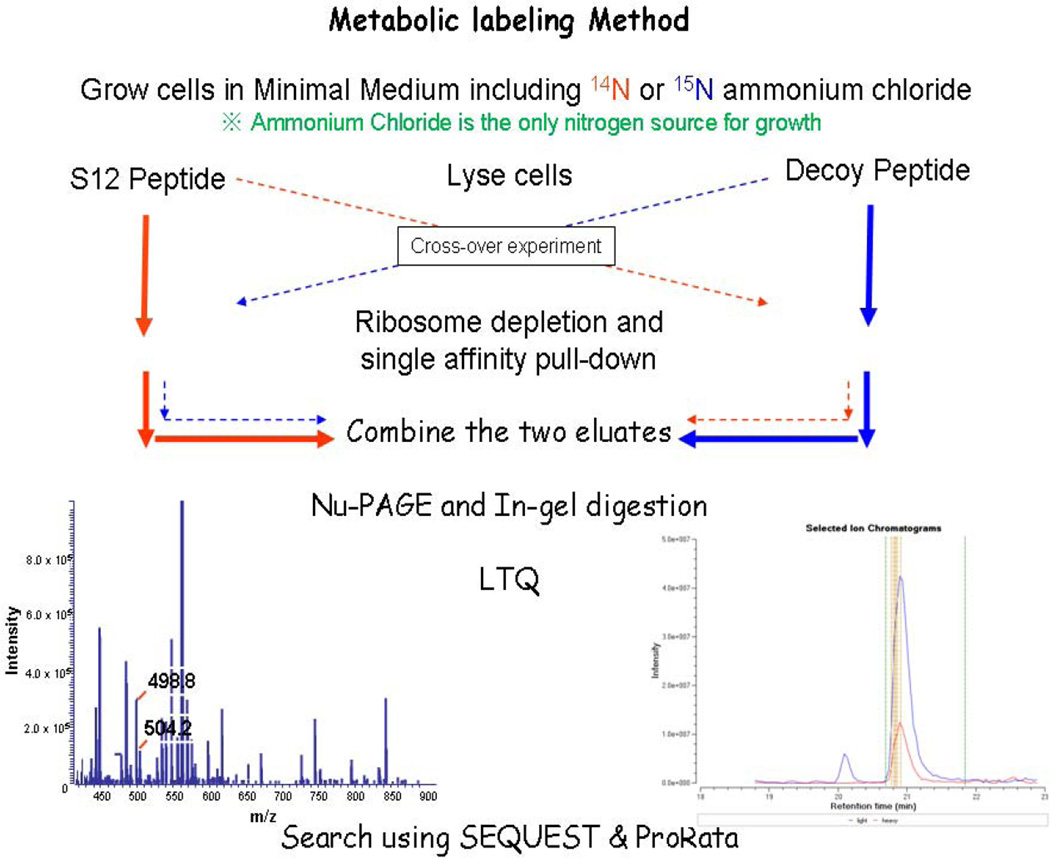
We next utilized isotope ratio proteomics by performing peptide affinity pull-downs using cell lysates from 14N and 15N metabolically labeled cell populations (see Figure 3). After 1D-LC/MS/MS analysis, extracted ion chromatograms and peptide abundance ratios were determined for all SEQUEST identified isotopologous peptide pairs using the software ProRata. Heavy and light versions of the same peptide were distinguishable with equal efficiency by mass spectrometry; the ion current ratio between two peptides, therefore, accurately reflected the ratio of the proteins in two populations. The total numbers of quantified proteins identified from pull-down and crossover experiments (see Table 5A) were fairly consistent although a small amount of variation was observed for background and decoy peptide specific protein interactors. All ProRata identifications meeting the criteria described in the experimental section are listed in Supplemental Table S6. Because no protein should specifically bind the decoy control peptide we used stringent abundance ratio criteria for determining S12 specific interactions. For each pull-down, background and non-specific binding proteins were those that were quantified to have log2 ratios < 2 in both pull-down experiments or were only identified in a single S12 peptide experiments (not found in crossover). A ratio of less than two indicates that a protein interacted equally to the S12 and decoy peptides. Proteins were, therefore, determined to be true S12 peptide interactors if they were identified with at least a 4 fold difference (corresponds to a log2 ratio of >2) in both pull-down and crossover experiments. We identified Hfq-1 specific interactions with the S12 peptide in both mid-log and stationary phase. Additionally we found RnaseR to interact with S12 in stationary phase (see table 5B). Both proteins have functions involving the ribosome and are S12 candidate interactors from SPA-S12 recombinant data listed in table 3B. Taken together, the correlation between datasets strongly suggests these proteins to interact with S12 at least partly via the D88 containing loop region.
Figure 3.
Schematic diagram representing peptide pull-down strategy. Synthetic S12 and decoy peptide pull-downs were incubated in PDE minimal media (14N or 15N). The combined eluates were subjected to PAGE/in gel digestions and then analyzed by 1D-LC/MS/MS. To reduce the variability associated with background protein binding, cross experiments were performed in which the S12 peptide was incubated in the lysate from 15N labeled cells in one pull-down (the scrambled peptide would be incubated in 14N labeled lysate) and 14N labeled lysate was in the next pull-down. After 1D-LC/MS/MS analysis, extracted ion chromatograms and peptide abundance ratios were determined for isotopologous peptide pairs. Heavy and light versions of the same peptide were distinguishable with equal efficiency; the ion current ratio therefore, reflected the ratio of the proteins in two populations.
Table 5.
| A. Total proteins identifications from S12 synthetic and scrambled decoy peptide pull-downs for mid-log and stationary phase cross-over experiments | ||
|---|---|---|
| Pull-down | Midlog Total Protein ids |
Stationary Total Protein ids |
| Pull-down | ||
| Total Quantified Protein ids | 185 | 112 |
| S12 peptide specific | 69 | 32 |
| Decoy peptide specific | 69 | 32 |
| Crossover | ||
| Total Quantified Protein ids | 119 | 113 |
| S12 peptide specific | 56 | 36 |
| Decoy peptide specific | 56 | 36 |
| B. S12 specific protein identifications from S12 synthetic peptide pull-downs for mid-log and stationary phase crossover experiments | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Acession | Protein Name | Pull-down | Crossover | ||||||
| No. peptides |
Cl* | log2 Ratio | Ratio* | No. peptides |
Cl* | log2 Ratio | Ratio | ||
| Mid-log phase | |||||||||
| P0A6X3 | Protein hfq | 7 | [4,5] | 4.5 | 23 | 5 | [2,3.2] | 2.6 | 6 |
| Stationary Phase | |||||||||
| P0A6X3 | Protein hfq | 5 | [1.8,2.8] | 2.3 | 5 | 10 | [2.1,3] | 2.6 | 6 |
| P21499 | Rinbonuclease R | 15 | [2.9,3.8] | 3.3 | 10 | 6 | [1.3,2.8] | 2 | 4 |
confidence interval widths for ProRata profile likelihood curves. A small CI width corresponds to a more accurate estimation of the ratio difference
Discussion
A primary goal of this research was to perform experiments aimed at testing the possibility that the D88 conserved loop region forms proteins-protein interactions. We therefore applied a complimentary proteomic approach using a combination of SPA-S12 pull-down and peptide affinity strategies to identify S12 binding proteins. Experimental strategies presented here revealed proteins that form direct interactions with the 30S subunit, and additionally elucidated which are likely to be S12 specific interactions via the conserved loop. Although many questions remain, this work provided evidence implicating the conserved loop region to be part of a protein binding interface.
Pull-downs from PDE samples localize several proteins to the ribosome small subunit
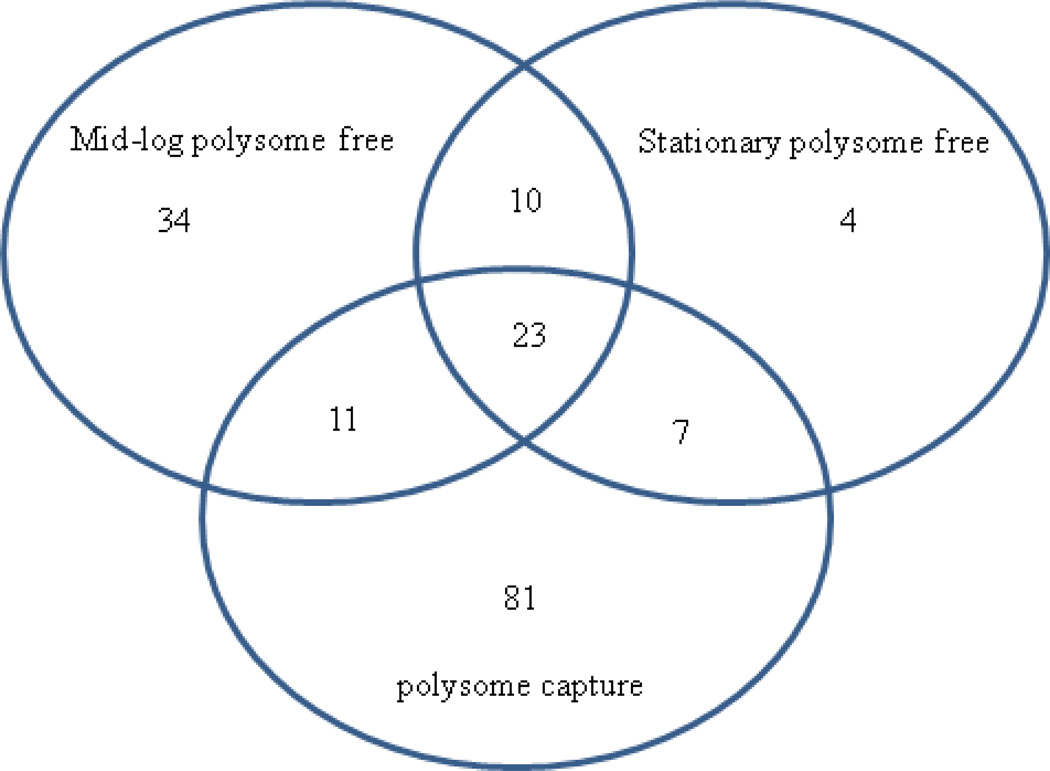
As illustrated in figure 4, the 100K centrifugal (removal) step significantly reduced the sample complexity observed in polysomal captures. Specifically, proteins unique to mid-log and stationary phase PDE pull-downs represented approximately 2 fold and 20 fold fewer identifications, respectively, than polysomal samples. Indeed, the PDE affinity capture excluded the identification of 50S large subunit proteins and many polysome associated proteins; a majority (30 out of 41) of the proteins listed in tables 2A and 2B are absent in tables 3A and 3B. The pull-down of small subunit complexes strongly suggests that proteins in table 3B were localized to the 30S subunit indicating a greater degree of resolution for subunit assignment than the polysomal dataset. To our surprise, both (mid-log and stationary phase) PDE pull-downs resulted in capturing nearly identical primary and secondary small subunit proteins. This suggests that these partially assembled particles were unique to this experimental condition and likely represent a low abundant complex in whole cell lysates (all 21 small subunit proteins were identified in whole cell lysate pull-downs). This is supported by the fact that several 30S specific proteins in table 3B were not identified in whole cell lysates, including the protein RimO, and the previously uncharacterized protein YcaO (which was among the most abundant as determined by spectra matched to peptides). These proteins were present in whole cell lysates but due to sample complexity were stochastically below the detection limit. On the other hand, under PDE pull-down conditions RimO and YcaO were enriched as reflected by their abundance in table 3B.
Figure 4.
Venn diagram representing total protein identifications from each SPA-S12 experimental condition. The whole cell lysate pull-downs resulted in capturing proteins associated with a heterogeneous population of isolated 30S subunits, 70S ribosomes and/or translationally active polysomes. Proteins unique to mid-log and stationary phase PDE pull-downs represented approximately 2 fold and 20 fold fewer identifications, respectively, than polysomal samples.
A large portion of proteins in table 3B have been reported to interact with the 30S small subunit and/or S12 thereby providing an additional level of confidence for assigning potential S12 interactors. Among those implicated to directly interact with S12 include the proteins Elongation factor-Tu, Elongation Factor G, the ribosome recycling factor and RimP (33). All three proteins are involved in either translation or recycling subunits. Interestingly, EF-Tu was negligibly identified in whole cell lysates but was the most abundantly identified in mid-log phase PDE as indicated by the number of spectra matched to peptides. Additionally, the above elongations factors, and RimO listed in table 3B were exclusively identified under mid-log phase conditions suggesting that these proteins may have reduced expression levels under stationary phase conditions. Based on our previous characterization regarding the role RimO and YcaO play in modifying S12 and the fact that these modifying enzyme interactions are likely to be transient, the data in 3B also suggest that RimO and YcaO likely modify S12 in this small subunit ribonucleoprotein particle in mid-log phase.
Identification of two proteins by peptide pull-down suggest the D88 containing loop to be part of a protein binding interface
Preliminary experiments using label free spectral counting were performed to compare differences in S12 and decoy peptide pull-downs (data not shown). While spectral counting provided preliminary data suggesting Rnase R and hfq-1 as potential interactors, the peptide pull-down strategy outlined in Figure 3 provided a more stringent complimentary approach (to SPA-S12 recombinant pull-downs) for identifying proteins that might specifically interact with the S12 D88 conserved loop. The use of synthetic peptides as bait can have potential limitations. First, a binding domain is often composed of several regions within the protein structure and peptide baits used outside of the context of the protein may reduce binding affinity. We did not identify RimO or YcaO in these pull-downs suggesting that they may require additional subunit interactions. Second, biologically specific protein interactions maybe masked by more abundant nonspecific interactions. The use of isotope ratio abundance measurements (log2 ratios), the decoy peptide control and cross-over pull-down experiments allowed a stringent method for filtering out these false positives. With this point being made, Rnase R and Hfq-1 were specifically captured with the S12 peptide and are therefore likely to form interactions with the S12 D88 conserved second loop region. Additionally, several factors found in both recombinant SPA-S12 and peptide pull-downs provided compelling evidence that Rnase R and Hfq-1 interacted with the S12 D88 second loop. These two proteins are likely to be specific to the S12 loop as they do not interact with the decoy control peptide. If both were merely engaging in non-specific binding we would have obtained ratios indicating equivalent binding (log2 ratios < 2; close to unity) to the decoy peptide. These proteins were also among the S12 candidates identified in SPA-S12 PDE data (and were abundant in the polysome datasets too). The correlation between two complementary approaches provides the strongest evidence that both proteins likely make direct contacts with S12 via the second loop region (and potentially other 30S elements). Interestingly, Rnase R has been shown to be involved in removing defective rRNAs from assembled subunits, facilitating ribosomal degradation during starvation (42, 43) and the selective decay of defective mRNAs to prevent translational stalling (on active polysomes) (44). The above studies aimed at understanding ribosomal degradation (and rRNA removal), collectively indicate that Rnase R may target individual subunits and may specifically make direct contacts with the exposed 30S subunit interface (S12 is located at the A site near the interface). Our results are consistent with this possibility by providing the first interaction data describing Rnase R contacts with a ribosomal subunit. It should also be noted Rnase R expression increases under stress conditions which may explain why we did not capture this protein in peptide pull-downs from mid-log phase conditions(45). Hfq has been shown to be involved in regulating the stability of various mRNAs by possibly competing with 30S subunits for binding to the mRNA template. This model suggests that Hfq does this by interactions with mRNA and blocking ribosome binding (46). The data we present in this report suggest that an interaction involving S12 may be important for Hfq function.
Conclusion
In a recent paper we reported data (from ΔrimO and ΔycaO genetic knockout studies) implicating a potential role for β-methylthiolation involving transcriptional regulation of anaerobic genes via enhancement of FNR and NarL mRNA translation (1). These results strongly suggested that if the PTM is involved in modulating translation, the D88 containing loop is likely part of a protein binding interface. While Rnase R and Hfq-1 both have been reported to play a role in mRNA maintenance stability future studies will need to be performed to determine if these potential interactions contribute to changes in FNR and NarL levels. An initial step toward this goal would be to identify how β-methylthiolated D88 affects the binding properties of this peptide. We recently synthesized β-methylthio-aspartic acid (are currently verifying that it is chemically equivalent to that on S12 D88) for making a modified version of the biotinylated peptide mimic. Using the same pull-down strategy we will quantify differences in binding properties for both peptides. The results from this work will potentially link this PTM to binding interactions and further support a model indicating the S12 conserved second loop to be part of a 30S subunit binding interface.
Supplementary Material
Acknowledgments
We would like to thank Jack Greenblatt (University of Toronto, Canada) for generously providing the SPA cassette used for generating SPA tagged genes. We would like to thank Mikhail Bunenko for his valuable comments and suggestions (National Cancer Institute/Frederick Cancer Research and Development Center, Frederick, MD). We would like to thank Chongle Pan for assistance in using ProRata software. We would like to thank Todd L. Mollan (CBER/FDA), Adele Blackler (NCI/NIMH) and Christopher Crutchfield (NICFID/NIH) for assistance with reading and/or editing this manuscript. M. B. Strader contributed as primary author and researcher. W. Hervey IV, N. Costantino, C. Chen, S. Fujigaki, C. Ihunnah and A. Makusky participated as supporting researchers. D. Court, S. Markey and J. Kowalak participated in the writing and/or editing of this manuscript. This work was supported by the Intramural Research Program of the National Institute of Mental Health (1ZIAMH000274).
References
- 1.Strader MB, Costantino N, Elkins CA, Chen CY, Patel L, Makusky AJ, Choy JS, Court DL, Markey SP, Kowalak JA. A proteomic and transcriptomic approach reveals new insight into beta- methylthiolation of Escherichia coli ribosomal protein S12. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.005199. M110 005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 3.Gustilo EM, Vendeix FA, Agris PF. tRNA's modifications bring order to gene expression. Curr Opin Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kowalak JA, Walsh KA. Beta-methylthio-aspartic acid: identification of a novel posttranslational modification in ribosomal protein S12 from Escherichia coli. Protein Sci. 1996;5:1625–1632. doi: 10.1002/pro.5560050816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 6.Carr JF, Hamburg DM, Gregory ST, Limbach PA, Dahlberg AE. Effects of streptomycin resistance mutations on posttranslational modification of ribosomal protein S12. J Bacteriol. 2006;188:2020–2023. doi: 10.1128/JB.188.5.2020-2023.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suh MJ, Hamburg DM, Gregory ST, Dahlberg AE, Limbach PA. Extending ribosomal protein identifications to unsequenced bacterial strains using matrix-assisted laser desorption/ionization mass spectrometry. Proteomics. 2005;5:4818–4831. doi: 10.1002/pmic.200402111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strader MB, Verberkmoes NC, Tabb DL, Connelly HM, Barton JW, Bruce BD, Pelletier DA, Davison BH, Hettich RL, Larimer FW, Hurst GB. Characterization of the 70S Ribosome from Rhodopseudomonas palustris using an integrated "top-down" and "bottom- up" mass spectrometric approach. J Proteome Res. 2004;3:965–978. doi: 10.1021/pr049940z. [DOI] [PubMed] [Google Scholar]
- 9.Gupta N, Tanner S, Jaitly N, Adkins JN, Lipton M, Edwards R, Romine M, Osterman A, Bafna V, Smith RD, Pevzner PA. Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17:1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Running WE, Reilly JP. Ribosomal proteins of Deinococcus radiodurans: their solvent accessibility and reactivity. J Proteome Res. 2009;8:1228–1246. doi: 10.1021/pr800544y. [DOI] [PubMed] [Google Scholar]
- 11.Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ. RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli. Proc NatlAcadSci USA. 2008;105:1826–1831. doi: 10.1073/pnas.0708608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arragain S, Garcia-Serres R, Blondin G, Douki T, Clemancey M, Latour JM, Forouhar F, Neely H, Montelione GT, Hunt JF, Mulliez E, Fontecave M, Atta M. Post-translational modification of ribosomal proteins: structural and functional characterization of RimO from Thermotoga maritima, a radical S-adenosylmethionine methylthiotransferase. J Biol Chem. 2010;285:5792–5801. doi: 10.1074/jbc.M109.065516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee KH, Saleh L, Anton BP, Madinger CL, Benner JS, Iwig DF, Roberts RJ, Krebs C, Booker SJ. Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily. Biochemistry. 2009;48:10162–10174. doi: 10.1021/bi900939w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierrel F, Hernandez HL, Johnson MK, Fontecave M, Atta M. MiaB protein from Thermotoga maritime. Characterization of an extremely thermophilic tRNA-methylthiotransferase. J Biol Chem. 2003;278:29515–29524. doi: 10.1074/jbc.M301518200. [DOI] [PubMed] [Google Scholar]
- 15.Pierrel F, Douki T, Fontecave M, Atta M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J Biol Chem. 2004;279:47555–47563. doi: 10.1074/jbc.M408562200. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez HL, Pierrel F, Elleingand E, Garcia-Serres R, Huynh BH, Johnson MK, Fontecave M, Atta M. MiaB, a bifunctional radical-S-adenosylmethionine enzyme involved in the thiolation and methylation of tRNA, contains two essential [4Fe-4S] clusters. Biochemistry. 2007;46:5140–5147. doi: 10.1021/bi7000449. [DOI] [PubMed] [Google Scholar]
- 17.Booker SJ, Cicchillo RM, Grove TL. Self-sacrifice in radical S-adenosylmethionine proteins. Curr Opin Chem Biol. 2007;11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, Richards D, Beattie B, Emili A, Greenblatt JF. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. JProteome Res. 2004;3:463–468. doi: 10.1021/pr034084x. [DOI] [PubMed] [Google Scholar]
- 19.Hervey WJt, Khalsa-Moyers G, Lankford PK, Owens ET, McKeown CK, Lu TY, Foote LJ, Asano KG, Morrell-Falvey JL, McDonald WH, Pelletier DA, Hurst GB. Evaluation of affinity-tagged protein expression strategies using local and global isotope ratio measurements. J Proteome Res. 2009;8:3675–3688. doi: 10.1021/pr801088f. [DOI] [PubMed] [Google Scholar]
- 20.Slotta DJ, McFarland MA, Markey SP. MassSieve: panning MS/MS peptide data for proteins. Proteomics. 2010;10:3035–3039. doi: 10.1002/pmic.200900370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan C, Kora G, McDonald WH, Tabb DL, VerBerkmoes NC, Hurst GB, Pelletier DA, Samatova NF, Hettich RL. ProRata: A quantitative proteomics program for accurate protein abundance ratio estimation with confidence interval evaluation. Anal Chem. 2006;78:7121–7131. doi: 10.1021/ac060654b. [DOI] [PubMed] [Google Scholar]
- 23.Pan C, Kim J, Chen L, Wang Q, Lee C. The HIV positive selection mutation database. Nucleic Acids Res. 2007;35:D371–D375. doi: 10.1093/nar/gkl855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Buul CP, van Knippenberg PH. Nucleotide sequence of the ksgA gene of Escherichia coli: comparison of methyltransferases effecting dimethylation of adenosine in ribosomal RNA. Gene. 1985;38:65–72. doi: 10.1016/0378-1119(85)90204-5. [DOI] [PubMed] [Google Scholar]
- 25.Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W. The native 3D organization of bacterial polysomes. Cell. 2009;136:261–271. doi: 10.1016/j.cell.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Del Campo M, Ofengand J, Malhotra A. Crystal structure of the catalytic domain of RluD, the only rRNA pseudouridine synthase required for normal growth of Escherichia coli. RNA. 2004;10:231–239. doi: 10.1261/rna.5187404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lesnyak DV, Osipiuk J, Skarina T, Sergiev PV, Bogdanov AA, Edwards A, Savchenko A, Joachimiak A, Dontsova OA. Methyltransferase that modifies guanine 966 of the 16 S rRNA: functional identification and tertiary structure. J Biol Chem. 2007;282:5880–5887. doi: 10.1074/jbc.M608214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basturea GN, Rudd KE, Deutscher MP. Identification and characterization of RsmE, the founding member of a new RNA base methyltransferase family. RNA. 2006;12:426–434. doi: 10.1261/rna.2283106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura S, Suzuki T. Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 2010;38:1341–1352. doi: 10.1093/nar/gkp1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purta E, O'Connor M, Bujnicki JM, Douthwaite S. YccW is the m5C methyltransferase specific for 23S rRNA nucleotide 1962. J Mol Biol. 2008;383:641–651. doi: 10.1016/j.jmb.2008.08.061. [DOI] [PubMed] [Google Scholar]
- 31.Gustafsson C, Persson BC. Identification of the rrmA gene encoding the 23S rRNA mlG745 methyltransferase in Escherichia coli and characterization of an mlG745-deficient mutant. J Bacteriol. 1998;180:359–365. doi: 10.1128/jb.180.2.359-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang L, Ku J, Pookanjanatavip M, Gu X, Wang D, Greene PJ, Santi DV. Identification of two Escherichia coli pseudouridine synthases that show multisite specificity for 23S RNA. Biochemistry. 1998;37:15951–15957. doi: 10.1021/bi981002n. [DOI] [PubMed] [Google Scholar]
- 33.Nord S, Bylund GO, Lovgren JM, Wikstrom PM. The RimP protein is important for maturation of the 30S ribosomal subunit. J Mol Biol. 2009;386:742–753. doi: 10.1016/j.jmb.2008.12.076. [DOI] [PubMed] [Google Scholar]
- 34.Lovgren JM, Wikstrom PM. Hybrid protein between ribosomal protein S16 and RimM of Escherichia coli retains the ribosome maturation function of both proteins. J Bacteriol. 2001;183:5352–5357. doi: 10.1128/JB.183.18.5352-5357.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang M, Sullivan SM, Walker AK, Strahler JR, Andrews PC, Maddock JR. Identification of novel Escherichia coli ribosome- associated proteins using isobaric tags and multidimensional protein identification techniques. J Bacteriol. 2007;189:3434–3444. doi: 10.1128/JB.00090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, Andrews PC, Maddock JR. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J Bacteriol. 2006;188:6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970;226:1214. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- 38.Held WA, Ballon B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 39.Gregory ST, Carr JF, Dahlberg AE. A signal relay between ribosomal protein S12 and elongation factor EF-Tu during decoding of mRNA. RNA. 2009;15:208–214. doi: 10.1261/rna.1355709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pai RD, Zhang W, Schuwirth BS, Hirokawa G, Kaji H, Kaji A, Cate JH. Structural Insights into ribosome recycling factor interactions with the 70S ribosome. J Mol Biol. 2008;376:1334–1347. doi: 10.1016/j.jmb.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulze WX, Mann M. A novel proteomic screen for peptide- protein interactions. J Biol Chem. 2004;279:10756–10764. doi: 10.1074/jbc.M309909200. [DOI] [PubMed] [Google Scholar]
- 42.Cheng ZF, Deutscher MP. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci USA. 2003;100:6388–6393. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zundel MA, Basturea GN, Deutscher MP. Initiation of ribosome degradation during starvation in Escherichia coli. RNA. 2009;15:977–983. doi: 10.1261/rna.1381309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richards J, Mehta P, Karzai AW. RNase R degrades non- stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol Microbiol. 2006;62:1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]
- 45.Chen C, Deutscher MP. Elevation of RNase R in response to multiple stress conditions. J Biol Chem. 2005;280:34393–34396. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- 46.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 47.Carter AP, demons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.