Abstract
To characterize microRNAs (miRNAs) involved in testicular toxicity in cynomolgus monkeys, miRNA profiles were investigated using next‐generation sequencing (NGS), microarray and reverse transcription‐quantitative real‐time‐PCR (RT‐qPCR) methods. First, to identify organ‐specific miRNAs, we compared the expression levels of miRNAs in the testes to those in representative organs (liver, heart, kidney, lung, spleen and small intestine) obtained from naïve mature male and female monkeys (n = 2/sex) using NGS analysis. Consequently, miR‐34c‐5p, miR‐202‐5p, miR‐449a and miR‐508‐3p were identified to be testicular‐specific miRNAs in cynomolgus monkeys. Next, we investigated miRNA profiles after testicular–hyperthermia (TH) treatment to determine which miRNAs are involved in testicular injury. In this experiment, mature male monkeys were divided into groups with or without TH‐treatment (n = 3/group) by immersion of the testes in a water bath at 43 °C for 30 min for 5 consecutive days. As a result, TH treatment induced testicular injury in all animals, which was characterized by decreased numbers of spermatocytes and spermatids. In a microarray analysis of the testis, 11 up‐regulated (>2.0 fold) and 13 down‐regulated (<0.5 fold) miRNAs were detected compared with those in the control animals. Interestingly, down‐regulated miRNAs included two testicular‐specific miRNAs, miR‐34c‐5p and miR‐449a, indicating their potential use as biomarkers for testicular toxicity. Furthermore, RT‐qPCR analysis revealed decreased expression levels of testicular miR‐34b‐5p and miR‐34c‐5p, which are enriched in meiotic cells, reflecting the decrease in pachytene spermatocytes and spermatids after TH treatment. These results provide valuable insights into the mechanism of testicular toxicity and potential translational biomarkers for testicular toxicity. Copyright © 2016 The Authors. Journal of Applied Toxicology published by John Wiley & Sons Ltd.
Keywords: microRNA, testicular toxicity, cynomolgus monkey, biomarker, next‐generation sequencing
Short abstract
In this study, we identified 4 testicular‐specific miRNAs, miR‐34c‐5p, miR‐202‐5p, miR‐449a, and miR‐508‐3p based on next‐generation sequencing of miRNAs from representative organs obtained from naïve mature cynomolgus monkeys. Next, miRNAs were profiled in a model of testicular injury induced by testicular hyperthermia. Microarray and PCR analyses revealed down‐regulation of miR‐34c‐5p in the testis, which is enriched in meiotic cells, reflecting decreased numbers of pachytene spermatocytes and spermatids by the treatment.
Introduction
Although testicular toxicity is one of the major concerns for drug development, the toxicity remains difficult to characterize and predict comprehensively using non‐clinical safety assessments for pharmaceuticals. The cynomolgus monkey has been recognized as the most relevant species for the characterization and prediction of testicular toxicity in humans because of strong similarities of spermatogenesis, sperm morphology and hormonal regulation to those in humans (Dreef et al., 2007; McLachlan et al., 2002). Despite the importance of utilizing monkeys in testicular toxicity assessments, few such investigations have been performed because of several limitations, including the number of animals and availability of assay systems for hormones and other biomarkers.
MicroRNAs (miRNAs) are 20 to 25 nucleotide‐long non‐coding RNAs that regulate functions in various organs by post‐transcriptional repression of messenger RNAs (Bartel, 2004). MiRNAs are highly conserved across species and relatively stable compared with messenger RNAs (Bartel, 2004; Gantier et al., 2011). The conditional knockout (KO) of a miRNA dicer, a prerequisite for the production of miRNAs, in Sertoli cells in mice resulted in infertility, suggesting the importance of miRNAs for spermatogenesis (Papaioannou et al., 2009). Additionally, double KO of miR‐34b/c and miR‐449 in mice resulted in severely disrupted spermatogenesis (Wu et al., 2014). Recently, we demonstrated that levels of testicular miR‐34b and/or miR‐449a were decreased in cynomolgus monkeys treated with ethylene glycol monomethyl ether (EGME), a representative testicular toxicant (Sakurai et al., 2015). Interestingly, decreases in these miRNAs were also seen in the testes of infertile men with histopathological abnormalities such as Sertoli cell‐only syndrome or mixed atrophy (Abu‐Halima et al., 2014). These characteristics and results provided the basis for the potential application of miRNAs as biomarkers for testicular toxicity, and the monkey model would be useful to investigate the mechanisms of testicular toxicity and the potential translational biomarkers in humans. Although testicular‐specific miRNAs have been identified in rats (Minami et al., 2014), miRNA profiling, using nucleotide sequencing and tissue distribution in cynomolgus monkeys, has not been studied adequately.
Lue et al. (2002) reported that testicular hyperthermia (TH)‐treated cynomolgus monkeys exhibited testicular injury characterized by apoptosis of pachytene spermatocytes and round spermatids. Notably, the TH model did not induce any other organ toxicities, unlike nitrobenzene‐ (Berner et al., 2009) or EGME‐induced testicular toxicity models (Johanson, 2000). For this reason, the TH model is considered suitable to investigate testicular toxicity‐specific translational biomarkers.
In the present study, we identified testicular‐specific miRNAs in cynomolgus monkeys using next‐generation sequencing (NGS). Furthermore, testicular toxicity‐related miRNAs were identified in a monkey model of TH‐induced testicular injury using miRNA microarray or reverse transcription‐quantitative real‐time‐PCR (RT‐qPCR) methods.
Materials and methods
Animals
Ten mature cynomolgus monkeys (eight males: six of Indonesian origin and two of Chinese origin; one female each of Indonesian origin and Chinese origin) were obtained from CLEA Japan, Inc. (Indonesian origin, Tokyo, Japan) or Hamri Co., Ltd. (Chinese origin, Ibaraki, Japan). The animals were between 5 and 11 years of age and weighed between 2.8 and 6.6 kg. Monkeys were housed individually in stainless‐steel cages (W 594 mm × D 870 mm × H 1015 mm) in an animal study room maintained at a temperature of 24 °C and humidity of 60%, and the lighting was on a 12‐h light (from 07:00 to 19:00) and 12‐h dark cycle. Commercial pellets for monkeys (PS‐A, Oriental Yeast Co., Ltd., Tokyo, Japan) were given to each animal (100 g day–1) in the morning (from 07:00 to 09:00). On the day of the TH treatment, half of an apple was also given to the animals. All animal procedures were performed in accordance with the guidelines of the Animal Care and Use Committee of Daiichi Sankyo Co., Ltd (Tokyo, Japan). The experimental protocols were approved by the Institutional Animal Care Committee of Daiichi Sankyo Co., Ltd.
NGS analysis
For NGS analysis, two males (Chinese origin) and two females (Chinese and Indonesian origin) were euthanized humanely under anesthesia with an intramuscular injection of ketamine hydrochloride (20 mg kg–1, Ketalar Intramuscular 500 mg; Daiichi Sankyo Co., Ltd) and xylazine (0.5 mg kg–1, Celactal; Bayer Yakuhin, Ltd., Osaka, Japan). The liver (left lateral lobe), heart (left ventricular wall), lung (left caudal lobe), kidney (cortex), spleen, small intestine (jejunum and ileum) and testis (left) were collected, snap‐frozen in liquid nitrogen, and stored at –80 °C. Necropsy was performed between 09:00 to 12:00 for each animal depending on the anesthetic condition of the animal and practical reason. Total RNA was extracted from 50 to 100 mg of each frozen tissue using miRNeasy mini kits (Qiagen, Venlo, the Netherlands) according to the manufacture's protocol followed by pooling the same amount of total RNA from four animals for each organ. The small RNA library construction and deep sequencing were carried out at Hokkaido System Science Co., Ltd. (Hokkaido, Japan). Briefly, 1 µg of total RNA including small RNA was prepared into a sequence library according to the instructions for the TruSeq small RNA sample preparation kit (Illumina, San Diego, CA, USA). Multiplex sequencing was performed using a HiSeq 2000 analyzer and 101‐bp single reads. From the sequenced reads, the adapter sequences were trimmed using the cutadapt program, version 1.1 (https://pypi.python.org/pypi/cutadapt/1.1). After the adapters had been trimmed, the reads for each unique sequence were counted using Linux commands. The reads of more than 16 bp were aligned to mature miRNA records present in miRBase (Release 19, http://www.mirbase.org/) using the BLAST program, version 2.2.26. The sum of the ‐3p and ‐5p counts was represented as the miRNA count. Count data were normalized by total sequenced nucleotides and expressed as reads per million (rpm). Testicular‐specific miRNAs were selected using the criterion that the proportion of the rpm for the testis accounted for more than 90% of the total rpm, with the rpm values for the testis being more than 500.
Experimental protocol for the TH treatment
Six mature males (Indonesian origin) were assigned to the TH treatment experiment. TH treatment was performed on three animals according to a previously reported method (Lue et al., 2002) with some modifications. In brief, the inferior gluteal region, including the scrota with the testes, was immersed in a 43 °C water bath for 30 min under anesthesia with an intramuscular injection of 10 mg kg–1 ketamine hydrochloride (Daiichi Sankyo Co., Ltd). After the treatment, the animals were dried with a clean towel and confirmed not to have any signs of burn injury. For the untreated control group, three animals were subject to a sham treatment involving immersion into an empty water bath at room temperature for 30 min. The TH treatment was performed once daily for 5 consecutive days; the first day of the TH treatment was designated as Day 1. Body weight measurement, blood sampling for miRNAs analysis, blood chemistry and testosterone measurements were conducted before TH treatment (Days –11 and –4) and on Days 12 and 25. Blood (c. 2 ml) was taken from the femoral vein, and the obtained whole blood was immediately placed in tubes (Microtainer®; Becton, Dickinson and Co., NJ, USA) containing EDTA‐2 K for miRNA analysis or heparin lithium for blood chemistry and testosterone measurements. Plasma samples were obtained by centrifugation at 4 °C and 11 190 g for 5 min and then stored in a freezer at –80 °C until analysis. Blood chemistry parameters were measured using an autoanalyzer (TBA‐200FR; Toshiba Medical Co., Ltd., Tochigi, Japan). The testosterone concentration was determined in duplicate by a fluoroimmunoassay with DELFIA testosterone reagents (R050‐101; PerkinElmer, Inc., MA, USA). On the day after the final blood collection (Day 26), animals were anesthetized with an intramuscular injection of 10 mg kg–1 ketamine hydrochloride, and the left testis was then removed by aseptic surgery under inhalation of isoflurane (1.0 to 1.5%, Escain®; Mylan Inc., Canonsburg, PA, USA). The surgical operation was done between 09:00 to 12:00. Each removed testis was immediately dissected in half, with one half used for histopathology analysis and the other for miRNA analysis.
Histopathology
The removed testis was fixed in formalin–sucrose–acetic acid and embedded in paraffin. Sections were stained with hematoxylin and eosin (H&E) for microscopic examination.
RNA extraction
Total RNA was extracted from plasma samples using miRNeasy mini kits (Qiagen) according to the manufacturer's instructions. For the testicular sample, frozen samples (50 to 100 mg) were homogenized with QIAzol lysis reagent (Qiagen). Then, the homogenate was used for RNA extraction according to the manufacture's protocol. The quality of extracted total RNA from the testis was checked using a 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA).
Microarray analysis of testicular samples
Total RNA from the testis was labeled with Hy5 using the miRCURY LNA Array miR labeling kit (Exiqon A/S, Vedbaek, Denmark) according to the manufacturer's protocol. Labeled RNAs were hybridized onto 3D‐Gene® Human miRNA Oligo chips (Human miRNA V20; TORAY Industries, Inc., Tokyo, Japan). After stringent washes of the samples, fluorescent signals were scanned with a 3D‐Gene® Scanner (TORAY Industries, Inc.) and analyzed using 3D‐Gene® Extraction software (TORAY Industries, Inc.).
The raw data for each spot were normalized by substitution with a mean intensity of the background signal, determined by the upper limit of the 95% confidence intervals of the signal intensities of all of the blank spots. Measurements of spots with signal intensities greater than two standard deviations (SD) of the background signal intensity were considered to be valid. The relative expression level of a given miRNA was calculated by comparing the signal intensities of the valid spots throughout the microarray experiments. The data were globally normalized per array, such that the median of the signal intensity was adjusted to 25. Up‐ and down‐regulated miRNAs in the testis induced by TH treatment were identified based on the criterion that the mean signal intensity was more than 2.0‐fold or less than 0.50‐fold of the control value.
RT‐qPCR analysis of the testis and plasma
Total RNA samples were reverse‐transcribed with a TaqMan® MicroRNA Reverse Transcription kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) to obtain cDNA samples. Using the results of the NGS and microarray analyses in this study and data in the literature (Dere et al., 2013; Fukushima et al., 2011; Lize et al., 2011; Sakurai et al., 2015; Smorag et al., 2012; Wu et al., 2014), the following miRNAs were examined for RT‐qPCR analysis: miR‐34b‐5p, miR‐34c‐5p and miR‐449a for the testis and miR‐34c‐5p and miR‐202‐5p for the plasma. Namely, plasma miR‐34c‐5p was selected as a representative miRNA of the miR‐34/449 family, and plasma miR‐202‐5p has been reported for its elevation in rat testicular toxicity model (Dere et al., 2013). These miRNAs were subjected to RT‐qPCR with a 7900HT Fast Real‐Time PCR System (Thermo Fisher Scientific Inc.). Although miR‐508‐3p was also a candidate miRNA because of its high specificity to the testis, the primer for mml‐miR‐508‐3p (Thermo Fisher Scientific Inc., Assay ID: #4440887, currently not available), which has the exact matched sequence to that of cynomolgus monkeys, did not work properly, probably because of its specific nucleotide sequence. The primers for hsa‐miR‐34b‐5p, hsa‐miR‐34c‐5p, hsa‐miR‐449a and hsa‐miR‐202‐5p (Thermo Fisher Scientific Inc., Assay IDs: #000427, #000428, #001030, and #002579, respectively) were selected based on homology of the sequence between our NGS data and each respective human sequence obtained from miRBase (http://www.mirbase.org/). The thermal cycling conditions were 50 °C for 2 min and 95 °C for 20 s followed by 40 cycles of 95 °C for 1 s and 60 °C for 20 s. The miRNA levels were normalized to the spiked‐in cel‐miR‐238 (Syn‐cel‐miR‐238 miScript miRNA Mimic; Qiagen) as an external control.
Statistical analysis
The signal intensity values from the microarray and RT‐qPCR analyzes are expressed as the mean ± SD. They were statistically analyzed using F‐tests to evaluate the homogeneity of variance, followed by the Student's t‐tests (for homogenous data) or Aspin–Welch's t‐tests (for heterogeneous data). These zanalyzes were performed using Microsoft® Office Excel 2010 (Microsoft Corporation, Redmond, WA, USA). A P‐value less than 0.05 was classified as a statistically significant change.
Results
Determination of testicular‐specific miRNAs by NGS analysis
A total of approximately 1100 mature miRNAs that had already been confirmed in humans were identified in monkeys. In total, 3 707 568 reads were obtained for the testis by sequencing. Among these mature miRNAs, the 30 most abundant miRNAs in the testis included miR‐508, miR‐34c, let‐7a, let‐7f and miR‐202 (Table 1), consistent with those in humans (Yang et al., 2013). The top hit sequences of miR‐508, miR‐34c and miR‐202 were annotated as mml‐miR‐508‐3p, hsa‐miR‐34c‐5p and hsa‐miR‐202‐5p, respectively. The nucleotide sequences of 29 of the 30 most abundant miRNAs except for miR‐508 perfectly matched to the corresponding miRNA sequences in humans, although there were isomiRs of each miRNA. In contrast, mml‐miR‐508‐3p, the top hit of miR‐508, had two mismatch sequences compared with humans (5′‐ugauuguagccuuuuggaguaga‐3′) based on miRBase (Release 19, http://www.mirbase.org/). When each rpm of the testicular miRNA was compared with those of the liver, heart, lung, kidney, spleen and small intestine, miR‐508‐3p, miR‐34c‐5p, miR‐202‐5p and miR‐449a were determined to be testicular‐specific miRNAs, although slight expression levels of miR‐34c‐5p and miR‐449a were detected in the lung (Fig. 1).
Table 1.
Top 30 most abundantly expressed testicular miRNAs in cynomolgus monkeys as determined by next generation sequencing
| No. | miRNA | Read count | Top hit sequence | Annotation |
|---|---|---|---|---|
| 1 | miR‐10b | 3707568 | UACCCUGUAGAACCGAAUUUGU | hsa‐miR‐10b‐5p |
| 2 | miR‐143 | 1362027 | UGAGAUGAAGCACUGUAGCU | hsa‐miR‐143‐3p |
| 3 | miR‐191 | 576073 | CAACGGAAUCCCAAAAGCAGCUG | hsa‐miR‐191‐5p |
| 4 | miR‐26a | 265785 | UUCAAGUAAUCCAGGAUAGGCU | hsa‐miR‐26a‐5p |
| 5 | miR‐508 | 262691 | AUUGUCGCCUUUUUGAGUAGA | mml‐miR‐508a ) |
| 6 | miR‐181a | 245703 | AACAUUCAACGCUGUCGGUGAG | hsa‐miR‐181a‐5p |
| 7 | miR‐22 | 237376 | AAGCUGCCAGUUGAAGAACUGU | hsa‐miR‐22‐3p |
| 8 | miR‐27b | 197483 | UUCACAGUGGCUAAGUUCUG | hsa‐miR‐27b‐3p |
| 9 | miR‐34c | 195393 | AGGCAGUGUAGUUAGCUGAUUGC | hsa‐miR‐34c‐5p |
| 10 | miR‐126 | 102445 | CAUUAUUACUUUUGGUACGCG | hsa‐miR‐126‐5p |
| 11 | miR‐92a | 97483 | UAUUGCACUUGUCCCGGCCUGU | hsa‐miR‐92a‐3p |
| 12 | let‐7a | 96729 | UGAGGUAGUAGGUUGUAUAGUU | hsa‐let‐7a‐5p |
| 13 | miR‐16 | 92968 | UAGCAGCACGUAAAUAUUGGCG | hsa‐miR‐16‐5p |
| 14 | let‐7f | 76400 | UGAGGUAGUAGAUUGUAUAGUU | hsa‐let‐7f‐5p |
| 15 | miR‐125a | 70066 | UCCCUGAGACCCUUUAACCUGU | hsa‐miR‐125a‐5p |
| 16 | miR‐30a | 59068 | UGUAAACAUCCUCGACUGGA | hsa‐miR‐30a‐5p |
| 17 | miR‐151a | 53252 | UCGAGGAGCUCACAGUCUAGU | hsa‐miR‐151a‐5p |
| 18 | miR‐148a | 53242 | UCAGUGCACUACAGAACUUUGU | hsa‐miR‐148a‐3p |
| 19 | miR‐202 | 51687 | UUCCUAUGCAUAUACUUCUUU | hsa‐miR‐202‐5p |
| 20 | miR‐99b | 49297 | CACCCGUAGAACCGACCUUGCG | hsa‐miR‐99b‐5p |
| 21 | miR‐127 | 48741 | UCGGAUCCGUCUGAGCUUGGCU | hsa‐miR‐127‐3p |
| 22 | miR‐21 | 44394 | UAGCUUAUCAGACUGAUGUUGA | hsa‐miR‐21‐5p |
| 23 | miR‐28 | 40298 | CACUAGAUUGUGAGCUCCUGGA | hsa‐miR‐28‐3p |
| 24 | miR‐204 | 39459 | UUCCCUUUGUCAUCCUAUGCCU | hsa‐miR‐204‐5p |
| 25 | let‐7 g | 38843 | UGAGGUAGUAGUUUGUACAGUU | hsa‐let‐7 g‐5p |
| 26 | miR‐449a | 32023 | UGGCAGUGUAUUGUUAGCUGGU | hsa‐miR‐449a |
| 27 | miR‐30d | 31543 | UGUAAACAUCCCCGACUGGA | hsa‐miR‐30d‐5p |
| 28 | miR‐486 | 28444 | UCCUGUACUGAGCUGCCCCGAGU | hsa‐miR‐486‐5p |
| 29 | miR‐25 | 28406 | CAUUGCACUUGUCUCGGUCUGA | hsa‐miR‐25‐3p |
| 30 | miR‐186 | 27971 | CAAAGAAUUCUCCUUUUGGGCU | hsa‐miR‐186‐5p |
Data of read count was obtained from a pooled sample of two mature male animals.
mml‐miR‐508 is a previous name of mml‐miR‐508‐3p.
Figure 1.
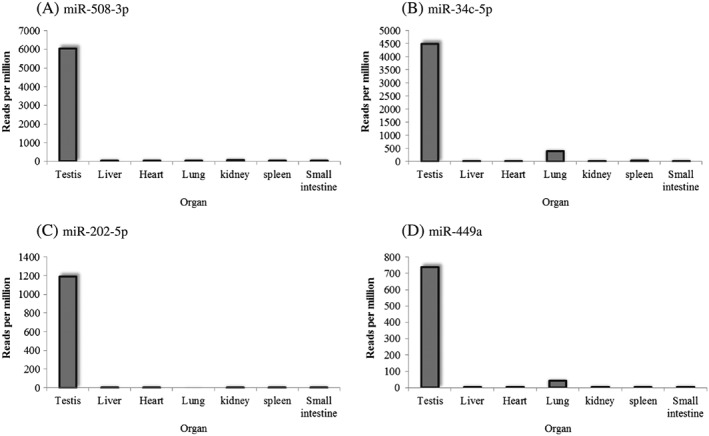
Tissue specificity of several miRNAs in cynomolgus monkeys. (A) miR‐508‐3p, (B) miR‐34c‐5p, (C) miR‐202‐5p and (D) miR‐449a. The y‐axis represents reads per million. Liver: Left lateral lobe, Heart: Left ventricular wall, Kidney: Cortex, Lung: Left caudal lobe, Small intestine: Jejunum and ileum.
Clinical observations, blood chemistry and testosterone measurement after the TH treatment
TH treatment for 5 consecutive days did not cause any abnormalities in general condition, body weight or blood chemistry parameters. Also, no noteworthy changes were observed in plasma testosterone levels (Fig. 2).
Figure 2.
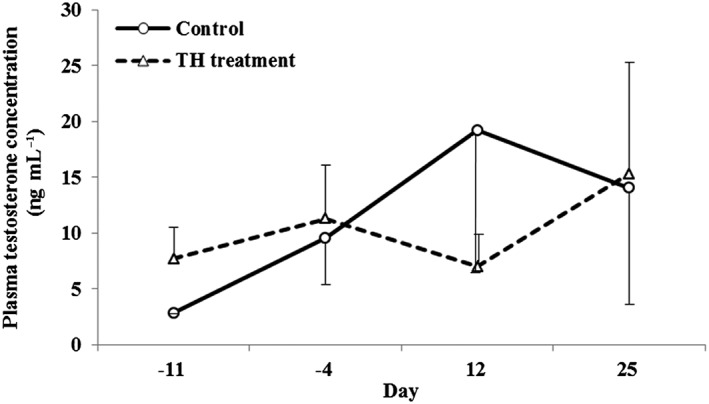
Effect of testicular hyperthermia on plasma testosterone levels in cynomolgus monkeys. Testosterone levels were measured before treatment (Days –11 and –4) and on Days 12 and 25. Data are shown as the mean ± SD (n = 3 per group).
Histopathology
In all the TH‐treated animals, a slight‐to‐moderate degeneration of spermatocytes/spermatids, slight‐to‐moderate decrease in the numbers of pachytene spermatocytes and round spermatids, and a severe decrease in the number of elongated spermatids were observed. No changes were observed in any of the control animals (Fig. 3).
Figure 3.
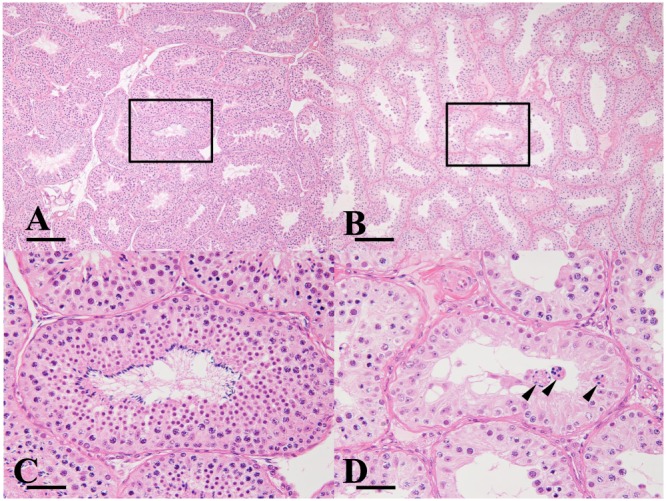
Testicular lesion in a cynomolgus monkey caused by testicular–hyperthermia treatment. (A) and (C): Control animal, (B) and (D): Testicular‐hyperthermia treated animal. (C) and (D) are higher magnified images of the areas surrounded by the rectangles in (A) and (B), respectively. In the testicular‐hyperthermia treated animal, elongated spermatids were nearly absent, the numbers of spermatocytes and round spermatids were decreased and degenerated spermatocytes/spermatids were observed. Arrowhead: degenerated spermatocytes/spermatids, Bar: 200 µm (A and B), 50 µm (C and D).
Microarray analysis of testicular miRNAs after TH treatment
In the comprehensive microarray analysis for human miRNAs, 24 testicular miRNAs were differentially expressed in the TH‐treated group relative to those in the control group. These included 11 up‐regulated miRNAs and 13 down‐regulated miRNAs (Table 2). The down‐regulated miRNAs included hsa‐miR‐34b‐3p, hsa‐miR‐34b‐5p, hsa‐miR‐34c‐3p, hsa‐miR‐34c‐5p, hsa‐miR‐449a and hsa‐miR‐449b‐5p, with 0.23 to 0.42‐fold lower expression than the control group. In contrast, the expression levels of the testicular‐specific hsa‐miR‐508‐3p and hsa‐miR‐202‐5p were not different compared with those in the control group.
Table 2.
Effect of testucular hyperthermia on testicular miRNAs in cynomolgus monkeys
| Testis | |||
|---|---|---|---|
| miRNA | Fold change | P‐value | |
| Up‐regulated miRNA | hsa‐miR‐6759‐5p | 2.84 | 0.022 |
| hsa‐miR‐6785‐5p | 2.40 | 0.023 | |
| hsa‐miR‐4433‐3p | 2.33 | 0.018 | |
| hsa‐miR‐557 | 2.31 | 0.045 | |
| hsa‐miR‐6742‐5p | 2.29 | 0.015 | |
| hsa‐miR‐4640‐5p | 2.27 | 0.027 | |
| hsa‐miR‐6840‐3p | 2.25 | 0.006 | |
| hsa‐miR‐4433b‐3p | 2.16 | 0.009 | |
| hsa‐miR‐6722‐3p | 2.13 | 0.005 | |
| hsa‐miR‐6737‐5p | 2.09 | 0.027 | |
| hsa‐miR‐199b‐5p | 2.02 | 0.016 | |
| Down‐regulated miRNA | hsa‐miR‐7162‐3p | 0.10 | <0.001 |
| hsa‐miR‐7153‐5p | 0.20 | 0.025 | |
| hsa‐miR‐34b‐3p | 0.23 | 0.004 | |
| hsa‐miR‐34c‐5p | 0.26 | 0.021 | |
| hsa‐miR‐34b‐5p | 0.28 | 0.015 | |
| hsa‐miR‐551b‐3p | 0.29 | 0.004 | |
| hsa‐miR‐200b‐3p | 0.29 | 0.003 | |
| hsa‐miR‐200a‐3p | 0.34 | <0.001 | |
| hsa‐miR‐34c‐3p | 0.39 | 0.010 | |
| hsa‐miR‐449b‐5p | 0.40 | 0.015 | |
| hsa‐miR‐449a | 0.42 | 0.008 | |
| hsa‐miR‐15b‐5p | 0.48 | 0.006 | |
| hsa‐miR‐4745‐5p | 0.48 | 0.013 | |
P‐values were calculated by Student's t‐test or Aspin–Welch's t‐test.
Fold changes were calculated based on the average values in the control group.
RT‐qPCR analysis of testicular and plasma miRNAs
Little variability was observed in the replicate samples using RT‐qPCR analysis. In the testis, miR‐34b‐5p and miR‐34c‐5p were significantly decreased, but miR‐449a did not differ compared with the control group (Fig. 4). Neither circulating miR‐34c‐5p nor miR‐202‐5p levels showed statistically significant changes on any examination day (Fig. 5).
Figure 4.
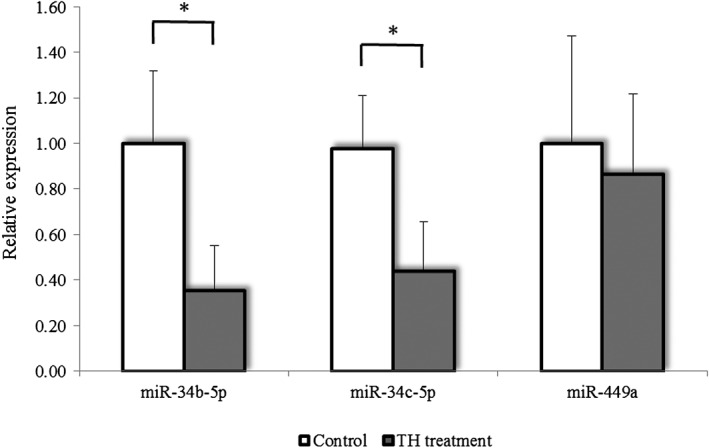
Effect of testicular hyperthermia on the expression levels of testicular‐specific miRNAs in cynomolgus monkeys. The expression level of each miRNA in the testicular‐hyperthermia treated group expressed relative to that of the control group (normalized as 1). Data are shown as the mean ± SD (n = 3 per group). The asterisks (*) indicate statistical significance (P‐value < 0.05) by t‐test.
Figure 5.
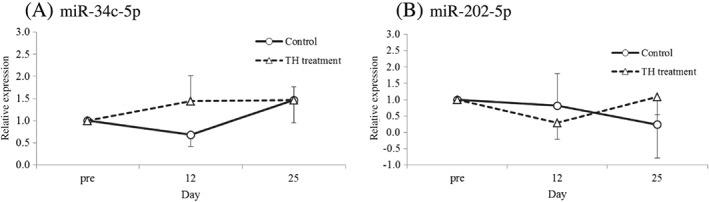
Effect of testicular hyperthermia on the expression levels of testicular‐specific plasma miRNAs in cynomolgus monkeys. (A) miR‐34c‐5p and (B) miR‐202‐5p. The expression level of each miRNA on Days 12 and 25 is expressed relative to the average of pre‐treatment values on Days –11 and –4 normalized as 1. Undetectable low values were regarded as 0. Data are shown as the mean ± SD (n = 3 per group). No statistically significant change was observed in the expression levels compared with those of the control group.
Discussion
MiRNAs have been recognized as novel biomarkers for detecting and monitoring organ toxicities in recent years (Starkey Lewis et al., 2011; Yamaura et al., 2012; Cheng et al., 2014; Nishimura et al., 2015) . Also, several miRNAs have been identified to be involved in testicular toxicity in rats (Dere et al., 2013) and humans (Abu‐Halima et al., 2014; Kotaja, 2014). However, only a few studies have investigated testicular miRNAs in cynomolgus monkeys despite the strong similarities in spermatogenesis and hormone regulation with those in humans (McLachlan et al., 2002; Dreef et al., 2007).
First, to identify organ‐specific miRNAs in cynomolgus monkeys, the 30 most abundant miRNAs in the testis of cynomolgus monkeys were identified using NGS. These miRNAs included let‐7a, let‐7f, miR‐34c, miR‐202 and miR‐508 (Table 1), which are highly expressed in the testis of humans (Yang et al., 2013). Furthermore, NGS analysis identified miR‐508‐3p, miR‐34c‐5p, miR‐202‐5p and miR‐449a as testicular‐specific miRNAs (Fig. 1). Among these miRNAs, miR‐34c and miR‐449a have been reported to have a critical role in spermatogenesis in mice (Wu et al., 2014). Furthermore, miR‐508‐3p has been reported to be abundant in the testis of humans as well (Yang et al., 2013). Interestingly, it has been reported that plasma miR‐202‐5p was elevated in rats with testicular toxicity induced by 1,3‐dinitrobenzene (1,3‐DNB) or carbendazim (CBZ) (Dere et al., 2013). Therefore, these four miRNAs are considered to be potential biomarkers for testicular toxicity even although the functional properties of miR‐508‐3p and miR‐202‐5p are not fully understood.
The cynomolgus monkey has been known to show genomic diversity depending on its origin (Kawamoto et al., 2008; Higashino et al., 2012). Although the effect of the origins on miRNA levels in cynomolgus monkeys has not been fully identified, our previous research did not show any obvious differences in testicular miRNAs levels in cynomolgus monkeys between Indonesian and Chinese origins (Sakurai et al., 2015).
The testicular injury was induced by our TH‐treatment protocol in all three animals without any changes in clinical signs, body weight, food consumption, blood chemistry parameters or plasma testosterone level. These phenotypic characteristics are consistent with a previous report on cynomolgus monkeys, except for a change in testosterone levels (Lue et al., 2002). In our model, histopathological analysis revealed a decreased number of pachytene spermatocytes, round spermatids and elongated spermatids (Fig. 3). Considering these findings, the spermatogenic cycle of monkeys (Dreef et al., 2007), and the findings of a previous report (Lue et al., 2002), the primary targets in this TH model are thought to be pachytene spermatocytes and round spermatids.
We next evaluated TH treatment‐dependent changes in miRNAs using microarray analysis, and we identified 11 up‐regulated miRNAs (>2.0 fold) and 13 down‐regulated miRNAs (<0.5 fold) in the testis of the TH‐treated group (Table 2). Although the functions of up‐regulated miRNAs are so far mostly unknown, down‐regulated miRNAs included several miR‐34/449 family members, such as miR‐34b‐3p, miR‐34b‐5p, miR‐34c‐3p, miR‐34c‐5p, miR‐449a and miR‐449b‐5p. The miR‐34/449 family has been reported to be preferentially expressed in the testis, especially in meiotic cells during spermatogenesis (Smorag et al., 2012). Additionally, miR‐449‐null male mice are known to show normal spermatogenesis with up‐regulation of miR‐34b/c (Bao et al., 2012), but miR‐34b/c and miR‐449 double KO mice exhibit severely disrupted spermatogenesis (Wu et al., 2014), suggesting that the miR‐34/449 family has a critical role in spermatogenesis. Taken together, decreased testicular miR‐34/449 expression in the TH‐treatment monkeys might reflect damage to meiotic cells such as pachytene spermatocytes.
Recently, miR‐34c was suggested to be involved in apoptosis and cell proliferation processes (Lize et al., 2011). Liang et al. (2012) demonstrated that miR‐34c enhanced germ cell apoptosis via activating transcription factor 1, and in contrast, inhibition of miR‐34c suppressed apoptosis with an increased B‐cell leukemia/lymphoma 2 (Bcl‐2) / Bcl‐2‐Associated X Protein (Bax) ratio. Thus, miR‐34c was suggested to be involved in apoptosis and cell proliferation through both pro‐apoptotic and anti‐proliferative functions (Liang et al., 2012). Interestingly, Jia et al. (2007) revealed that TH treatment in cynomolgus monkeys for 2 consecutive days increased testicular Bcl‐2 levels, but Bcl‐2 was inactivated as a result of phosphorylation at serine 70; therefore, the anti‐apoptotic activity through this pathway may be inactivated in this TH model. In the present study, decreased levels of testicular miR‐34b‐5p and miR‐34c‐5p were confirmed by RT‐qPCR. MiR‐34b‐5p and miR‐34c‐5p are known to be preferentially expressed in pachytene spermatocytes and round spermatids (Bouhallier et al., 2010; Smorag et al., 2012). Collectively, considering that decreased numbers of pachytene spermatocytes and round spermatids were observed in our TH‐treated monkey model, the down‐regulation of miR‐34b‐5p and miR‐34c‐5p may simply reflect the depletion of highly expressed germ cells.
In this study, a human microarray analysis was applied for cynomolgus monkey miRNAs, as this is a genetically similar species to humans. According to the genome sequence of the rhesus monkey, a congeneric species with cynomolgus monkey, both the nucleotide and amino acid sequences of rhesus monkey have 97.5% homology with those of humans (Gibbs et al., 2007). Moreover, many miRNAs are highly conserved among different mammalian species. Our NGS results also showed high homology to humans, namely, the sequences of 29 out of 30 testicular miRNAs perfectly matched those in humans. These data support the application of human microarray analysis to cynomolgus monkey samples.
Plasma miR‐34c‐5p and miR‐202‐5p levels in the TH‐treated group did not change compared with those in the control groups. Various miRNAs are present in the blood, which indicates that they are constitutively released from organs or tissues. Once the organs or tissues are injured by drugs, organ‐specific miRNAs could increase in plasma as is the case with hepatotoxicity (Starkey Lewis et al., 2011). However, the results of the present study indicated that the leakage of miRNAs from damaged testes did not occur or had little impact on their circulating levels because of small amounts of leaked miRNAs. Elkin et al. (2010) suggested that blood–testis barrier (BTB) integrity was a key factor in determining whether a molecule leaked from the germinal epithelium into the blood. In that study, cadmium induced the leakage of a 76‐kDa molecule via a damaged BTB, whereas methoxy acetic acid or 1,3‐DNB did not damage the BTB or induce leakage of a 15‐kDa molecule. The molecular weight of a miRNA is less than 15 kDa (approximately 5 to 8 kDa); however, circulating miRNAs are packed in an exosome or bound to proteins (Zhang et al., 2015). Therefore, the BTB would still prevent the miRNAs from leaking into circulation in this TH model because the TH‐treatment has been reported to not disturb BTB integrity (Turner et al., 1996). Alternatively, leakage of miRNAs from the testes might have little impact on the circulating levels because of their small size compared to other large organs such as the liver, heart or gastrointestinal tract. Indeed, plasma miR‐202‐5p was elevated only in some animals with testicular lesions induced by 1,3‐DNB or CBZ (Dere et al., 2013). Furthermore, recent studies have shown that miRNAs in exosomes might be useful potential biomarkers if exosomes are leaked, as is the case with the hepatotoxicity (Bala et al., 2012; Momen‐Heravi et al., 2015), but under our current assay condition, it might be difficult to measure exosomal miRNAs owing to quite low expressions of our target miRNAs (miR‐34c‐5p and miR‐202‐5p) in plasma in this study. Therefore, further studies are necessary to establish a circulating miRNA that can be used as a testicular toxicity biomarker.
In conclusion, NGS data showed that miR‐34b‐5p, miR‐34c‐5p, miR‐202‐5p and miR‐508‐3p were testicular‐specific miRNAs in cynomolgus monkeys. Furthermore, testicular miR‐34b‐5p and miR‐34c‐5p are suggested to be involved in testicular damage, including decreased pachytene spermatocytes and spermatids, caused by TH treatment. Our results would be useful for investigating the mechanisms of testicular toxicity in not only monkeys but also humans, considering the physiological similarity between monkeys and humans. Although further investigations are necessary to clarify the application of miRNAs as translational biomarkers for testicular toxicity in humans, our results provide valuable insights into the miRNAs involved in testicular toxicity.
Conflict of interest
The authors did not report any conflicts of interest.
Acknowledgments
The authors thank Drs. Naoki Kiyosawa and Kenji Watanabe for the NGS analysis and Takashi Yamaguchi for the technical assistance with the clinical pathology analysis.
Sakurai, K. , Mikamoto, K. , Shirai, M. , Iguchi, T. , Ito, K. , Takasaki, W. , and Mori, K. (2016) MicroRNA profiles in a monkey testicular injury model induced by testicular hyperthermia. J. Appl. Toxicol., 36: 1614–1621. doi: 10.1002/jat.3326.
References
- Abu‐Halima M, Backes C, Leidinger P, Keller A, Lubbad AM, Hammadeh M, Meese E. 2014. MicroRNA expression profiles in human testicular tissues of infertile men with different histopathologic patterns. Fertil. Steril. 101(78–86): e72 DOI:10.1016/j.fertnstert.2013.09.009. [DOI] [PubMed] [Google Scholar]
- Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, Alao H, Kodys K, Szabo G. 2012. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug‐induced, and inflammatory liver diseases. Hepatology 56: 1946–1957. DOI:10.1002/hep.25873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Li D, Wang L, Wu J, Hu Y, Wang Z, Chen Y, Cao X, Jiang C, Yan W, Xu C. 2012. MicroRNA‐449 and microRNA‐34b/c function redundantly in murine testes by targeting E2F transcription factor‐retinoblastoma protein (E2F‐pRb) pathway. J. Biol. Chem. 287: 21686–21698. DOI:10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Berner T, Dannan GA, Hawkins B, Hogan K, Holder JW, Ching‐Hung Hsu CH, Kopylev L, Stedeford T, Zodrow J. 2009. Toxicological Review of Nitrobenzene In Support of Summary Information on the Integrated Risk Information System (IRIS). US Environmental Protection Agency (US EPA). [Google Scholar]
- Bouhallier F, Allioli N, Lavial F, Chalmel F, Perrard MH, Durand P, Samarut J, Pain B, Rouault JP. 2010. Role of miR‐34c microRNA in the late steps of spermatogenesis. RNA 16: 720–731. DOI:10.1261/rna.1963810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Wang Q, You W, Chen M, Xia J. 2014. MiRNAs as biomarkers of myocardial infarction: a meta‐analysis. PLoS One 9: e88566 DOI:10.1371/journal.pone.0088566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dere E, Anderson LM, Coulson M, McIntyre BS, Boekelheide K, Chapin RE. 2013. SOT Symposium Highlight: Translatable Indicators of Testicular Toxicity: Inhibin B, MicroRNAs, and Sperm Signatures. Toxicol. Sci. 136: 265–273. DOI:10.1093/toxsci/kft207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreef HC, Van Esch E, De Rijk EP. 2007. Spermatogenesis in the cynomolgus monkey (Macaca fascicularis): a practical guide for routine morphological staging. Toxicol. Pathol. 35: 395–404. DOI:10.1080/01926230701230346. [DOI] [PubMed] [Google Scholar]
- Elkin ND, Piner JA, Sharpe RM. 2010. Toxicant‐induced leakage of germ cell‐specific proteins from seminiferous tubules in the rat: relationship to blood‐testis barrier integrity and prospects for biomonitoring. Toxicol. Sci. 117: 439–448. DOI:10.1093/toxsci/kfq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Taki K, Ise R, Horii I, Yoshida T. 2011. MicroRNAs expression in the ethylene glycol monomethyl ether‐induced testicular lesion. J. Toxicol. Sci. 36: 601–611. [DOI] [PubMed] [Google Scholar]
- Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, Williams BR. 2011. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 39: 5692–5703. DOI:10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, Batzer MA, Bustamante CD, Eichler EE, Hahn MW, Hardison RC, Makova KD, Miller W, Milosavljevic A, Palermo RE, Siepel A, Sikela JM, Attaway T, Bell S, Bernard KE, Buhay CJ, Chandrabose MN, Dao M, Davis C, Delehaunty KD, Ding Y, Dinh HH, Dugan‐Rocha S, Fulton LA, Gabisi RA, Garner TT, Godfrey J, Hawes AC, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Kirkness EF, Cree A, Fowler RG, Lee S, Lewis LR, Li Z, Liu YS, Moore SM, Muzny D, Nazareth LV, Ngo DN, Okwuonu GO, Pai G, Parker D, Paul HA, Pfannkoch C, Pohl CS, Rogers YH, Ruiz SJ, Sabo A, Santibanez J, Schneider BW, Smith SM, Sodergren E, Svatek AF, Utterback TR, Vattathil S, Warren W, White CS, Chinwalla AT, Feng Y, Halpern AL, Hillier LW, Huang X, Minx P, Nelson JO, Pepin KH, Qin X, Sutton GG, Venter E, Walenz BP, Wallis JW, Worley KC, Yang SP, Jones SM, Marra MA, Rocchi M, Schein JE, Baertsch R, Clarke L, Csuros M, Glasscock J, Harris RA, Havlak P, Jackson AR, Jiang H, Liu Y, Messina DN, Shen Y, Song HX, Wylie T, Zhang L, Birney E, Han K, Konkel MK, Lee J, Smit AF, Ullmer B, Wang H, Xing J, Burhans R, Cheng Z, Karro JE, Ma J, Raney B, She X, Cox MJ, Demuth JP, Dumas LJ, Han SG, Hopkins J, Karimpour‐Fard A, Kim YH, Pollack JR, Vinar T, Addo‐Quaye C, Degenhardt J, Denby A, Hubisz MJ, Indap A, Kosiol C, Lahn BT, Lawson HA, Marklein A, Nielsen R, Vallender EJ, Clark AG, Ferguson B, Hernandez RD, Hirani K, Kehrer‐Sawatzki H, Kolb J, Patil S, Pu LL, Ren Y, Smith DG, Wheeler DA, Schenck I, Ball EV, Chen R, Cooper DN, Giardine B, Hsu F, Kent WJ, Lesk A, Nelson DL, O'Brien WE, Prufer K, Stenson PD, Wallace JC, Ke H, Liu XM, Wang P, Xiang AP, Yang F, Barber GP, Haussler D, Karolchik D, Kern AD, Kuhn RM, Smith KE, Zwieg AS. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316: 222–234. DOI:10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Higashino A, Sakate R, Kameoka Y, Takahashi I, Hirata M, Tanuma R, Masui T, Yasutomi Y, Osada N. 2012. Whole‐genome sequencing and analysis of the Malaysian cynomolgus macaque (Macaca fascicularis) genome. Genome Biol. 13: R58 DOI:10.1186/gb-2012-13-7-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Hikim AP, Lue YH, Swerdloff RS, Vera Y, Zhang XS, Hu ZY, Li YC, Liu YX, Wang C. 2007. Signaling pathways for germ cell death in adult cynomolgus monkeys (Macaca fascicularis) induced by mild testicular hyperthermia and exogenous testosterone treatment. Biol. Reprod. 77: 83–92. DOI: 10.1095/biolreprod.106.058594. [DOI] [PubMed] [Google Scholar]
- Johanson G. 2000. Toxicity review of ethylene glycol monomethyl ether and its acetate ester. Crit. Rev. Toxicol. 30: 307–345. DOI:10.1080/10408440091159220. [DOI] [PubMed] [Google Scholar]
- Kawamoto Y, Kawamoto S, Matsubayashi K, Nozawa K, Watanabe T, Stanley MA, Perwitasari‐Farajallah D. 2008. Genetic diversity of longtail macaques (Macaca fascicularis) on the island of Mauritius: an assessment of nuclear and mitochondrial DNA polymorphisms. J. Med. Primatol. 37: 45–54. DOI:10.1111/j.1600-0684.2007.00225.x. [DOI] [PubMed] [Google Scholar]
- Kotaja N. 2014. MicroRNAs and spermatogenesis. Fertil. Steril. 101: 1552–1562. DOI:10.1016/j.fertnstert.2014.04.025. [DOI] [PubMed] [Google Scholar]
- Liang X, Zhou D, Wei C, Luo H, Liu J, Fu R, Cui S. 2012. MicroRNA‐34c enhances murine male germ cell apoptosis through targeting ATF1. PLoS One 7: e33861 DOI:10.1371/journal.pone.0033861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lize M, Klimke A, Dobbelstein M. 2011. MicroRNA‐449 in cell fate determination. Cell Cycle 10: 2874–2882. [DOI] [PubMed] [Google Scholar]
- Lue YH, Lasley BL, Laughlin LS, Swerdloff RS, Hikim AP, Leung A, Overstreet JW, Wang C. 2002. Mild testicular hyperthermia induces profound transitional spermatogenic suppression through increased germ cell apoptosis in adult cynomolgus monkeys (Macaca fascicularis). J. Androl. 23: 799–805. [PubMed] [Google Scholar]
- McLachlan RI, O'Donnell L, Meachem SJ, Stanton PG, de Kretser DM, Pratis K, Robertson DM. 2002. Identification of specific sites of hormonal regulation in spermatogenesis in rats, monkeys, and man. Recent Prog. Horm. Res. 57: 149–179. [DOI] [PubMed] [Google Scholar]
- Minami K, Uehara T, Morikawa Y, Omura K, Kanki M, Horinouchi A, Ono A, Yamada H, Ohno Y, Urushidani T. 2014. miRNA expression atlas in male rat. Sci. Data 1: 140005 DOI:10.1038/sdata.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen‐Heravi F, Saha B, Kodys K, Catalano D, Satishchandran A, Szabo G. 2015. Increased number of circulating exosomes and their microRNA cargos are potential novel biomarkers in alcoholic hepatitis. J. Transl. Med. 13: 261 DOI:10.1186/s12967-015-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y, Kondo C, Morikawa Y, Tonomura Y, Torii M, Yamate J, Uehara T. 2015. Plasma miR‐208 as a useful biomarker for drug‐induced cardiotoxicity in rats. J. Appl. Toxicol. 35: 173–180. DOI:10.1002/jat.3044. [DOI] [PubMed] [Google Scholar]
- Papaioannou MD, Pitetti JL, Ro S, Park C, Aubry F, Schaad O, Vejnar CE, Kuhne F, Descombes P, Zdobnov EM, McManus MT, Guillou F, Harfe BD, Yan W, Jegou B, Nef S. 2009. Sertoli cell Dicer is essential for spermatogenesis in mice. Dev. Biol. 326: 250–259. DOI:10.1016/j.ydbio.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Mikamoto K, Shirai M, Iguchi T, Ito K, Takasaki W, Mori K. 2015. MicroRNA profiling in ethylene glycol monomethyl ether‐induced monkey testicular toxicity model. J. Toxicol. Sci. 40: 375–382. DOI:10.2131/jts.40.375. [DOI] [PubMed] [Google Scholar]
- Smorag L, Zheng Y, Nolte J, Zechner U, Engel W, Pantakani DV. 2012. MicroRNA signature in various cell types of mouse spermatogenesis: evidence for stage‐specifically expressed miRNA‐221, ‐203 and ‐34b‐5p mediated spermatogenesis regulation. Biol. Cell 104: 677–692. DOI:10.1111/boc.201200014. [DOI] [PubMed] [Google Scholar]
- Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DG, Antoine DJ, French NS, Dhaun N, Webb DJ, Costello EM, Neoptolemos JP, Moggs J, Goldring CE, Park BK. 2011. Circulating microRNAs as potential markers of human drug‐induced liver injury. Hepatology 54: 1767–1776. DOI:10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- Turner KJ, McKinnell C, McLaren TT, Qureshi SJ, Saunders PT, Foster PM, Sharpe RM. 1996. Detection of germ cell‐derived proteins in testicular interstitial fluid: potential for monitoring spermatogenesis in vivo. J. Androl. 17: 127–136. [PubMed] [Google Scholar]
- Wu J, Bao J, Kim M, Yuan S, Tang C, Zheng H, Mastick GS, Xu C, Yan W. 2014. Two miRNA clusters, miR‐34b/c and miR‐449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc. Natl. Acad. Sci. U.S.A. 111: E2851–E2857. DOI:10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaura Y, Nakajima M, Takagi S, Fukami T, Tsuneyama K, Yokoi T. 2012. Plasma microRNA profiles in rat models of hepatocellular injury, cholestasis, and steatosis. PLoS One 7: e30250 DOI:10.1371/journal.pone.0030250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Hua J, Wang L, Xu B, Zhang H, Ye N, Zhang Z, Yu D, Cooke HJ, Zhang Y, Shi Q. 2013. MicroRNA and piRNA profiles in normal human testis detected by next generation sequencing. PLoS One 8: e66809 DOI:10.1371/journal.pone.0066809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. 2015. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 13: 17–24. DOI:10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


