Abstract
The biosynthesis of the class II lanthipeptide geobacillin II was reconstituted in vitro. The purified precursor peptide was modified by the lanthipeptide synthetase GeoM at temperatures ranging between 37 °C and 80 °C demonstrating the thermostability of the enzyme. Geobacillin II shares with cytolysin, haloduracin, and carnolysin a DhxDhxXxxXxxCys motif (Dhx =dehydroalanine or dehydrobutyrine) as precursor to its N-terminal A-ring. Like in these other three lantibiotics, the lanthionine in the A-ring of geobacillin II had the ll stereochemical configuration as shown by chiral gas chromatography/mass spectrometry. Various analogues of geobacillin II were produced using co-expression of mutants of the precursor peptide GeoAII and the synthetase GeoM in Escherichia coli. The findings in this study suggest that the stereochemical outcome of the A-ring in geobacillin II is not solely dependent on the peptide sequence as previously suggested for haloduracin β and cytolysin.
Keywords: antibiotics, lantibiotics, thermophile, stereochemistry, RiPP
Graphical abstract
INTRODUCTION
The recent surge in the availability of genome sequences from a wide variety of bacterial sources has led to the rapid discovery of ribosomally-synthesized and post-translationally modified peptides (RiPPs).1 The biosynthetic machinery of RiPPs offers a platform for the discovery and engineering of peptidic natural products. Although rapid advances have been made in elucidating biosynthetic pathways generating various RiPPs, the understanding of the molecular details of enzyme catalysis is still limited. One class of RiPPs is the lantibiotics,2 lanthipeptides with antimicrobial activity. The unifying posttranslational modifications (PTMs) found in lanthipeptides are thioether-crosslinked bis amino acids called lanthionine and methyl lanthionine. These structures are formed via the dehydration of serine and threonine residues to dehydroalanine (Dha) and dehydrobutyrine (Dhb) residues, respectively, followed by Michael-type addition of cysteines to the dehydrated residues.3 These PTMs are introduced in the C-terminal core region of the precursor peptide with the N-terminal leader region of the peptide acting as a guide for the lanthipeptide biosynthetic enzymes.4–6 Many lantibiotics have useful properties that have led to commercial use or that are in current development. Nisin is used in the food industry to combat food-borne pathogens7 and for treatment of bovine mastitis,8–10 a semi-synthetic analog of actagardine has been in clinical development for treatment of C. difficile,11–13 duramycin has completed phase II trials for cystic fibrosis as it promotes chloride secretion in lung epithelial cells,14–17 microbisporicin (NAI-107) is in development as an antibiotic,18–20 labyrinthopeptin has activity against neuropathic pain,21–23 and mutacin 1140 is in development against gram-positive multi-drug resistant pathogens.24
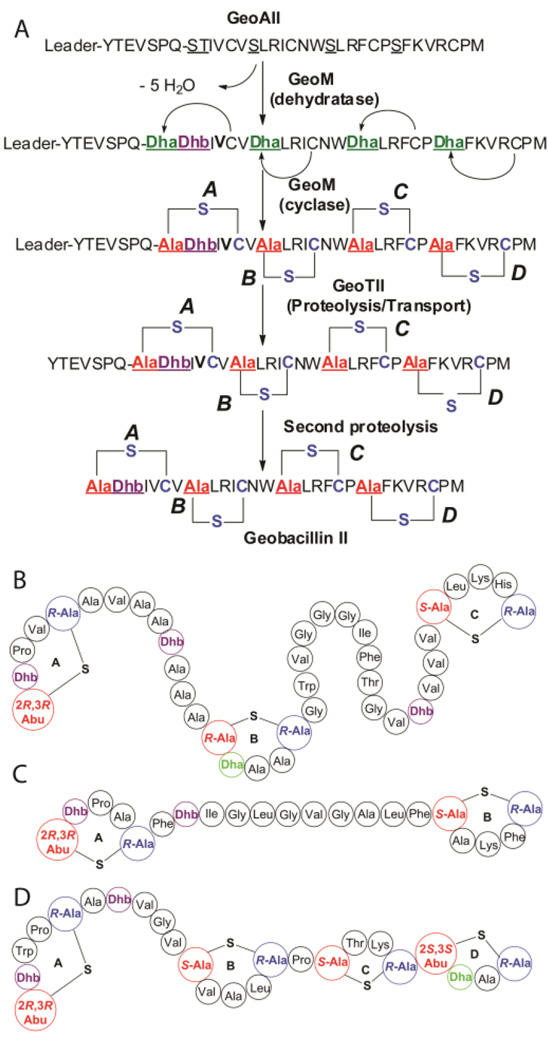
Recently, we reported the biosynthetic gene clusters and structures of two lantibiotics, geobacillin I and geobacillin II from Geobacillus thermodenitrificans NG80-2,25 a thermophilic bacterium with an optimal growth temperature of 65 °C.26 The lantibiotic geobacillin II is generated from the precursor peptide GeoAII and contains four thioether bridges, which are enzymatically synthesized by the lanthipeptide synthetase GeoM. This enzyme is a bifunctional class II lanthipeptide synthetase that catalyzes both the dehydration and cyclization reactions.25 The N-terminal domain of this class of enzymes catalyzes the phosphorylation of Ser/Thr followed by elimination of phosphate yielding the Dha/Dhb residues.27–31 Recent structural studies have shown that this domain has typical features associated with mammalian lipid kinases, and catalyzes both the phosphorylation and elimination reactions in the same active site.31 The C-terminal domain of the bifunctional lanthipeptide synthetases displays structural and sequence homology with class I lanthipeptide cyclases that catalyze the formation of the thioether crosslinks by reaction of the Cys side chains with the Dha/Dhb residues.32,33 Removal of the leader peptide by the bifunctional protease/transporter GeoTII and a second proteolysis step by an unknown protease25 results in the production of bioactive geobacillin II containing four lanthionine rings and one Dhb residue (Fig. 1A).
Figure 1.
(A) Predicted biosynthesis of geobacillin II. The steps leading to incorporation of post-translational modifications in the biosynthesis of lantibiotic geobacillin II. The dehydration and cyclization reactions are catalyzed by the bifunctional enzyme, GeoM. GeoATII is a bifunctional transporter/protease catalyzing transport of modified GeoAII followed by cleavage of the leader peptide. A second proteolysis step catalyzed by an unknown enzyme leads to the synthesis of fully modified geobacillin II. (B)–(D) Structures of lantibiotics with unusual stereochemistry. (B) CylLL", (C) CylLS", and (D) haloduracin β. Sequence of the leader peptide that is not shown: MKGGIQMEKQEQTFVSKISEEELKKLAGG. Treatment with endoproteinase GluC results in cleavage after Glu−5, leaving the tetrapeptide VSPQ derived from the leader peptide at the N-terminus of geobacillin II.
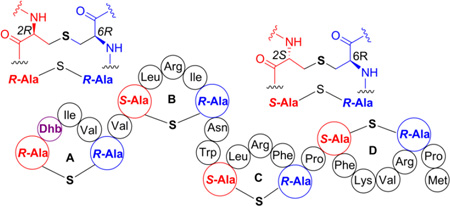
The stereochemistry of Lan and MeLan residues of select lanthipeptides including nisin,34 epidermin,35 subtilin,36, Pep5,37 mersacidin,38 lactocin S,39 and some prochlorosins40 was found to be 2S,6R for lanthionine and 2S,3S,6R for methyllanthionine (abbreviated hereafter as dl stereoisomers, d at the α-carbon of the former Ser/Thr and l at the former Cys). Recently, a different stereochemical configuration was reported for some of the thioether rings of the two-component lantibiotics cytolysin, haloduracin, and carnolysin.41,42 These compounds were shown to contain lanthionines with 2R,6R configuration and methyllanthionines with the 2R,3R,6R configuration, hereafter abbreviated as ll stereoisomers (Fig. 1B).43 The methyllanthionine A-ring and lanthionine B-ring of CylLL" (Fig. 1C) and the methyllanthionine A-ring of CylLS" (Fig. 1C), as well as the corresponding rings in the two peptides of the structurally related carnolysin, all had the ll stereochemical configuration. The LL stereochemistry was also found in the the A-ring of Halβ (Fig. 1D). Based on studies performed on cytolysin and haloduracin, it was suggested that the presence of a Dhx-Dhx-Xxx-Xxx-Cys sequence motif in the precursor peptide, where Dhx is Dha or Dhb, and Xxx is any amino acid except Ser/Thr/Cys, may be dictating the formation of the ll stereochemical configuration of the thioether crosslinks.44 Since such a motif is also present in the precursor peptide for geobacillin II, we investigated herein the stereochemistry of the resulting lanthionine as well as the determinants that govern the stereochemical outcome of the reaction. This study also presents the first in vitro investigation of a lanthipeptide synthetase from a thermophile.
RESULTS AND DISCUSSION
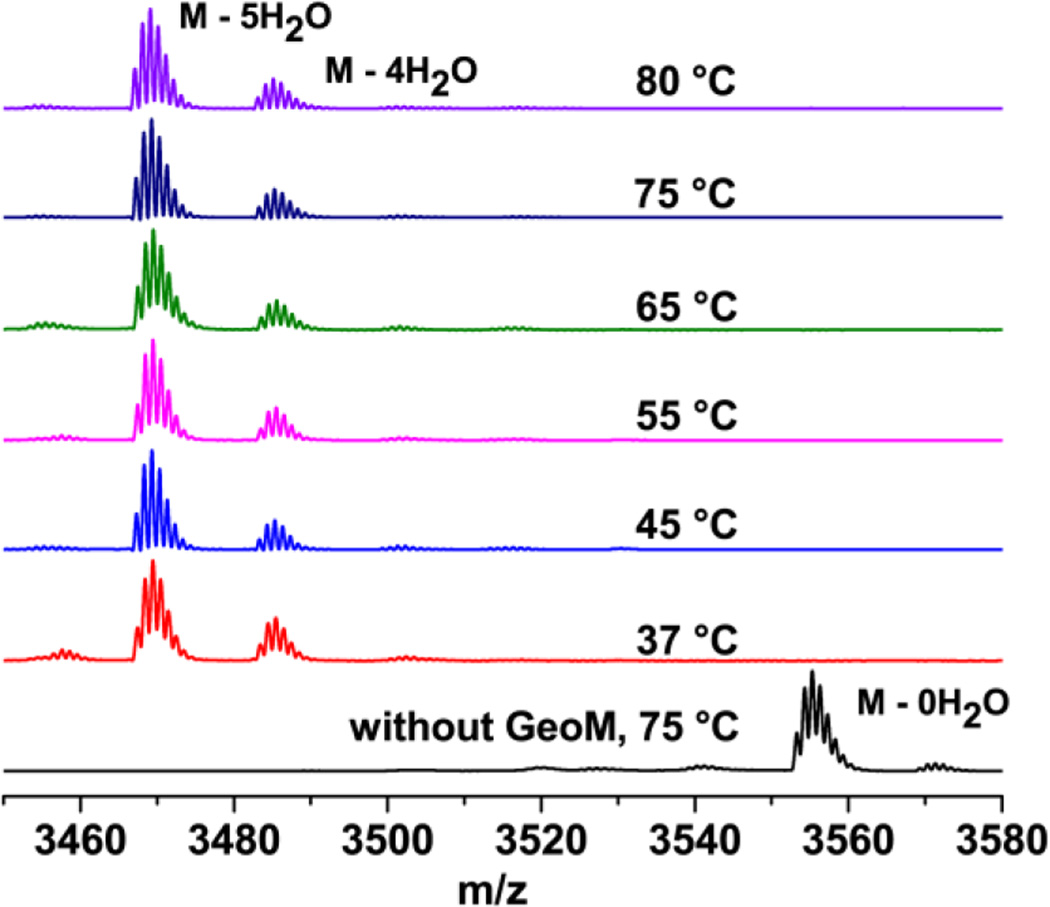
In addition to the interest in the factors that determine the stereochemistry, the geobacillin II biosynthetic enzymes are potentially valuable as they constitute the first such proteins from a thermophilic organism. As such, its lanthipeptide synthetases may provide promising candidates for generation of cyclic peptide libraries if they have increased thermostability. Towards this aim, we sought to characterize the enzymatic activity of GeoM. The genes encoding the precursor peptide GeoAII and the modification enzyme GeoM were heterologously expressed as recombinant His6-tagged fusion proteins in Escherichia coli. The precursor peptide was purified using nickel affinity chromatography followed by reversed phase high performance liquid chromatography (RP-HPLC). N-terminally His6-tagged GeoM was purified using nickel affinity chromatography followed by gel filtration chromatography. In vitro activity assays were carried out at 37 °C, 45 °C, 55 °C, 65 °C, 75 °C, and 80 °C for 2 h in the presence of 20 µM GeoAII, ATP, Mg2+, tris(2-carboxyethyl)phosphine (TCEP), and 5 µM GeoM. Commercial GluC protease was used for the removal of most of the leader peptide, cleaving after the −5 position and leaving a four-amino-acid overhang at the N-terminus of the core peptide. The GluC cleavage product was analyzed by matrix assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS), demonstrating that GeoAII was dehydrated five times by GeoM in 2 h at all temperatures, thus illustrating the thermostability of the enzyme (Figure 2). At the same time, the data demonstrate that the enzyme still has good activity at lower temperatures unlike other enzymes from thermophiles that because of their rigid structures, often exhibit poor activity at low temperature.45 These findings also explain why geobacillin II could be obtained previously by co-expression of GeoAII with GeoM in E. coli at 37 °C.25 The efficiency of modification of GeoAII by GeoM was investigated by monitoring the time-dependence of product formation at 37 °C, 55 °C, and 70 °C with 1 µM enzyme (a five-fold decrease compared to Figure 2). The reaction was analyzed at 30 min, 60 min, and 2 h for each temperature using MALDI-TOF MS (Figure S1), demonstrating that the enzymatic efficiency increased with temperature. Full conversion was observed by 2 h at 55 °C and 60 min at 70 °C whereas a significant amount of starting material was left after 2 h at 37 °C.
Figure 2. In vitro reconstitution of GeoM activity.
MALDI-MS spectra of His6-GeoAII (20 µM) incubated with His6-GeoM (5 µM), ATP (2.5 mM), Mg2+ (10 mM), and TCEP (2 mM) in HEPES pH 7.5 and cleaved with endoproteinase GluC.
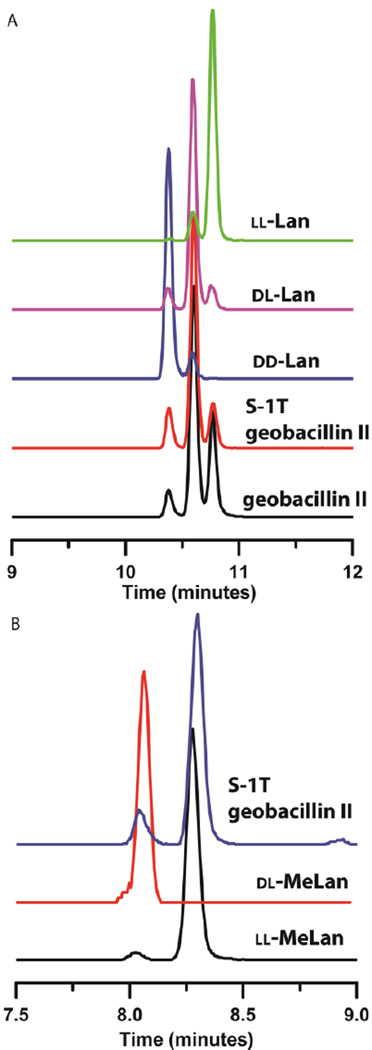
Like cytolysin and haloduracin β, the A-ring of geobacillin II is also formed from a Dhx-Dhx-Xxx-Xxx-Cys motif. In order to produce sufficient quantities of geobacillin II to determine the stereochemistry of the lanthionines, a hexa-histidine tagged precursor peptide mutant (GeoAII-G−8K) was co-expressed with the non-tagged tailoring enzyme GeoM in E. coli.25,46 Mutation of glycine at position −8 to lysine increased the yield and solubility of the peptide without affecting the post-translational modification by GeoM. The modified precursor peptide thus obtained was hydrolyzed and the resulting amino acids were derivatized for gas chromatography/mass spectrometry (GC/MS) analysis. As predicted based on the previous studies on cytolysin, haloduracin β, and carnolysin, and the fact that geobacillin II contains four lanthionines, the analysis demonstrated a mixture of dl and ll stereoisomers of lanthionine suggesting that at least one of the lanthionines has ll stereochemistry (Figure 3A). To determine unambiguously the position of the lanthionine(s) with ll stereochemical configuration, the Ser residue at position 1 was mutated to Thr using site directed mutagenesis such that ring A would contain a unique methyllanthionine. Co-expression of the precursor peptide GeoAII-S1T with GeoM resulted in five-fold dehydrated product. The modified GeoAII-S1T was hydrolyzed, derivatized, and the derivatized amino acids were analyzed by GC/MS using a chiral stationary phase. The A-ring, now methyllanthionine, was found to have the ll configuration and the lanthionine rings B, C, and D had the dl configuration (Figure 3A and B). Thus, we conclude that the ring A of authentic geobacillin II formed from the amino acid sequence Dha-Dhb-Ile-Val-Cys had the ll configuration. This is the first example of a lanthipeptide containing the ll stereochemistry that is not part of a two-component system.
Figure 3. Stereochemical configuration of the thioether bridges of wild type geobacillin II and geobacillin II S1T.
(A) GC-MS traces of hydrolyzed and derivatized Lan residues from GeoM-modified His6-GeoAII-G−8K (black), His6-GeoAII-S1T (red), dd-Lan standard (blue), dl-Lan standard (pink), and ll- Lan standard (green). (B) GC-MS traces of hydrolyzed and derivatized MeLan residues from GeoM-modified His6GeoAII-S1T (blue), dl-MeLan standard (red), and ll-MeLan standard (black). The small shoulders on the Lan peaks result from partial epimerization during hydrolysis and/or derivatization as previously reported.39 The experiments in panels A and B were conducted with different columns, which led to different retention times.
Previous studies have used chimeric peptides consisting of the leader peptides of haloduracin β, lacticin 3147 A2, or prochlorosin 3.2 fused to the core peptide of cytolysin CylLS. These studies showed that the modification enzymes HalM2, LtnM2, and ProcM all catalyzed the formation of the cytolysin A-ring with the ll configuration.44 This finding suggested that it is the property of the sequence of the core peptide and not the modification enzyme that dictates the stereochemical outcome of ring formation. To investigate the importance of the suggested motif in determining the stereochemical outcome for GeoM, analogues were produced with the amino acid composition of the precursor to the ring A changed to Dha-Gly-Ile-Val-Cys and Dha-Ala-Ile-Val-Cys. Co-expression of the corresponding precursor peptides GeoAII-G−8K/T2G and GeoAII-G−8K/T2A with GeoM resulted in four-fold dehydrated products (compared to five-fold in wild type GeoAII), as anticipated as the Thr at position 2 was replaced. The modified GeoAII-G−8K/T2G and GeoAII-G−8K/T2A peptides were hydrolyzed, derivatized, and the derivatized amino acids were analyzed by GC/MS. The stereochemistry of the lanthionine rings in these analogues was found to be a mixture of dl and ll stereoisomers that was similar to that of the wild type sequence (Fig. S2 and S3). Thus, it appeared that, unlike the previous findings for haloduracin β,44 the second Dhx in the sequence motif Dhx-Dhx-Xxx-Xxx-Cys is not essential for a stereochemical outcome that generates the ll configuration of the A-ring of geobacillin II. Thus, for geobacillin II, additional factors apart from the sequence of core peptide seem to play a role in determining the stereochemistry of ring formation.
The presence of three additional Lan with the dl stereochemistry in geobacillin II and the partial epimerization that is typically observed during hydrolysis39 and derivatization of Lan structures confounds the analysis of the stereochemistry of the A-ring. In order to clearly establish the stereochemical configuration of the ring A formed from a precursor peptide lacking the Dhx-Dhx-Xxx-Xxx-Cys motif, a GeoA variant (GeoAII-S1T/T2A) was used for coexpression with GeoM in E. coli which would result in the sequence Dhb-Ala-Ile-Val-Cys for the A-ring prior to cyclization. This mutant again allowed us to distinguish the derivatized MeLan residue of ring A from derivatized Lan residues arising from the B, C and D rings. The derivatized methyllanthionine residue originating from ring A showed a mixture of DL and LL configuration with the LL isomer as the major product but the dl isomer enriched compared to the wild type sequence (Figure S4, red trace). Interestingly, the GeoAII-S1T mutant resulted in a product in which the ll-isomer was favoured more than the GeoAII-S1T/T2A variant (Figure S4, blue trace). This observation suggests that the second Dhx in the Dhx-Dhx-Xxx-Xxx-Cys motif increases the amount of ll product formed but may not be the only governing factor for a stereochemical outcome of ll configuration by GeoM.
The B, C and D rings of geobacillin II all have a positively charged amino acid residue while ring A does not. To investigate whether the absence of a charged amino acid is (in part) responsible for the observed alternate stereochemistry of the A-ring, we constructed a precursor to ring A with amino acid sequence Dhb-Ala-Arg-Val-Cys (GeoAII-S1T/T2A/I3R). However, instead of increasing the amount of the dl methyllanthionine isomer compared to what was observed for the Dhb-Ala-Ile-Val-Cys variant, the abundance of the dl isomer in the GC-MS trace was decreased, showing that a charged amino acid does not favour the formation of the dl stereochemistry (Figure S4, black trace). As expected, the stereochemistry of all lanthionine rings (i.e. rings B-D) in these analogues was dl (Figure S5). Thus, in light of the results obtained in this study, the presence of the sequence Dhx-Dhx-Xxx-Xxx-Cys motif is not required to produce the ll stereoisomer of the thioether bridge in geobacillin II.
In order to investigate whether GeoM can catalyze the formation of ll stereoisomers at positions other than the A-ring, each of the B, C, and D lanthionine rings was first converted to methyllanthionine rings by mutation of the Ser involved in ring formation to Thr; as discussed above, this procedure allows clean analysis of the stereochemistry of these rings without complications from the other rings. Subsequently, we attempted to access a Dhx-Dhx-Xxx-Xxx-(Xxx)-Cys motif in these rings by introducing a second Thr residue (Thr-Thr-Arg-Ile-Cys for ring B, Thr-Thr-Arg-Phe-Cys for ring C, and Thr-Thr-Lys-Val-Arg-Cys for ring D, Figures S6A, S7A, and S8A). The GeoAII peptide variants were co-expressed with GeoM in E. coli. Unfortunately, following co-expression, five-fold dehydrated peptides were obtained for the mutants GeoAII-G−8K/S7T/L8T (ring B mutant), GeoAII-G−8K/S14T/L15T (ring C mutant), and GeoAII-G−8K/S20T/F21T (ring D mutant) as opposed to the desired six-fold dehydrated peptides. Although we were not able to determine which of the two adjacent Thr residues was not dehydrated, a likely explanation is that GeoM was unable to catalyze the conversion of the newly introduced, second Thr in the Thr-Thr-Xxx-Xxx-(Xxx)-Cys motif to Dhb. Thus, the modified amino acid sequences of the B, C and D ring analogues are Dhb-Thr-Xxx-Xxx-Cys instead of Dhb-Dhb-Xxx-Xxx-Cys. The lack of dehydration may possibly occur because of premature closure of the thioether bridge by the GeoM cyclization domain preventing access of the introduced threonine residue to the enzyme active site involved in catalyzing the dehydration reaction. Such alternating dehydration and cyclization activities have been reported previously for the class II lanthipeptide synthetase HalM2.47 Although we were unable to test how introduction of two consecutive dehydro amino acids in these rings would affect the stereochemistry, we did determine the stereochemistry of the B, C and D ring analogues that were obtained as described above. All newly introduced methyllanthionines in these analogues had the dl configuration (Figures S6B, S7B, and S8B). Similar to wild type geobacillin II, the lanthionines in the unperturbed rings in these analogues contained a mixture of ll and dl stereochemical configuration (Figures S6C, S7C, and S8C).
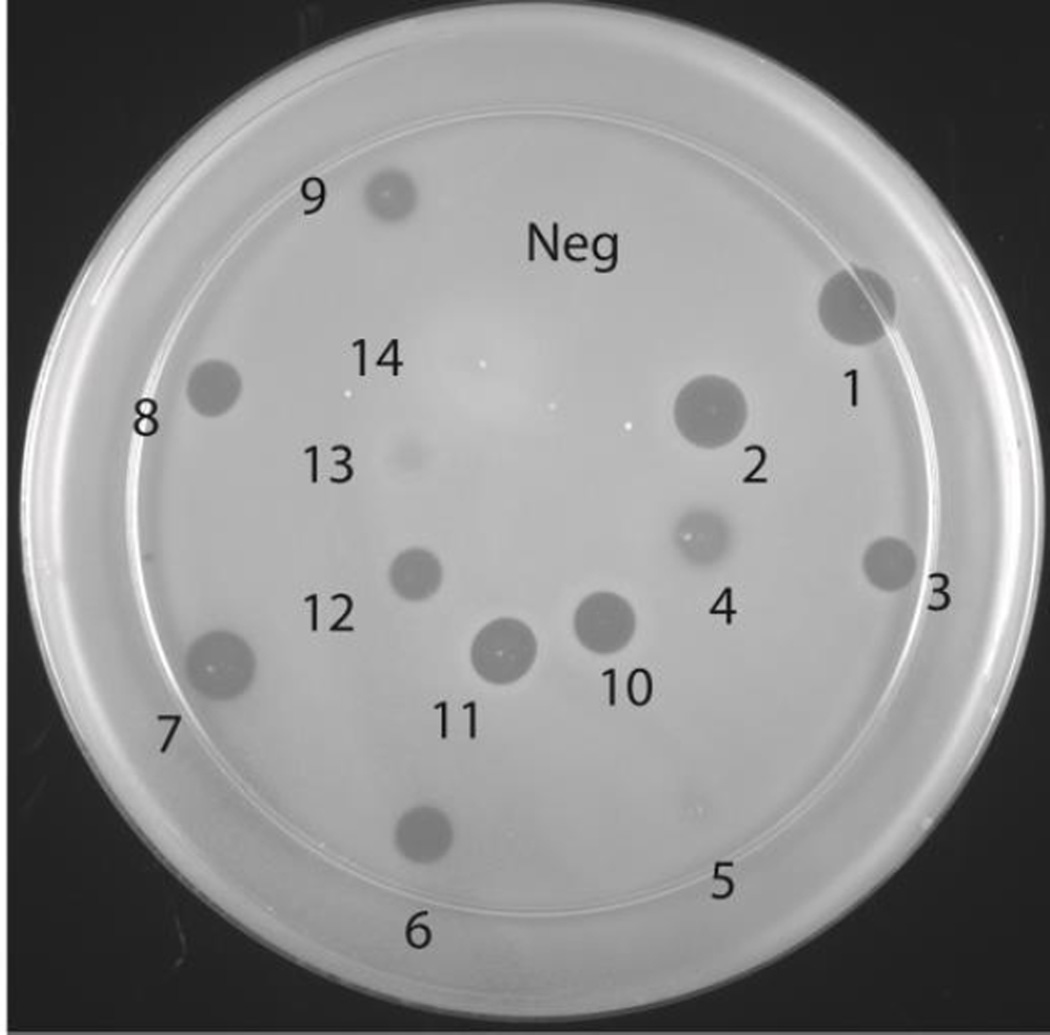
The bioactivity of the geobacillin II analogues produced in this study was tested using an agar diffusion growth inhibition assay, as we did not obtain sufficient quantities of material for determination of minimal inhibitory concentrations in liquid media. The purified modified peptides were incubated with GluC in order to remove the leader peptide (see Fig. S9 and S10 and Table S2 for mass spectra and Table 1 for a summary of the biological activity of all mutants) and used for antimicrobial activity. GluC cleaved at the −5 position leaving a four amino acid overhang at the N-terminus of core peptide, but this overhang does not inhibit the antibacterial activity of wild type geobacillin II.25 All analogues resulted in comparable activity as wild type geobacillin II treated in the same manner, except the A-ring analogues formed from precursors with amino acid compositions Dhb-Ala-Ile-Val-Cys (zone 13, Figure 4) and Dhb-Ala-Arg-Val-Cys (zone 14) and the two B-ring analogues constructed in this study (zone 5, and less obviously in zone 4 in Figure 4).
Table 1. Summary of qualitative agar inhibition assays conducted for the wild type sequence and the mutants generated in this study.
All peptides contained the four-amino acid sequence VSPQ at the N-terminus that derives from the leader peptide
| Peptide sequence | Ring position | Biological activity |
|---|---|---|
| DhaDhbIVC (wt) | A | wild type |
| DhbLRIC | B | decreased |
| DhbTRIC | B | decreased |
| DhbLRFC | C | unchangeda |
| DhbTRFC | C | unchanged |
| DhbFKVRC | D | unchanged |
| DhbTKVRC | D | unchanged |
| DhaAIVC | A | unchanged |
| DhaGIVC | A | unchanged |
| DhbDhbIVC | A | unchanged |
| DhbAIVC | A | decreasedb |
| DhbARVC | A | decreasedb |
Unchanged means a qualitatively similar size of the zone of growth inhibition (Figure 4).
The decrease in activity appears to be caused by the presence of about 50% uncyclized material that antagonizes the activity of the cyclized compound. See text.
Figure 4. Agar diffusion growth inhibition assay of geobacillin II analogues produced in this study with B. subtilis ATCC6633.
For a summary of the bioactivities, see Table 1. Zone 1: nisin (10 µL of 75 µM), Zone 2: nisin (10 µL of 50 µM). Zones 3–14 show growth inhibition by geobacillin II and the analogues produced in this study (20 µL of 150 µM solutions from GluC cleavage of each modified precursor peptide). Zone 3: wild type geobacillin II, Zone 4: ring B formed from Dhb-Leu-Arg-Ile-Cys, Zone 5: ring B formed from Dhb-Thr-Arg-Ile-Cys, Zone 6: ring C formed from Dhb-Leu-Arg-Phe-Cys, Zone 7: ring C formed from Dhb-Thr-Arg-Phe-Cys, Zone 8: ring D formed from Dhb-Phe-Lys-Val-Arg-Cys, Zone 9: ring D formed from Dhb-Thr-Lys-Val-Arg-Cys, Zone 10: ring A formed from Dha-Ala-Ile-Val-Cys, Zone 11: ring A formed from Dha-Gly-Ile-Val-Cys, Zone 12: ring A formed from Dhb-Dhb-Ile-Val-Cys, Zone 13: ring A formed from Dhb-Ala-Ile-Val-Cys, and Zone 14: ring A formed from Dhb-Ala-Arg-Val-Cys.
Interestingly, the GeoM-modified core peptides of the two methyllanthionine A-ring analogues that did not show bioactivity in Figure 4 (formed from Dhb-Ala-Arg-Val-Cys and Dhb-Ala-Ile-Val-Cys) both eluted as two separate peaks whereas the core peptides for the other modified geobacillin II analogues produced in this study eluted as a single peak (Figures S11 and S12). In order to further investigate their identity, the two compounds giving rise to the peaks observed in RP-HPLC after GluC cleavage of the modified GeoAII-S1T/T2A/I3R were collected separately and lyophilized. The lyophilized peptides were analysed by RP-HPLC to determine their purity (Figure S13). The isolated peptides were then hydrolyzed and derivatized for GC/MS analysis. Peak 1 contained a mixture of dl and ll stereoisomers of MeLan whereas only a very small signal for MeLan was observed in the analysis of peak 2 suggesting ring A had not been formed (Figure S14). Indeed, incomplete cyclization of ring A was confirmed by alkylation of free cysteines in these peptides with iodoacetamide (IAA) using a previously published protocol.28 MALDI-TOF MS spectra showed no carbamidomethyl adducts were formed in the reaction containing the peptide corresponding to peak 1 whereas one carbamidomethyl adduct was present in the reaction containing the peptide corresponding to peak 2 (Figure S15). Similar results upon incubation with IAA were obtained for the geobacillin II analogue GeoAII-S1T/T2A that also consisted of two peaks by HPLC. The observed incomplete cyclization upon mutation of these residues in the A-ring suggests that the wild type sequence of this ring is important for efficient modification by GeoM.
For both analogues, the cyclized material (peak 1) inhibited the growth of B. subtilis whereas the uncyclized material (peak 2) resulted in no inhibition (Figure S16). When the cyclized and uncyclized material were applied together, no zone of inhibition was observed (zone 4 and 5, Fig. S16). This observation suggests that the uncyclized material serves as antagonist of the fully cyclized peptides when the mixture of both peptides was tested (Figure 4, zones 13 and 14). In turn, this finding suggests formation of an oligomeric active complex involving multiple geobacillin II molecules may be required for antibacterial activity. For instance, the lantibiotic nisin has been shown to form a multimeric complex in the cell membrane upon lipid II binding.48 Formation of an inactive complex between fully cyclized and partially cyclized compounds, and potentially a target molecule, may explain the lack of activity of geobacillin II S1T/T2A/I3R and geobacillin II S1T/T2A. Since the ring B analogues had identical stereochemistry as geobacillin II and eluted as single peaks, the reduction in biological activity observed in zones 4 and 5 (Figure 4) may arise from mutation of the Dha residue to a Dhb residue. This mutation may reduce binding to the target, which may be optimal with a Lan at this position. Alternatively, a Lan B-ring may be important for interactions between geobacillin II molecules. Deeper investigation of these results requires additional mechanism-of-action studies.
In conclusion, the lantibiotic synthetase GeoM is a thermostable enzyme and is active up-to temperatures of at least 80 °C. Similar to cytolysin and haloduracin β, the A-ring of geobacillin II has unusual ll stereochemistry. Unlike haloduracin β, the stereochemical outcome at the A-ring of the analogues generated in this study does not depend only on the Dhx-Dhx-Xxx-Xxx-Cys motif. The findings in this study suggest more complex factors such as amino acid sequence and/or differential binding in the enzyme active site, and perhaps even some features of the leader peptide might play a concerted role in determining the stereochemistry of the thioether bridges synthesized by GeoM. A role of the leader peptide is suggested by the position of nearly all (methyl)lanthionines with LL-stereochemistry at the N-termini of the mature lantibiotics.
MATERIALS AND METHODS
General materials and methods
The synthetic genes were purchased from GeneArt (Invitrogen). The codon optimized gene sequences are given in the supporting information. Restriction enzymes, Phusion DNA polymerase, and T4 DNA ligase were purchased from New England Biolabs. The culture media was purchased from BD Biosciences. The oligonucleotide primers were purchased from Integrated DNA Technologies. DNA manipulation was performed in E. coli DH5α as host and E. coli BL21 (DE3) and E. coli BL21 Rossetta 2 was used as expression host for purification of modified peptides and protein, respectively. The endoproteinase GluC was purchased from Roche Biosciences. Unless otherwise mentioned, chemicals were obtained from Sigma-Aldrich.
Purification of His6GeoM
The gene encoding codon-optimized GeoM for expression in E. coli (for sequence see the Supporting Information) was purchased from GeneArt inserted between NdeI and HindIII restriction sites in a pMK vector. The plasmid was digested with these restriction enzymes and the geoM gene was inserted into a pET28b vector digested with the same enzymes. Single colony transformants of E. coli Rossetta2 cells containing pET28/His6-GeoM were grown in a 37 °C shaker for 12–15 h in 30 mL of LB medium supplemented with 50 µg/mL kanamycin and 25 µg/mL chloramphenicol. These cultures were used to inoculate 3 L of LB media and grown at 37 °C until the OD600 was ~ 0.6. IPTG was added to a final concentration of 0.1 mM and the cultures were grown at 18 °C for an additional 12 h. Cells were harvested by centrifugation at 11,900×g for 25 min at 4 °C. The cell paste was resuspended in 20 mM Tris-HCl, 1 mM NaCl, and 10% glycerol, pH 8.0. The cells were lysed using a C3 Homogenizer (Avestin Inc.). The lysed cells were pelleted by centrifugation at 23,700 × g for 25 min. Purification by nickel column chromatography was accomplished using a 5 mL HiTrap chelating HP nickel affinity column (GE Healthcare) on an AKTA FPLC system (GE Healthcare). The supernatant from the centrifugation step was applied to the column using a 50 mL superloop. The protein was detected by absorbance at 280 nm. The column was washed with buffer A containing 20 mM Tris-HCl, pH 8.0, 1 M NaCl, and 30 mM imidazole to remove any non-specifically bound proteins until a flat baseline was observed. A gradient of 0–100%B over 40 min was used to elute the bound proteins where buffer B contained 1 M NaCl, 20 mM Tris-HCl, pH 8.0, and 200 mM imidazole. The most concentrated protein fraction (4 mL) was loaded onto an S-200 gel filtration column (Amersham Biosciences) equilibrated in 20 mM HEPES pH 7.5, 300 mM NaCl, and 10% glycerol.
Purification of His6-GeoAII
The gene encoding GeoAII was cloned in a pET15b vector between restriction sites NdeI and XhoI. Electrocompetent E. coli BL21 (DE3) cells were transformed with pET15b/His6-GeoAII. Single colony transformants were picked and grown in LB media overnight. The culture after centrifugation and resuspension in fresh LB media was used to inoculate 12 L of LB media containing 100 mg/L ampicillin. The culture was shaken at 200 rpm at 37 °C until the OD600 reached between 0.6 and 0.8. At this point, the culture was induced with 1 mM IPTG. The induced cells were shaken continually at 37 °C for an additional 2 h, and the cells were harvested by centrifugation (11,900×g for 10 min, Beckman JLA-10.500 rotor). The cell pellet was resuspended in 50 mL of start buffer (20 mM Tris, pH 8.0, 500 mM NaCl, and 10% glycerol), and cell lysis was carried out using a MultiFlex C3 homogenizer (Avestin). The lysed cells were centrifuged at 23,700×g for 30 min at 4 °C. The supernatant was loaded onto a 5 mL HiTrap HP nickel affinity column (GE Healthcare) pre-equilibrated with start buffer. The column was washed with wash buffer (start buffer containing 30 mM imidazole), and the peptide was eluted from the column with elution buffer (start buffer containing 1 M imidazole). To increase the yield, GeoAII was also recovered from the insoluble fraction after cell lysis. The pellet was homogenized using a sonicator (35% amplitude, 4.4 s pulse, 9.9 s pause for 20 min total) in start buffer to remove any soluble proteins. The suspension was centrifuged and the pellet was homogenized in 30 mL of denaturing buffer (4 M guanidine hydrochloride, 20 mM NaH2PO4, 500 mM NaCl, pH 7.5). The supernatant after centrifugation was loaded onto a HiTrap HP nickel affinity column, the column was washed with denaturing buffer containing 30 mM imidazole, and the product was eluted with 15 mL of denaturing buffer containing 1 M imidazole. Small aliquots of eluent were desalted using a ZipTip (µC18) and analyzed for the presence of GeoAII using MALDI-TOF MS. For large scale purification, the eluent was desalted using preparative scale RP-HPLC using a Waters Delta-pak C4 15 µm; 300 Å; 25×100 mm PrepPak Cartridge. Following injection, the column was kept at 2% solvent A (solvent A = 0.086% TFA in 2% ACN/98% water) for 2 min followed by a gradient of 2–100% of solvent B (0.086% TFA in 80% ACN/20% water) over 45 min. The fractions containing GeoAII were lyophilized.
In vitro GeoM assay
The activity assay mixture contained 20 µM His6-GeoAII, 5 µM His6-GeoM, 20 mM HEPES pH 7.5, 10 mM MgCl2, 2 mM TCEP, and 2.5 mM ATP. The assay mixtures were incubated at various temperatures (37 °C, 45 °C, 55 °C, 65 °C, 75 °C, and 80 °C) for 2 h, desalted using µC18 Ziptip devices, and analyzed by MALDI-TOF MS. In order to determine the effect of temperature on enzyme activity, time dependent in vitro assays were carried out at 37 °C, 55 °C, and 70 °C with 1 µM GeoM. The reactions (20 µL) were quenched with 5 µL of 3% TFA at 30 min, 60 min, and 2 h. The reaction mixture obtained at different time points was desalted using µC18 Ziptip and analyzed by MALDI-TOF MS The mass spectrometry data were acquired using a Bruker Ultraflex TOF/TOF mass spectrometer.
Production of geobacillin II and analogues using co-expression
The construction of a pRSFDuet-1 vector carrying the genes encoding GeoAII and GeoM was described previously.25 Site directed mutagenesis was carried out on this construct to generate vectors encoding analogues of geobacillin II. The list of primers used is given in Table S1. The modified geobacillin II and analogues were obtained using co-expression as described previously.25
Chiral gas chromatography/mass spectrometry (GC/MS) analysis
The stereochemistry of (methyl)lanthionine residues was determined by acid hydrolysis of geobacillin II and chemical derivatization of the resulting amino acids as previously described.39,40,49 The derivatized material was then analyzed by GC/MS. Briefly, the modified peptides with leader attached (1 mg) were dissolved in 3 mL of 6 M HCl. The mixture was heated at 110 °C in a sealed tube for 20 h. The reaction was cooled and dried under nitrogen overnight. Methanol (5 mL) was chilled in an ice-water bath, and acetyl chloride (1.5 mL) was added dropwise. This solution was added to the hydrolyzed peptides and heated at 110 °C for 1 h. The sample was cooled and dried under a stream of nitrogen. CH2Cl2 (3 mL) and pentafluoropropionic anhydride (1 mL) was added and the material was heated at 110 °C for 1 h. The reaction was allowed to cool and dried under a stream of nitrogen. The hydrolyzed and derivatized peptides were taken up in methanol and dried under nitrogen. This step was repeated twice to ensure complete removal of residual pentafluoropropionic anhydride. The derivatized amino acids from modified peptides, as well as synthetic (methyl)lanthionine standards treated the same way were analyzed by GC/MS using an Agilent 7890A gas chromatograph equipped with an Agilent 5975C Inert XL EI/CI MS detector and a Varian CP-Chirasil-L-Val fused silica column (25 m × 250 µm × 0.12 µm). The samples were introduced to the instrument via a pulsed split injection at an inlet temperature of 200 °C and flow rate of 2 mL/min helium gas. The temperature gradient used was 160 °C for 5 min, then 160 °C to 200 °C at 3 °C/min. The MS was operated in simultaneous scan/single-ion monitoring (SIM) mode, monitoring at fragment masses of 365 for lanthionine and 379 for methyllanthionine.
Agar diffusion growth inhibition assay
Lyophilized peptides were dissolved in 50 mM HEPES pH 7.5 to give stock solutions of 150 µM. The dissolved peptides (200 µL) were incubated with 3 µL of GluC (1 mg/mL, catalog number: 10791156001, Roche Biosciences) for 5 h. The cleavage products were analyzed using a Bruker Ultraflex TOF/TOF mass spectrometer and complete cleavage was observed in 5 h. Bacillus subtilis ATCC 6633 was used as indicator strain to determine biological activity of the analogues produced in this study. LB agar media (25 mL) was melted in a microwave and was cooled in a 45 °C water bath for 10 min. The overnight culture of B. subtilis ATCC 6633 was diluted to an optical density at 600 nm of 0.1 and mixed with molten agar. The molten agar containing cells was poured in a petri dish, allowed to solidify and 20 µL of cleaved peptides were spotted on the plate.
Supplementary Material
Acknowledgments
Contract grant sponsor: National Institutes of Health
Contract grant number: R01 GM058822
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA. Nat Prod Rep. 2013;30:108. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cotter PD, Hill C, Ross RP. Curr Protein Pept Sci. 2005;6:61. doi: 10.2174/1389203053027584. [DOI] [PubMed] [Google Scholar]
- 3.Knerr PJ, van der Donk WA. Annu Rev Biochem. 2012;81:479. doi: 10.1146/annurev-biochem-060110-113521. [DOI] [PubMed] [Google Scholar]
- 4.Oman TJ, van der Donk WA. Nat Chem Biol. 2010;6:9. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plat A, Kuipers A, Rink R, Moll GN. Curr Protein Pept Sci. 2013 doi: 10.2174/1389203711314020001. [DOI] [PubMed] [Google Scholar]
- 6.Yang X, van der Donk WA. Chem Eur J. 2013;19:7662. doi: 10.1002/chem.201300401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delves-Broughton J, Blackburn P, Evans RJ, Hugenholtz J. Antonie van Leeuwenhoek. 1996;69:193. doi: 10.1007/BF00399424. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez L, Delgado S, Herrero H, Maldonado A, Rodriguez JM. J Hum Lact. 2008;24:311. doi: 10.1177/0890334408317435. [DOI] [PubMed] [Google Scholar]
- 9.Cao LT, Wu JQ, Xie F, Hu SH, Mo Y. J Dairy Sci. 2007;90:3980. doi: 10.3168/jds.2007-0153. [DOI] [PubMed] [Google Scholar]
- 10.Wu J, Hu S, Cao L. Antimicrob Agents Chemother. 2007;51:3131. doi: 10.1128/AAC.00629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson MJ, Scott RW. Curr Opin Pharmacol. 2012;12:545. doi: 10.1016/j.coph.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowther GS, Baines SD, Todhunter SL, Freeman J, Chilton CH, Wilcox MH. J Antimicrob Chemother. 2013;68:168. doi: 10.1093/jac/dks359. [DOI] [PubMed] [Google Scholar]
- 13.Mathur H, O'Connor PM, Hill C, Cotter PD, Ross RP. Antimicrob Agents Chemother. 2013;57:2882. doi: 10.1128/AAC.00261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliynyk I, Varelogianni G, Roomans GM, Johannesson M. APMIS. 2010;118:982. doi: 10.1111/j.1600-0463.2010.02680.x. [DOI] [PubMed] [Google Scholar]
- 15.Jones AM, Helm JM. Drugs. 2009;69:1903. doi: 10.2165/11318500-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 16.McNulty MJ, Hutabarat RH, Findlay JW, Devereux K, Knick VC, Harvey RJ, Molina L. Xenobiotica. 2003;33:197. doi: 10.1080/0049825021000022320. [DOI] [PubMed] [Google Scholar]
- 17.Steiner I, Errhalt P, Kubesch K, Hubner M, Holy M, Bauer M, Muller M, Hinterberger S, Widmann R, Mascher D, Freissmuth M, Kneussl M. Naunyn Schmiedebergs Arch Pharmacol. 2008;378:323. doi: 10.1007/s00210-008-0293-8. [DOI] [PubMed] [Google Scholar]
- 18.Jabés D, Brunati C, Candiani G, Riva S, Romanó G, Donadio S. Antimicrob Agents Chemother. 2011;55:1671. doi: 10.1128/AAC.01288-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castiglione F, Lazzarini A, Carrano L, Corti E, Ciciliato I, Gastaldo L, Candiani P, Losi D, Marinelli F, Selva E, Parenti F. Chem Biol. 2008;15:22. doi: 10.1016/j.chembiol.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Lepak AJ, Marchillo K, Craig WA, Andes DR. Antimicrob Agents Chemother. 2015;59:1258. doi: 10.1128/AAC.04444-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seibert G, L V, Wink J, Winkler I, Süssmuth RD, Sheldrick GM, Meindl K, Broenstrup M, Hoffmann H, Guehring H, Toti L. Sanofi-aventis (174, A. d. F., 75013 Paris, FR), Ed. 2008 [Google Scholar]
- 22.Meindl K, Schmiederer T, Schneider K, Reicke A, Butz D, Keller S, Guhring H, Vertesy L, Wink J, Hoffmann H, Bronstrup M, Sheldrick GM, Süssmuth RD. Angew Chem Int Ed. 2010;49:1151. doi: 10.1002/anie.200905773. [DOI] [PubMed] [Google Scholar]
- 23.Férir G, Petrova MI, Andrei G, Huskens D, Hoorelbeke B, Snoeck R, Vanderleyden J, Balzarini J, Bartoschek S, Brönstrup M, Süssmuth RD, Schols D. PLoS One. 2013;8:e64010. doi: 10.1371/journal.pone.0064010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith L, Hillman J. Curr Opin Microbiol. 2008;11:401. doi: 10.1016/j.mib.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garg N, Tang W, Goto Y, Nair SK, van der Donk WA. Proc Natl Acad Sci U S A. 2012;109:5241. doi: 10.1073/pnas.1116815109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng L, Wang W, Cheng J, Ren Y, Zhao G, Gao C, Tang Y, Liu X, Han W, Peng X, Liu R, Wang L. Proc Natl Acad Sci U S A. 2007;104:5602. doi: 10.1073/pnas.0609650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You YO, van der Donk WA. Biochemistry. 2007;46:5991. doi: 10.1021/bi602663x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. Proc Natl Acad Sci USA. 2006;103:17243. doi: 10.1073/pnas.0606088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. Science. 2004;303:679. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 30.Chatterjee C, Miller LM, Leung YL, Xie L, Yi M, Kelleher NL, van der Donk WA. J Am Chem Soc. 2005;127:15332. doi: 10.1021/ja0543043. [DOI] [PubMed] [Google Scholar]
- 31.Dong SH, Tang W, Lukk T, Yu Y, Nair SK, van der Donk WA. eLife. 2015;4:e07607. doi: 10.7554/eLife.07607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B, Yu J-PJ, Brunzelle JS, Moll GN, van der Donk WA, Nair SK. Science. 2006;311:1464. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- 33.Paul M, Patton GC, van der Donk WA. Biochemistry. 2007;46:6268. doi: 10.1021/bi7000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross E, Morell JL. J Am Chem Soc. 1971;93:4634. doi: 10.1021/ja00747a073. [DOI] [PubMed] [Google Scholar]
- 35.Allgaier H, Jung G, Werner RG, Schneider U. Angew Chem, Int Ed. 1985;24:1051. [Google Scholar]
- 36.Gross E, Kiltz HH, Nebelin E. Hoppe-Seyler's Z Physiol Chem. 1973;354:810. [PubMed] [Google Scholar]
- 37.Kellner R, Jung G, Josten M, Kaletta C, Entian KD, Sahl HG. Angew Chem. 1989;101:618. [Google Scholar]
- 38.Chatterjee S, Chatterjee S, Lad SJ, Phansalkar MS, Rupp RH, Ganguli BN, Fehlhaber HW, Kogler H. J Antibiot. 1992;45:832. doi: 10.7164/antibiotics.45.832. [DOI] [PubMed] [Google Scholar]
- 39.Ross AC, Liu H, Pattabiraman VR, Vederas JC. J Am Chem Soc. 2010;132:462. doi: 10.1021/ja9095945. [DOI] [PubMed] [Google Scholar]
- 40.Li B, Sher D, Kelly L, Shi Y, Huang K, Knerr PJ, Joewono I, Rusch D, Chisholm SW, van der Donk WA. Proc Natl Acad Sci U S A. 2010;107:10430. doi: 10.1073/pnas.0913677107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang W, van der Donk WA. Nat Chem Biol. 2013;9:157. doi: 10.1038/nchembio.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohans CT, Li JL, Vederas JC. J Am Chem Soc. 2014;136:13150. doi: 10.1021/ja5070813. [DOI] [PubMed] [Google Scholar]
- 43.Chatterjee C, Paul M, Xie L, van der Donk WA. Chem Rev. 2005;105:633. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- 44.Tang W, Jiménez-Osés G, Houk KN, van der Donk WA. Nat Chem. 2015;7:57. doi: 10.1038/nchem.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siddiqui KS. Biotechnol Adv. 2015;33:1912. doi: 10.1016/j.biotechadv.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Shi Y, Yang X, Garg N, van der Donk WA. J Am Chem Soc. 2011;133:2338. doi: 10.1021/ja109044r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thibodeaux CJ, Ha TJ, van der Donk WA. J Am Chem Soc. 2014;136:17513. doi: 10.1021/ja5089452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hasper HE, de Kruijff B, Breukink E. Biochemistry. 2004;43:11567. doi: 10.1021/bi049476b. [DOI] [PubMed] [Google Scholar]
- 49.Liu W, Chan AS, Liu H, Cochrane SA, Vederas JC. J Am Chem Soc. 2011;133:14216. doi: 10.1021/ja206017p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.