Abstract
An increase in hepatic triglyceride (TG) contents usually results in non-alcoholic fatty liver disease (NAFLD) and related metabolic diseases. However, the mechanisms underlying perturbations of hepatic TG homeostasis remain largely unknown. Here, we showed that MicroRNA-124 was up-regulated in the livers of C57BL/6 mice fed a short-term high-fat-diet (HFD). Adenoviral overexpression of miR-124 in C57BL/6 mice led to accumulation of excessive triglycerides and up-regulation of lipogenic genes in the liver. We further identified tribbles homolog 3 (TRB3) as a direct target of miR-124. AKT signaling, which is negatively regulated by TRB3, was enhanced by miR-124 overexpression. Moreover, restoration of TRB3 expression markedly abolished the effect of miR-124 on hepatic TG metabolism. Therefore, our findings revealed that miR-124 played a role in mediating high-fat-diet induced TG accumulation in the liver.
In mammals, liver is a metabolic organ that plays an important role in the regulation of triglyceride (TG) homeostasis. The homeostasis of hepatic TG metabolism is attributed to the precise regulation between TG acquisition and removal. Liver TG accumulates when an increase in de novo lipogenesis (DNL) or uptake of fatty acid cannot be offset by fatty acid oxidation or very low-density lipoprotein (VLDL) secretion1,2. Hepatic lipogenesis is primarily controlled by a transcription factor named sterol regulatory element binding protein 1c (SREBP-1c), which transcriptionally up-regulates most of lipogenic genes, including fatty acid synthase (Fasn), acetyl-CoA carboxylase (ACC) and stearoyl-CoA desaturase 1 (SCD-1)3. As a result, increased expression of SREBP-1c is closely associated with TG accumulation in the liver4,5. Of note, SREBP-1c expression can be regulated by multiple pathways, involving AKT, STAT3 and mTOR signaling6,7. Our previous studies demonstrated that SREBP-1c could also be regulated by a nuclear receptor cascade involving farnesoid X receptor (FXR) and small heterodimer partner (SHP) in obesity and aging8,9.
MicroRNAs (miRNAs), a class of small non-coding RNAs, regulate gene expression post-transcriptionally through binding to complementary regions in the 3′ untranslated regions (3′ UTRs) of target mRNA, which results in mRNA degradation or attenuated translation10,11. Several studies have shown that miRNAs may play a pathological role in the hepatic TG homeostasis. For instance, miR-122a-deficient mice developed hepatosteatosis as a consequence of dysfunctional expression of genes involved in TG synthesis, storage and secretion12,13. miR-33 affected TG metabolism in concert with their host genes SREBF through targeting key genes of fatty acid oxidation14,15. miR-378 could ameliorate hepatic steatosis in obese mice by directly targeting p110α subunit, a core component of insulin signaling16.
miR-124, which is abundantly expressed in the brain, plays a key role in neuronal differentiation and brain development17,18. miR-124 also participated in tumorigenesis by targeting oncogenic proteins including cyclin-dependent kinase 6 (CDK6), signal transducerand activator of transcription 3 (STAT3), rho-kinase2 (ROCK2) and enhancer of the zeste homologue 2 (EZH2)19,20,21. Consistently, downregulation of miR-124 was observed in several types of human cancers20,21,22,23,24,25. Moreover, miR-124 was shown to regulate pancreatic islet development through suppression of the forkhead box protein A2 (FoxA2) transcription factor and Rab27a26,27. However, the role of miR-124 in the regulation of hepatic TG homeostasis remains unknown.
In the present study, we identified a critical role of miR-124 in the regulation of SREBP-1c and hepatic TG homeostasis, which might help us to understand the pathogenesis of hepatic metabolic disorders.
Results
Short term high-fat-diet increases hepatic triglyceride contents in mice
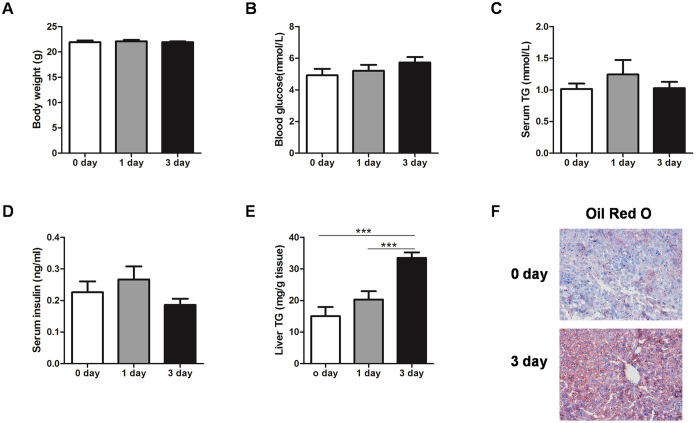
Obese mice, such as ob/ob and db/db mice, and long-term high-fat-diet (HFD) fed mice, are usually used to investigate the mechanisms of hepatic TG homeostasis. However, these mice are characterized by many metabolic disorders, including obesity, insulin resistance and hyperglycemia, which may contribute as confounders to hepatic TG disorders1,28,29,30,31. Therefore, we employed short term (1 and 3 day) HFD mice to investigate hepatic TG homeostasis. As a result, there were no significant differences in body weight, blood glucose, serum TG or insulin levels among 0, 1 and 3 day HFD mice (Fig. 1A–D). However, hepatic TG contents were markedly increased in mice fed a HFD for 3 days (Fig. 1E), which was also confirmed by Oil Red O staining (Fig. 1F).
Figure 1. Short term HFD increases hepatic TG contents in mice.
(A–E) Body weight (A), blood glucose (B), serum TG levels (C), insulin levels (D) and liver TG contents (E) in C57BL/6 mice fed a normal diet or high-fat-diet for 1 or 3 days. (F) Oil Red O staining showing TG accumulation in the liver. Original magnification, x200. ***P < 0.001.
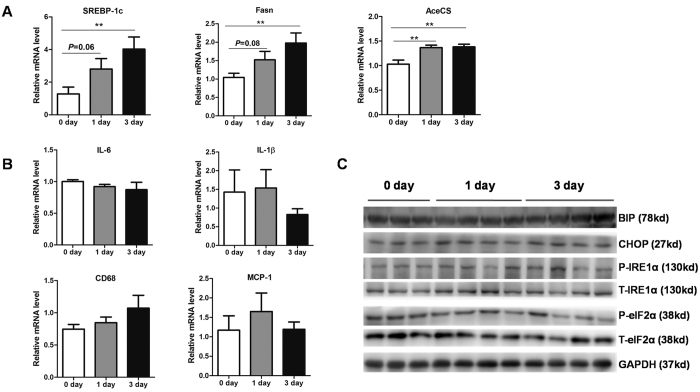
To explore the molecular basis for hepatic TG accumulation, expression levels of those genes involved in TG homeostasis were determined by quantitative real-time PCR (qPCR) using livers of mice fed a HFD for 0 day, 1 day and 3 days. As a result, we found that lipogenic genes such as SREBP-1c and its down-stream target genes (Fasn and AceCS) were elevated in the livers of mice fed a HFD for 1 day and 3 days (Fig. 2A), suggesting that up-regulation of lipogenic genes contributed to hepatic TG accumulation in short-term HFD mice.
Figure 2. Short-term HFD promotes hepatic TG lipogenesis in mice.
(A) Relative mRNA expression of lipogenic genes (SREBP-1C, Fasn and AceCS) in the liver from mice fed a HFD 3 days (n = 7–8). (B) Relative mRNA expression of pro-inflammatory cytokine (IL 6, IL-1β, CD68 and MCP-1) in the liver from mice fed a HFD for 1 day or 3 days (n = 7–8). (C) Western blot analysis of key molecules (BIP, CHOP, p-IRE1α and p-eIF2α) in ER stress signaling in the liver from mice fed a HFD for 1 day or 3 days (n = 4). **P < 0.01.
It has been well-established that inappropriate activation of inflammation and endoplasmic reticulum (ER) stress could promote hepatic TG accumulation in long term HFD obese mice29,30,31. Therefore, we assessed whether inflammation and ER stress were activated in livers of short-term HFD mice. mRNA levels of pro-inflammatory factors, including IL-6, IL-1β, CD68 and MCP-1 were not changed (Fig. 2B). Besides, protein levels of BIP, CHOP, phosphorylated IRE1α and eIF2α, markers of ER stress, remained unaffected (Fig. 2C). Taken together, our data indicate that the mechanism for hepatic TG accumulation and lipogenesis in short term HFD mice may be different from that in obese mice.
miR-124 expression is elevated in short term HFD mice
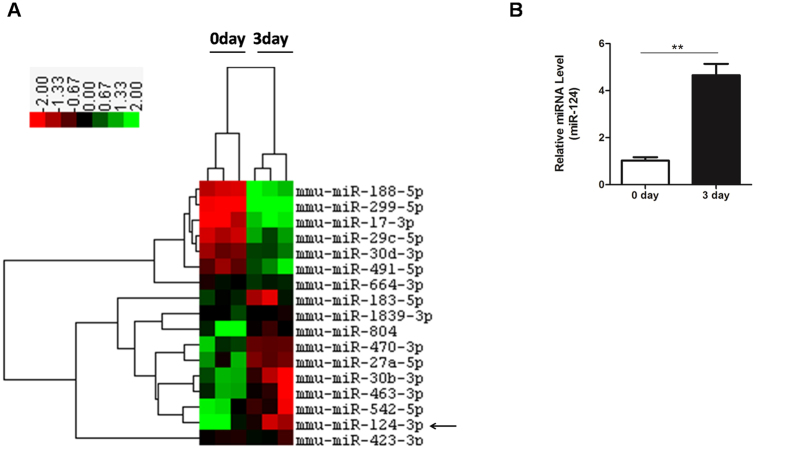
In order to identify the factors that promote hepatic lipogenesis in short-term HFD mice, we performed a clustering analysis of MicroRNA arrays using livers of mice fed a normal diet or high-fat-diet for 3 days. Here, the time 0 day in the heat map represents normal diet (ND), while the time 3 day represents HFD for 3 days (Fig. 3A). As a result, we found that a number of miRNAs were significantly altered in the livers of mice of two groups (P < 0.05). Our screen revealed a pronounced up-regulation of miR-124 in the livers of 3 days HFD mice compared to ND mice (Fig. 3A), which was further confirmed by qPCR (Fig. 3B). Here, we focus on miR-124 because up-regulation of miR-124 was one of the most pronounced changes among differential expressed miRNAs (Fig. 3A). Besides, miR-124 has been shown play a critical role in neuronal differentiation, pancreatic islet development and tumorigenesis17,18,20,21,22,23,24,25,26,27. However, its role in the regulation of hepatic TG homeostasis remains uncharacterized. Therefore, miR-124 was chosen for further experiments.
Figure 3. Up-regulation of miR-124 in short term HFD mice.
(A) Cluster analysis of miRNA expression in livers from C57BL/6 mice aged 8 weeks fed with a normal diet (ND) or high-fat diet (HFD) for 3 days. Cluster in red indicates upregulation and in green indicates downregulation. (B) Comparative real-time PCR analysis of miR-124 expression in the liver of 3 days HFD mice. **P < 0.01.
miR-124 promotes hepatic TG accumulation
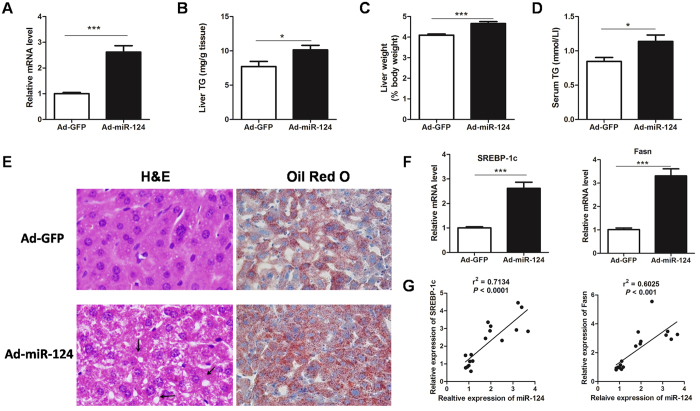
Next, to elucidate the role of miR-124 in the liver, adenovirus containing miR-124 or negative control (NC) were administered into C57BL/6 mice via tail vein injection. As shown in Fig. 4A, miR-124 expression levels were elevated in livers of mice infected with adenoviral miR-124 (Ad-miR-124). As expected, overexpression of miR-124 resulted in a marked increase in TG contents and liver weights (Fig. 4B,C), as well as serum TG levels (Fig. 4D). The observed alteration in hepatic TG content was further confirmed by H&E and Oil red O staining (Fig. 4E). However, the body weight, blood glucose, serum insulin and cholesterol levels were not changed (Fig. S1A–S1D). In agreement, mRNA levels of lipogenic genes, SREBP-1C and Fasn, were elevated and correlated well with the expression of miR-124 (Fig. 4F,G).
Figure 4. Overexpression of miR-124 increases hepatic lipogenesis.
(A) The expression of miR-124 was determined in the liver of C57BL/6 mice infected with adenovirus (Ad-) containing GFP or miR-124 (n = 8–10). (B–D) Liver TG content (B) and weight (C) and serum TG levels (D) in mice infected with Ad-GFP Ad-miR-124 (n = 8–10). (E) Representative histology (H&E, left) or Oil Red O (right) staining showing TG accumulation in livers from mice injected with miR-124 versus GFP adenovirus. Original magnification, x200. (F) Relative expression of SREBP-1c and Fasn in two groups of mice. (G) Pearson R and P value for normalized SREBP-1c and Fasn mRNA levels versus miR-124 expression in livers of mice (n = 17). *P < 0.05, **P < 0.01, ***P < 0.001.
TRB3 is a direct target of miR-124
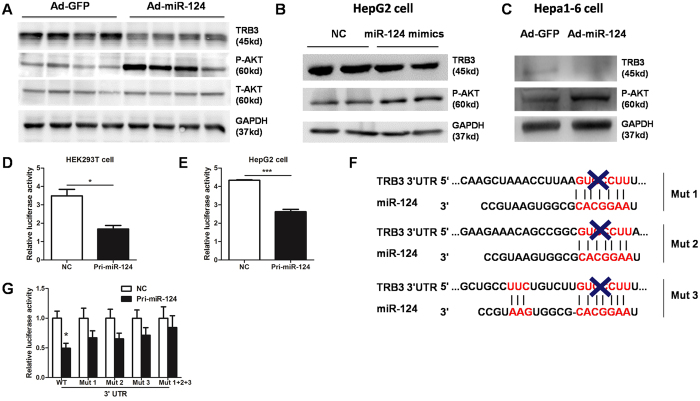
Using a stringent bioinformatics approach, we identified several putative murine miR-124 target genes, among which the gene encoding tribbles homolog 3 (TRB3) harboured a miR-124 binding site. Previous studies have shown that TRB3 is a pseudokinase that impaires insulin signaling by binding to Akt and blocking its activation32. Given that activation of Akt signaling could promote lipogenesis and hepatic TG accumulation33, we hypothesized that miR-124 may regulate TG metabolism through a TRB3-AKT pathway. As predicted, protein levels of TRB3 were decreased in the liver overexpressing miR-124 (Fig. 5A). Consistently, phosphorylation of Akt was increased as well (Fig. 5A). Overexpression of miR-124 also led to a decrease in TRB3 expression and an increase in Akt phosphorylation in HepG2 and Hepa1-6 cells (Fig. 5B,C).
Figure 5. miR-124 regulates TRB3 expression.
(A) Western blot analysis of TRB3 and phospho-Akt in the liver of mice infected with Ad-GFP or Ad-miR-124. (B) Western blot analysis of TRB3 and phospho-Akt in HepG2 cells treated with miR-124 mimics (100 nM) or vehicle control for 48 hours. (C) Western blot analysis of TRB3 and phospho-Akt in Hepa1-6 cells treated with Ad-GFP or Ad-miR-124 (100MOI) for 72 hours. (D,E) Relative luciferase activity of the firefly reporter containing the 3′UTR of TRB3 was detected in HEK293T cells (D) and HepG2 cells (E) co-transfected with pri-miR-124. (F) Sequence alignment of the 3′UTRs of TRB3 and miR-124. (G) Relative luciferase activity of reporters containing either the wild-type or mutated 3′UTRs of TRB3 gene (Mut1, Mut2, Mut3 and Mut1 + 2 + 3) were also analyzed in HEK293T cells. *P < 0.05, ***P < 0.001.
To further determine whether TRB3 is a direct target gene of miR-124, we constructed a luciferase reporter containing the TRB3 3′UTR and co-transfected with pri-miR-124 or the control into HEK293T and HepG2 cells. As expected, miR-124 markedly repressed the luciferase reporter activity in both cells (Fig. 5D,E). Besides, there are three potential binding sites in the TRB3 3′UTR (Fig. 5F). Each mutation attenuated the reduction of luciferase activity by miR-124 (Fig. 5G), while mutation of all three binding sites resulted in abolished repression in luciferase activity (Fig. 5G). Thus, our data clearly indicate that TRB3 is a direct target gene of miR-124 in the liver.
TRB3 restoration abolishes the effect of miR-124
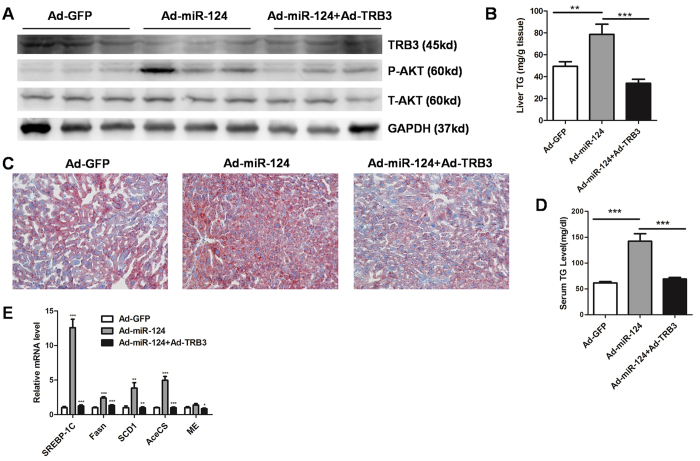
To clarify whether TRB3 mediates the effect of miR-124 on hepatic lipogenesis, we restored TRB3 (Ad-TRB3) expression in the liver in which adenoviral miR-124 was overexpressed (Fig. 6A). As a result, TRB3 restoration completely ameliorated hepatic triglyceride accumulation (Fig. 6B,C), lowered serum TG levels and down-regulated hepatic lipogenic genes expression (Fig. 6D,E). Therefore, our data support the notion that miR-124 mediated TRB3 down-regulation plays a critical role in hepatic lipogenesis and TG accumulation.
Figure 6. TRB3 overexpression alleviates miR-124 induced TG retention.
(A) Western blot analysis of TRB3 and phospho-Akt in the liver of mice infected with Ad-miR-124 or both Ad-miR-124 and Ad-TRB3. (B) Liver TG levels in mice infected with Ad-miR-124 or both Ad-miR-124 and Ad-TRB3 (n = 8–9). (C) Oil Red O staining showing TG accumulation in the liver from mice infected with Ad-miR-124 or both Ad-miR-124 and Ad-TRB3. Original magnification, x200. (D) Serum TG levels in mice infected with Ad-miR-124 or both Ad-miR-124 and Ad-TRB3 (n = 8–9). (E) Relative mRNA expression of lipogenic genes in the liver from mice infected with Ad-miR-124 or both Ad-miR-124 and Ad-TRB3 (n = 8–9). *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In the current study, we for the first time identified miR-124 as a regulator in hepatic TG homeostasis. We demonstrated that miR-124 directly targeted TRB3, and led to activation of AKT and up-regulation of lipogenic genes. Thus, our findings suggest that manipulating miR-124 expression might provide an alternative approach for amelioration of hepatic TG accumulation.
At the molecular level, we functionally validate TRB3 as a miR-124 target. It has been shown that TRB3 inhibited insulin signaling by binding directly to Akt and blocking its activation32. Besides, specific overexpression or knockdown of TRB3 in livers promoted or improved insulin resistance, respectively34,35. However, whole-body deletion of TRB3 displayed normal hepatic insulin signaling and glucose homeostasis, including unchanged Akt activation36. Although the reason for this inconsistence remains unknown, we speculate that compensatory function of TRB3 in other metabolic tissues such as skeletal muscle and adipose tissues should be considered.
It has been well established that Akt is a critical node to promote lipogenesis37. This is supported by several evidences. First, Akt enhances post-translational processing of SREBP-1c through phosphorylating SREBP-1c precursor38. Second, Akt increases SREBP-1c activity via mammalian target of rapamycin complex 1 (mTORC1)32. Third, Akt prevents degradation of mature SREBP-1c through inhibiting glycogen synthase kinase 3 (GSK3), a kinase that promotes the proteasomal degradation of SREBP-1c39,40. Therefore, hepatic overexpression of constitutively active Akt induced marked steatosis and up-regulation of lipogenic genes, including SREBP-1c, Fasn and SCD-133. Consistently, our data show that overexpression of miR-124 causes increased expression of SREBP1-c and TG accumulation in the liver. Of note, restoration of hepatic TRB3 expression almost completely abrogated the effect of miR-124 on lipids homeostasis, indicating that TRB3 might be the primary target of miR-124 in the regulation of liver TG metabolism.
Taken together, our study uncovers a novel miRNA, miR-124, that is responsible for early hepatic TG accumulation, preceding the systemic metabolic disorders. Our findings may expand our knowledge of mechanisms underlying the disturbance of hepatic TG metabolism and provide a promising therapeutic target for related diseases.
Methods
Animal experiments
Male C57BL/6 mice at age of 8–12 weeks were purchased from the Shanghai Laboratory Animal Company (SLAC, Shanghai, China). All mice were housed at 21 ± 1 °C with a humidity of 55 ± 10% and a12-hour light/12-hour dark cycle with free access to food and water. The high-fat diet (HFD) (D12492, Research Diets, New Brunswick, New Jersey, USA) contained 60% kcal from fat, 20% kcal from carbohydrate, and 20% kcal from protein. The normal diet (ND) contained 10% kcal from fat, 70% kcal from carbohydrate, and 20% kcal from protein. All animal protocols were approved by the Animal Care Committee of Shanghai Jiao Tong University School of Medicine and adhere to the criteria outlined in the “Guide for the Care and Use of Laboratory Animals”.
Adenovirus preparation
Recombinant adenovirus expressing mouse miR-124 and TRB3 (Ad-miR-124, Ad-TRB3) was generated using the pAd-Easy system according to the manufacturer’s instructions. Viruses were diluted in PBS and administered via a tail vein injection using 2*109 plaque-forming units per mouse.
Microarray analysis
Total RNA was isolated from hepatic tissues using the standard TRIzol method according to the manufacturer’s instructions (Invitrogen, Shanghai). After having passed RNA quantity measurement using the NanoDrop2000, the samples were labeled using the miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek, Denmark) and hybridized on the miRCURY™ LNA Array (v.18.0, Exiqon). Following washing steps the slides were scanned using the Axon GenePix 4000B microarray scanner. Scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged and miRNAs that intensities > = 30 in all samples were chosen for calculating normalization factor. Expressed data were normalized using the Median normalization. After normalization, significant differentially expressed miRNAs were identified through Volcano Plot filtering, which displays unstandardized signal (e.g. log(2) fold-change) against noise-adjusted/standardized signal (e.g. log(10) (p-value)).
Hepatic and cellular TG measurement
Liver tissues (weighed ∼100 mg) were harvested and homogenized in chloroform/methanol (2:1 v/v) using a Polytron tissue grinder (Kinematica AG, Luzern, Switzerland). Lipid extracts were prepared by the classical Folch method. Extracts were dried under N2 flow and dissolved in isopropanol. Cellular TG contents were measured using commercial kits (Biovision, USA) according to the manufacturer’s instructions.
Cell culture and luciferase assays
HepG2 and Hepa1-6 cell lines were purchased from the Cell Bank of Type Culture Collection, Chinese Academy of Sciences (CAS, Shanghai) and cultured in DMEM (Gibco, Shanghai, China) containing 10% FBS (Gibco), 100 IU/ml penicillin and 100 ug/ml streptomycin (Gibco). The primary miR-124 (pri-miR-124) sequence was amplified from the mouse genomic DNA and inserted into pcDNA3.1 vector (Invitrogen, USA). The complete 3′ UTR of murine TRB3 containing either the wildtype or mutated miR-124 binding sites was cloned and inserted into pRL-null vector (Promega, USA). For luciferase assays, cells were co-transfected in 24-well plates in duplicate wells with TRB3 reporter vectors together with pri-miR-124 plasmids and empty vectors using Lipofectamine 2000 (Invitrogen, USA). Luciferase activities were measured consecutively by using the Dual Luciferase Reporter Assay System (Promega, USA).
RNA isolation and real-time PCR
Total RNA of cells or tissues were extracted using standard TRIzol method according to the manufacturer’s instructions (Invitrogen, Shanghai). qRT-PCR was performed using a SYBR Green Premix Ex Taq (Takara, Japan) on Light Cycler480 (Roche, Switzerland). For miRNA detection, total RNA was reverse-transcribed using miScript II RT Kit (QIGEN, Shanghai). Subsequently, qRT-PCR was measured using miScript SYBR Green PCR Kit (QIGEN, Shanghai). U6 was used for normalization of miR-124 expression.
Western blot
Hepatic tissues and cells prepared with radioimmunoprecipitation (RIPA) buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5 mM MgCl2, 2 mM EDTA, 1 mM NaF, 1% NP40 and 0.1% SDS, supplemented with protease and phosphatase inhibitors (Millipore, USA). 50 μg lysates were loaded onto 10% SDS-PAGE and transferred to polyvinylidenedifluoride (PVDF) membranes (Millipore, USA). The proteins were visualized with Immobilon Western Chemiluminescent HRP Substrate (Millipore, USA) according to the manufacturer’s protocol. Western blots were performed using antibodies against rabbit anti-p-AKT (Cell Signaling, #4060, 1:1000), rabbit anti-t-AKT (Cell Signaling, #9272, 1:2000), goat anti-TRB3 (Santa Cruz, sc-34211, 1:500), mouse anti-CHOP (Beyotime, #AC532, 1:1000), rabbit anti-BIP (Beyotime, #AB310, 1:1000), rabbit anti-p-eIF2α (Cell Signaling, #9721, 1:1000), rabbit anti-t-eIF2α (Cell Signaling, #9722, 1:1000), rabbit anti-p-IRE1α (Novus Biologicals, #NB100-2323, 1:1000), rabbit anti-t-IRE1α (Cell Signaling, #3294, 1:1000) and rabbit anti-GAPDH (Cell Signaling, #5174, 1:1000).
Histological analysis
For hematoxylin and eosin staining (H&E staining), liver tissues were fixed in 10% neutral buffered formalin and embedded in paraffin. Sections were subjected to standard H&E staining. For oil red O staining, liver tissues were fixed in 4% paraformaldehyde in PBS, embedded in optimum cutting temperature compound (OCT), and cryosectioned. Frozen liver sections were stained with 0.15% oil red O according to standard procedures. Pictures were taken using an Olympus BX-51 microscope.
Statistical analysis
All data were expressed as mean ± standard error of mean (SEM). Statistical differences were determined by a Student t test. Statistical significance is displayed as *p < 0.05, **p < 0.01 or ***p < 0.001.
Additional Information
How to cite this article: Liu, X. et al. MicroRNA-124 promotes hepatic triglyceride accumulation through targeting tribbles homolog 3. Sci. Rep. 6, 37170; doi: 10.1038/srep37170 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This study is supported by grants from Natural Science Foundation of China (Nos 31530033, 81361128009, 81570769, 81500617 and 81300700) and the Doctoral Innovation Fund Projects from Shanghai Jiao Tong University School of Medicine (BXJ201416).
Footnotes
Author Contributions Y.Lu. and X.Y.L. conceived the project; Y.Lu. and X.L. designed experiments; X.L., J.J.Z., Q.L., X.X.X., Z.J.Z., Y.J., B.L. and Y.Li. performed experiments; X.L. drafted the manuscript and performed the statistical analysis; Y.Lu. and X.Y.L. handled funding and supervision. All authors reviewed the manuscript.
References
- Cohen J. C., Horton J. D. & Hobbs H. H. Human fatty liver disease: old questions and new insights. Science. 332, 1519–1523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano Y. & Cohen D. E. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol. 48, 434–441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. et al. Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. PNAS. 98, 13607–13612 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. D., Goldstein J. L. & Brown M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 109, 1125–1131 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura I., Bashmakov Y. & Horton J. D. Increased Levels of Nuclear SREBP-1c Associated with Fatty Livers in Two Mouse Models of Diabetes Mellitus. THE Journal Of Biological Chemistry. 274, 30028–30032 (1999). [DOI] [PubMed] [Google Scholar]
- Krycer J. R., Sharpe L. J., Luu W. & Brown A. J. The Akt-SREBP nexus: cell signaling meets lipid metabolism. Trends Endocrinol Metab. 21, 268–276 (2010). [DOI] [PubMed] [Google Scholar]
- Elam M. B. et al. Dysregulation of sterol regulatory element binding protein-1c in livers of morbidly obese women is associated with altered suppressor of cytokine signaling-3 and signal transducer and activator of transcription-1 signaling. Metabolism. 59, 587–598 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. et al. Yin Yang 1 promotes hepatic steatosis through repression of farnesoid X receptor in obese mice. Gut. 63, 170–178 (2012). [DOI] [PubMed] [Google Scholar]
- Xiong X. L. et al. Hepatic steatosis exacerbated by endoplasmic reticulum stress-mediated downregulation of FXR in aging mice. Journal of Hepatology. 60, 847–854 (2014). [DOI] [PubMed] [Google Scholar]
- Rottiers V. & Naar A. M. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 13, 239–250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli S., Panera N., Gnani D. & Nobili V. Dual role of microRNAs in NAFLD. Int J Mol Sci. 14, 8437–8455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W. C. et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 122, 2884–2897 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S. H. et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver.J Clin Invest. 122, 2871–2883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerin I. et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation.J Biol Chem. 285, 33652–33661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dávalosa A. et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. PNAS. 108, 9232–9237 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. et al. Hepatic miR-378 targets p110 alpha and controls glucose and lipid homeostasis by modulating hepatic insulin signalling. Nat Commun. 5, 5684 (2014). [DOI] [PubMed] [Google Scholar]
- Sonntag K. C., Woo T. U. & Krichevsky A. M. Converging miRNA functions in diverse brain disorders: a case for miR-124 and miR-126. Exp Neurol. 235, 427–435 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun A. X., Crabtree G. R. & Yoo A. S. MicroRNAs: regulators of neuronal fate. Curr Opin Cell Biol. 25, 215–221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J. et al. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 73, 3913–3926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre X. et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 69, 4443–4453 (2009). [DOI] [PubMed] [Google Scholar]
- Zheng F. et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 61, 278–289 (2012). [DOI] [PubMed] [Google Scholar]
- Cheng Y. et al. STAT3 is involved in miR-124-mediated suppressive effects on esophageal cancer cells. BMC Cancer. 15, 306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T. et al. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 124, 2367–2374 (2009). [DOI] [PubMed] [Google Scholar]
- Wilting S. M. et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer. 9, 167 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X. B. et al. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene. 32, 4130–4138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroukh N. et al. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem. 282, 19575–19588 (2007). [DOI] [PubMed] [Google Scholar]
- Lovis P., Gattesco S. & Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol Chem. 389, 305–312 (2008). [DOI] [PubMed] [Google Scholar]
- Reddy J. K. & Rao M. S. Lipid Metabolism and Liver Inflammation.II. Fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol. 290, G852–G858 (2006). [DOI] [PubMed] [Google Scholar]
- Shoelson S. E., Lee J. & Goldfine A. B. Inflammation and insulin resistance. J Clin Invest. 116, 1793–1801 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-Kent C. et al. Cytokines in the pathogenesis of fatty liver and disease progression to steatohepatitis: implications for treatment. Am J Gastroenterol. 103, 1036–1042 (2008). [DOI] [PubMed] [Google Scholar]
- Kammoun H. M. et al. GRP78 expression inhibits insulin and ER stress–induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest. 119, 1201–1215 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K., Herzig S., Kulkarni R. N. & Montminy M. TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science. 300, 1574–1577 (2003). [DOI] [PubMed] [Google Scholar]
- Ono H. et al. Hepatic Akt activation induces marked hypoglycemia, hepatomegaly, and hypertriglyceridemia with sterol regulatory element binding protein involvement. Diabetes. 52, 2905–2913 (2003). [DOI] [PubMed] [Google Scholar]
- Koo S. H. et al. PGC-1 promotes insulin resistance in liver through PPAR-α-dependent induction of TRB-3. Nature Medicine. 10, 530–534 (2004). [DOI] [PubMed] [Google Scholar]
- Yu J. J. et al. Hepatic phosphoserine aminotransferase 1 (PSAT1) regulates insulin sensitivity in mice via tribbles homolog 3 (TRB3). Diabetes. 64, 1591–1602 (2015). [DOI] [PubMed] [Google Scholar]
- Okamoto H. et al. Genetic deletion of Trb3, the mammalian Drosophila tribbles homolog, displays normal hepatic insulin signaling and glucose homeostasis. Diabetes. 56, 1350–1356 (2007). [DOI] [PubMed] [Google Scholar]
- Wong R. H. & Sul H. S. Insulin signaling in fatty acid and fat synthesis: a transcriptional perspective. Curr Opin Pharmacol. 10, 684–691 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellaturu C. R. et al. Insulin enhances post-translational processing of nascent SREBP-1c by promoting its phosphorylation and association with COPII vesicles. J Biol Chem. 284, 7518–7532 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punga T., Bengoechea-Alonso M. T. & Ericsson J. Phosphorylation and ubiquitination of the transcription factor sterol regulatory element-binding protein-1 in response to DNA binding. J Biol Chem. 281, 25278–25286 (2006). [DOI] [PubMed] [Google Scholar]
- Bengoechea-Alonso M. T. & Ericsson J. A phosphorylation cascade controls the degradation of active SREBP1. J Biol Chem. 284, 5885–5895 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.