Abstract
Clostridium difficile is a major cause of intestinal infection and diarrhoea in individuals following antibiotic treatment. Recent studies have begun to elucidate the mechanisms that induce spore formation and germination and have determined the roles of C. difficile toxins in disease pathogenesis. Exciting progress has also been made in defining the role of the microbiome, specific commensal bacterial species and host immunity in defence against infection with C. difficile. This Review will summarize the recent discoveries and developments in our understanding of C. difficile infection and pathogenesis.
ToC blurb
Treating infection with Clostridium difficile and post-antibiotic disease can be difficult. In this Review, Abt, McKenney and Pamer show how insights into spore germination, virulence and interactions with the host and microbiota can help to combat this pathogen.
Clostridium difficile-induced colitis is the most common and costly healthcare-associated infection with an estimate of nearly half a million cases and approximately 29,000 deaths occurring annually in the United States1. Disease associated with C. difficile infection ranges from mild diarrhoea to pseudomembranous colitis, which was first shown to be caused by C. difficile 40 years ago2. A pristine intestinal microbiota provides resistance against C. difficile infection and disruption of the microbiota (for example, through antibiotic treatment), allows the bacterium to proliferate in the gut. C. difficile is a Gram-positive, spore-forming, obligate anaerobic bacterium. The formation of spores enables C. difficile to survive in oxic conditions, which contributes to transmission in healthcare settings and maybe also in the community (BOX 1). Once inside the gastrointestinal tract, pathogenesis is tightly linked to spore germination and the production of toxins. In this Review, we highlight factors that regulate the spore-forming life cycle of C. difficile, virulence and mechanisms that are mediated by the host and the microbiota that contribute to protection from disease. A more comprehensive understanding of C. difficile pathogenesis is emerging that may lead to new and innovative therapeutic and diagnostic options, which are urgently required to treat infection with this bacterium (BOX 2).
Box 1. Genomic insights into the taxonomy and transmission of Clostridium difficile.
Clostridium difficile was first isolated from the intestines of infants and was shown to cause colitis in guinea pigs and rabbits156. It was assigned to the genus Clostridium because of its morphology, ability to from spores and inability to undergo vegetative growth in the presence of oxygen. The Clostridium genus is complex and contains many distinct and dissimilar species that were grouped together based on traditional microbiological methods. Recent analyses, mostly based on sequences of 16S rRNA and ribosomal protein genes, place C. difficile in the Peptostreptococcaceae family and its genus name, therefore, has been changed to Peptoclostridium157. The number of infections with C. difficile is increasing and the repeated emergence of new and evolved epidemic strains, such as the notorious BI/NAP1/027 strain, raises the possibility that new strains are spreading into human populations from environmental sources that are yet to be defined. Although healthcare and chronic care facilities have been typical places in which patients acquire C. difficile infections, in the past decade, more patients are presenting with community-acquired C. difficile infections158. New evidence indicates that the transmission of C. difficile between patients who are hospitalized has been overestimated159, which suggests that many patients who develop an infection with C. difficile did not acquire it during hospitalization and presumably harboured the organism asymptomatically. Pigs, horses and a wide range of other mammals can be colonized with C. difficile and viable C. difficile spores have been detected in various locations160. A deeper understanding of the environmental distribution of C. difficile strains will provide important insights into the epidemiology of human infections.
Box 2. Clinical dilemmas regarding C. difficile infection.
The diagnosis and treatment of Clostridium difficile infection in clinical settings are far from straightforward. Many clinical studies have been carried out to address these complex topics and our understanding has certainly improved. However, the development of more sensitive diagnostic tests, the application of whole-genome sequencing, the introduction of new antibiotics and greater acceptance of faecal microbial transplants have led to many new questions that require further investigation.
Patients who are hospitalized and have diarrhoea are generally tested for C. difficile infection. The recent introduction of a fast and highly sensitive PCR test that detects tcdB has led to higher rates of detection but it also potentially identifies patients colonized with C. difficile whose diarrhoea is caused by other factors. The concern about false-positivity is particularly important in patient populations with diarrhoea that is associated with laxative administration or following irradiation for haematopoietic stem cell transplantation. Rates of C. difficile carriage in the overall population are variable, but in some cases can be quite high — for example, in infants in which carriage rates are very high but are not associated with colitis. The administration of antibiotics following a false-positive test in a patient who is asymptomatically colonized with C. difficile has the potential to disrupt the protective gut microbiota and induce colitis.
The high recurrence rate of C. difficile infection in patients following conventional treatment with antibiotics is probably the result of collateral damage to the microbiota induced by metronidazole or oral vancomycin161. Indeed, these two antibiotics, which are used to treat C. difficile infection, are destructive to the commensal microbiota and render the host highly susceptible to reinfection. A recent addition to the treatment armamentarium is fidaxomycin, a non-absorbable antibiotic that is less toxic to obligate anaerobic commensal bacteria. Recurrence of C. difficile infection caused by some, but not all, strains is decreased following treatment with fidaxomycin, when compared with other antibiotics162.
The most rapidly evolving and certainly most effective treatment for recurrent infection is faecal microbial transplant. Donor selection, route of transplantation, timing of transplantation and screening for potentially transmissible pathogens remain important issues that, with continued studies, should be resolved. It is likely that faecal microbial transplant will be replaced with precision bacteriotherapy with defined consortia of commensal bacteria.
C. difficile sporulation
The lumen of the human colon is anoxic, which enables obligate anaerobic bacteria such as C. difficile to survive and, if the conditions are suitable, to proliferate, produce toxins and damage the intestinal epithelium. C. difficile forms spores that are resistant to heat, oxygen and common disinfectants, such as ethanol-based hand sanitizers, which facilitates spread3–5. Efforts to determine the mechanism of C. difficile sporulation benefit from more than 50 years of work on sporulation in Bacillus subtilis and, mechanistically, much is conserved between these two members of the Firmicutes phylum6.
Formation of the forespore
In culture, sporulation occurs at stationary phase when nutrients become limiting. At one pole of a C. difficile (or a B. subtilis) cell a septum is constructed that results in asymmetric division and the creation of two unequally sized compartments. The smaller compartment — the forespore — will develop into the spore, whereas the larger compartment — the mother cell — will prepare the forespore for dormancy. The forespore matures into a desiccated, stress-resistant chromosome-storage vessel, which is released into the environment through lysis of the mother cell. Spores can germinate and produce new vegetative cells when conditions become favourable again (FIG. 1). The transcription factors that are responsible for sporulation seem to be well conserved, which enables sporulation in B. subtilis to act as a template for understanding the regulation of sporulation in C. difficile6. The development of increasingly sophisticated tools for reverse genetics7,8 and forward genetics9, combined with transcriptional profiling, has enabled the identification of hundreds of genes that are involved in sporulation in C. difficile10.
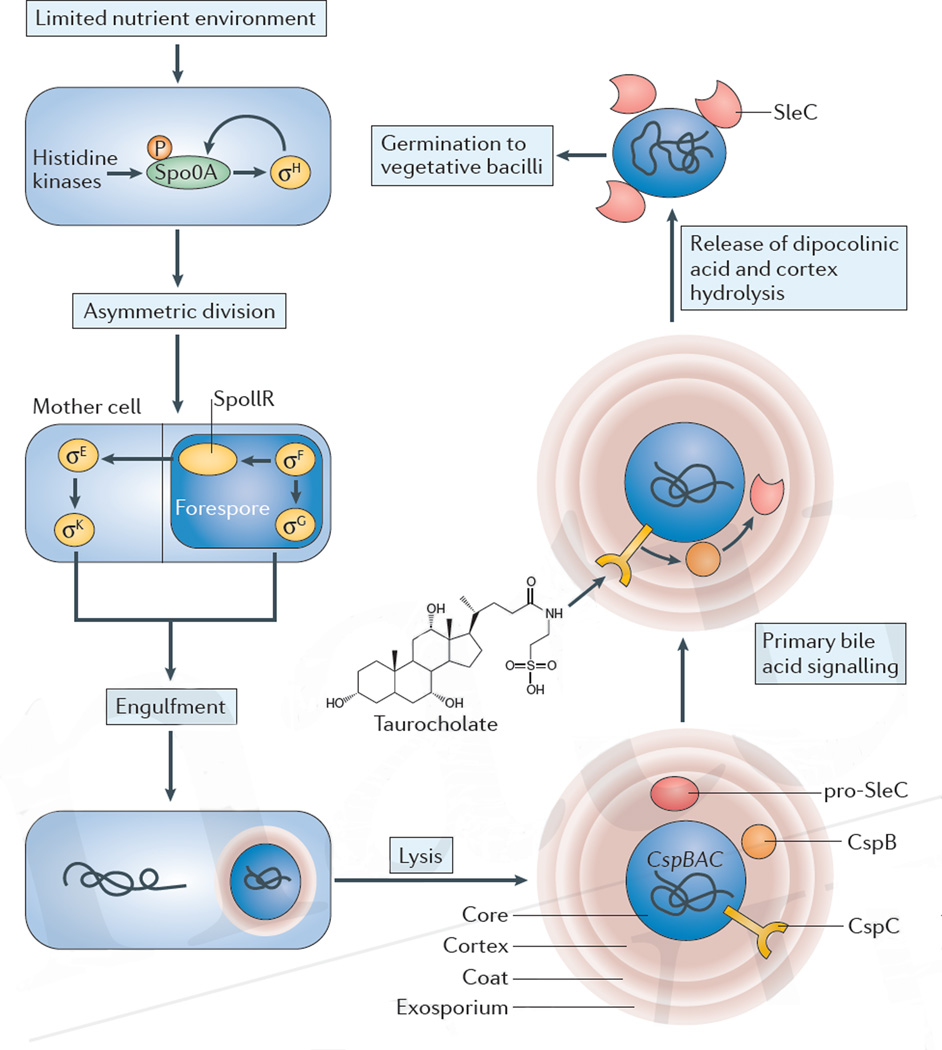
Figure 1. Sporulation and germination of C. difficile.
A limited nutrient environment induces sporulation. The transcription factor stage 0 sporulation protein A (Spo0A) is phosphorylated by histidine kinases, activating a cascade of signalling and morphological events that create a forespore within the mother cell of the bacterium. After lysis, the spore is released into the environment. The core of the spore, which contains the condensed chromosome, is encapsulated by three protective layers: the cortex, coat and exosporium. Germination of the spore can be initiated by bile acids, such as taurocholate, which signal through the CspC receptor. Activation of the SleC enzyme by CspB leads to the degradation of the cortex of the spore and eventually leads to outgrowth of a new vegetative cell. P, phosphate.
Transcriptional regulation of sporulation
Although the exact environmental signals that trigger sporulation in C. difficile have remained elusive, the molecular circuitry that results in the formation of spores is now well described. In C. difficile, sporulation is initiated by signalling through sensor histidine kinases that ultimately results in the phosphorylation and activation of the transcription factor stage 0 sporulation protein A (Spo0A)11. Five such kinases have been identified in C. difficile strain CD630 (REF. 12) and one kinase (CD1579) directly phosphorylates Spo0A in vitro13. Phosphorylated Spo0A drives the sporulation regulatory network and directly regulates dozens of genes14,15. Spo0A is essential for C. difficile sporulation and mutants that lack this gene only exist as vegetative cells. The inability of Spo0A mutants to form spores profoundly decreases the spread of the organism from infected mice to susceptible, uninfected mice16. Spo0A induces the expression of the first sporulation-specific sigma factor, σH, which forms a positive feed-forward loop with Spo0A17.
Expression levels of Spo0A are also controlled by two transcription factors, CodY and CcpA, which may integrate nutritional state into the decision to form spores. CodY is mainly a transcriptional repressor that binds to DNA in the presence of GTP and branched-chain amino acids and represses the transcription of dozens of operons in C. difficile18, including the toxin locus19 and two regulators of sporulation20. CcpA, a global regulator of carbon catabolite repression, directly represses spo0A21 and leads to decreased toxin production when C. difficile is cultured in the presence of glucose22. Finally, deletion of the oligopeptide permeases opp and app increases the expression of Spo0A, spore production and virulence in a hamster model23. Deletion of opp and app may decrease the ability of C. difficile to harvest nutrients from peptides in the local environment, which leads to the starvation of cells and accelerates the transition to stationary phase. Taken together, these data suggest that C. difficile represses sporulation in the presence of nutrients.
Downstream of Spo0A is a transcriptional programme that is driven by the sequential activation of four compartment-specific alternative sigma factors — σF, σE, σG and σK. In B. subtilis, σF is activated in the forespore following asymmetric division24. Inter-compartmental signalling across the division septum then activates σE in the mother cell, followed by σG in the forespore and finally σK in the mother cell24. The activation of these four sporulation sigma factors in C. difficile differs from that described for B. subtilis10,25 (FIG. 1). Specifically, the checkpoints that ensure the sequential, inter-compartmental progression of sigma factor activation in B. subtilis seem to be missing from late-stage sporulation in C. difficile. In both systems, σF is activated only in the forespore, which results in the production of the SpoIIR signalling protein and the subsequent activation of σE in the mother cell10,26. Individual deletion of sigma factors, complemented by morphological characterization of C. difficile sporangia, demonstrated that σF induces the activation of σG in the forespore, whereas σE promotes the activity of σK in the mother cell. However, in contrast to B. subtilis, the activity of σG does not depend on σE and σK is independent of σG, which suggests that inter-compartmental signalling for the activation of sigma factors is not conserved (FIG. 1). In addition, the SpoIIIA–SpoIIQ ‘feeding tube’ channel, which is necessary for the activation of σG in B. subtilis, is dispensable for the activation of σG in C. difficile27,28. A recent study reported a transposon mutant library in C. difficile that identified 404 candidate genes that are involved in sporulation, including known sporulation-associated genes, such as spo0A, but also genes such as splA the contribution of which to sporulation is unknown9.
The spore envelope
The spore core is encased in three protective layers: the peptidoglycan cortex, a coat that predominantly consists of proteins, and a third and outermost layer, the exosporium, which predominantly consists of glycoproteins29. Only 25% of the more than 70 proteins that comprise the spore coat are conserved between B. subtilis and C. difficile30, which suggests that the spore surface may be an important source of evolutionary adaptation among members of the Firmicutes6. Deletion of the major spore coat morphogenetic protein SpoIVA in C. difficile caused the coat to fail to localize to the spore surface31. Screening for envelope proteins from extracts of uncoated spores has uncovered a high degree of enzymatic activity among C. difficile spore coat proteins including a catalase with superoxide dismutase activity32, and CdeC, a cysteine-rich protein that localizes to the exosporium and enhances resistance to heat, lysozyme and ethanol33. The C. difficile exosporium contains three collagen-like glycoproteins, BclA1, BclA2 and BclA3, which are conserved with the exosporium of Bacillus anthracis. Deletion of bclA1 results in a slight decrease in virulence, but an increase in both spore germination and adherence, which suggests that changes to the structure of the spore envelope may affect disease34,35.
C. difficile germination and vegetative growth
Germination of C. difficile spores and the growth of vegetative forms occurs only in the lower gastrointestinal tract, in part, because oxygen concentration at this site is negligible36. Substances that are present in the intestine, most notably bile acids, induce germination of the spore into an actively replicating vegetative cell, a process that is controlled by the cspBAC gene locus.
The role of bile acids in spore germination
An early study demonstrated that the addition of taurocholate, a conjugated primary bile acid that is present in the small intestine, to culture media greatly increased the growth of colonies from clostridial spores that were isolated from human, calf and rat faeces37. Subsequent studies demonstrated that the addition of taurocholate to cycloserine cefoxitin fructose agar (CCFA) media, which is used to culture C. difficile, increased the recovery of colonies from spores38. Importantly, bile acids differ in their ability to promote germination and to influence vegetative growth39. Thus, whereas taurocholate together with the amino acid glycine act as co-germinants of spores without affecting the vegetative growth of C. difficile, the secondary bile acid deoxycholate promotes germination but markedly suppresses vegetative growth39,40. Interestingly, chenodeoxycholate, another primary bile acid, inhibits spore germination in C. difficile and acts both as a competitive inhibitor of taurocholate and a suppressor of vegetative growth in liquid culture41,42. The potential role of the intestinal bile acid pool in determining C. difficile colonization following exposure to spores was supported by ex vivo studies of intestinal extracts from antibiotic-treated and untreated mice, which revealed a correlation between the capacity to support the growth of C. difficile and decreased levels of secondary bile acids43,44. These observations led to the hypothesis that the production of secondary bile acids by the commensal microbiota, and their ablation by antibiotic treatment, modulates susceptibility to C. difficile colitis (BOX 3).
Box 3. Microbiota-mediated primary bile acid conversion.
Bile acids are produced in the liver and secreted into the small intestine. Most are reabsorbed in the terminal ileum; however, approximately 5% of bile acids flow into the large intestine in which a subset of anaerobic bacteria converts them into secondary bile acids163. Primary bile acids, such as glycocholate and taurocholate, are deconjugated to yield cholate by bile salt hydrolases that are expressed on the surface of a wide range of bacterial species in the gut164. A smaller subset of bacterial species that reside in the caecum and colon, such as Clostridium scindens165, take up cholate and chenodeoxycholate and dehydroxylate the 7α carbon of the bile acid backbone to produce deoxycholate or lithocholate, respectively166. The benefit of this dehydroxylation step for the bacteria that carry it out remains unclear, but bile acids may simply represent an electron acceptor in the anaerobic environment of the lower intestinal tract. The association between the levels of secondary bile acids and colon cancer suggests that their production is deleterious to the host. Recent studies have determined that another common intestinal commensal bacterium, Ruminococcus gnavus, detoxifies secondary bile acids by converting them into iso-bile acids167. Balancing the potential negative effects of secondary bile acids to the host is their ability to inhibit spore germination and suppress the vegetative growth of C. difficile. Indeed, loss of secondary bile acids in the caecum and colon following antibiotic treatment is strongly associated with susceptibility to infection with C. difficile, and recovery from recurrent C. difficile infection following microbiota reconstitution by faecal transplantation is highly correlated with the recovery of secondary bile acid levels115. For reasons that remain unclear, the effect of bile acids on spore germination and growth in C. difficile varies between strains, with some strains inhibited by chenodeoxycholate, whereas other strains are not168. Bile acids vary somewhat between mammalian species. For example, muricholic acids that are produced by mice inhibit the germination of C. difficile spores169, although it remains unclear whether this applies to spores of all strains. Ursodeoxycholate, which is present in human bile and is also used therapeutically to treat cholestatic liver disease, inhibits both germination and vegetative growth of C. difficile170.
Degradation of the spore cortex
Bile acid-induced germination leads to cortex degradation, the release of calcium and dipocolinic acid and rehydration of the spore, which are important early steps in the germination process45,46 (FIG. 1). The receptor for bile acids on C. difficile spores, CspC, was recently identified by screening ethyl methanesulfonate mutagenized bacteria47. CspC is encoded by the cspBAC locus and is similar in sequence to subtilisin-family proteases in Clostridium perfringens that have been associated with the activation of SleC, a lytic enzyme that is essential for spore germination47, but lacks the catalytic triad that is required for proteolytic activity47. Deletion of CspC renders C. difficile spores unresponsive to taurocholate. Notably, a single amino acid substitution of glycine to arginine at residue 457 of CspC alters the germination-inhibitory effect of chenodeoxycholate to a germination-stimulatory effect, markedly supporting the notion that CspC directly associates with primary bile acids47. Also encoded by the cspBAC locus is a hybrid protein, CspB–CspA, which consists of the subtilisin-family proteins CspB and CspA. Biochemical studies have demonstrated that the CspA pseudoprotease domain regulates the level of the CspC receptor in mature spores48. The YabG protease functions to cleave both the CspBA fusion protein and full-length SleC during sporulation, releasing CspB and pro-SleC, which become components of the spore coat48,49. Only CspB contains an enzymatically active catalytic triad; however, structural analysis of CspB from C. perfringens revealed that its prodomain, even after autocleavage, remains tightly bound, thereby occluding the active site47,49. CspB is required to complete the proteolytic activation of pro-SleC into mature and active SleC, which degrades the dense proteoglycan cortex during spore germination48,49,50 (FIG. 1). It remains unclear how the association of taurocholate with CspC activates CspB and subsequently SleC, and it is possible that additional factors are required for the activation of germination. Indeed, recent studies have demonstrated that the lipoprotein GerS is essential for the full enzymatic activity of SleC following CspB-mediated cleavage of pro-SleC27. Once initiated, germination of C. difficile spores is a complex process that, as determined by microarray analyses, involves the upregulation or downregulation of more than 500 genes51. The efficiency of germination varies between clinical strains of C. difficile, and whether this correlates with virulence remains an active area of investigation52,53.
Virulence factors
The ability of C. difficile to cause colitis depends on a range of virulence factors, including toxins, which are encoded in the pathogenicity locus54, and adherence and motility factors. In response to limited nutrient availability, C. difficile produces toxins that primarily target intestinal epithelial cells. Following toxin endocytosis and activation in the cytosol, epithelial cells undergo necrosis, which leads to loss of intestinal membrane integrity, host exposure to intestinal microorganisms and activation of the host inflammatory response.
TcdA and TcdB toxins
The pathogenicity locus generally encodes five proteins and, in most strains, is localized at a specific site in the C. difficile chromosome55, although a recent study characterized unusual strains in which the pathogenicity locus was localized in atypical regions56. The major toxins that are encoded by the pathogenicity locus are TcdA (also known as ToxA) and TcdB (also known as ToxB), which are two large secreted proteins that contain four structurally homologous domains57. TcdA and TcdB contain RHO and RAC glucosyl transferase domains (GTDs) at the amino terminus and mediate toxicity by glycosylating and thereby inactivating host RHO and RAC GTPases in the cytosol of targeted cells (FIG. 2a). This disrupts the cytoskeleton and leads to the disassociation of tight junctions between colonic epithelial cells and the loss of epithelial integrity55.
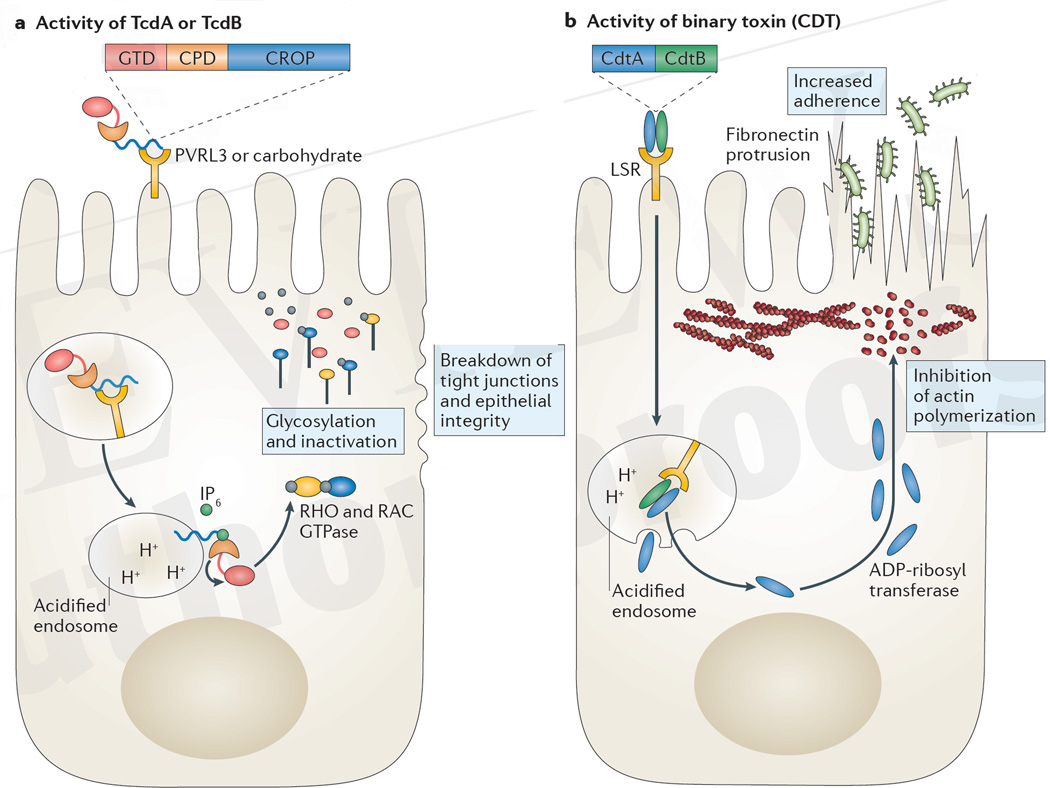
Figure 2. Mechanism of action of C. difficile toxin in epithelial cells.
a | The combined repetitive oligopeptide repeat (CROP) domain of TcdA binds to carbohydrates on the apical surface of epithelial cells, whereas TcdB binds to poliovirus receptor-like 3 (PVRL3) expressed on colonic epithelial cells. Toxin is internalized and acidification of the endosome enables the CROP domain to embed into the endosomal membrane and the subsequent transport of the cysteine protease domain (CPD) and the glucosyl transferase domain (GTD) into the cytosol. Inositol hexakisphosphate (IP6) activates the cysteine protease to cleave and release the toxin glycotransferase. Glycosylation and thereby inactivation of RHO or RAC GTPases ultimately results in the breakdown of tight junctions and epithelial integrity. b | Binary toxin (also known as Clostridium difficile transferase (CDT)) binds to the lipolysis-stimulated lipoprotein receptor (LSR) and is internalized. The CdtB subunit creates pores in the acidified endosome that enable the release of the CdtA subunit into the cytosol. The ADP-ribosyl transferase activity of CdtA inhibits actin polymerization near the cell membrane, enabling fibronectin microtubules to elongate and protrude through microvilli, which increases C. difficile adherence to the epithelium through type IV pili.
Adjacent to the GTD, TcdA and TcdB contain a cysteine protease domain (CPD) that autocatalytically cleaves the glucosyl transferase in the cytosol of eukaryotic cells on association with inositol hexakisphosphate (myo-inositol)58,59. More efficient autoprocessing by the cysteine protease has been associated with increased virulence in C. difficile strains60. A potent small-molecule inhibitor of the toxin cysteine protease was recently described that blocked the release of the glucosyl transferase and abrogated toxicity61. Administration of this drug reduced intestinal epithelial damage in mice that were infected with C. difficile and offers a potential therapeutic avenue to limit toxin-mediated damage.
The other two domains of TcdA and TcdB consist of a hydrophobic protein sequence that is involved in host membrane insertion and the combined repetitive oligopeptide repeat (CROP) domains that are hypothesized to bind to cell surface receptors before TcdA and TcdB endocytosis and internalization. The CROP domain of TcdA contains up to 38 repeats62 with a structure that enables the binding of carbohydrates63 and associates with gp96 on the apical surface of colonocytes64. Conversely, the CROP domain of TcdB has fewer repeats and associates with the N-terminal, extracellular domain of chondroitin sulfate proteoglycan 4, although the expression of this receptor in intestinal epithelial cells has not yet been reported65. TcdB has also been shown to bind to poliovirus receptor-like 3 (PVRL3), which is expressed on the surface of colonic epithelial cells, although the association of TcdB with PVRL3 does not involve the CROP domain66.
The relative contribution of TcdA and TcdB to in vivo pathogenesis has been investigated by deletion mutagenesis and infection of antibiotic-treated hamsters67–69. The combined deletion of TcdB and TcdA completely abolished in vivo virulence68,69. Single deletion of TcdA did not alter C. difficile virulence in the hamster model, which indicates that TcdB alone is capable of mediating colitis. The potential role of TcdA is more controversial, with one report suggesting that C. difficile mutants that lack TcdB but express TcdA do not cause colitis67, whereas two other reports showed TcdA-mediated disease68,69. These inconsistent findings probably reflect differences in C. difficile strains, experimental animals (mice versus hamsters) and their microbiota composition and antibiotic sensitivity. Indeed, a carefully carried out, multi-laboratory follow-up study demonstrated that TcdB is the major virulence factor that mediates colonic epithelial damage, inflammation and mortality in the murine model, whereas TcdA is a relatively minor driver of inflammation in mice and is slightly more toxic in hamsters54.
Remarkable sequence diversity in genes that encode TcdB exists among different C. difficile strains and it has been hypothesized that sequence differences in toxins may contribute to heterogeneity in virulence between strains12,70. Studies with purified TcdB from the epidemic BI/NAP1/027 strain of C. difficile demonstrated 4-fold-greater toxicity in mice compared with TcdB from typical C. difficile strains71. It remains unresolved whether environmental factors promote toxin diversification and whether the host environment selects for increased or decreased virulence.
TcdR, TcdC and TcdE
The pathogenicity locus also encodes three other proteins, TcdR, TcdC and TcdE. TcdR is an alternative sigma factor that facilitates the binding of RNA polymerase to the promoters of the tcdA and tcdB genes and also, in a positive feedback loop, its own promoter72. On reaching stationary growth, the transcription of tcdA and tcdB is driven by TcdR in C. difficile72. During exponential growth, C. difficile expresses higher levels of TcdC, which has been hypothesized to act as an anti-sigma factor and thus suppresses the transcription of tcdA and tcdB73. In contrast to typical anti-sigma factors, which associate with sigma factors to inhibit transcription, TcdC may directly associate with single-stranded DNA to inhibit tcdA and tcdB transcription74. Hypervirulence has been associated with deletions in the tcdC sequence, which supports the potential role of TcdC in limiting toxin expression by C. difficile75. However, neither genetically engineered tcdC mutant strains nor restoration of tcdC expression in a hypervirulent 027 ribotype strain altered levels of toxin production when grown in culture media8,76. These more recent studies cast doubt on the exact role of TcdC in negatively regulating the expression of tcdA and tcdB.
The fifth gene in the pathogenicity locus is tcdE, which encodes a holin-like protein that is believed to facilitate the secretion of TcdA and TcdB, which lack conventional secretion signal sequences. As with other proteins in C. difficile, controversy regarding the necessity of TcdE for toxin secretion exists, with one publication demonstrating its requirement for secretion77 and another showing toxin secretion in its absence78. More recent work suggests that TcdE is involved in the secretion of TcdA and TcdB in strains of C. difficile that secrete high amounts of toxin79.
Binary toxin (CDT)
Some strains of C. difficile, in particular the hypervirulent BI/NAP1/027 strain, also express another toxin, referred to as binary toxin or C. difficile transferase (CDT), which may enhance virulence and is not encoded in the pathogenicity locus80. The role of CDT in virulence remains unproven, although there is an association between its presence and higher mortality in patients80. CDT is composed of two proteins, CdtA, an ADP-ribosyl transferase that ribosylates actin in eukaryotic cells, and CdtB, which forms pores in acidified endosomes and facilitates the transfer of CdtA to the cytosol. The cellular receptor for CDT is the lipolysis-stimulated lipoprotein receptor (LSR)81. Ribosylation interferes with polymerization of the actin meshwork that underlies the cell membrane, which results in cellular protrusions that are formed by microtubules and enhanced fibronectin delivery to the cell surface, thereby enhancing C. difficile adhesion to targeted cells82 (FIG. 2b).
Non-toxin virulence factors
The regulation of genes that control motility and adherence is an important factor that contributes to colonization efficiency and the virulence of C. difficile83,84. Flagellar expression is highly variable among C. difficile strains85 and lack of flagella has been linked to impaired adherence to the intestinal epithelium86,87. Interestingly, mutant strains that lack components of the flagellar machinery exhibit dysregulated toxin expression and corresponding altered virulence in vivo, which suggests a link between flagellar expression and toxin regulation87,88. In C. difficile, flagellum expression is regulated by the intracellular second messenger cyclic dimeric guanosine monophosphate (c-di-GMP)89,90. c-di-GMP, which is synthesized from GTP by diguanylate cyclase, acts as a specific ligand and binds to a riboswitch upstream of the flgB operon, which is crucial in early flagellum formation, and terminates the transcription of flgB90. High levels of intracellular c-di-GMP repress flagellum expression and thereby motility91, and also repress the synthesis of TcdA and TcdB92. Concurrently, c-di-GMP activates another riboswitch that induces the expression of type IV pili that interact with the intestinal epithelium and contribute to C. difficile aggregation and biofilm formation93,94. In this manner, c-di-GMP acts as a key signal that can switch C. difficile between a highly motile, toxin-producing state and a strongly adherent biofilm-producing state. In addition, the adhesin fibronectin binding protein A, cell wall proteins such as Cwp66, S-layer protein A and its modifying protease Cwp84, and even Spo0A (the master regulator for sporulation), all contribute to C. difficile adherence and have a role in biofilm formation95–97. The extracellular matrix of the biofilm is composed of proteins, polysaccharides and free DNA from dead cells, which insulate vegetative cells from oxidative stress, antibodies and antibiotics, creating a protected niche for sporulation98.
Microbiota-mediated resistance to C. difficile
Antibiotic treatment is the main risk factor for the development of C. difficile colitis through the disruption of colonization resistance. Restoration of the microbiota, for example, through faecal microbiota transplant, re-establishes resistance mechanisms that inhibit the growth of C. difficile.
The role of the microbiota in infection with C. difficile
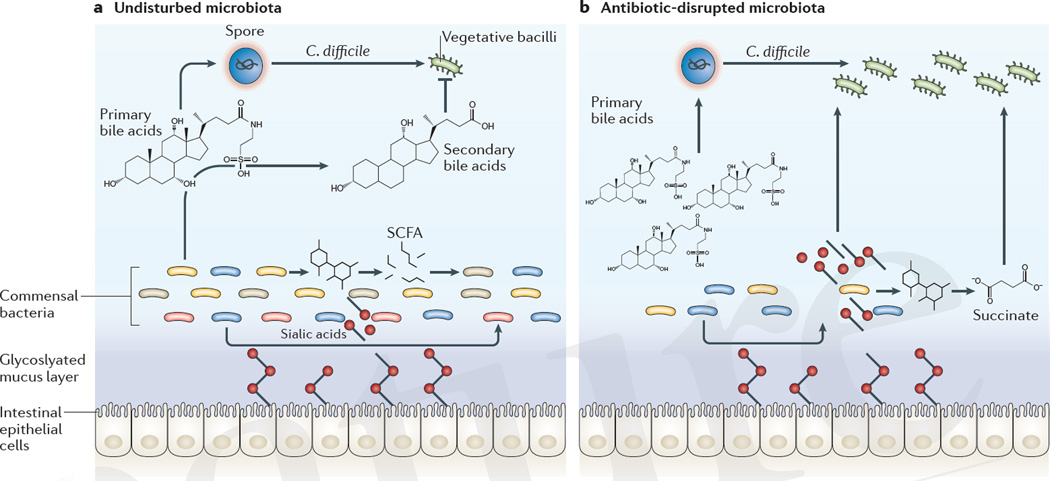
Antibiotic treatment that precedes C. difficile infection substantially alters the intestinal metabolome, creating a more hospitable environment for the growth of C. difficile99. In the colon, sialidase-producing commensal bacteria cleave sugars from glycosylated proteins that are bound to the epithelial cell membrane, which releases free sialic acid into the lumen100. Primary fermenters break down complex carbohydrates into short-chain fatty acids101. Both of these metabolites are rapidly consumed as energy sources by commensal bacteria. However, antibiotic treatment can deplete competing commensal bacteria, which leads to an abundance of sialic acid and succinate, a short-chain fatty acid that is produced during fermentation. C. difficile has genes for both sialic acid catabolism and succinate transporters, which enables it to use the excess sialic acid and succinate for growth102,103 (FIG. 3). In addition to bacteria-derived metabolites, direct interactions between bacteria can limit the expansion of C. difficile. A limited number of bacteriocins have been identified that exhibit antimicrobial activity against Gram-positive pathogens such as C. difficile104,105. These bacteriocins can target C. difficile while causing minimal disruption to intestinal microbial communities and could be used therapeutically as an alternative, or in addition, to standard antibiotic treatment.
Figure 3. Microbiota-mediated defences against C. difficile.
a | The intact microbiota converts primary bile acids into secondary bile acids, several derivatives of which inhibit the growth of Clostridium difficile through detergent-induced toxicity to vegetative bacilli. Commensal bacteria that express sialidases cleave sugars that are attached to epithelial cells and release sialic acid into the intestinal lumen. Fermenting commensal bacterial species convert carbohydrates into short-chain fatty acids (SCFAs), such as succinate. Bystander commensal bacterial populations can consume these metabolites as energy sources. b | Antibiotic-mediated disruption of the microbiota depletes primary bile acid converters, which enables C. difficile sporulation and growth. Antibiotics can also deplete competing sialic acid and succinate consumers, liberating an energy source for C. difficile.
A recent study characterized the composition of the microbiota of a set of antibiotic-treated mice that exhibited a range of susceptibilities to C. difficile colitis and used mathematical modelling to identify commensal bacterial species that were significantly associated with resistance to the development of infection106. The same approach was also used to characterize intestinal members of the microbiota that were associated with resistance to the development of C. difficile colitis in patients who were undergoing haematopoietic stem cell transplantation106,107. Combining the human and murine datasets identified the commensal species Clostridium scindens as most highly associated with resistance to C. difficile infection106. C. scindens has a bile acid inducible (bai) operon that encodes dehydroxylating enzymes that are necessary to convert primary bile acids into secondary bile acids108. The inhibitory effect of C. scindens on C. difficile was negated through the addition of the bile acid sequestrant cholestyramine to cultures, which indicated that secondary bile acids were probably mediators of C. difficile growth inhibition (FIG. 3). Bile acids are probably not the only metabolites present in the intestinal lumen that inhibit the expansion of C. difficile. Through both direct and indirect mechanisms intestinal commensal microbial communities have a central role in determining whether C. difficile successfully colonizes the large intestine.
Faecal microbial transplants
Resolution of antibiotic-associated diarrhoea through the transfer of normal faecal microbiota to patients was demonstrated even before C. difficile was implicated as its cause109. Anecdotal case reports and small uncontrolled trials that were conducted after the failure of antibiotic therapy demonstrated the effectiveness of faecal microbial transplants as a treatment for recurrent C. difficile colitis110. Scepticism among many clinicians limited the adoption of faecal microbial transplants as a routine treatment until a randomized clinical trial of faecal microbial transplant versus conventional antibiotic treatment clearly demonstrated the superiority of faecal microbial transplants to resolve recurrent episodes of disease associated with C. difficile infection111.
Currently, faecal microbial transplant is only available for patients who experience recurrent C. difficile infection. Efficacy of treating primary C. difficile infection or patients who are actively receiving antibiotics with a faecal microbial transplant has not been carefully studied. Furthermore, an important concern about faecal microbial transplants is that the complete composition of the faeces cannot be determined. Thus, uncharacterized viruses and bacterial species may be transferred and the consequences are unpredictable to some extent, particularly in patients who have compromised immune systems. Admittedly, the short-term complications of faecal microbial transplants have been minimal, especially when balanced with its remarkable effectiveness. Nevertheless, several studies have demonstrated that consortia of a small number of commensal bacteria can resolve symptoms and infection in patients with C. difficile colitis. The first of these studies112 demonstrated that administration of a mixture of 10 commensal bacterial species cured patients with recurrent C. difficile infection. For reasons that remain unclear, bacteriotherapy for C. difficile languished for several decades and only 25 years later did a follow-up paper, using mice, identify a smaller consortium of bacterial species that also reduced the severity of C. difficile infection113. Another study, using germ-free mice, demonstrated that administration of a single bacterial species in the Lachnospiraceae family could decrease the density of C. difficile growth in vitro and ameliorate disease severity114. The mechanism of clearance remained undefined although metabolomics studies suggested that clearance correlated with the re-establishment of normal secondary bile acid concentrations in the lower gastrointestinal tract115. Two separate phase II clinical trials of patients recovering from C. difficile infection following initial antibiotic treatment, reported that administration of a non-toxigenic strain of C. difficile116 or a consortia of spore-forming commensal species117 significantly decreased the recurrence of disease. These clinical trials demonstrate the potential of a targeted approach to inhibit toxigenic C. difficile; however, the mechanism of action of these bacterial therapeutics remains undefined.
Host response to C. difficile infection
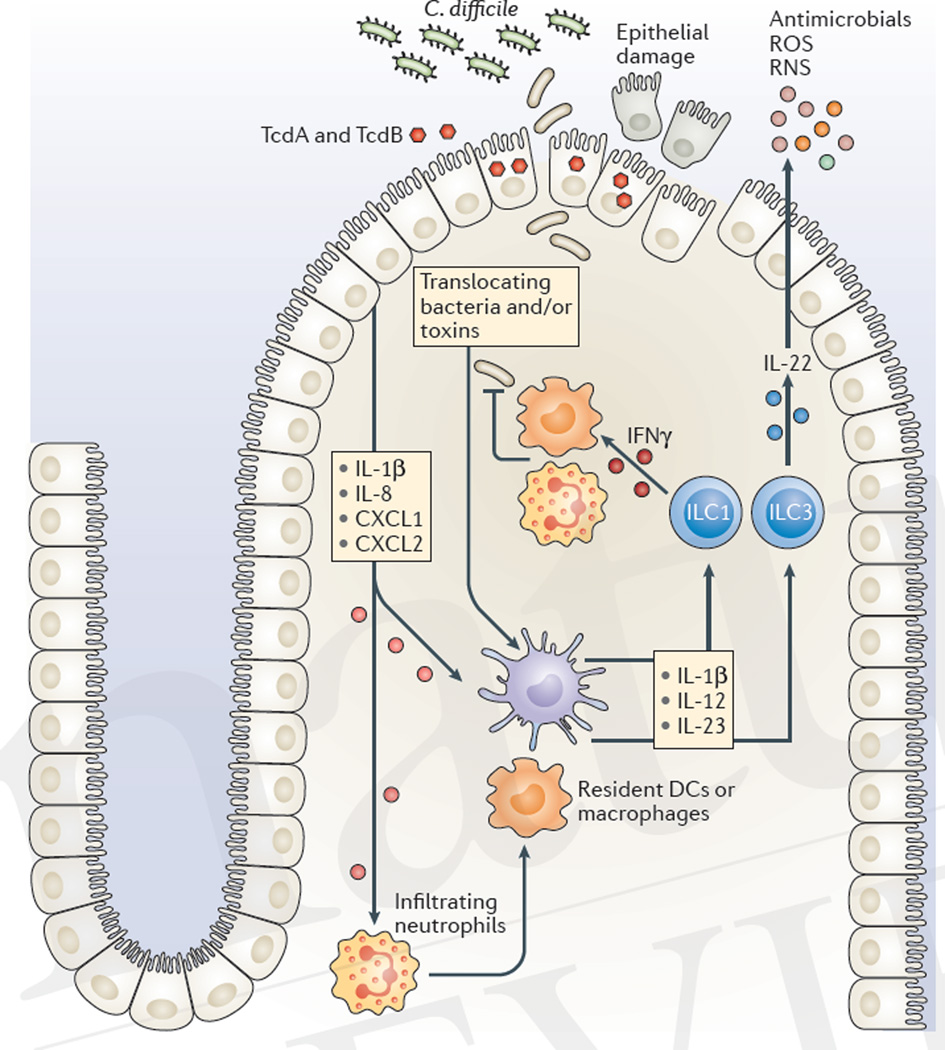
The immune system of the host rapidly responds to microbial molecules that traverse the epithelial barrier, a process that goes into overdrive in the setting of C. difficile toxin-mediated damage to the colonic epithelium. Loss of epithelial integrity results in increased intestinal permeability and the translocation of bacteria from the gut lumen into deeper tissues118. In response, resident immune cells and intoxicated epithelial cells release pro-inflammatory cytokines and chemokines that recruit circulating innate and adaptive immune cells and drive the expression of antimicrobial peptides, and the production of reactive nitrogen species (RNS) and reactive oxygen species (ROS)119,120 (FIG. 4). Although C. difficile has evolved resistance mechanisms against ROS and some antimicrobial peptides121–123, these effector molecules limit the translocation of other intestinal bacteria. Furthermore, S-nitrosylation of the cysteine protease domain of TcdA and TcdB by RNS inhibits the release of the GTD into the cytosol, thereby attenuating toxin potency124. Inflammatory responses are essential for host survival following C. difficile infection, but overly robust inflammation can be detrimental. For example, ROS that are produced in response to a toxin can exacerbate epithelial damage125 and mice that lack the pro-inflammatory cytokine interleukin-23 (IL-23) show improved survival compared with wild-type mice126. Furthermore, clinical studies demonstrate that the magnitude of the inflammatory response, as measured by faecal cytokine levels, correlates more closely with severity and duration of infection than with the C. difficile burden127.
Figure 4. Innate immune-mediated defences against C. difficile.
The acute host response to Clostridium difficile is initiated by toxin-mediated damage, loss of epithelial integrity and the detection of translocating bacteria. Intestinal epithelial cells and resident innate immune cells secrete pro-inflammatory chemokines (such as chemokine C-X-C motif ligand 1 (CXCL1), CXCL2, and interleukin-8 (IL-8)) and pro-inflammatory cytokines (such as IL-1β, IL-12 and IL-23), which leads to the recruitment of neutrophils and the activation of innate lymphoid cells (ILCs). IL-12 signalling drives the expression of interferon-γ (IFNγ), whereas IL-1β and IL-23 signalling induces the production of IL-22. The effector cytokines IFNγ and IL-22 induce defence mechanisms such as increased phagocytic activity of macrophages and neutrophils and the production of antimicrobial peptides and enzymes that synthesize reactive oxygen species (ROS) and reactive nitrogen species (RNS). These defence mechanisms limit bacterial dissemination, attenuate toxin activity and repair epithelial damage. DCs, dendritic cells.
Inflammation and cytokine production
TcdA or TcdB in C. difficile can activate nuclear factor-κB (NF-κB) and activator protein 1 (AP-1) signalling pathways in intestinal epithelial cells through the phosphorylation of mitogen-activated protein kinases (MAPK), which leads to the transcription of pro-inflammatory chemokines, such as IL-8, chemokine C-X-C motif ligand 1 (CXCL1; also known as GROα), CXCL2 and CC-chemokine ligand 2 (CCL2), and the recruitment of innate immune cells128–132. Furthermore, TcdB glycosylation and the resulting inactivation of RHO GTPases in epithelial cells are detected by the intracellular pyrin receptor, which binds to apoptosis-associated speck-like protein containing a CARD (ASC), which leads to inflammasome formation and the secretion of IL-1β133. The pattern-recognition receptors nucleotide-binding oligomerization domain-containing 1 (NOD1)134, Toll-like receptor 4 (TLR4)135 and downstream signalling proteins, such as myeloid differentiation primary response protein 88 (MYD88)136 and ASC137 support early host defence against C. difficile infection and mice that lack these proteins have increased mortality following infection. Notably however, Asc−/− mice show decreased intestinal inflammation following direct instillation of TcdA or TcdB in the ileum138, which exemplifies how differences in experimental protocols can lead to seemingly contrary results. The use of different antibiotic pre-treatment regimens to induce susceptibility to C. difficile139,140 or the composition of the infecting inoculum (vegetative bacilli versus spores)141 also affect C. difficile virulence in the murine infection model. These variables can provide a useful spectrum of disease severity to gain insights into when pro-inflammatory mediators have beneficial or deleterious effects on C. difficile pathogenesis.
Cellular responses
Resolution of diarrhoea and recovery of lost weight in mice infected with C. difficile are independent of adaptive immune defences as Rag1−/− mice, which lack T cells and B cells, recover from acute infection similarly to wild-type mice141,142. Intestinal innate lymphoid cells (ILCs) respond to IL-1β, IL-12 and IL-23, which are pro-inflammatory cytokines that are upregulated during the acute phase of C. difficile infection133,143,144, by producing effector cytokines, such as IL-22, IL-17a, interferon-γ (IFNγ) and tumour necrosis factor (TNF). The production of these effector cytokines by ILCs activates recruited neutrophils and macrophages and induces the expression of antimicrobial peptides, the production of RNS and ROS, and repair mechanisms in epithelial cells145. In contrast to Rag1−/− mice, Rag1−/−Il2rg−/− mice, which lack ILCs (in addition to T cells and B cells), fail to upregulate IFNγ, IL-22 or downstream effector molecules and exhibit high mortality following C. difficile infection141. Two ILC subsets, T-bet-expressing ILC1s, which produce IFNγ, and retinoic acid-related orphan receptor-γt (Rorγt)-expressing ILC3s, which produce IL-22, are activated during acute C. difficile infection and contribute to host defence141. IL-22 induces the production of antimicrobial peptides in the gut146 immediately following C. difficile infection, and activates the complement pathway in the lung and liver to clear translocated bacteria142. IFNγ produced by ILC1s can increase phagocytic mechanisms and the expression of ROS-producing and RNS-producing enzymes in the colonic mucosa.
Neutrophils are crucial in the early host defence against C. difficile-mediated damage as depletion of these cells in mice results in acute mortality following infection136. The pro-inflammatory cytokines IL-23 and granulocyte–macrophage-colony stimulating factor (GM-CSF) contribute to neutrophil migration to the site of infection by augmenting the expression of neutrophil chemotactic factors CXCL1 and CXCL2 (REFS 147,148). Once in the intestinal mucosa, neutrophils have several host protective functions, including the production of ROS in response to the activation of the N-formyl peptide receptor by TcdB149 and the secretion of IFNγ150, potentially acting in concert with ILC1s to enhance phagocytosis and bacterial killing by macrophages.
Although T cells and B cells do not contribute to the resolution of the acute phase of C. difficile infection in mice, clinical data indicate that adaptive immune responses can have protective effects. Severity of disease is inversely correlated with the presence of toxin-specific immunoglobulin A (IgA) and IgG antibodies151,152. Furthermore, a randomized, placebo-controlled trial demonstrated that patients who received humanized monoclonal antibodies that were specific for C. difficile TcdA and TcdB had reduced recurrences of disease153. The role of T cells is less well understood. Major histocompatibility complex class II (MHC II)-deficient mice, which lack CD4+ T cells, exhibit unaltered acute infection154, but toxin-specific antibody production and protection against secondary challenge with C. difficile is decreased compared with wild-type mice154. Interestingly, even in fully immunocompetent mice, C. difficile colonization can persist following initial infection and recovery140,154,155, which suggests that the main function of the immune system is to limit and repair C. difficile-mediated damage, not to sterilely clear C. difficile from the intestine.
Conclusions
Our understanding of the pathogenesis of C. difficile infection has increased markedly during the past decade. Deciphering the genes and proteins that are involved in sporulation, germination and toxin production has the potential to lead to new approaches to treatment (BOX 1). Dissection of the interactions between commensal bacterial species and C. difficile and the identification of commensal bacteria-derived inhibitory mechanisms may yield interventions that prevent or ameliorate C. difficile colitis. Greater understanding of host immune defence mechanisms may enable clinicians to reduce deleterious inflammatory responses while enhancing protective immune defences. As effective as infection control efforts have been to reduce transmission and protect patients from C. difficile infection, it is likely that new approaches, such as administration of protective probiotic bacterial strains, will ultimately be more effective at decreasing the incidence of infection. Because faecal microbial transplant is indisputably effective in curing C. difficile infections, C. difficile will be the first infection to be treated with bacteriotherapy.
Key points.
Disease that is associated with infection by Clostridium difficile represents an urgent public health threat. The severity of C. difficile infection is determined by strain virulence, interactions with intestinal commensal microbial communities, and the host immune response to damage of the intestinal epithelium that is induced by C. difficile.
The ability to sporulate and germinate is essential to C. difficile virulence. Hundreds of genes that are involved in sporulation and germination have been identified as well as a bile acid receptor that induces germination.
C. difficile secretes toxin proteins that are internalized by host cells through receptor-mediated endocytosis and cause disruption to cytoskeletal architecture, which leads to cell death. Toxin-mediated cell death results in the loss of intestinal barrier integrity and the translocation of bacteria into underlying tissues.
The intestinal microbiota provides colonization resistance against C. difficile infection. Commensal bacteria that are capable of converting primary bile acids to secondary bile acids inhibit the growth of C. difficile by depriving C. difficile spores of an important germinant and by increasing the concentration of secondary bile acids in the intestinal lumen, which are toxic to the vegetative form of C. difficile.
Toxin-mediated damage to the epithelium activates the host inflammatory immune response. The role of the immune system is to limit epithelial damage and the dissemination of intestinal bacteria into the circulation. However, an overly robust inflammatory response can be damaging to the host and contribute to disease pathology.
Acknowledgments
The Pamer Laboratory is supported by the US National Cancer Institute (NCI; core grant P30 CA008748) and the US National Institutes of Health (NIH; grants RO1 AI042135 and AI095706). M.C.A. is supported by the NIH (grant K99 AI125786). P.T.M. was supported by the NIH immunology training grant (T32CA009149).
Glossary
- Germination
The transformation of a dormant spore to an actively replicating bacterial cell.
- Reverse genetics
Targeted alterations of the genome. In Clostridium difficile, tools include ClosTron and allelic replacement.
- Forward genetics
Untargeted alterations of the genome that are achieved by chemical mutagens or transposable elements.
- Sensor histidine kinases
Signal-sensing proteins that pass phosphate to response regulator transcription factors that alter gene expression in response to extracellular stimuli.
- Sigma factor
The DNA-binding subunit of RNA polymerase, each sigma factor binds to a distinct consensus sequence.
- Catabolite repression
The regulation of gene expression such that preferred carbon sources are metabolized first.
- Sporangia
A cell of a spore-forming bacterium that has completed asymmetric division.
- Vegetative growth
Normal exponential growth of bacteria in rich media. Clostridium difficile switches between vegetative growth and sporulation.
- Prodomain
A peptide sequence at the amino terminus of a protein that is cleaved for the protein to be active and fully functional.
- Riboswitch
A secondary structure of mRNA, typically in the 5′-untranslated region, that binds to small molecules and regulates transcription and/or translation in cis.
- Type IV pili
Polymer filaments on the surface of Gram-positive and Gram-negative bacteria that facilitate motility or adhesion.
- Bacteriocins
Ribosomally synthesized antimicrobial peptides that are produced by bacteria that can selectively act against specific bacterial species or exhibit broad-spectrum activity.
- Inflammasome
A cytosolic multiprotein complex that detects pathogen- associated molecular patterns. The detection of these ‘danger signals’ activates transcription of pro-inflammatory cytokine genes.
- Innate lymphoid cells
(ILCs). Haematopoietic-derived innate immune cells that are capable of producing effector cytokines tailored to coordinate the early host response against distinct classes of pathogen.
- Complement pathway
A series of interactions between plasma-derived proteins that lead to the opsonization of a pathogen and activation of the inflammatory immune response.v
Biographies
Michael C. Abt received his Ph.D. from the University of Pennsylvania, Philadelphia, USA, in the field of immunology. His current research in the laboratory of Eric Pamer focuses on understanding the role of the host immune response and intestinal microbiota in modulating the severity of Clostridium difficile-associated disease.
Peter T. McKenney studied Bacillus subtilis spore morphogenesis and received his Ph.D. from New York University (NYU), USA, where he worked under Patrick Eichenberger. He is now a postdoctoral researcher in the laboratory of Eric Pamer, where he works on bile acid-mediated resistance to Gram-positive pathogens.
Eric Pamer received his M.D. from Case Western Reserve University, Cleveland, Ohio, USA. He is an infectious disease clinician scientist at the Memorial Sloan Kettering Cancer Center, New York, USA, and research in his laboratory focuses on defence against antibiotic-resistant pathogens.
Footnotes
Competing interests statement
The authors declare no competing interests.
Subject categories
Biological sciences / Immunology / Infectious diseases / Clostridium difficile [URI /631/250/255/1911]
Biological sciences / Microbiology / Bacteria / Bacterial pathogenesis [URI /631/326/41/2531]
Biological sciences / Microbiology / Bacteria / Bacterial toxins [URI /631/326/41/1319]
Biological sciences / Microbiology / Bacteria / Bacterial host response [URI /631/326/41/2533]
Biological sciences / Microbiology / Microbial communities / Microbiome [URI /631/326/2565/2134]
References
- 1. Lessa FC, et al. Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 2015;372:825–834. doi: 10.1056/NEJMoa1408913. This study provides a comprehensive assessment of the effect of C. difficile-associated disease on the current healthcare system in the United States.
- 2.George RH, et al. Identification of Clostridium difficile as a cause of pseudomembranous colitis. Br. Med. J. 1978;1:695. doi: 10.1136/bmj.1.6114.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawley TD, et al. Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl. Environ. Microbiol. 2010;76:6895–6900. doi: 10.1128/AEM.00718-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson LF, Valiente E, Donahue EH, Birchenough G, Wren BW. Hypervirulent Clostridium difficile PCR-ribotypes exhibit resistance to widely used disinfectants. PLoS ONE. 2011;6:e25754. doi: 10.1371/journal.pone.0025754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Palacios A, Lejeune JT. Moist-heat resistance, spore aging, and superdormancy in Clostridium difficile. Appl. Environ. Microbiol. 2011;77:3085–3091. doi: 10.1128/AEM.01589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Hoon MJ, Eichenberger P, Vitkup D. Hierarchical evolution of the bacterial sporulation network. Curr. Biol. 2010;20:R735–R745. doi: 10.1016/j.cub.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods. 2007;70:452–464. doi: 10.1016/j.mimet.2007.05.021. This paper introduces the ClosTron technology — a substantial step forward in Clostridium genetics.
- 8.Cartman ST, Kelly ML, Heeg D, Heap JT, Minton NP. Precise manipulation of the Clostridium difficile chromosome reveals a lack of association between the tcdC genotype and toxin production. Appl. Environ. Microbiol. 2012;78:4683–4690. doi: 10.1128/AEM.00249-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dembek M, et al. High-throughput analysis of gene essentiality and sporulation in Clostridium difficile. mBio. 2015;6:e02383. doi: 10.1128/mBio.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fimlaid KA, et al. Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet. 2013;9:e1003660. doi: 10.1371/journal.pgen.1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edwards AN, McBride SM. Initiation of sporulation in Clostridium difficile: a twist on the classic model. FEMS Microbiol. Lett. 2014;358:110–118. doi: 10.1111/1574-6968.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebaihia M, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 2006;38:779–786. doi: 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 13.Underwood S, et al. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J. Bacteriol. 2009;191:7296–7305. doi: 10.1128/JB.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pettit LJ, et al. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics. 2014;15:160. doi: 10.1186/1471-2164-15-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenbusch KE, Bakker D, Kuijper EJ, Smits WK. C. difficile 630Δerm Spo0A regulates sporulation, but does not contribute to toxin production, by direct high-affinity binding to target DNA. PLoS ONE. 2012;7:e48608. doi: 10.1371/journal.pone.0048608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deakin LJ, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect. Immun. 2012;80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J. Bacteriol. 2011;193:3186–3196. doi: 10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dineen SS, McBride SM, Sonenshein AL. Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 2010;192:5350–5362. doi: 10.1128/JB.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 2007;66:206–219. doi: 10.1111/j.1365-2958.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- 20.Nawrocki KL, Edwards AN, Daou N, Bouillaut L, McBride SM. CodY-dependent regulation of sporulation in Clostridium difficile. J. Bacteriol. 2016;198:2113–2130. doi: 10.1128/JB.00220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antunes A, et al. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res. 2012;40:10701–10718. doi: 10.1093/nar/gks864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antunes A, Martin-Verstraete I, Dupuy B. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol. Microbiol. 2011;79:882–899. doi: 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- 23.Edwards AN, Nawrocki KL, McBride SM. Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect. Immun. 2014;82:4276–4291. doi: 10.1128/IAI.02323-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 25.Pereira FC, et al. The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet. 2013;9:e1003782. doi: 10.1371/journal.pgen.1003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saujet L, et al. Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet. 2013;9:e1003756. doi: 10.1371/journal.pgen.1003756. Together with references 10, 17 and 25, this study provides a comprehensive catalogue of sporulation genes and regulatory dynamics.
- 27.Fimlaid KA, et al. Identification of a novel lipoprotein regulator of Clostridium difficile spore germination. PLoS Pathog. 2015;11:e1005239. doi: 10.1371/journal.ppat.1005239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serrano M, et al. The SpoIIQ–SpoIIIAH complex of Clostridium difficile controls forespore engulfment and late stages of gene expression and spore morphogenesis. Mol. Microbiol. 2016;100:204–228. doi: 10.1111/mmi.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 2014;22:406–416. doi: 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henriques AO, Moran CP. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 31.Putnam EE, Nock AM, Lawley TD, Shen A. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J. Bacteriol. 2013;195:1214–1225. doi: 10.1128/JB.02181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Permpoonpattana P, et al. Functional characterization of Clostridium difficile spore coat proteins. J. Bacteriol. 2013;195:1492–1503. doi: 10.1128/JB.02104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barra-Carrasco J, et al. The Clostridium difficile exosporium cysteine (CdeC)-rich protein is required for exosporium morphogenesis and coat assembly. J. Bacteriol. 2013;195:3863–3875. doi: 10.1128/JB.00369-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phetcharaburanin J, et al. The spore-associated protein BclA1 affects the susceptibility of animals to colonization and infection by Clostridium difficile. Mol. Microbiol. 2014;92:1025–1038. doi: 10.1111/mmi.12611. [DOI] [PubMed] [Google Scholar]
- 35.Pizarro-Guajardo M, et al. Characterization of the collagen-like exosporium protein, BclA1, of Clostridium difficile spores. Anaerobe. 2014;25:18–30. doi: 10.1016/j.anaerobe.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Koenigsknecht MJ, et al. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect. Immun. 2015;83:934–941. doi: 10.1128/IAI.02768-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raibaud P, Ducluzeau R, Muller M-C, Sacquet E. Le taurocholate de sodium, facteur de germination in vitro et in vivo dans le tube digestif d’animaux “gnotoxeniques”, pour les spores de certaines bacteries anaerobies strictes isolee de feces humaines et animales. Ann. Microbiol. (Inst. Pasteur) 1974;125B:381–391. (in French) [PubMed] [Google Scholar]
- 38.Wilson KH, Kennedy MJ, Fekety FR. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 1982;15:443–446. doi: 10.1128/jcm.15.3.443-446.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson KH. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J. Clin. Microbiol. 1983;18:1017–1019. doi: 10.1128/jcm.18.4.1017-1019.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 2009;191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sorg JA, Sonenshein AL. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 2010;192:4983–4990. doi: 10.1128/JB.00610-10. Together with references 40 and 41, this is the first investigation of the effects of individual bile acids on the growth of C. difficile.
- 43.Giel JL, Sorg JA, Sonenshein AL, Zhu J. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS ONE. 2010;5:e8740. doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theriot CM, Bowman AA, Young VB. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 2016;1:e00045–e00015. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Francis MB, Allen CA, Sorg JA. Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J. Bacteriol. 2015;197:2276–2283. doi: 10.1128/JB.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, Shen A, Setlow P, Li YQ. Characterization of the dynamic germination of individual Clostridium difficile spores using raman spectroscopy and differential interference contrast microscopy. J. Bacteriol. 2015;197:2361–2373. doi: 10.1128/JB.00200-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Francis MB, Allen CA, Shrestha R, Sorg JA. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog. 2013;9:e1003356. doi: 10.1371/journal.ppat.1003356. This paper identifies CspC as the receptor that recognizes bile acids and induces C. difficile spore germination.
- 48.Kevorkian Y, Shirley DJ, Shen A. Regulation of Clostridium difficile spore germination by the CspA pseudoprotease domain. Biochimie. 2016;122:243–254. doi: 10.1016/j.biochi.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams CM, Eckenroth BE, Putnam EE, Doublie S, Shen A. Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog. 2013;9:e1003165. doi: 10.1371/journal.ppat.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burns DA, Heap JT, Minton NP. SleC is essential for germination of Clostridium difficile spores in nutrient-rich medium supplemented with the bile salt taurocholate. J. Bacteriol. 2010;192:657–664. doi: 10.1128/JB.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dembek M, Stabler RA, Witney AA, Wren BW, Fairweather NF. Transcriptional analysis of temporal gene expression in germinating Clostridium difficile 630 endospores. PLoS ONE. 2013;8:e64011. doi: 10.1371/journal.pone.0064011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore P, Kyne L, Martin A, Solomon K. Germination efficiency of clinical Clostridium difficile spores and correlation with ribotype, disease severity and therapy failure. J. Med. Microbiol. 2013;62:1405–1413. doi: 10.1099/jmm.0.056614-0. [DOI] [PubMed] [Google Scholar]
- 53.Carlson PE, Jr, et al. Variation in germination of Clostridium difficile clinical isolates correlates to disease severity. Anaerobe. 2015;33:64–70. doi: 10.1016/j.anaerobe.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carter GP, et al. Defining the roles of TcdA and TcdB in localized gastrointestinal disease, systemic organ damage, and the host response during Clostridium difficile infections. mBio. 2015;6:e00551. doi: 10.1128/mBio.00551-15. This multicentre study uses two different animal models to compare the relative roles of TcdA and TcdB in C. difficile pathogenesis.
- 55.Hunt JJ, Ballard JD. Variations in virulence and molecular biology among emerging strains of Clostridium difficile. Microbiol. Mol. Biol. Rev. 2013;77:567–581. doi: 10.1128/MMBR.00017-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monot M, et al. Clostridium difficile: new insights into the evolution of the pathogenicity locus. Sci. Rep. 2015;5:15023. doi: 10.1038/srep15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pruitt RN, Chambers MG, Ng KK, Ohi MD, Lacy DB. Structural organization of the functional domains of Clostridium difficile toxins A and B. Proc. Natl Acad. Sci. USA. 2010;107:13467–13472. doi: 10.1073/pnas.1002199107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reineke J, et al. Autocatalytic cleavage of Clostridium difficile toxin B. Nature. 2007;446:415–419. doi: 10.1038/nature05622. [DOI] [PubMed] [Google Scholar]
- 59.Egerer M, Giesemann T, Jank T, Satchell KJ, Aktories K. Auto-catalytic cleavage of Clostridium difficile toxins A and B depends on cysteine protease activity. J. Biol. Chem. 2007;282:25314–25321. doi: 10.1074/jbc.M703062200. [DOI] [PubMed] [Google Scholar]
- 60.Lanis JM, Hightower LD, Shen A, Ballard JD. TcdB from hypervirulent Clostridium difficile exhibits increased efficiency of autoprocessing. Mol. Microbiol. 2012;84:66–76. doi: 10.1111/j.1365-2958.2012.08009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bender KO, et al. A small-molecule antivirulence agent for treating Clostridium difficile infection. Sci. Transl Med. 2015;7:306ra148. doi: 10.1126/scitranslmed.aac9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dingle T, et al. Functional properties of the carboxy-terminal host cell-binding domains of the two toxins, TcdA and TcdB, expressed by Clostridium difficile. Glycobiology. 2008;18:698–706. doi: 10.1093/glycob/cwn048. [DOI] [PubMed] [Google Scholar]
- 63.Greco A, et al. Carbohydrate recognition by Clostridium difficile toxin A. Nat. Struct. Mol. Biol. 2006;13:460–461. doi: 10.1038/nsmb1084. [DOI] [PubMed] [Google Scholar]
- 64.Na X, Kim H, Moyer MP, Pothoulakis C, LaMont JT. gp96 is a human colonocyte plasma membrane binding protein for Clostridium difficile toxin A. Infect. Immun. 2008;76:2862–2871. doi: 10.1128/IAI.00326-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan P, et al. Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res. 2015;25:157–168. doi: 10.1038/cr.2014.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. LaFrance ME, et al. Identification of an epithelial cell receptor responsible for Clostridium difficile TcdB-induced cytotoxicity. Proc. Natl Acad. Sci. USA. 2015;112:7073–7078. doi: 10.1073/pnas.1500791112. This study identifies PVRL3 expressed on colonic epithelial cells as a receptor for TcdB; this interaction is necessary for toxin-mediated cytotoxicity.
- 67.Lyras D, et al. Toxin B is essential for virulence of Clostridium difficile. Nature. 2009;458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuehne SA, et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 69.Kuehne SA, et al. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J. Infect. Dis. 2014;209:83–86. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stabler RA, et al. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 2006;188:7297–7305. doi: 10.1128/JB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lanis JM, Heinlen LD, James JA, Ballard JD. Clostridium difficile 027/BI/NAP1 encodes a hypertoxic and antigenically variable form of TcdB. PLoS Pathog. 2013;9:e1003523. doi: 10.1371/journal.ppat.1003523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mani N, Dupuy B. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc. Natl Acad. Sci. USA. 2001;98:5844–5849. doi: 10.1073/pnas.101126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matamouros S, England P, Dupuy B. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol. Microbiol. 2007;64:1274–1288. doi: 10.1111/j.1365-2958.2007.05739.x. [DOI] [PubMed] [Google Scholar]
- 74.van Leeuwen HC, Bakker D, Steindel P, Kuijper EJ, Corver J. Clostridium difficile TcdC protein binds four-stranded G-quadruplex structures. Nucleic Acids Res. 2013;41:2382–2393. doi: 10.1093/nar/gks1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spigaglia P, Mastrantonio P. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 2002;40:3470–3475. doi: 10.1128/JCM.40.9.3470-3475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bakker D, Smits WK, Kuijper EJ, Corver J. TcdC does not significantly repress toxin expression in Clostridium difficile 630Δerm. PLoS ONE. 2012;7:e43247. doi: 10.1371/journal.pone.0043247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Govind R, Dupuy B. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog. 2012;8:e1002727. doi: 10.1371/journal.ppat.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Olling A, et al. Release of TcdA and TcdB from Clostridium difficile cdi 630 is not affected by functional inactivation of the tcdE gene. Microb. Pathog. 2012;52:92–100. doi: 10.1016/j.micpath.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 79.Govind R, Fitzwater L, Nichols R. Observations on the role of TcdE isoforms in Clostridium difficile toxin secretion. J. Bacteriol. 2015;197:2600–2609. doi: 10.1128/JB.00224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gerding DN, Johnson S, Rupnik M, Aktories K. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes. 2014;5:15–27. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Papatheodorou P, et al. Lipolysis-stimulated lipoprotein receptor (LSR) is the host receptor for the binary toxin Clostridium difficile transferase (CDT) Proc. Natl Acad. Sci. USA. 2011;108:16422–16427. doi: 10.1073/pnas.1109772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwan C, et al. Clostridium difficile toxin CDT hijacks microtubule organization and reroutes vesicle traffic to increase pathogen adherence. Proc. Natl Acad. Sci. USA. 2014;111:2313–2318. doi: 10.1073/pnas.1311589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Awad MM, Johanesen PA, Carter GP, Rose E, Lyras D. Clostridium difficile virulence factors: insights into an anaerobic spore-forming pathogen. Gut Microbes. 2014;5:579–593. doi: 10.4161/19490976.2014.969632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vedantam G, et al. Clostridium difficile infection: toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes. 2012;3:121–134. doi: 10.4161/gmic.19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pituch H, et al. Variable flagella expression among clonal toxin A−/B+ Clostridium difficile strains with highly homogeneous flagellin genes. Clin. Microbiol. Infect. 2002;8:187–188. doi: 10.1046/j.1469-0691.2002.00394.x. [DOI] [PubMed] [Google Scholar]
- 86.Tasteyre A, Barc MC, Collignon A, Boureau H, Karjalainen T. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 2001;69:7937–7940. doi: 10.1128/IAI.69.12.7937-7940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baban ST, et al. The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS ONE. 2013;8:e73026. doi: 10.1371/journal.pone.0073026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aubry A, et al. Modulation of toxin production by the flagellar regulon in Clostridium difficile. Infect. Immun. 2012;80:3521–3532. doi: 10.1128/IAI.00224-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bordeleau E, Burrus V. Cyclic-di-GMP signaling in the Gram-positive pathogen Clostridium difficile. Curr. Genet. 2015;61:497–502. doi: 10.1007/s00294-015-0484-z. [DOI] [PubMed] [Google Scholar]
- 90.Sudarsan N, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 2012;194:3307–3316. doi: 10.1128/JB.00100-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R. The second messenger cyclic di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J. Bacteriol. 2013;195:5174–5185. doi: 10.1128/JB.00501-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bordeleau E, et al. Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. J. Bacteriol. 2015;197:819–832. doi: 10.1128/JB.02340-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Purcell EB, McKee RW, Bordeleau E, Burrus V, Tamayo R. Regulation of type IV pili contributes to surface behaviors of historical and epidemic strains of Clostridium difficile. J. Bacteriol. 2015;198:565–577. doi: 10.1128/JB.00816-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ethapa T, et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 2013;195:545–555. doi: 10.1128/JB.01980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dawson LF, Valiente E, Faulds-Pain A, Donahue EH, Wren BW. Characterisation of Clostridium difficile biofilm formation, a role for Spo0A. PLoS ONE. 2012;7:e50527. doi: 10.1371/journal.pone.0050527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barketi-Klai A, Hoys S, Lambert-Bordes S, Collignon A, Kansau I. Role of fibronectin-binding protein A in Clostridium difficile intestinal colonization. J. Med. Microbiol. 2011;60:1155–1161. doi: 10.1099/jmm.0.029553-0. [DOI] [PubMed] [Google Scholar]
- 98.Dapa T, Unnikrishnan M. Biofilm formation by Clostridium difficile. Gut Microbes. 2013;4:397–402. doi: 10.4161/gmic.25862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Theriot CM, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014;5:3114. doi: 10.1038/ncomms4114. This paper reports a link between antibiotic treatment, subsequent susceptibility to C. difficile and a distinct metabolomic profile in the intestine.
- 100.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 101.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006;40:235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 102. Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. This paper shows that sialic acids that are cleaved from glycoproteins of epithelial cells by commensal bacteria are consumed as an energy source by C. difficile.
- 103.Ferreyra JA, et al. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rea MC, et al. Effect of broad- and narrowspectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl Acad. Sci. USA. 2011;108(Suppl. 1):4639–4644. doi: 10.1073/pnas.1001224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Trzasko A, Leeds JA, Praestgaard J, Lamarche MJ, McKenney D. Efficacy of LFF571 in a hamster model of Clostridium difficile infection. Antimicrob. Agents Chemother. 2012;56:4459–4462. doi: 10.1128/AAC.06355-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. This paper demonstrates that reconstitution of primary bile acid-converting bacteria restores colonization resistance against C. difficile.
- 107.Kinnebrew MA, et al. Early Clostridium difficile infection during allogeneic hematopoietic stem cell transplantation. PLoS ONE. 2014;9:e90158. doi: 10.1371/journal.pone.0090158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ridlon JM, Hylemon PB. Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium. J. Lipid Res. 2012;53:66–76. doi: 10.1194/jlr.M020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Eiseman B, Silen W, Bascom GS, Kauvar AJ. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery. 1958;44:854–859. [PubMed] [Google Scholar]
- 110.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 2011;53:994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 111. van Nood E, Dijkgraaf MG, Keller JJ. Duodenal infusion of feces for recurrent Clostridium difficile. N. Engl. J. Med. 2013;368:2145. doi: 10.1056/NEJMc1303919. This is the first placebo-controlled double-blind study to demonstrate the efficacy of faecal microbiota transplant therapy to cure recurrent C. difficile disease.
- 112.Tvede M, Rask-Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet. 1989;1:1156–1160. doi: 10.1016/s0140-6736(89)92749-9. [DOI] [PubMed] [Google Scholar]
- 113.Lawley TD, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect. Immun. 2012;80:3786–3794. doi: 10.1128/IAI.00647-12. Together with reference 11, this paper identifies specific commensal bacterial species that protect the host from C. difficile infection.
- 115.Weingarden AR, et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol. 2014;306:G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gerding DN, et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: a randomized clinical trial. JAMA. 2015;313:1719–1727. doi: 10.1001/jama.2015.3725. This phase II clinical trial reports that administration of non-toxigenic C. difficile spores to patients who are recovering from C. difficile infection can prevent recurrence of disease.
- 117.Khanna S, et al. A novel microbiome therapeutic increases gut microbial diversity and prevents recurrent Clostridium difficile infection. J. Infect. Dis. 2016;214:173–181. doi: 10.1093/infdis/jiv766. [DOI] [PubMed] [Google Scholar]
- 118.Naaber P, Mikelsaar RH, Salminen S, Mikelsaar M. Bacterial translocation, intestinal microflora and morphological changes of intestinal mucosa in experimental models of Clostridium difficile infection. J. Med. Microbiol. 1998;47:591–598. doi: 10.1099/00222615-47-7-591. [DOI] [PubMed] [Google Scholar]
- 119.Madan R, Petri WA. Immune responses to Clostridium difficile infection. Trends Mol. Med. 2012;18:658–666. doi: 10.1016/j.molmed.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Solomon K. The host immune response to Clostridium difficile infection. Ther. Adv. Infect. Dis. 2013;1:19–35. doi: 10.1177/2049936112472173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.McBride SM, Sonenshein AL. The dlt operon confers resistance to cationic antimicrobial peptides in Clostridium difficile. Microbiology. 2011;157:1457–1465. doi: 10.1099/mic.0.045997-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Girinathan BP, Braun SE, Govind R. Clostridium difficile glutamate dehydrogenase is a secreted enzyme that confers resistance to H2O2. Microbiology. 2014;160:47–55. doi: 10.1099/mic.0.071365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ho TD, Ellermeier CD. PrsW is required for colonization, resistance to antimicrobial peptides, and expression of extracytoplasmic function sigma factors in Clostridium difficile. Infect. Immun. 2011;79:3229–3238. doi: 10.1128/IAI.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Savidge TC, et al. Host S-nitrosylation inhibits clostridial small molecule-activated glucosylating toxins. Nat. Med. 2011;17:1136–1141. doi: 10.1038/nm.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fradrich C, Beer LA, Gerhard R. Reactive oxygen species as additional determinants for cytotoxicity of Clostridium difficile toxins A and B. Toxins (Basel) 2016;8:E25. doi: 10.3390/toxins8010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Buonomo EL, et al. Role of interleukin 23 signaling in Clostridium difficile colitis. J. Infect. Dis. 2013;208:917–920. doi: 10.1093/infdis/jit277. This paper finds that mice that are deficient in the pro-inflammatory cytokine IL-23 exhibit improved survival following infection with C. difficile, which suggests a detrimental role of overactive inflammatory responses.
- 127.El Feghaly RE, et al. Markers of intestinal inflammation, not bacterial burden, correlate with clinical outcomes in Clostridium difficile infection. Clin. Infect. Dis. 2013;56:1713–1721. doi: 10.1093/cid/cit147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jefferson KK, Smith MF, Bobak DA. Roles of intracellular calcium and NF-κB in the Clostridium difficile toxin A-induced up-regulation and secretion of IL-8 from human monocytes. J. Immunol. 1999;163:5183–5191. [PubMed] [Google Scholar]
- 129.Lee JY, et al. Effects of transcription factor activator protein-1 on interleukin-8 expression and enteritis in response to Clostridium difficile toxin A. J. Mol. Med. (Berl.) 2007;85:1393–1404. doi: 10.1007/s00109-007-0237-7. [DOI] [PubMed] [Google Scholar]
- 130.Warny M, et al. p38 MAP kinase activation by Clostridium difficile toxin A mediates monocyte necrosis, IL-8 production, and enteritis. J. Clin. Invest. 2000;105:1147–1156. doi: 10.1172/JCI7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Castagliuolo I, et al. Clostridium difficile toxin A stimulates macrophage-inflammatory protein-2 production in rat intestinal epithelial cells. J. Immunol. 1998;160:6039–6045. [PubMed] [Google Scholar]
- 132.Kim JM, et al. NF-κB activation pathway is essential for the chemokine expression in intestinal epithelial cells stimulated with Clostridium difficile toxin A. Scand. J. Immunol. 2006;63:453–460. doi: 10.1111/j.1365-3083.2006.001756.x. [DOI] [PubMed] [Google Scholar]
- 133.Xu H, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 134.Hasegawa M, et al. Nucleotide-binding oligomerization domain 1 mediates recognition of Clostridium difficile and induces neutrophil recruitment and protection against the pathogen. J. Immunol. 2011;186:4872–4880. doi: 10.4049/jimmunol.1003761. [DOI] [PubMed] [Google Scholar]
- 135.Ryan A, et al. A role for TLR4 in Clostridium difficile infection and the recognition of surface layer proteins. PLoS Pathog. 2011;7:e1002076. doi: 10.1371/journal.ppat.1002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jarchum I, Liu M, Shi C, Equinda M, Pamer EG. Critical role for MyD88-mediated neutrophil recruitment during Clostridium difficile colitis. Infect. Immun. 2012;80:2989–2996. doi: 10.1128/IAI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hasegawa M, et al. Protective role of commensals against Clostridium difficile infection via an IL-1β-mediated positive-feedback loop. J. Immunol. 2012;189:3085–3091. doi: 10.4049/jimmunol.1200821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ng J, et al. Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology. 2010;139:542–552. doi: 10.1053/j.gastro.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 139.Chen X, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008;135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 140.Buffie CG, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Abt MC, et al. Innate immune defenses mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe. 2015;18:27–37. doi: 10.1016/j.chom.2015.06.011. This paper identifies that IFNγ-producing type 1 innate lymphoid cells are crucial for host protection during acute C. difficile infection.
- 142. Hasegawa M, et al. Interleukin-22 regulates the complement system to promote resistance against pathobionts after pathogen-induced intestinal damage. Immunity. 2014;41:620–632. doi: 10.1016/j.immuni.2014.09.010. This paper observes that IL-22 systemically activates the complement pathway as a defence against disseminating bacteria that translocate across the epithelial barrier.
- 143.Cowardin CA, et al. Inflammasome activation contributes to interleukin-23 production in response to Clostridium difficile. mBio. 2015;6:e02386–e02314. doi: 10.1128/mBio.02386-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sadighi Akha AA, et al. Acute infection of mice with Clostridium difficile leads to eIF2α phosphorylation and pro-survival signalling as part of the mucosal inflammatory response. Immunology. 2013;140:111–122. doi: 10.1111/imm.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat. Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sadighi Akha AA, et al. Interleukin-22 and CD160 play additive roles in the host mucosal response to Clostridium difficile infection in mice. Immunology. 2015;144:587–597. doi: 10.1111/imm.12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McDermott AJ, et al. Role of GM-CSF in the inflammatory cytokine network that regulates neutrophil influx into the colonic mucosa during Clostridium difficile infection in mice. Gut Microbes. 2014;5:476–484. doi: 10.4161/gmic.29964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McDermott AJ, et al. Interleukin-23 (IL-23), independent of IL-17 and IL-22, drives neutrophil recruitment and innate inflammation during Clostridium difficile colitis in mice. Immunology. 2016;147:114–124. doi: 10.1111/imm.12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Goy SD, Olling A, Neumann D, Pich A, Gerhard R. Human neutrophils are activated by a peptide fragment of Clostridium difficile toxin B presumably via formyl peptide receptor. Cell. Microbiol. 2015;17:893–909. doi: 10.1111/cmi.12410. [DOI] [PubMed] [Google Scholar]
- 150.El-Zaatari M, et al. Tryptophan catabolism restricts IFNγ-expressing neutrophils and Clostridium difficile immunopathology. J. Immunol. 2014;193:807–816. doi: 10.4049/jimmunol.1302913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Johnson S, Gerding DN, Janoff EN. Systemic and mucosal antibody responses to toxin A in patients infected with Clostridium difficile. J. Infect. Dis. 1992;166:1287–1294. doi: 10.1093/infdis/166.6.1287. [DOI] [PubMed] [Google Scholar]
- 152.Kyne L, Warny M, Qamar A, Kelly CP. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N. Engl. J. Med. 2000;342:390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 153.Lowy I, et al. Treatment with monoclonal antibodies against Clostridium difficile toxins. N. Engl. J. Med. 2010;362:197–205. doi: 10.1056/NEJMoa0907635. [DOI] [PubMed] [Google Scholar]
- 154.Johnston PF, Gerding DN, Knight KL. Protection from Clostridium difficile infection in CD4 T Cell- and polymeric immunoglobulin receptor-deficient mice. Infect. Immun. 2014;82:522–531. doi: 10.1128/IAI.01273-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lawley TD, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hall IC, O’Toole E. Intestinal flora in new-born infants, with a description of a new pathogenic anaerobe Bacillus dificillis. Am. J. Dis. Child. 1935;49:390–402. [Google Scholar]
- 157.Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ. Microbiol. 2013;15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chitnis AS, et al. Epidemiology of community-associated Clostridium difficile infection, 2009 through 2011. JAMA Intern. Med. 2013;173:1359–1367. doi: 10.1001/jamainternmed.2013.7056. [DOI] [PubMed] [Google Scholar]
- 159. Eyre DW, et al. Diverse sources of C. difficile infection identified on whole-genome sequencing. N. Engl. J. Med. 2013;369:1195–1205. doi: 10.1056/NEJMoa1216064. This study details whole-genome sequencing on strains that were isolated from patients infected with C. difficile during a three year period and found a high degree of diversity among strains, which suggests community sources may be as prevalent as direct hospital transmission for pathogen acquisition.
- 160.Janezic S, et al. International Clostridium difficile animal strain collection and large diversity of animal associated strains. BMC Microbiol. 2014;14:173. doi: 10.1186/1471-2180-14-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lewis BB, et al. Loss of microbiota-mediated colonization resistance to Clostridium difficile infection with oral vancomycin compared with metronidazole. J. Infect. Dis. 2015;212:1656–1665. doi: 10.1093/infdis/jiv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Venugopal AA, Johnson S. Fidaxomicin: a novel macrocyclic antibiotic approved for treatment of Clostridium difficile infection. Clin. Infect. Dis. 2012;54:568–574. doi: 10.1093/cid/cir830. [DOI] [PubMed] [Google Scholar]
- 163.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 164.Joyce SA, Shanahan F, Hill C, Gahan CG. Bacterial bile salt hydrolase in host metabolism: potential for influencing gastrointestinal microbe–host crosstalk. Gut Microbes. 2014;5:669–674. doi: 10.4161/19490976.2014.969986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Morris GN, Winter J, Cato EP, Ritchie AE, Bokkenheuser VD. Clostridium scindens sp. nov., a human intestinal bacterium with desmolytic activity on corticoids. Int. J. Syst. Bacteriol. 1985;35:478–481. [Google Scholar]
- 166.Kang DJ, Ridlon JM, Moore DR, Barnes S, Hylemon PB. Clostridium scindens baiCD and baiH genes encode stereo-specific 7α/7β-hydroxy-3-oxo-Δ4-cholenoic acid oxidoreductases. Biochim. Biophys. Acta. 2008;1781:16–25. doi: 10.1016/j.bbalip.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 2015;11:685–690. doi: 10.1038/nchembio.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Heeg D, Burns DA, Cartman ST, Minton NP. Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS ONE. 2012;7:e32381. doi: 10.1371/journal.pone.0032381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Francis MB, Allen CA, Sorg JA. Muricholic acids inhibit Clostridium difficile spore germination and growth. PLoS ONE. 2013;8:e73653. doi: 10.1371/journal.pone.0073653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Weingarden AR, et al. Ursodeoxycholic acid inhibits Clostridium difficile spore germination and vegetative growth, and prevents the recurrence of ileal pouchitis associated with the infection. J. Clin. Gastroenterol. 2016;50:624–630. doi: 10.1097/MCG.0000000000000427. [DOI] [PMC free article] [PubMed] [Google Scholar]