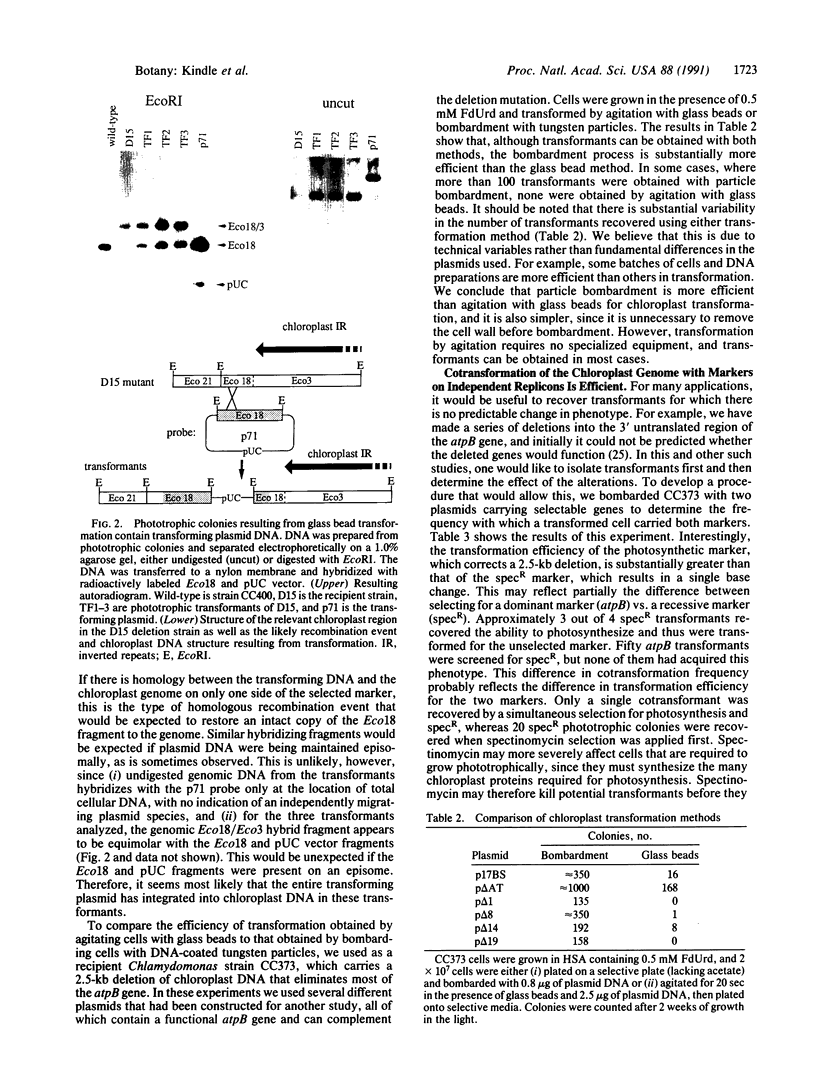
Abstract
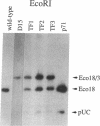
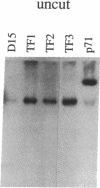
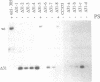
Chloroplast transformation of Chlamydomonas reinhardtii has been accomplished by agitating cell wall-deficient cells in the presence of glass beads and DNA. By using the atpB gene as the selected marker and cells grown in 0.5 mM 5-fluorodeoxyuridine, we have recovered up to 50 transformants per microgram of DNA. This method is easy and does not require specialized equipment, although it is not as efficient as the tungsten particle bombardment method [Boynton, J. E., Gillham, N. W., Harris, E. H., Hosler, J. P., Johnson, A. M., Jones, A. R., Randolph-Anderson, B. L., Robertson, D., Klein, T. M., Shark, K. B. & Sanford, J. C. (1988) Science 240, 1534-1537]. By using particle bombardment, we have developed a cotransformation approach in which spectinomycin-resistant 16S rRNA-encoding DNA is the selected marker, and we have demonstrated that cotransformation of an unselected marker on an independent replicon is very efficient. We have used this strategy (i) to recover transformants with partially deleted atpB genes that could not otherwise have been selected since they did not restore photosynthetic capability to a recipient carrying a more extensive atpB deletion and (ii) to generate specific deletion mutations in a wild-type recipient. This methodology should allow the introduction of any desired change into the chloroplast genome, even in the absence of phenotypic selection, and thus a detailed functional analysis of any chloroplast DNA sequence should be possible.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blowers A. D., Bogorad L., Shark K. B., Sanford J. C. Studies on Chlamydomonas chloroplast transformation: foreign DNA can be stably maintained in the chromosome. Plant Cell. 1989 Jan;1(1):123–132. doi: 10.1105/tpc.1.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton J. E., Gillham N. W., Harris E. H., Hosler J. P., Johnson A. M., Jones A. R., Randolph-Anderson B. L., Robertson D., Klein T. M., Shark K. B. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988 Jun 10;240(4858):1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde M. F., Boynton J. E., Gillham N. W., Harris E. H., Tingle C. L., Wang W. L. Chloroplast genes in Chlamydomonas affecting organelle ribosomes. Genetic and biochemical analysis of analysis of antibiotic-resistant mutants at several gene loci. Mol Gen Genet. 1975 Oct 3;140(3):183–220. doi: 10.1007/BF00334266. [DOI] [PubMed] [Google Scholar]
- Debuchy R., Purton S., Rochaix J. D. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 1989 Oct;8(10):2803–2809. doi: 10.1002/j.1460-2075.1989.tb08426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener D. R., Curry A. M., Johnson K. A., Williams B. D., Lefebvre P. A., Kindle K. L., Rosenbaum J. L. Rescue of a paralyzed-flagella mutant of Chlamydomonas by transformation. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5739–5743. doi: 10.1073/pnas.87.15.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Harris E. H., Burkhart B. D., Gillham N. W., Boynton J. E. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: correlation of genetic and physical maps of the chloroplast genome. Genetics. 1989 Oct;123(2):281–292. doi: 10.1093/genetics/123.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K. L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle K. L., Schnell R. A., Fernández E., Lefebvre P. A. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989 Dec;109(6 Pt 1):2589–2601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield S. P., Kindle K. L. Stable nuclear transformation of Chlamydomonas reinhardtii by using a C. reinhardtii gene as the selectable marker. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2087–2091. doi: 10.1073/pnas.87.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers A. M., Grant D. M., Rabert D. K., Harris E. H., Boynton J. E., Gillham N. W. Mutants of Chlamydomonas reinhardtii with physical alterations in their chloroplast DNA. Plasmid. 1982 Mar;7(2):133–151. doi: 10.1016/0147-619x(82)90073-7. [DOI] [PubMed] [Google Scholar]
- Shepherd H. S., Boynton J. E., Gillham N. W. Mutations in nine chloroplast loci of Chlamydomonas affecting different photosynthetic functions. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1353–1357. doi: 10.1073/pnas.76.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell W. J., Eskue W. A., Buchanan M. J. Regulated secretion of a serine protease that activates an extracellular matrix-degrading metalloprotease during fertilization in Chlamydomonas. J Cell Biol. 1989 Oct;109(4 Pt 1):1689–1694. doi: 10.1083/jcb.109.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. MITOTIC REPLICATION OF DEOXYRIBONUCLEIC ACID IN CHLAMYDOMONAS REINHARDI. Proc Natl Acad Sci U S A. 1960 Jan;46(1):83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z., Hajdukiewicz P., Maliga P. Stable transformation of plastids in higher plants. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner J. P., Gillham N. W., Boynton J. E. The sequence of the chloroplast atpB gene and its flanking regions in Chlamydomonas reinhardtii. Gene. 1986;44(1):17–28. doi: 10.1016/0378-1119(86)90038-7. [DOI] [PubMed] [Google Scholar]