Abstract
The adenovirus E1A 243R oncoprotein targets TRRAP, a scaffold protein that assembles histone acetyltransferase (HAT) complexes, such as the NuA4/Tip60 complex which mediates transcriptional activity of the proto-oncogene MYC and helps determine the cancer cell phenotype. How E1A transforms cells through TRRAP remains obscure. We performed proteomic analysis with the N-terminal transcriptional repression domain of E1A 243R (E1A 1-80) and showed that E1A 1-80 interacts with TRRAP, p400, and three other members of the NuA4 complex - DMAP1, RUVBL1 and RUVBL2 - not previously shown to associate with E1A 243R. E1A 1-80 interacts with these NuA4 components and MYC through the E1A TRRAP-targeting domain. E1A 243R association with the NuA4 complex was demonstrated by co-immunoprecipitation and analysis with DMAP1, Tip60, and MYC. Significantly, E1A 243R promotes association of MYC/MAX with the NuA4/Tip60 complex, implicating the importance of the MYC/NuA4 pathway in cellular transformation by both MYC and E1A.
Keywords: E1A 1-80, 243R, NuA4, TRRAP, DMAP1, RUVBL1, RUVBL2, Tip60
Graphical Abstract
Introduction
Histone acetylation plays important roles in gene activation by relaxation of chromatin structure, allowing recognition of promoter regulatory sequences by the transcription machinery. Histone acetyltransferases (HATs) are often recruited to promoters by transcription activators such as the proto-oncogene product MYC (Cowling and Cole, 2006; Dang et al., 2006; Meyer and Penn, 2008). Among HATs, p300 is enzymatically proficient by itself, whereas Tip60, GCN5 and PCAF are assembled into multi-subunit complexes to enhance enzymatic activity or promoter recruitment (for a review, see (Lee and Workman, 2007). These HAT complexes are assembled by the transformation/transcription domain associated protein, TRRAP. For example, the human NuA4/Tip60 HAT complex contains TRRAP, Tip60, DNA methlytransferase 1 associated protein (DMAP1), E. coli RuvB-like 1 (RUVBL1), and RUVBL2, in addition to other components (Doyon et al., 2004; Lee and Workman, 2007). This complex is thought to represent a fusion between two yeast complexes, the NuA4 complex which has HAT activity, and the SWR1 complex which has ATP-dependent chromatin remodeling activity (Auger et al., 2008; Doyon et al., 2004).
The NuA4/Tip60 complex is involved in regulation of transcription, DNA repair, and growth control (Doyon et al., 2004; Lee and Workman, 2007; Squatrito et al., 2006). It is required for stem cell functions since RNAi screening has identified shRNAs against several members of the complex that induce phenotypic changes in embryonic stem (ES) cells (Fazzio et al., 2008). Its critical roles in cancer have been suggested by the observation that the proto-oncogene product MYC and the NuA4 complex co-activate a set of genes (the MYC module) that are expressed in both the mouse ES cells and human cancer cells (Kim et al., 2010). In mouse ES cells, expression of both the MYC module and the stem cell Core module is essential to maintain ES cell identity. In human cancers, however, only the MYC module is expressed (Kim et al., 2010), suggesting that the MYC/NuA4 pathway may be responsible for the similarities between cancer cells and stem cells. Of interest, in human breast cancers, higher expression of the MYC module is correlated with a shorter time span for tumors to become metastatic (Kim et al., 2010).
E1A 243R is an adenoviral oncogene product that targets several major cellular pathways for transformation (for a review, see (Berk, 2005; Pelka et al., 2008)). Our previous studies show that the N-terminal 80 amino acid region of E1A 243R, E1A 1-80, can function as a transcriptional repressor by targeting p300 and TBP (Green et al., 2008; Song et al., 1995a; Song et al., 1995b). More recently, we showed that E1A 1-80 represses MYC transcription by targeting both p300 and TRRAP, thus inhibiting the acetylation of H3K18 and H4K16 on the MYC promoter (Zhao et al., 2016). Interaction with TRRAP is essential for cellular transformation by E1A 243R (Deleu et al., 2001). Additionally, the TRRAP-binding domain of MYC can be functionally substituted by the E1A 243R TRRAP-targeting domain for cellular transformation (Deleu et al., 2001). Recently, MYC was shown to be part of an E1A 243R-TRRAP complex (Vijayalingam et al., 2015), suggesting that MYC and E1A 243R may functionally cooperate in cellular transformation (Chakraborty and Tansey, 2009). While these studies suggest the importance of the TRRAP pathway for E1A 243R function, it remains unclear what happens subsequent to E1A 243R interaction with TRRAP that leads to cellular transformation.
To further understand E1A 243R interaction with cellular regulatory pathways, as described here, we performed a proteomic analysis with Flag-tagged E1A 1-80, and found that E1A 1-80 interacts with TRRAP and p400, as well as three other members of the NuA4 complex: DMAP1, RUVBL1 and RUVBL2, which have not been shown previously to interact with E1A 243R. These proteins interact with E1A 243R as shown by co-immunoprecipitation with antibody against the C-terminal domain of E1A 243R, as well as by co-immunoprecipitation with DMAP1-Flag, Flag-Tip60 and Flag-HA-MYC. Significantly, E1A 243R enhances the association of MYC with the NuA4/Tip60 complex, and we propose that the NuA4/Tip60 complex is the platform for the cooperation of E1A 243R with MYC in cellular transformation.
Materials and Methods
Lentiviral and adenoviral constructs
pLenti6-E1A 1-80FH: the Ad2 E1A 1-80 region was PCR-amplified with the following primers: G1-E1AN5″: CACCATGAGACATATTATCTGCCACGGAGG, and G3-E1AN3′-FH: GCTTACTCGAGGCTGGCGTAATCGGGGACGTCATAAGGGTAAGCCTTGTCATCGTCATCCTTG TAGTCGCCTTCTCTGCCTAAGTCAATCCCTTCCTGCACCGC. PCR reactions were carried out with the Platinum High Fidelity PCR kit following manufacturer’s recommendations (Invitrogen). The PCR DNA was cloned first into the pENTR-SD/D-TOPO vector, and then transferred to the pLenti6/capTEV-CT-DEST vector to generate pLenti6-E1A 1-80FH following the supplier’s protocol (the Gateway System, Invitrogen).
pLenti6-FH-GFP: the GFP open reading frame was PCR-amplified with the following primers: G5-FH-GFP5′: CACCATGGACTACAAGGATGACGATGACAAGGCTTACCCTTATGAC-GTCCCCGATTACGCCAGCCTCGAGATGGTGAGCAAGGGCGAGGAGCTG, and G6-GFP3′: GCTTACTTGTACAGCTCGTCCATGCCGAG. The PCR DNA was cloned the same way as for pLenti6-E1A 1-80FH.
pLenti6-DMAP1-Flag: the DMAP1 open reading frame was PCR amplified from RNA prepared from SKBR3 cells with the following primers: G153-DMAP1-A: ACTGCCGAATTCCATGGCTACGGGCGCGGATGTACGG, and G156-DMAP1-D: GCTTACGGATCCTTAAGCCTTGTCATCGTCATCCTTGTAGTCGCCTTCTCTGCCCTCGAGCGG CTTCTTGGCTTTCTTCACGGA. The PCR DNA was digested with EcoRI and BamHI and cloned into the EcoRI/BamHI sites of pLenti6Δ (a derivative of the pLenti6/capTEV-CT-DEST vector with a deletion of the XbaI fragment) to generate pLenti6-DMAP1-Flag.
pLenti6-Flag-Tip60: The Tip60 open reading frame was PCR-amplified from pBABE-FH-Tip60 (a gift of G. Chinnadurai) with the following primers: G199-Flag-Tip60: ACATCGTCTAGAACCATGGACTACAAGGATGACGATGACAAGGCTGGAGAATTCGGACGGGA AGGTATGGCGGAGGTGGGGGAGATAATC, and G202-Tip60-3′Rev: GTCAGCGGATCCTCACCACTTCCCCCTCTTGCTCCA. The PCR DNA was digested with XbaI and BamHI, and ligated into the XbaI/BamHI sites of pLenti6-DMAP1-Flag to generate pLenti6-Flag-Tip60.
pCDH-FH-MYC: a synthetic double-stranded DNA encoding Flag-HA tag with the following sequence (top-strand): CTAGAACCATGGACTACAAGGATGACGATG-ACAAGGCTTACCCTTATGACGTCCCCGATTACGCCGGCGGAGGTGGCCTCGAGG was inserted into the XbaI/EcoRI sites of pCDH-MYC (Addgene plasmid # 46970) (Cheng et al., 2013) to generate pCDH-FH-MYC.
All constructs described above except pLenti6-FH-GFP are confirmed by sequence analysis. pLenti6-FH-GFP was confirmed by detection of green fluorescence in lentivirus infected cells and by immunoprecipitation with Flag antibody.
Ad-E1A 1-80 C+, Ad-E1A 243R, Ad-E1A 1-80FH, Ad-E1A 1-80(Δ2-11)FH, and Ad-E1A 1-80(Δ26-35)FH have been described previously (Loewenstein and Green, 2011; Zhao et al., 2016)
Lentivirus generation
293T cells were plated in a 6-well plate (6 × 106 cells/well) the day before transfection, and transfected with JetPrime reagent following manufacturer’s guideline (Polyplus-transfection). Briefly, 0.5 μg each of pCMV-VSV-G and the packaging plasmid pCMV-dR8.2 dvpr (Stewart et al., 2003), and 1.5 μg of a lentivirus expression construct were combined in 100 μl of the transfection buffer, and mixed with 100 μl of the buffer with 5 μl of JetPrime reagent for DNA precipitates to form. DNA precipitates were incubated with cells for 5 h, after which cells were washed and maintained for 42 h prior to use of culture supernatant for transduction.
Lentiviral transduction, Ad viral infection and immunoprecipitation
HeLa cells (2 × 106 cells) were plated in a T180 flask and transduced 5 h later with 2 ml of a lentiviral vector overnight in 20 ml medium with 8 ng/ml polybrene. The next day, cells were washed with medium and selected with 1.0 μg/ml of puromycin (for FH-MYC) or 6 μg/ml of blasticidin (for DMAP1-Flag and Flag-Tip60) for 3 days. After drug selection, 4 × 106 cells were plated in a T180 flask and infected 24 h later with an Ad vector for 24 h (with 20 PFU/cell of Ad-lacZ or Ad-E1A 1-80 C+, or 40 PFU/cell of Ad-E1A 243R due to its lower expression level compared to E1A 1-80 C+). Cells were harvested and processed for immunoprecipitation with the Flag antibody beads as described (Zhao et al., 2014).
Proteomic analysis
HeLa cells in six T180 flasks (Greiner) were transduced with lentiviral particles and selected with blasticidin (6 μg/ml) for 3 days, after which cells were washed and cultured for one more day in complete DMEM medium. Cell lysis and immunoprecipitation with Flag antibody were carried out as previously described (Zhao et al., 2014). The Flag antibody beads with bound proteins were submitted to the Donald Danforth Plant Science Center Proteomics and Mass Spectrometry facility for Liquid chromatography-tandem MS (LC-MS/MS) and protein identification.
Antibodies
p300: sc-584 (Santa Cruz Biotechnology, SCBT); DMAP1: sc-135244 (SCBT); RUVBL1: sc-48798 (SCBT); RUVBL2: 06-1300 (Millipore); p400, A300-541A (Bethyl Laboratory); TRRAP: A301-132A (Bethyl Laboratory); Tip60: DR1041 (Calbiochem); MYC: sc-40 (SCBT); MAX: sc-197 (SCBT); HA: sc-805 (SCBT); E1A 243R: sc-58658 (SCBT); E1A 1-80: rabbit polyclonal antisera.
Results and Discussion
Proteomic analysis: E1A 1-80 interacts with DMAP1, RUVBL1, and RUVBL2 in addition to TRRAP and p400
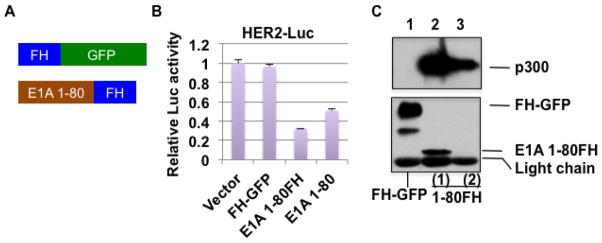
E1A 1-80 is capable of targeting TRRAP (Zhao et al., 2016), an activity essential for E1A 243R to transform cells (Deleu et al., 2001). To understand further the mechanism of E1A 1-80-mediated transcriptional repression and to gain a better understanding of the mechanism of cellular transformation by E1A 243R, we performed proteomic analysis to identify cellular proteins that specifically interact with E1A 1-80. Lentiviral expression vectors were constructed expressing Flag-HA-tagged E1A 1-80 (E1A 1-80FH) and FH-GFP (green fluorescence protein) as a control (Figure 1A). Lentiviral vectors were chosen for a longer-term and more moderate level of expression compared to using Ad vectors. To first confirm the functional activity of E1A 1-80FH as a transcriptional repressor, a co-transfection/luciferase reporter assay was performed with pHER2-Luc (Figure 1B), which expresses firefly luciferase under the control of the E1A-repressible HER2 promoter (Loewenstein and Green, 2011). As shown, E1A 1-80FH represses the HER2 promoter as well as does E1A 1-80 (Loewenstein and Green, 2011), whereas FH-GFP does not. Thus, E1A 1-80FH retains the repression activity of E1A 1-80.
Figure 1. E1A 1-80FH is active in transcriptional repression and interaction with p300.
A. Lentiviral expression constructs for E1A 1-80FH and FH-GFP. B. Luciferase reporter assays with pHER2-Luc and the lentiviral constructs. HeLa cells were co-transfected with pHER2-Luc, phRL-0 (renilla luciferase), and the lentiviral constructs as indicated, and dual luciferase assays performed as described previously (Loewenstein and Green, 2011). Data plotted represent average of three experiments. C. Interaction of E1A 1-80FH with p300. Lentiviral particles from the constructs in A were used to infect HeLa cells to establish blasticidin-resistant cells. Cell lysates were co-immunoprecipitated with Flag antibody beads, and the bound proteins eluted and examined by Western blots for p300 (upper panel) or for FH-GFP and E1A 1-80FH using HA antibody (lower panel). Lanes 2 and 3 represent two samples of eluted E1A 1-80FH complexes with high (lane 2) and low (lane 3, below detection limit in this Western blot) quantities of E1A 1-80FH.
For proteomic analysis, E1A 1-80FH and FH-GFP were expressed in HeLa cells using lentiviral transduction. Transduced cells were selected with blasticidin and used to make lysates which were then immunoprecipitated using Flag antibody beads. A small portion of the eluted proteins were examined by Western blot with p300 antibody (Figure 1C, upper panel) or HA antibody for tagged proteins (lower panel). The two samples of E1A 1-80FH used for this analysis had different quantities of E1A 1-80FH as shown (lanes 2 and 3; lane 3 is below the detection limit in this blot). E1A 1-80FH bound p300 strongly (lanes 2 and 3), whereas the control FH-GFP did not (lane 1). Both E1A 1-80FH samples and the control FH-GFP were subjected to proteomic analysis by mass spectrometry (see Experimental Procedures).
Analysis revealed eight proteins of interest that are specific to E1A 1-80FH (Table 1). The use of two E1A 1-80FH samples with high and low quantities of E1A 1-80FH provides additional confidence of the specificity of the interaction. While some of the identified proteins in Table 1, such as p300 and TRRAP, are known to interact with E1A 243R, three proteins are novel: DMAP1, RUVBL1, and RUVBL2. These three proteins, together with TRRAP and p400, are important members of the NuA4/Tip60 complex (Doyon et al., 2004). Human NuA4/Tip60 complex reportedly consists of up to 20 subunits (Lee and Workman, 2007). In our proteomic analysis, not all the commonly-reported components of the NuA4 complex are identified, including the HAT enzyme Tip60. Similarly, E1A 243R has been shown to associate with a p400 complex that lacks Tip60 (Fuchs et al., 2001). Likewise, in a study which defined a MYC gene regulatory module that is co-regulated by MYC and NuA4, proteomic analysis of the MYC complex also identified only some members of the NuA4 complex and lacked Tip60 (Kim et al., 2010). It is possible that the assembly of the NuA4 complex is a dynamic process and not all the components are present in the NuA4/Tip60 complex at all times. In addition, HeLa cells are known to express the E6 and E7 proteins of the human papillomavirus (HPV), and HPV E6 has been shown to down-regulate Tip60 (Jha et al., 2010), which may contribute to the lack of Tip60 detection in our proteomic analysis.
Table 1.
Proteins identified by proteomic analysis with E1A 1-80FH
| Number of identified peptides | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| # | Identified Proteins | Accession Number | Molecular Weight | FH- GFP | E1A 1-80FH 1 | E1A 1-80FH 2 |
| 1 | CBP | gi|119943104 | 265 kDa | 0 | 68 | 41 |
| 2 | p300 | gi|50345997 | 264 kDa | 0 | 51 | 20 |
| 3 | RUVBL1* | gi|197692395 | 50 kDa | 1 | 10 | 7 |
| 4 | RUVBL2* | gi|119572814 | 47 kDa | 0 | 8 | 3 |
| 5 | p400 | gi|119618940 | 335 kDa | 0 | 11 | 7 |
| 6 | TRRAP | gi|119597101 | 444 kDa | 0 | 12 | 2 |
| 7 | DMAP1* | gi|13123776 | 53 kDa | 0 | 2 | 2 |
| 8 | MYC | gi|127619 | 49 kDa | 0 | 1 | 1 |
Novel proteins that have not been shown previously to interact with E1A 243R.
Sample with higher level of E1A 1-80FH.
Sample with lower level of E1A 1-80FH.
Interaction of E1A 1-80 with the NuA4/Tip60 complex through the E1A TRRAP-targeting domain
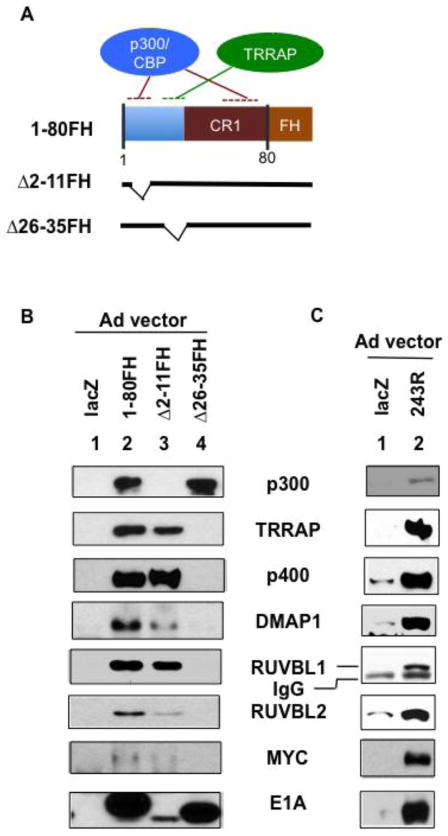
To determine which domains in E1A 1-80 are responsible for interaction with the proteins identified by proteomic analysis, we used Ad vectors to express higher levels of E1A 1-80FH or two deletion mutants deficient in ability to interact with either p300 (Δ2-11FH) or TRRAP (Δ26-35FH) (Figure 2A) (Zhao et al., 2016). HeLa cells were infected with the indicated Ad vectors, and cell lysates prepared for immunoprecipitation with Flag antibody beads (Figure 2B). As shown by Western blot (Figure 2B), E1A 1-80FH associated with all seven of the proteins identified by proteomic analysis (lane 2). The Δ2-11FH mutant associated with all the examined proteins except p300 (lane 3) as expected. In contrast, the Δ26-35FH mutant associated with p300 but not any of the other proteins (lane 4). Significantly, E1A 1-80FH, but not Δ26-35FH, also bound MYC, consistent with a previous report showing association of E1A 243R with MYC (Vijayalingam et al., 2015). Thus, association of E1A 1-80 with TRRAP, p400, DMAP1, RUVBL1, RUVBL2 and MYC occurs through the TRRAP-targeting domain of E1A 1-80. Importantly, all of the proteins requiring E1A amino acids 26-35 for interaction, except MYC, belong to the NuA4/Tip60 complex (Doyon et al., 2004). TRRAP is a component of several HAT complexes, including the GCN5 complex, PCAF complex and the NuA4/Tip60 complex (Lee and Workman, 2007), each being an assembly of multiple components. However, our proteomic analysis with E1A 1-80 did not identify subunits of the TRRAP-GCN5 and TRRAP-PCAF complexes, suggesting that E1A 1-80 preferentially interacts with the NuA4/Tip60 complex.
Figure 2. Interaction of E1A 1-80 and E1A 243R with NuA4 components through the TRRAP targeting domain.
A. Illustration of expression constructs. B. Adenoviral-expressed E1A 1-80FH interacts with components of the NuA4 complex identified by proteomic analysis. HeLa cells were infected with Ad expression vectors as indicated, and cell lysates co-immunoprecipitated with Flag antibody beads. Bound proteins were identified by Western blots with indicated antibodies. E1A Western blot analysis was with a rabbit polyclonal antibody. The apparent low quantity of E1A Δ2-11FH in the E1A Western blot analysis was possibly due to weak binding of the protein to the PVDF membrane during the assay. E1A Western blot analysis with Flag and HA antibodies revealed the same result (not shown). C. Adenoviral-expressed E1A 243R interacts with the identified components of the NuA4 complex. HeLa cells were infected with Ad-lacZ (lane 1) or Ad-E1A 243R (lane 2), and co-immunoprecipitation was with E1A M73 antibody beads (SCBT, Inc.). Western blot analysis was as in B, except E1A 243R Western blot analysis was with M58 antibody (SCBT, Inc.)
E1A 243R associates with DMAP1, RUVBL1 and RUVBL2 of the NuA4/Tip60 complex as does E1A 1-80
DMAP1, RUVBL1 and RUVBL2 were identified through our proteomic analysis with E1A 1-80. It is likely that the full-length E1A 243R also interacts with these proteins. To examine this possibility, we infected HeLa cells with Ad-lacZ or Ad-E1A 243R, and immunoprecipitated cell lysates with antibody against the C-terminal region of E1A (M73). These immunoprecipitates were analyzed by Western blot with antibodies as indicated (Figure 2C). As shown, E1A 243R interacted with TRRAP, p400 and MYC as expected. Importantly, it also interacted with DMAP1, RUVBL1 and RUVBL2, indicating that the interaction of E1A 1-80 with these proteins reflects their interaction with E1A 243R.
HPV E6 and E7 which are constituently expressed in HeLa cells are transforming proteins that target p300 and Rb, respectively (for a review, see (Moody and Laimins, 2010)), and appear to be in competition with E1A 243R in this regard. However, the HPV E6 and E7 proteins have not been shown to interact with the NuA4/Tip60 complex and thus would seem unlikely to directly compete with 243R for access to this complex. Further, E1A 1-80(Δ2-11)FH, which does not interact with p300 or Rb, remains capable of interaction with the NuA4/Tip60 components (Figure 2B), suggesting that interaction of E1A 243R and E1A 1-80 with the NuA4/Tip60 complex does not require interaction with p300 or Rb, and may not be impacted directly by E6 or E7.
While TRRAP-targeting by E1A is required for E1A 243R to induce cellular transformation (Deleu et al., 2001), the exact role of TRRAP-targeting in E1A 243R function is unclear. It was demonstrated that the TRRAP-targeting domain of E1A 243R can be substituted for the TRRAP-interaction domain of MYC to maintain MYC transformation activity (Deleu et al., 2001), suggesting that MYC and E1A 243R target the same pathway for transformation. Since the NuA4/Tip60 complex and MYC co-regulate a set of genes that help define the cancer cell phenotype (Kim et al., 2010), it is possible that the NuA4/Tip60 complex is also targeted by E1A 243R for transcriptional regulation as well as cellular transformation.
Co-immunoprecipitation with Flag-tagged DMAP1 confirms interaction of E1A 1-80 and E1A 243R with the NuA4/Tip60 complex
To more thoroughly examine E1A 1-80 and 243R interaction with the identified components of the NuA4/Tip60 complex, we attached a Flag tag to the C-terminus of DMAP1, and expressed DMAP1-Flag in HeLa cells using a lentivirus vector. The cells were then infected with Ad-lacZ, Ad-E1A 1-80 C+, or Ad-E1A 243R. Cell lysates were immunoprecipitated with Flag antibody beads and examined by Western blot analysis with the indicated antibodies (Figure 3B). As shown, DMAP1-Flag associated with TRRAP, p400, RUVBL1 and RUVBL2 regardless of expression of lacZ, E1A 1-80 C+ or E1A 243R (Figure 3B, lanes 1–4), whereas the control FH-GFP did not interact with any protein of the NuA4/Tip60 complex (lanes 5 and 6).
Figure 3. E1A 243R and E1A 1-80 promote association of MYC/MAX with the NuA4/Tip60 complex.
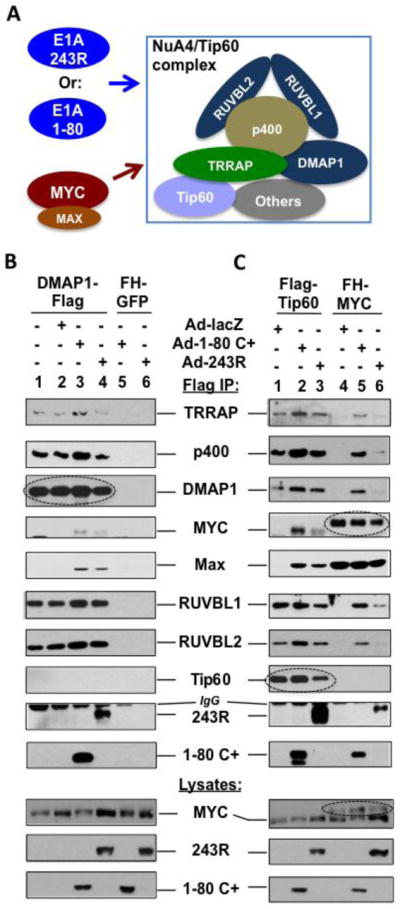
A. Experimental model. The relative position of each protein in the NuA4/Tip60 complex remains to be elucidated. B. Flag-DMAP1 interacts with E1A 1-80 and E1A 243R as well as components of the NuA4 complex identified by proteomic analysis. Flag-DMAP1 was expressed in HeLa cells using a lentivirus vector (lanes 1–4). The cells were then infected with Ad-lacZ (lane 2), Ad-E1A 1-80 C+ (lane 3), or Ad-E1A 243R (lane 4) as indicated. Cell lysates were immunoprecipitated with Flag antibody beads and immunoprecipitates examined by Western blots with indicated antibodies. Lanes 5 and 6 are controls with HeLa cells expressing FH-GFP. Expressed DMAP1-Flag immunoprecipitated by the Flag antibody is indicated by a dashed oval. Expression of E1A 1-80 C+ or E1A 243R recruited MYC/MAX into the NuA4 complex (lanes 3 and 4). C. E1A 243R and E1A 1-80 promote the association of MYC/MAX with the NuA4/Tip60 complex. Experimental conditions are the same as in B, except that Flag-Tip60 (lanes 1–3) and FH-MYC (lanes 4–6) are expressed by lentivirus vectors. When the cells were not infected with an Ad vector (not shown), Flag-Tip60 and FH-MYC had the same patterns of binding as when Ad-lacZ was used (lanes 1 and 4, respectively). Expressed Flag-Tip60 or FH-MYC immunoprecipitated are indicated by a dashed oval to differentiate from cellular endogenous proteins. Expression of both E1A 1-80 C+ and E1A 243R recruited MYC/MAX into the NuA4/Tip60 complex when Flag-Tip60 was used for analysis (lanes 2 and 3). When FH-MYC was immunoprecipitated for analysis, E1A 1-80 and E1A 243R both enhanced the association of FH-MYC with the examined components of the NuA4/Tip60 complex (lanes 5 and 6).
E1A 1-80 and E1A 243R associated with DMAP1-Flag (Figure 3B, lanes 3 and 4, respectively), whereas control FH-GFP did not co-immunoprecipitate E1A 1-80 C+ (lane 5) or E1A 243R (lane 6). Both E1A proteins were well expressed in the cell lysates (see lanes 5 and 6, lysates). We conclude that interaction of E1A 1-80 and E1A 243R with DMAP1 as well as the other members of the NuA4/Tip60 complex is specific, and propably through TRRAP (Figure 2B). Since the same proteins are co-immunoprecipitated whether E1A 243R (Figure 2C), Flag-tagged E1A 1-80 (Figure 2B), or Flag-tagged DMAP1 (Figure 3B) is used for immunoprecipitation, it is likely that the identified components of the NuA4/Tip60 complex interact with E1A 1-80 or E1A 243R as a complex. Of importance, MYC and MAX were co-immunoprecipitated with DMAP1-Flag only in the presence of E1A 1-80 (Figure 3B, lane 3) or E1A 243R (lane 4), suggesting that MYC is recruited together with MAX into the NuA4/Tip60 complex by E1A 1-80 and E1A 243R.
E1A 1-80 and E1A 243R promote the association of MYC/MAX with the NuA4/Tip60 complex
Our proteomic analysis did not detect all of the components of the NuA4/Tip60 complex (Table 1), e.g., the HAT enzyme Tip60. This may reflect instability of the complex or a low sensitivity of detection. We have noted that detection of Tip60 expression in several human breast cancer cell lines and HeLa cells has a low sensitivity (unpublished observations). Since MYC/MAX is recruited to the NuA4/Tip60 complex by E1A 1-80 and E1A 243R (Figure 3B), we examined whether MYC/MAX is also recruited into the NuA4/Tip60 complex when Flag-tagged Tip60 is expressed in cells followed by co-immunoprecipitation. We first expressed Flag-Tip60 in HeLa cells using a lentivirus vector. These cells were then infected with Ad-lacZ, Ad-E1A 1-80 C+, or Ad-E1A 243R (Figure 3C). Cell lysates were immunoprecipitated with Flag antibody beads and immunoprecipitates examined by Western blot analysis with the indicated antibodies. Flag-Tip60 associated with TRRAP, p400, DMAP1, RUVBL1 and RUVBL2, but not with MYC (lane 1). Most important, MYC/MAX was detected in the Flag-Tip60 immunoprecipitates only in the presence of E1A 1-80 C+ or 243R (lanes 2 and 3), similar to results observed with DMAP1-Flag (Figure 3B, lanes 3 and 4). Flag-Tip60 also associated with E1A 1-80 and E1A 243R (lanes 2 and 3). Thus, although Tip60 is not identified in our proteomic analysis (Table 1), Flag-Tip60 is capable of incorporating into the NuA4 complex and associating with E1A 243R.
To further examine MYC interaction with the NuA4/Tip60 complex, we expressed FH-MYC in HeLa cells, followed by infection with Ad vectors expressing lacZ, E1A 1-80 C+ or 243R (Figure 3C). MYC interaction with the NuA4 components was examined by Flag co-IP followed by Western blot. As shown, FH-MYC was not found to associate with the NuA4 components when Ad-lacZ was used for infection (Figure 3C, lane 4). Significantly, expression of E1A 1-80 C+ dramatically increased MYC association with these proteins (lane 5). E1A 243R also enhanced MYC recruitment, as all the examined components of the NuA4/Tip60 complex were detected in the FH-MYC immunoprecipitates (lane 6). In contrast, neither E1A 1-80 C+ (lane 5) nor E1A 243R (lane 6) affected FH-MYC protein level or FH-MYC association with MAX in the immunoprecipitates. FH-MYC also associated with E1A 1-80 and E1A 243R (lanes 5 and 6). Since MYC and MAX were always detected together in our analyses, it appears that the MYC/MAX heterodimer is recruited by E1A 1-80 and E1A 243R to the NuA4/Tip60 complex.
Several lines of evidence suggest that the enhanced association of MYC/MAX with the NuA4/Tip60 complex in the presence of E1A 243R (Figure 3B and 3C) is likely mediated by TRRAP. First, both MYC and E1A 243R can interact with TRRAP (Deleu et al., 2001; McMahon et al., 1998). Second, the sequences in TRRAP responsible for interaction with MYC and E1A 243R are located in broad regions of TRRAP (Deleu et al., 2001; Park et al., 2001), rendering it feasible that MYC and E1A 243R interact with different domains in TRRAP and function cooperatively. Third, recent studies suggest that MYC interacts with E1A 243R indirectly through TRRAP (Vijayalingam et al., 2015). Since MYC interaction with TRRAP and the NuA4/Tip60 complex is inefficient (Figure 3C, lane 4), it is likely that E1A 243R interacts with TRRAP and facilitates its interaction with MYC/MAX. Because TRRAP is critical for the assembly of the NuA4/Tip60 complex, the more efficient interaction of MYC/MAX with TRRAP mediated by E1A 243R can lead to more efficient MYC/MAX interaction with the NuA4/Tip60 complex.
Implications of E1A 243R interaction with MYC and the NuA4/Tip60 complex for the induction of cellular transformation
From yeast to humans, several types of chromatin-remodeling and histone acetyltransferase complexes have been discovered and their functions are beginning to be understood (Doyon et al., 2004). The human NuA4/Tip60 HAT complex is proposed to represent a fusion of the yeast SWR1 chromatin remodeling complex and the yeast NuA4 HAT complex (Auger et al., 2008; Doyon et al., 2004), and may be a highly effective transcriptional activator due to both activities. Our results suggest that MYC may interact more efficiently with TRRAP in the E1A 243R - NuA4/Tip60 complex and form a putative E1A 243R - NuA4/Tip60 - MYC supercomplex.
We suggest two possible mechanisms by which such a putative supercomplex could engage in transcriptional regulation and cellular transformation. First, MYC may normally interact with the NuA4/Tip60 complex poorly, thus avoiding over-activation of the MYC/NuA4/Tip60 pathway that could lead to cellular transformation. Enhanced association of MYC with the NuA4/Tip60 complex in the presence of E1A 243R may drive MYC transcriptional activity of genes with critical roles in cellular transformation. There may be E1A-like cellular proteins that regulate MYC interaction with the NuA4/Tip60 complex. Second, since MYC can recruit both TRRAP-GCN5 and TRRAP-Tip60 to promoters (Frank et al., 2003; Liu et al., 2003; McMahon et al., 2000), MYC may preferrentially recruit the NuA4/Tip60 complex for gene regulation in the presence of E1A 243R. It has been reported that E1A 243R interacts with GCN5 (Kulesza et al., 2002; Lang and Hearing, 2003), presumably through targeting TRRAP. However, since our proteomic analysis has not identified any component of the TRRAP-GCN5 complex, it seems likely that E1A 243R favors the NuA4/Tip60 complex.
In summary, our finding that E1A 243R specifically enhances the association of MYC with the NuA4/Tip60 complex through the E1A N-terminal domain may facilitate our understanding of the functional regulation of the NuA4/Tip60 complex as well as its role in cellular transformation by MYC and E1A 243R. These studies may guide cancer therapies based on targeting the MYC/NuA4/Tip60 pathway.
Conclusions
We demonstrate that the adenoviral oncogene product E1A 243R, as well as the E1A N-terminal repression domain, E1A 1-80, interacts with components of the NuA4 complex, including DMAP1, RUVBL1, RUVBL2, which have not been shown to interact with E1A 243R previously. E1A 243R and E1A 1-80 interact with the NuA4 complex through a TRRAP-targeting domain, and enhance MYC association with the NuA4 complex. Since TRRAP is key for the assembly of the NuA4 complex and is required for transformation by both MYC and E1A 243R, we propose that MYC targeting of the NuA4 complex is regulated and critical for cellular transformation by MYC and E1A.
Highlights.
E1A 243R interacts with DMAP1, RUVBL1, RUVBL2, TRRAP, p400, and MYC.
These cellular proteins, except MYC, are part of the NuA4/Tip60 complex.
E1A 243R interaction with the NuA4/Tip60 complex promotes its association with MYC.
The NuA4/Tip60 complex may be critical for cellular transformation by MYC and E1A.
Acknowledgments
Funding sources: MG is supported by Lifetime Research Career Award (RCA #AI-04739) from the NIH. The Lottie Caroline Hardy Charitable Trust and Saint Louis University Cancer Research Fund have provided support for the project.
The authors thank Dr. W. Wold for critical reading of the manuscript, and are grateful to Drs. G. Chinnadurai (Saint Louis University) for the gift of pBABE-FH-Tip60 and J. Wang (Vanderbilt University Medical Center) for the gift of pCDH-MYC plasmid (Addgene.com). Proteomic analysis was performed by the Donald Danforth Plant Science Center Proteomics and Mass Spectrometry facility.
The authors declare no conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auger A, Galarneau L, Altaf M, Nourani A, Doyon Y, Utley RT, Cronier D, Allard S, Cote J. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol. 2008;28:2257–2270. doi: 10.1128/MCB.01755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk AJ. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene. 2005;24:7673–7685. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- Chakraborty AA, Tansey WP. Adenoviral E1A function through Myc. Cancer Res. 2009;69:6–9. doi: 10.1158/0008-5472.CAN-08-3026. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, Thompson RC, Muller S, Knapp S, Wang J. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling VH, Cole MD. Mechanism of transcriptional activation by the Myc oncoproteins. Semin Cancer Biol. 2006;16:242–252. doi: 10.1016/j.semcancer.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Deleu L, Shellard S, Alevizopoulos K, Amati B, Land H. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene. 2001;20:8270–8275. doi: 10.1038/sj.onc.1205159. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Cote J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- Green M, Panesar NK, Loewenstein PM. The transcription-repression domain of the adenovirus E1A oncoprotein targets p300 at the promoter. Oncogene. 2008;27:4446–4455. doi: 10.1038/onc.2008.85. [DOI] [PubMed] [Google Scholar]
- Jha S, Vande Pol S, Banerjee NS, Dutta AB, Chow LT, Dutta A. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol Cell. 2010;38:700–711. doi: 10.1016/j.molcel.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesza CA, Van Buskirk HA, Cole MD, Reese JC, Smith MM, Engel DA. Adenovirus E1A requires the yeast SAGA histone acetyltransferase complex and associates with SAGA components Gcn5 and Tra1. Oncogene. 2002;21:1411–1422. doi: 10.1038/sj.onc.1205201. [DOI] [PubMed] [Google Scholar]
- Lang SE, Hearing P. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene. 2003;22:2836–2841. doi: 10.1038/sj.onc.1206376. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Liu X, Tesfai J, Evrard YA, Dent SY, Martinez E. c-Myc transformation domain recruits the human STAGA complex and requires TRRAP and GCN5 acetylase activity for transcription activation. J Biol Chem. 2003;278:20405–20412. doi: 10.1074/jbc.M211795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewenstein PM, Green M. Expression of the Adenovirus Early Gene 1A Transcription-Repression Domain Alone Downregulates HER2 and Results in the Death of Human Breast Cancer Cells Upregulated for the HER2 Proto-Oncogene. Genes Cancer. 2011;2:737–744. doi: 10.1177/1947601911426570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- McMahon SB, Wood MA, Cole MD. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol. 2000;20:556–562. doi: 10.1128/mcb.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Park J, Kunjibettu S, McMahon SB, Cole MD. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 2001;15:1619–1624. doi: 10.1101/gad.900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka P, Ablack JN, Fonseca GJ, Yousef AF, Mymryk JS. Intrinsic structural disorder in adenovirus E1A: a viral molecular hub linking multiple diverse processes. J Virol. 2008;82:7252–7263. doi: 10.1128/JVI.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CZ, Loewenstein PM, Green M. Repression in vitro, by human adenovirus E1A protein domains, of basal or Tat-activated transcription of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1995a;69:2907–2911. doi: 10.1128/jvi.69.5.2907-2911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CZ, Loewenstein PM, Toth K, Green M. Transcription factor TFIID is a direct functional target of the adenovirus E1A transcription-repression domain. Proc Natl Acad Sci U S A. 1995b;92:10330–10333. doi: 10.1073/pnas.92.22.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayalingam S, Subramanian T, Zhao LJ, Chinnadurai G. The cellular protein complex associated with a transforming region of E1A contains c-MYC. J Virol. 2015 doi: 10.1128/JVI.02039-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LJ, Loewenstein PM, Green M. The adenoviral E1A N-terminal domain represses MYC transcription in human cancer cells by targeting both p300 and TRRAP and inhibiting MYC promoter acetylation of H3K18 and H4K16. Genes Cancer. 2016 doi: 10.18632/genesandcancer.99. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LJ, Subramanian T, Vijayalingam S, Chinnadurai G. CtBP2 proteome: Role of CtBP in E2F7-mediated repression and cell proliferation. Genes Cancer. 2014;5:31–40. doi: 10.18632/genesandcancer.2. [DOI] [PMC free article] [PubMed] [Google Scholar]