Abstract
To make insight into the cytological basis of reproductive isolation, we examined chromosome synapsis and recombination in sterile male and female hybrids between Microtus arvalis and M. levis. These sibling species differ by a series of chromosomal rearrangements (fusions, inversions, centromere shifts and heterochromatin insertions). We found that meiosis in male hybrids was arrested at leptotene with complete failure of chromosome pairing and DNA double-strand breaks repair. In the female hybrids meiosis proceeded to pachytene; however, the oocytes varied in the degree of pairing errors. Some of them demonstrated almost correct chromosome pairing, while most of them contained a varying number of univalents and multivalents with extensive regions of asynapsis and non-homologous synapsis. Variation between oocytes was probably caused by stochasticity in the ratio of homologous to non-homologous pairing initiations. We suggest that substantial chromosomal and genetic divergence between the parental species affects preliminary alignment of homologues, homology search and elimination of ectopic interhomologue interactions that are required for correct homologous pairing. Apparently, pairing failure in male and aberrant synapsis in female vole hybrids followed by meiotic silencing of unsynapsed chromatin cause apoptosis of gametocytes and sterility.
Homologous chromosome recombination in meiotic prophase is required for orderly chromosome segregation. Recombination is preceded by chromosome prealignment and the scheduled formation of DNA double-strand breaks (DSBs), followed by a RAD51-mediated search for homologous DNA sequences and the formation of heteroduplexes involving DNA strands of homologous chromosomes at early stages of meiotic prophase (leptotene and zygotene). Polymerisation of the synaptonemal complex (SC), a meiotic-specific proteinaceous structure, stabilises these connections and completes homologous chromosome synapsis. A small proportion of DSBs (at least one per chromosome pair) is repaired as crossovers (reciprocal exchanges between homologues). The sites of crossing over can be visualised in mid-meiotic prophase (pachytene) as late recombination nodules containing MLH1 (mismatch repair protein), and at diplotene-diakinesis as chiasmata. Sister chromatid cohesion beyond the chiasmata holds homologues together at metaphase-I, ensuring proper orientation and orderly segregation1,2.
These complex and highly coordinated processes are thoroughly checked by natural selection at each meiosis, and so the genetic unity of the species is preserved. The evolution of geographically isolated populations, however, leads to the fixation of novel chromosomal rearrangements and a divergence of the factors controlling DSB formation and the DNA sequences involved in homology search. In hybrids, karyotypic and genetic divergence can result in meiotic aberrations and variable degrees of infertility due to germ-cell death or the formation of unbalanced gametes.
Although mammalian hybrids have been known for centuries3, studies of the genetic and cellular bases of hybrid sterility in mammals are surprisingly scarce. Several genes causing male sterility in hybrids between karyotypically identical species of the house mouse (Mus musculus x M. domesticus)5,6,7,8 and felines (Felis catus x Profelis serval, F. catus x Prionailurus bengalensis)9 have been localised and sequenced. One of them, PRDM9 (referred to as a speciation gene), is involved in the control of DSB formation10. Male sterility or reduced fertility accompanied by synaptic aberrations of autosomes and sex chromosomes has been widely reported in hybrids of chromosomally divergent mammalian species, subspecies and local populations3,11,12,13,14,15,16,17,18,19.
Sterility in mammalian hybrids is in a good agreement with Haldane’s rule that “when in the F1 offspring of a cross between two animal species or races one sex is absent, rare, or sterile, that sex is always the heterozygous sex”20. Several hypotheses have been proposed to explain the genetic basis of this rule. Among them, the dominance and “faster male” hypotheses are considered the most plausible21,22,23,24,25. The dominance model ascribes the predominant inviability/sterility of the heterogametic sex to the alternative fixation of X-linked recessive mutations. The higher the ratio between recessive and dominant mutations of incompatibility, the larger the time lag between hetero- and homogametic sexes in the evolution of hybrid unfitness26. The “faster male” hypothesis suggests that male fertility alleles evolve and diverge faster due to divergent sexual selection. At the same time, strong sperm selection leads to increase of the stringency of male meiotic checkpoints25,27. In mammals, male meiosis is very sensitive to genetic and chromosomal aberrations. Meiotic mutants and knockouts affect meiosis earlier and more severely in males than in females28 and this amplifies the effects of genetic and chromosomal incompatibility on the fertility of male hybrids.
Haldane’s rule usually applies only to early stages of speciation. Building reproductive barriers is a snowball process29, or, as Darwin (1866) put it, a series of “graduated steps from very slightly lessened fertility to utter and absolute sterility”30. Therefore, at advanced stages of reproductive isolation, both sexes are sterile. We are currently unclear, however, as to whether males and females proceed to complete sterility by the same or by different routes. Sex differences in meiotic disruption within hybrids, then, can tell us about the genetic and cytological bases of these steps. Unfortunately, due to technical difficulties female meiosis in the hybrids has rarely been analysed. We are aware of only one such study31, which found that female hybrids between Mus musculus and M. domesticus were fertile, even though their oocytes displayed the same pairing abnormalities as male hybrids - with half the frequency, though.
To make insight into the cytological basis of hybrid sterility, we examined chromosome synapsis and recombination in male and female sterile hybrids between two sibling species of the grey vole, Microtus arvalis (dams) and M. levis (sires). These species diverged from 0.5 to 4.3 MYA32,33, differ by a series of chromosomal rearrangements33,34, yet remain morphologically indistinguishable35. In nature, hybrids occur at the zone of sympatry in the Urals, and can easily be produced in laboratory settings35,36,37,38,39. Reciprocal hybrids between M. arvalis and M. levis are completely sterile; however, males differ from females in the stage of reproductive collapse38. In males, testis mass is severely reduced and no sperm is found in the epididymis. Male meiosis arrests at prophase I but occasional nuclei reach diakinesis-metaphase I, where they mainly show univalents. More advanced stages of spermatogenesis have not been detected40. In female hybrids, oocyte growth and development was described as normal. However, follicular atresia was detected in hybrids even at the primordial follicle stage, and, at all stages, was more pronounced than in females of the parental species. Hybrid females display abnormal ovulation without follicle wall breakage. Mature oocytes move into Graafian follicles where they undergo the second meiotic division. No corpus luteum was detected in hybrid females41. According to Gileva et al.37 backcross progeny may be produced very rarely, but in our hybridisation experiments none occurred. Thus, these hybrids provide a promising model for studying cellular mechanisms of male and female hybrid sterility.
We analysed key events of chromosome synapsis and recombination. DSBs were detected by immunolocalisation of RAD51, a marker for single-stranded DNA ends42, and γH2A.X, a phosphorylated form of histone H2A.X43. Polymerisation of the lateral elements of SC and the formation of its central element were visualised with antibodies to SYCP3 and SYCP143. The number and distribution of recombination events were estimated by immunolocalisation of MLH144. This analysis allowed us to detect meiotic aberrations leading to hybrid sterility.
Results
Parental species
Chromosome pairing and recombination in male M. arvalis and M. levis have been described previously45,46,47. Both species showed asynapsis of the X and Y chromosomes (Fig. 1a,b, Supplementary Fig. S1) which underwent meiotic sex chromosome inactivation and were labeled by γH2A.X antibodies (Fig. 1c, Fig. S1). This feature is characteristic of the entire arvalis lineage of the genus Microtus47. Asynapsis of autosomes at pachytene stage, when most chromosomes contained MLH1 foci, was very rare (less than 2% in both sexes) and always affected small chromosomes. M. arvalis and M. levis males did not differ from each other in total autosomal SC length (t212,1 = 2.6, P = 0.09) or MLH1 focus number per cell (t212,1 = 1.1, P = 0.29) (Table 1). The number of MLH1 foci was usually restricted to one per autosomal bivalent in spermatocytes, as was in oocytes (Fig. 1d, Table 1). Assuming at least one focus on the XX bivalent, no significant sex differences in recombination rates can therefore be seen in M. levis.
Figure 1.
Pachytene spermatocytes of M. levis (a,c) and M. аrvalis (b) and an oocyte of M. levis (d). Arrowheads indicate unpaired sex chromosomes. Bar – 5 μm.
Table 1. The total length of SC and the number of MLH1 foci (mean ± S.D.) per cell in M. arvalis, M. levis and their F1 hybrids.
| Genotype | n | SC length, μm | n | MLH1 foci number |
|---|---|---|---|---|
| M. arvalis, males | 150 | 144.7 ± 14.8 | 150 | 27.6 ± 1.2 |
| M. levis, males | 64 | 150.9 ± 17.6 | 64 | 27.8 ± 1.4 |
| M. levis, females | 50 | 163.6 ± 21.2 | 58 | 28.8 ± 1.4 |
| F1 hybrids, males | 30 | 170.4 ± 24.2 | 50 | 0 |
| F1 hybrids, females | 30 | 168.7 ± 23.5 | 40 | 24.6 ± 2.9 |
n - number of cells examined.
By contrast, a drastic sex difference in chromosome behavior is observed in hybrids, in which oocytes show almost normal progression until late pachytene while most spermatocytes are arrested at leptotene.
Female hybrids
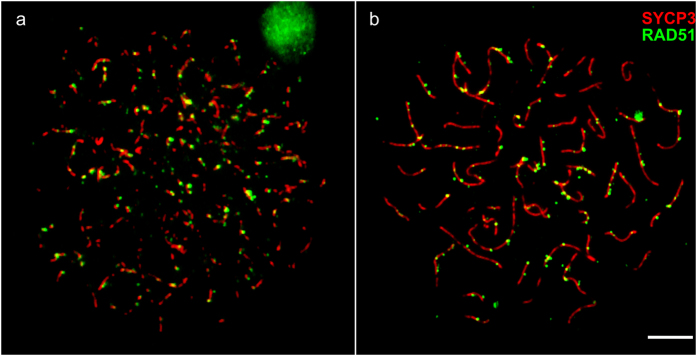
Leptotene oocytes of hybrids appeared normal with assembling lateral elements of SC labeled by RAD51 (Fig. 2a). At zygotene, lateral elements established contacts with each other while the central elements formed asynchronously. Thus, while some lateral elements were already paired, others displayed extensive asynapsis. Asynaptic regions were often intertwined and labeled by RAD51 and γH2A.X (Fig. 2b,c, Fig. S2). Some regions kept these marks until pachytene when most autosomes were synapsed (Fig. S2).
Figure 2.
Leptotene (a) and zygotene (b,c) oocytes of F1 hybrids between M. аrvalis and M. levis. Bar – 5 μm. (a) Assembling lateral elements of the SC (revealed by SYCP3 antibodies) accompanied by extensive RAD51 labeling. (b,c) Asynchronous formation of central elements of the SC. At completely pared SC blue signal of SYCP3 is co-localised with red signal of SYCP1. Regions with delayed synapsis are marked with RAD51 and γH2A.X antibodies.
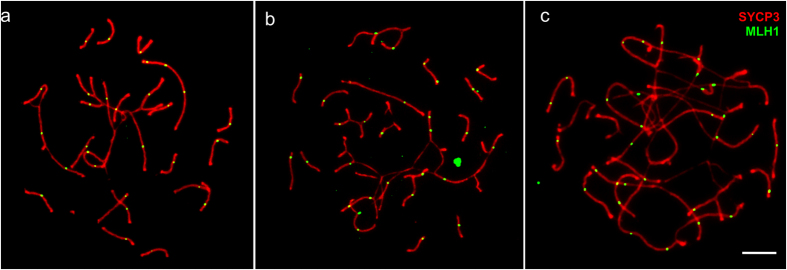
Pachytene oocytes were very variable in their appearance, containing asynaptic and heterosynaptic configurations. However, about 10% of oocytes had 23 “correctly” paired configurations, with four large trivalents, twelve heteromorphic and seven homomorphic bivalents (six acrocentric and one metacentric) (Fig. 3a). The heteromorphic bivalents comprised the X pair with long arms of different length and misaligned centromeres, and one large and ten smaller bivalents with the two centromere signals shifted. This is in a good agreement with pairing expected from comparative metaphase analysis of the parents (Fig. 3b). Parental karyotypes differ by at least four tandem fusions (producing trivalents in F1 hybrids), one paracentric and six pericentric inversions (which may produce inversion loops or straight bivalents with misaligned centromeres), four to seven putative centric transpositions (producing bivalents with misaligned centromeres), and an insertion of a large heterochromatic block in the X of M. levis33,34.
Figure 3.
A microphotograph (a) and a schematic (b) of a completely paired SC complement in a pachytene oocyte of an F1 hybrid between M. arvalis and M. levis. Bar – 5 μm. (a) Red and blue layers are slightly shifted in the merged image to show complete co-localisation of SYCP1 (central element) and SYCP3 (lateral elements of SC) in all synaptic configurations, except the heterochromatic block of the M. levis X chromosome. (b) Diagram of M. arvalis- (red) and M. levis- (blue) derived lateral elements, suggested by comparative metaphase analysis of the parental karyotypes. Arrows show bivalents with centromere signals shifted from each other, and an arrowhead shows a bivalent with centromere signals at both ends.
Although good correspondence between expected and observed synaptic configurations was found, there were two discordances. One heteromorphic bivalent showed centromere signals on its both ends (Fig. 3b, arrowhead). A trivalent comprising metacentric chromosome 1 of M. arvalis and twin acrocentrics of M. levis contained three misaligned centromeres. Lemskaya et al.34 proposed that chromosome 1 of M. arvalis resulted from tandem fusion of the proximal end of chromosome 3 and the distal end of chromosome 2 of M. levis. However, the synaptic configurations observed in these F1 hybrids indicate that chromosome 1 of M. arvalis was generated by fusion of the distal ends of the acrocentrics followed by inactivation of their centromeres, and re-activation of the centromere at the point of fusion.
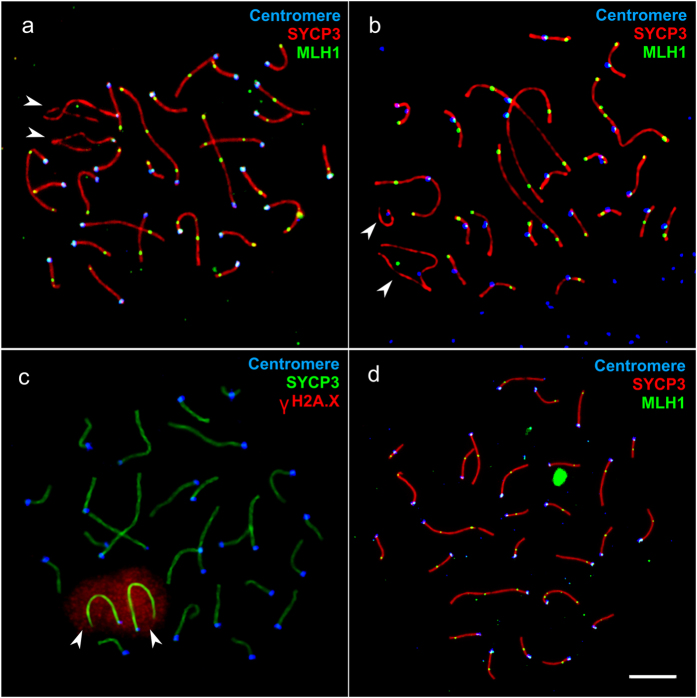
More than 90% of pachytene cells in female hybrids contained synaptic aberrations. Supplementary Fig. S2 shows examples of these aberrations. The most common were multivalents containing more than three lateral elements (from four to twenty, 6.5 ± 3.4 on average) (Figs 4 and 5). The number of multivalents varied from one to four per cell (1.7 ± 0.9 on average) and most multivalents were composed of large chromosomes. The probability for large metacentric chromosomes 1 to 4 of M. arvalis and the acrocentric pairs of M. levis to be involved in multivalent formation was 0.60 ± 0.06. This probability was also high for the X chromosomes (0.42 ± 0.11) and submetacentric chromosome 5 of M. arvalis with homologous acrocentric chromosome 1 of M. levis (0.32 ± 0.11). The probability for medium sized and small chromosomes, however, was very low (0.05 ± 0.01).
Figure 4. Synaptic aberrations in pachytene oocytes of F1 hybrids between M. аrvalis and M. levis.
Univalents (arrowheads) and unpaired regions in multivalents (arrows) contain unrepaired DSBs, revealed by antibodies to γH2A.X (a,b) and RAD51 (c,d). Bar – 5 μm.
Figure 5.
Immunolocalisation of MLH1 and SCs in pachytene oocytes of F1 hybrids between M. аrvalis and M. levis, containing one complex multivalent (a), several multivalents (b), and long asynaptic regions (c). Bar – 5 μm.
Univalents were also seen, varying in number from one to nine (3.0 ± 2.4 on average). In many cells odd numbers of univalents were found, indicating that some of their partners were involved in non-homologous synapsis in a multivalent. For the same reason the average number of observed trivalents was lower than the expected four (2.5 ± 1.4 on average). The number of bivalents in oocytes was lower than the expected 19 (15.4 ± 1.5 on average), again due to involvement of one or both partners in non-homologous synapsis in multivalents. Asynapsis was frequently observed around pairing partner switch points and at the ends of multivalent chains. Unpaired regions were labeled with γH2A.X (Fig. 4a,b, Fig. S2) and RAD51 (Fig. 4c,d, Fig. S2), indicating that they contained unrepaired DSBs.
Figure 5 and Fig. S2 show immunolocalisation of MLH1 at the SCs in pachytene oocytes of F1 hybrids between M. аrvalis and M. levis. The mean number of MLH1 foci was significantly lower in hybrid oocytes than in oocytes of M. levis (t96,1 = 9.6, P < 0.001: Table 1), while there was no difference in total SC length (t78,1 = 1.0, P = 0.32: Table 1). The decrease in hybrids was mainly due to a lack of foci in univalents. There was no substantial decrease in recombination between the homologous segments in homomorphic and heteromorphic bivalents, trivalents or multivalents. Most paired arms had at least one MLH1 focus. Even in the cells containing very complex multivalents (Fig. 5a), a series of multivalents (Fig. 5b), or long asynaptic regions (Fig. 5c) we did not observed a substantial decrease in the number of MLH1 foci.
Male hybrids
Male meiosis in hybrids was generally arrested at leptotene. The majority of spermatocytes carried only fragments of the axial elements (Fig. 6a, Fig. S2), indicating incomplete assembly. The most advanced spermatocytes contained almost normal axial elements, but even in these cells no initiation of synapsis was seen (Fig. 6b). The halved total length of the lateral elements in the male hybrids at this stage was about 10% longer than the total SC length of the male M. levis at pachytene (t92,1 = 4.4, P < 0.001: Table 1). Hybrid spermatocytes showed heavy labeling with RAD51 antibodies indicating DSB presence.
Figure 6. Leptotene spermatocytes of F1 hybrids between M. аrvalis and M. levis.
Bar – 5 μm. (a) A typical spermatocyte with incompletely assembled axial elements. (b) One of the most advanced spermatocytes with almost completely assembled axial elements.
Discussion
This study supports breeding records and histological assessments showing the sterility of hybrids between M. arvalis and M. levis35,36,37,38,39,41. Hybrids of both sexes are sterile; however, males, in accordance with Haldane’s rule, are “more sterile” than females. Male meiosis is uniformly arrested at leptotene, while female meiosis is affected at pachytene with wide variation between oocytes in the degree of disturbances of chromosome pairing. This might be due to the sex difference in degree of methylation on the meiotic onset. Studies in mice demonstrated that female germ cells enter meiosis in a demethylated state, while the genome of male germ cells is heavily remethylated after mitotic arrest at the beginning of meiosis48,49,50. A faster evolution of male hybrid sterility in the grey voles might also be accelerated by constitutive asynapsis and total lack of recombination between XY in this phylogenetic lineage47.
Several features of meiotic disturbances in female hybrids between M. arvalis and M. levis also characterise male meiosis in many cases of male-only hybrid sterility. The zygotene-pachytene transition is mainly affected, asynapsis and heterosynapsis occur in both heteromorphic and homomorphic chromosomes, while cells vary widely in the degree of disturbances and the number of chromosomes affected11,12,51,52,53.
Despite the sex differences in the stage and degree of meiotic disturbances, the cause of these disturbances is probably the same. This is failure of a homologous synapsis between homologous chromosome regions, which is complete in males and partial and sporadic in females. The majority of oocytes contains multiple regions of asynapsis and non-homologous synapsis. The establishment and extension of non-homologous synapsis leads to multivalent and univalent formation. Thus, when one homologue is involved in a multivalent, the other either remains univalent or pairs non-homologously with another chromosome region in the same or a different multivalent.
Homologous pairing involves several steps: preliminary DSB-independent pairing, DSB-mediated homology search with formation of interhomologue interactions, and elimination of unwanted contacts between nonhomologous chromosomes1.
DSB-independent pairing probably depends on associations of centromeres and/or telomeres at early prophase. It restricts the searching area for homologous recognition and alignment2. This process may be impaired in grey vole hybrids, which are heterozygous for at least 15 chromosomal rearrangements33,34. Although heterozygosity for a single fusion, pericentric inversion or centromeric shift does not usually lead to pairing abnormalities45,54,55,56, a high number of such heterozygosities may impede or delay presynaptic alignment of homologous regions57. It has been shown that multiple heterozygosity for a series of Robertsonian translocations in mice alters the nuclear architecture characteristic of the telocentric karyotype, changes patterns of centromere clustering and other interactions between chromosomal domains and leads to ectopic associations58. Similar problems in centromere clustering should occur in multiple heterozygotes for pericentric inversions and centromeric shifts.
Insufficient pre-DSB coalignment in the hybrids may enhance the probability of entanglements between chromosomes and increase the occurrence of ectopic recombinational intermediates between nearly homologous sites of nonhomologous chromosomes. Because the sequence homology matching in these intermediates is apparently low, they should be less stable. The unwanted DNA connections tend to be eliminated by the mismatch repair system59 and active chromosome movements62. We propose that the balance in stability between “wanted” and “unwanted” connections is impaired in hybrids, due to a decrease of sequence homology between the homologous regions of the parental species. This leads to variability between oocytes in the number and size of regions involved in multivalents.
These requirements of normal synapsis between homologous chromosomes are apparently not met in the hybrids. Multiple heterozygosity for chromosomal rearrangements hinders preliminary DSB-independent pairing between homologous chromosomes and increases the incidence of non-homologous associations. Divergence between parental genomes decreases homology at the sequence level, the stability of homologous heteroduplexes, and affects the efficiency of discrimination between correct and ectopic interhomologue interactions. In hybrids, the wide variation between genetically and chromosomally identical oocytes in the ratio of homologously paired to non-homologously paired regions indicates that the choice between homologous and non-homologous synapsis at each of these steps is random. Detailed molecular mechanisms of these processes remain to be elucidated, and interspecies hybrids, such as those reported here, provide an excellent model for future studies.
The sex difference observed in this study can be categorised as an example of “graduated steps of sterility”30 from advanced in females to complete in males. Genetic and chromosomal incompatibility is probably amplified in the male hybrids by the well known vulnerability of spermatogenesis to pairing aberrations28. We observed stochastic variation in “degree of sterility” even between oocytes of the same F1 genotype. Those containing “correctly” paired configurations are probably able to produce viable balanced oocytes. The more asynapsed regions an oocyte contain, the larger part of its genome undergoes meiotic silencing of unsynapsed chromatin (MSUC)64,65, and the higher the chance for the oocyte to be directed to apoptosis. The results of this study indicate that reproductive isolation based on hybrid sterility may be built up in a gradual mode. A gradual genetic divergence and the sequential fixation of different chromosome rearrangements in isolated populations increase the probability of pairing errors followed by MSUC and apoptosis in the hybrid gametocytes.
Materials and Methods
Seven adult male and 12 newborn female hybrids between M. arvalis (dams) and M. levis (sires) were examined, as well as three adult male M. arvalis, four adult male and three newborn female M. levis. Captive-bred colonies of the parental species were established from individuals trapped in Leningrad district (M. arvalis) and Novosibirsk district (M. levis) and maintained in the animal house of the Institute of Cytology and Genetics. Maintenance, handling and euthanasia of animals followed protocols approved by the Animal Care and Use Committee of the Institute of Cytology and Genetics. Experiments described in this manuscript were carried out in accordance with the approved national guidelines for the care and use of laboratory animals.
Chromosome spreads were prepared from spermatocytes or embryonic oocytes according to Peters et al.66. Cell spreads were treated as described in Anderson et al.44 using rabbit polyclonal anti-SYCP3 (1:500; Abcam), mouse monoclonal anti-SYCP3 (1:100; Abcam), rabbit polyclonal anti-SYCP1 (1:500; Abcam), mouse monoclonal anti-MLH1 (1:50; Abcam), rabbit polyclonal anti-RAD51 (1:200; Calbiochem), mouse monoclonal anti-γH2A.X (1:500; Abcam), rabbit polyclonal anti-γH2A.X (1:500; Abcam) and human anticentromere (ACA) (1:100; Antibodies Inc) primary antibodies. The secondary antibodies used were Cy3-conjugated goat anti-rabbit (1:500; Jackson ImmunoResearch), Alexa450-conjugated goat anti-rabbit (1:100; Invitrogen), FITC-conjugated donkey anti-rabbit (1:200; Jackson ImmunoResearch), FITC-conjugated goat anti-mouse (1:50; Jackson ImmunoResearch), AMCA-conjugated donkey anti-human (1:100; Jackson ImmunoResearch), and Cy3-conjugated goat anti-human (1:100; Jackson ImmunoResearch) antibodies. Antibodies were diluted in PBT (3% bovine serum albumin and 0.05% Tween 20 in phosphate-buffered saline). A solution of 10% PBT was used for blocking. Primary antibody incubations were performed overnight in a humid chamber at 37 °C; and secondary antibody incubations, for 1 h at 37 °C. Slides were mounted in Vectashield antifade mounting medium (Vector Laboratories) to reduce fluorescence fading.
Preparations were visualised with an Axioplan 2 microscope (Carl Zeiss) equipped with a CCD camera (CV M300, JAI), CHROMA filter sets and an ISIS4 image processing package (MetaSystems GmbH).
Centromeres were identified by ACA foci. MLH1 signals were scored only if they were localised on the SC. In the parental species the length of the SC of all bivalents was measured in micrometers using MicroMeasure 3.367. To estimate the total SC length in the hybrids, whose pachytene cells contained partially or completely unpaired chromosomes, we measured the lateral elements of SC and then divided the sum by two.
Statistica 6.0 software package (StatSoft, Tulsa, OK, USA) was used for descriptive statistics. For the comparisons of SC length and MLH1 foci number between parental species and their F1 hybrids, Students’ t-tests (two-sided) were performed.
Additional Information
How to cite this article: Torgasheva, A. A. and Borodin, P. M. Cytological basis of sterility in male and female hybrids between sibling species of grey voles Microtus arvalis and M. levis. Sci. Rep. 6, 36564; doi: 10.1038/srep36564 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Ms. Antonina Zhelezova for breeding the hybrids, Mrs. Maria Rodionova and Mrs. Olesya Klimova for help in chromosome preparations, Prof. John Parker for editing the manuscript, and the Microscopic Center of the Siberian Branch of the Russian Academy of Sciences for granting access to microscopic equipment. This work was supported by the Federal Agency of Scientific Organisations via the Institute of Cytology and Genetics (project # 0324-2015-0003).
Footnotes
Author Contributions A.T. and P.B. analysed all of the data in this study and wrote the manuscript; A.T. performed all of the experiments, P.B. conceived and designed the study.
References
- Zickler D. & Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harbor perspectives in biology 7, 10.1101/cshperspect.a016626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. & Kleckner N. A few of our favorite things: Pairing, the bouquet, crossover interference and evolution of meiosis. Seminars in cell & developmental biology 54, 135–148, 10.1016/j.semcdb.2016.02.024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benirschke K. Sterility and fertility of interspecific mammalian hybrids. (Springer, 1967). [Google Scholar]
- Gray A. P. Mammalian hybrids. A check-list with bibliography. Technical Communications. Commonwealth Bureau of Animal Breeding and Genetics (1972). [Google Scholar]
- Flachs P. et al. Prdm9 incompatibility controls oligospermia and delayed fertility but no selfish transmission in mouse intersubspecific hybrids. PloS one 9, e95806 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O., Trachtulec Z., Vlcek C., Schimenti J. C. & Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323, 373–375, 10.1126/science.1163601 (2009). [DOI] [PubMed] [Google Scholar]
- Oka A. et al. Disruption of genetic interaction between two autosomal regions and the x chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics 175, 185–197, 10.1534/genetics.106.062976 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A. & Shiroishi T. Regulatory divergence of X-linked genes and hybrid male sterility in mice. Genes and Genetic Systems 89, 99–108 (2014). [DOI] [PubMed] [Google Scholar]
- Davis B. W. et al. Mechanisms underlying mammalian hybrid sterility in two feline interspecies models. Molecular biology and evolution 32, 2534–2546, 10.1093/molbev/msv124 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver P. L., Goodstadt L., Bayes J. J., Birtle Z. & Roach K. C. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS genetics 5, e1000753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodin P., Barreiros-Gomez S., Zhelezova A., Bonvicino C. & D’Andrea P. Reproductive isolation due to the genetic incompatibilities between Thrichomys pachyurus and two subspecies of Thrichomys apereoides (Rodentia, Echimyidae). Genome 49, 159–167 (2006). [DOI] [PubMed] [Google Scholar]
- Borodin P. M., Rogatcheva M. B., Zhelezova A. I. & Oda S. Chromosome pairing in inter-racial hybrids of the house musk shrew (Suncus murinus, Insectivora, Soricidae). Genome 41, 79–90 (1998). [DOI] [PubMed] [Google Scholar]
- Castiglia R. & Capanna E. Contact zone between chromosomal races of Mus musculus domesticus. 2. Fertility and segregation in laboratory-reared and wild mice heterozygous for multiple Robertsonian rearrangements. Heredity 85, 147–156 (2000). [DOI] [PubMed] [Google Scholar]
- Chandley A., Jones R., Dott H., Allen W. & Short R. Meiosis in interspecific equine hybrids. Cytogenetic and genome research 13, 330–341 (1974). [DOI] [PubMed] [Google Scholar]
- Chandley A., Short R. & Allen W. Cytogenetic studies of three equine hybrids. Journal of reproduction and fertility. Supplement, 356–370 (1975). [PubMed] [Google Scholar]
- Hauffe H. C. & Searle J. B. Chromosomal heterozygosity and fertility in house mice (Mus musculus domesticus) from Northern Italy. Genetics 150, 1143–1154 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishishita S., Tsuboi K., Ohishi N., Tsuchiya K. & Matsuda Y. Abnormal pairing of X and Y sex chromosomes during meiosis I in interspecific hybrids of Phodopus campbelli and P. sungorus. Scientific reports 5, 9435, 10.1038/srep09435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. K. et al. Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proceedings of the National Academy of Sciences 111, 13415–13420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong E. & Fan G. The variety of sterility and gradual progression to fertility in hybrids of the horse and donkey. Heredity 62, 393–406 (1989). [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S. Sex ratio and unisexual sterility in hybrid animals. Journal of Genetics 12, 101–109 (1922). [Google Scholar]
- Coyne J. A. The genetic basis of Haldane’s rule. Nature 314, 736–738, 10.1038/314736a0 (1985). [DOI] [PubMed] [Google Scholar]
- Hurst L. D. & Pomiankowski A. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics 128, 841–858 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A. Haldane’s rule has multiple genetic causes. Nature 361, 532–533, 10.1038/361532a0 (1993). [DOI] [PubMed] [Google Scholar]
- Orr H. A. & Turelli M. Dominance and Haldane’s rule. Genetics 143, 613–616 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. I. & Davis A. W. Evolution of postmating reproductive isolation: the composite nature of Haldane’s rule and its genetic bases. American naturalist 142, 187–212, 10.1086/285534 (1993). [DOI] [PubMed] [Google Scholar]
- Turelli M. & Orr H. A. The dominance theory of Haldane’s rule. Genetics 140, 389–402 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. I., Johnson N. A. & Palopoli M. F. Haldane’s rule and its legacy: Why are there so many sterile males? Trends in ecology & evolution 11, 281–284, 10.1016/0169-5347(96)10033-1 (1996). [DOI] [PubMed] [Google Scholar]
- Hunt P. A. & Hassold T. J. Sex matters in meiosis. Science 296, 2181–2183 (2002). [DOI] [PubMed] [Google Scholar]
- Coyne J. A. & Orr H. A. Speciation. (Sinauer Associates, 2004). [Google Scholar]
- Darwin C. The origin of species. (Oxford University Press, 1996). [Google Scholar]
- Bhattacharyya T. et al. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proceedings of the National Academy of Sciences 110, E468–E477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz S. A., Bininda-Emonds O. R. & Purvis A. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecology letters 12, 538–549, 10.1111/j.1461-0248.2009.01307.x (2009). [DOI] [PubMed] [Google Scholar]
- Mazurok N. A. et al. Comparative chromosome and mitochondrial DNA analyses and phylogenetic relationships within common voles (Microtus, Arvicolidae). Chromosome research 9, 107–120 (2001). [DOI] [PubMed] [Google Scholar]
- Lemskaya N. A. et al. Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). III. Karyotype relationships of ten Microtus species. Chromosome research 18, 459–471 (2010). [DOI] [PubMed] [Google Scholar]
- Malygin V. & Sablina O. In Common vole: sibling species (eds Sokolov V. E. & Bashenina N. V.) 7–25 (Nauka, 1994). [Google Scholar]
- Gileva E. A., Bolshakov V. N., Polyavina O. V. & Cheprakov M. I. Microtus arvalis and M. rossiaemeridionalis in the Urals: hybridization in the wild. Dokl Biol Sci 370, 134–137 (2000). [PubMed] [Google Scholar]
- Gileva E. A., Bolshakov V. N. & Yalkovskaya L. E. Voles of the Microtus arvalis group in zones of ecological risk: Interspecies hybridization. Dokl Biol Sci 381, 567–569 (2001). [DOI] [PubMed] [Google Scholar]
- Meyer M. In Common vole: sibling species (eds Sokolov V. E. & Bashenina N. V.) 26–32 (Nauka, 1994). [Google Scholar]
- Meyer M. N., Golenishchev F. N., Radjably S. I. & Sablina O. V. Voles (subgenus Microtus Schrank) of Russia and adjacent territories. (Zoological Institute of RAS, 1996). [Google Scholar]
- Meyer M., Radjabli S., Bulativa N. & Golenishchev F. Karyological features and possible relations of the voles of “arvalis” group (Rodentia, Cricetidae). Zoologitcheskii zhurnal 64, 417–428 (1985). [Google Scholar]
- Zybina E. & Sukhol E. Peculiarities of oogenesis in the hybrids betweeen sibling species of the grey vole. Tsytologia 14, 433–437 (1972). [PubMed] [Google Scholar]
- MacQueen A. J. Catching a (double-strand) break: The RAD51 and DMC1 strand exchange proteins can co-occupy both ends of a meiotic DNA double-strand break. PLoS genetics 11, e1005741, 10.1371/journal.pgen.1005741 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turinetto V. & Giachino C. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res 43, 2489–2498, 10.1093/nar/gkv061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. K., Reeves A., Webb L. M. & Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localisation of MLH1 protein. Genetics 151, 1569–1579 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basheva E. A., Torgasheva A. A., Golenischev F. N., Frisman L. V. & Borodin P. M. Chromosome synapsis and recombination in the hybrids between chromosome races of the common vole Microtus aravalis: “arvalis” and “obscurus”. Dokl Biol Sci 456, 206–208, 10.1134/S0012496614030144 (2014). [DOI] [PubMed] [Google Scholar]
- Borodin P. M. et al. Immunofluorescence and electron microscopic analysis of meiotic chromosome pairing and recombination in four species of voles (genus Microtus; Arvicolinae, Rodentia). Vavilovskii zhurnal genetiki i selektsii 14, 89–95 (2010). [Google Scholar]
- Borodin P. M. et al. Multiple independent evolutionary losses of XY pairing at meiosis in the grey voles. Chromosome research 20, 259–268, 10.1007/s10577-011-9261-0 (2012). [DOI] [PubMed] [Google Scholar]
- Popp C. et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463, 1101–1105 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y., Lees-Murdock D. J., Xu G.-L. & Walsh C. P. Timing of establishment of paternal methylation imprints in the mouse. Genomics 84, 952–960 (2004). [DOI] [PubMed] [Google Scholar]
- Kelly T. & Trasler J. Reproductive epigenetics. Clinical genetics 65, 247–260 (2004). [DOI] [PubMed] [Google Scholar]
- Forejt J. Hybrid sterility in the mouse. Trends in genetics 12, 412–417 (1996). [DOI] [PubMed] [Google Scholar]
- Forejt J. & Gregorova S. Meiotic studies of translocations causing male sterility in the mouse. I. Autosomal reciprocal translocations. Cytogenetics and cell genetics 19, 159–179 (1977). [DOI] [PubMed] [Google Scholar]
- Forejt J. & Ivanyi P. Genetic studies on male sterility of hybrids between laboratory and wild mice (Mus musculus L.). Genetical research 24, 189–206 (1974). [DOI] [PubMed] [Google Scholar]
- Basheva E. A. et al. Chromosome synapsis and recombination in simple and complex chromosomal heterozygotes of tuco-tuco (Ctenomys talarum: Rodentia: Ctenomyidae). Chromosome research 22, 351–363, 10.1007/s10577-014-9429-5 (2014). [DOI] [PubMed] [Google Scholar]
- Everett C. A., Searle J. B. & Wallace B. M. A study of meiotic pairing, nondisjunction and germ cell death in laboratory mice carrying Robertsonian translocations. Genetical research 67, 239–247 (1996). [DOI] [PubMed] [Google Scholar]
- Mary N. et al. Meiotic recombination analyses in pigs carrying different balanced structural chromosomal rearrangements. PloS one 11, e0154635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garagna S., Page J., Fernandez-Donoso R., Zuccotti M. & Searle J. B. The Robertsonian phenomenon in the house mouse: mutation, meiosis and speciation. Chromosoma 123, 529–544, 10.1007/s00412-014-0477-6 (2014). [DOI] [PubMed] [Google Scholar]
- Berrios S. et al. Robertsonian chromosomes and the nuclear architecture of mouse meiotic prophase spermatocytes. Biological research 47, 1–16, 10.1186/0717-6287-47-16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borts R. H., Chambers S. & Abdullah M. F. The many faces of mismatch repair in meiosis. Mutation research/Fundamental and molecular mechanisms of mutagenesis 451, 129–150 (2000). [DOI] [PubMed] [Google Scholar]
- Hoffmann E. R. & Borts R. H. Meiotic recombination intermediates and mismatch repair proteins. Cytogenetics Genome Research 107, 232–248 (2004). [DOI] [PubMed] [Google Scholar]
- Radman M. & Wagner R. Mismatch recognition in chromosomal interactions and speciation. Chromosoma 102, 369–373 (1993). [DOI] [PubMed] [Google Scholar]
- Goldman A. S. & Lichten M. Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proceedings of the National Academy of Sciences 97, 9537–9542 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R. & Kleckner N. Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends in cell biology 19, 716–724 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. M., Mahadevaiah S. K., Ellis P. J., Mitchell M. J. & Burgoyne P. S. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell 10, 521–529 (2006). [DOI] [PubMed] [Google Scholar]
- Cloutier J. M., Mahadevaiah S. K., ElInati E., Tóth A. & Turner J. Mammalian meiotic silencing exhibits sexually dimorphic features. Chromosoma 125, 215–226, 10.1007/s00412-015-0568-z (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A. H., Plug A. W., van Vugt M. J. & de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome research 5, 66–68 (1997). [DOI] [PubMed] [Google Scholar]
- Reeves A. MicroMeasure: a new computer program for the collection and analysis of cytogenetic data. Genome 44, 439–443 (2001). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.