Abstract
Human parvovirus B19 is the only parvovirus known to be a human pathogen. The structure of recombinant B19-like particles has been determined to ≈3.5-Å resolution by x-ray crystallography and, to our knowledge, represents the first near-atomic structure of an Erythrovirus. The polypeptide fold of the major capsid protein VP2 is a “jelly roll” with a β-barrel motif similar to that found in many icosahedral viruses. The large loops connecting the strands of the β-barrel form surface features that differentiate B19 from other parvoviruses. Although B19 VP2 has only 26% sequence identity to VP3 of adeno-associated virus, 72% of the Cα atoms can be aligned structurally with a rms deviation of 1.8 Å. Both viruses require an integrin as a coreceptor, and conserved surface features suggest a common receptor-binding region.
Human parvovirus B19, a member of the Parvoviridae family, is a common human pathogen associated with a wide variety of diseases. Most commonly, it causes a mild childhood rash, Erythema infectiosum (1), but in some cases more serious symptoms can be linked to B19, such as acute or persistent arthropathies (2), critical failures of red cell production (3), Hydrops fetalis (4), fetal loss (5), myocarditis (6), or hepatitis (2).
Parvovirus B19 is currently the only accepted member of the genus Erythrovirus, which is characterized by a high tropism for erythroid progenitor cells (7). Nevertheless, some other human viruses have been discovered that can tentatively be considered to be erythroviruses. Human erythroviruses cluster into three genotypes, B19-, LaLi-, and V9-related viruses, based on nucleotide sequence comparisons (8). Simian parvovirus (SPV) (9), pig-tailed macaque parvovirus (PtPV), and rhesus macaque parvovirus (RhPV) (10) have also been proposed to be members of the Erythrovirus genus. Little is known about the molecular basis of infection and pathogenesis of these viruses. The glycosphingolipid globoside has been suggested to be the cellular receptor of B19 (11) but is not sufficient for viral infection (12). Recently, α5β1 integrin was described as a cellular coreceptor of B19 that is required for viral entry into human cells (13).
The single-stranded DNA genome of B19 is packaged into a nonenveloped, icosahedral protein shell of ≈280 Å in diameter (14). The capsid consists of 60 structural subunits, ≈95% of which are the major viral protein VP2 (58 kDa) (15). The other structural protein, VP1, differs from VP2 only in a N-terminal “unique region” composed of 227 additional amino acids, which are mostly located outside the virion and, therefore, accessible to antibody binding (16, 17).
The three-dimensional capsid structures of canine parvovirus (18, 19), feline panleukopenia virus (FPV) (20), minute virus of mice (21), Galleria mellonella densovirus (22), adeno-associated virus 2 (AAV-2) (23), and porcine parvovirus (24) were previously determined to near atomic resolution. An eight-stranded, antiparallel β-barrel (“jelly roll”) comprises most of the internal portion of the protein shell. The surface of the virion is formed by large insertions connecting the strands of the β-barrel, thereby creating features that govern antigenicity and receptor binding. The loop between the β-strands G and H and its symmetry-related equivalents form protrusions at or around the icosahedral threefold axes. Other common characteristics of parvoviral surfaces are depressions on the twofold axes and canyons surrounding the fivefold axes. In canine parvovirus, a glycine-rich motif near the N terminus of VP2 is located in the channel along the fivefold axes of the capsid (18, 19). The conservation of the glycine-rich motif in parvoviruses suggests its function for the externalization of the unique region at the icosahedral fivefold axes.
Cell culture systems that are susceptible for B19 infection are not suitable for a large-scale propagation of the virus. Therefore, it has been necessary to use recombinant, empty protein shells that are self-assembled from either VP2 or VP2 and VP1 for structural and biochemical studies. Recombinant capsids are morphologically and immunologically similar to infectious virions (25). Nevertheless, there are small differences in the immune response against recombinant VP2 capsids compared with particles that, like infectious virions, also contain some VP1 subunits (25, 26), suggesting that there may be minor structural differences between the structure reported here and the infectious virus. Previous studies on recombinant B19 VP2 particles by x-ray crystallography were limited to 8-Å resolution and showed differences on the viral surface, in particular on the threefold icosahedral axes, compared with other parvoviruses (14).
The neutralizing effect of antibodies can be achieved by interference of the antibody with the attachment of the virion to the cell surface or subsequent processes. The neutralization might be accomplished by physically blocking the receptor binding site or by an antibody-induced conformational change of the virus structure. The mapping of the antigenic surface of a virus, therefore, might indicate functional areas that govern receptor recognition, binding, or cell entry. Although the immune response against B19 is mainly elicited by the VP1 unique region (27), several antigenic target regions for neutralizing antibodies have been described for VP2 (26, 28–31). The major area of enhanced antigenicity corresponds to the C-terminal half of VP2, situated mostly around the threefold axes of the capsid.
We report here the three-dimensional structure of recombinant, empty parvovirus B19 VP2 particles at ≈3.5-Å resolution and show that, of the available parvoviral structures, it is structurally most similar to AAV-2. Although these viruses share a common host, their low sequence similarity suggests a host-independent evolution (32, 33). The structure of B19 will provide a basis for the mutational analysis of nonhuman primate erythroviruses that might help to elucidate the mechanisms of infection and pathogenesis of B19 in an animal model.
Methods
Recombinant VP2 capsids were purified from insect cells by a modification of the procedure described by Kajigaya et al. (25). The capsids were crystallized by using the hanging drop vapor diffusion technique. Aliquots (1.5 μl) of the purified capsid (9 mg·ml–1 in 10 mM Tris·HCl, pH 7.5) were mixed with an equal volume of reservoir solution (16–22% 2-methyl-2,4-pentanediol in 0.1 M sodium cacodylate buffer, pH 7.4), and the droplets were equilibrated against 750 μl of reservoir solution at 20°C. X-ray diffraction data were collected from frozen crystals at the Advanced Photon Source (Table 1). The diffraction images were integrated and scaled by using the hkl data processing system Version 1.97.3 (34). The crystals belonged to space group P213. Most of the several hundred crystals tested diffracted from 8- to 5-Å resolution with cell dimensions of ≈358 Å. Only a few crystals of one batch extended to higher resolution. One of these crystals gave weak diffraction to ≈3.5 Å and had unit cell dimensions of 351.4 Å. All reflections between 3.84 and 3.81 Å were omitted to avoid interference from ice crystal reflections (Table 1). The space group and crystal packing arrangement are essentially the same as previously observed on crystals that diffracted to only 8-Å resolution (14).
Table 1. Data collection and structure refinement.
| Data collection and processing | |
| Beam spot size, mm2 | 0.15 × 0.15 |
| Detector | marCCD 165 |
| Exposure time per frame | 5 min |
| No. of crystals (no. of frames) | 1 (50) |
| Oscillation angle, ° | 0.2 |
| Sample-to-detector distance, mm | 240 |
| Wavelength, Å | 1.107 |
| Space group (cell dimensions, Å) | P213 (a=351.4) |
| Mosaicity, ° | 0.18 |
| Resolution range processed (omitted range), Å | 20-3.5 (3.84-3.81) |
| No. of unique reflections | 123,626 |
| 〈I〉/〈σ(I)〉 | 7.21 (1.38)* |
| Rsym† (I > 0) | 0.098 (0.436)* |
| Redundancy (I > 0) | 1.6 |
| Completeness (I > 0) | 62.4% (52.6%)* |
| Completeness (I > 3σ) | 43.8% (12.8%)* |
| Structure refinement | |
| R factor (F > 0σ) | 0.309 (0.525)* |
| R factor (F > 3σ) | 0.262 (0.450)* |
Values in parentheses are for the highest-resolution shell (3.62-3.50 Å).
Rsym = Σ|(I - 〈I〉)|/Σ(I); σ cutoff 0.0 after partial addition.
The structure of the B19 VP2 particle was determined by standard procedures. An electron density map of recombinant B19 particles with 10-Å resolution obtained by cryoelectron microscopy (data not shown) was used as a model for molecular replacement. The structure was built by using FPV and AAV-2 as starting models with the o package (35) and refined with cns by using noncrystallographic symmetry constraints alternated with model rebuilding. The map was further improved by a translation, libration, and screw rotation refinement with the program refmac (36). The axis of greatest rotational displacement almost matched the threefold crystallographic axis. This rotational disorder is probably the cause of the anisotropic smearing that had been observed in the electron density map. The final R factors for the model are shown in Table 1. Of the non-glycine, non-proline residues, 95.8% were within the limits of the generously allowed regions of the Ramachandran plot. The bond lengths and angles deviated from idealized values by 0.01 Å and 1.9°, respectively.
Results
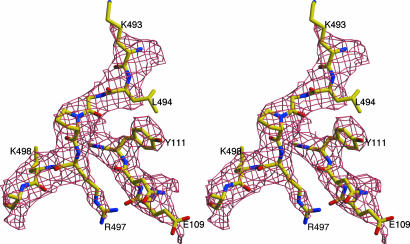
The crystal structure of parvovirus B19, to our knowledge the first near-atomic structure of an Erythrovirus, has been solved by using data to 3.5-Å resolution (Table 1). The high degree of noncrystallographic redundancy helped to produce an interpretable electron density map for the protein shell (Fig. 1), although the data beyond 3.7-Å resolution were weak. Most of the 554 residues of the B19 VP2 capsid protein were traceable in the electron density, except for 18 N-terminal residues and 13 amino acids (amino acids 301–313) that form the tip of a surface loop adjacent to a threefold icosahedral axis. Regions with the highest temperature factors coincide with exposed loops (Fig. 2). Additional ordered density was found close to the internal surface of the capsid, which can be interpreted as being partially ordered nonviral DNA, but the density is not sufficiently well defined to permit tracing of the nucleic acid backbone. This density is in contact with the protein shell in the vicinity of the threefold axes, where the inner particle surface is positively charged.
Fig. 1.
Stereodiagram of a section of the B19 electron density and its structural interpretation for residues comprising amino acids 493–499 and amino acids 109–111.
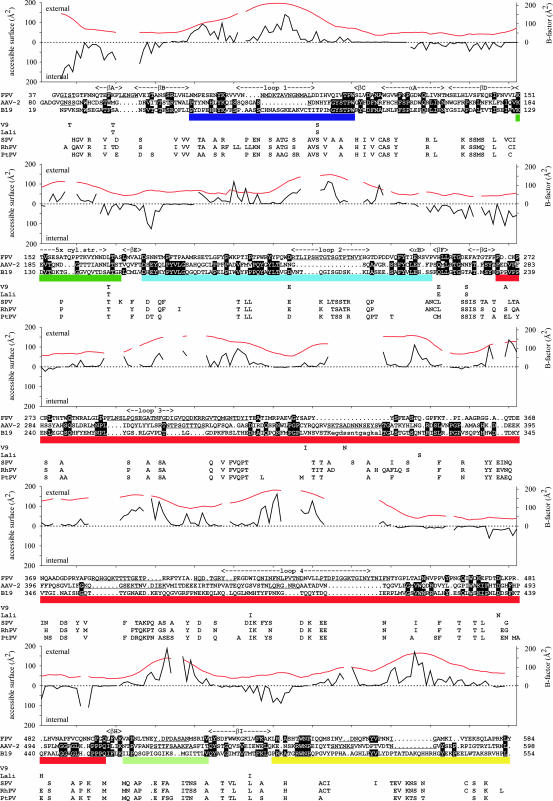
Fig. 2.
Structural alignment of B19 VP2 with the capsid proteins of FPV (PDB ID code 1C8F) and AAV-2 (PDB ID code 1LP3). Secondary structural elements of FPV are labeled above the sequence alignment. Aligned identical residues are white with a black background. Residues of AAV-2 or FPV that could not be structurally equivalenced to B19 are underlined. The bars below the sequence alignment highlight the surface loops, with colors according to Fig. 3a. Plotted above the sequence alignments are the external and internal accessible surface area (black) for the capsid. Shown also is the averaged temperature factor for each residue (red). Additionally, amino acids that are not identical to the B19 VP2 sequence are shown for the major capsid proteins of nonhuman primate erythroviruses SPV (accession no. AAA74975), RhPV (accession no. AAF61211), and PtPV (accession no. AAF61214) and human erythroviruses LaLi (accession no. AAK95571) and V9 (accession no. NP_694865).
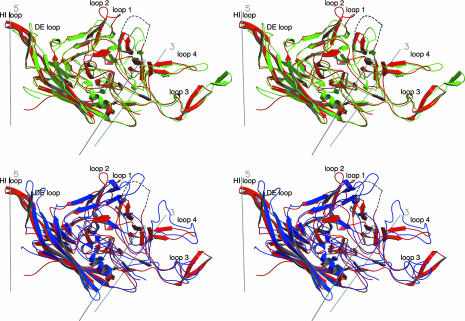
The B19 VP2 structure contains an eight-stranded, antiparallel β-barrel consisting of two β-sheets in the standard BIDG and CHEF arrangement (37) (Fig. 3a). This motif is in approximately the same position relative to the icosahedral symmetry axes as in other mammalian parvoviruses. About two-thirds of the parvoviral structure are made up of insertions connecting the strands of the β-barrel. The capsid appears to be stabilized by the interlocking of neighboring VP2 molecules, where loops 3 and 4 of the GH loop of one subunit are closely intertwined with loop 1 in the BC loop and a portion of the GH loop of a threefold symmetry-related subunit (see Fig. 3a for nomenclature). Although most of the C-terminal 50 residues of VP2 are accessible on the particle surface, the final four amino acids are buried within the protein shell, close to the inner capsid surface. The N terminus of the ordered VP2 structure (amino acid 19) is located on the inside of the capsid, close to a fivefold axis. The elaborate interstrand loops contain secondary structural elements, which comprise the viral surface (Fig. 3b). Compared with all other known parvovirus structures, B19 has the closest structural similarity to AAV-2, a Dependovirus that also replicates in human cells (Figs. 2 and 4). The backbone of B19 (523 ordered residues) superimposes onto AAV-2 (519 ordered residues) with a rms deviation (RMSD) of 1.8 Å for 400 equivalenced Cα atoms (or 77% of the structure), whereas the RMSD for FPV (548 ordered residues) is 2.0 Å for 324 Cα atoms (or 62% of the structure). The structural similarity of B19 to AAV-2 is especially apparent in that the structure of the large insertion (209 ordered residues), between strands G and H of the β-barrel, superimposes with an RMSD of 2.0 Å for 168 Cα atoms (80%) in the case of AAV-2 and 2.2 Å for 102 Cα atoms (49%) for FPV. Between AAV-2 and B19, 28% of the structurally aligned residues are identical, whereas only 17% are identical between FPV and B19 (Fig. 2).
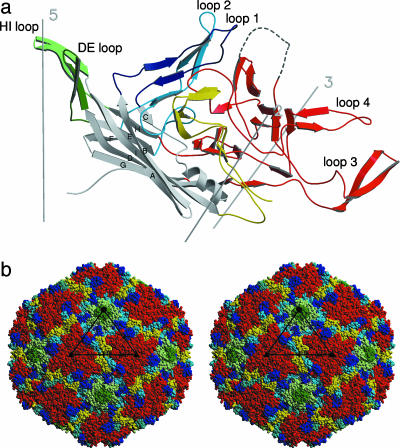
Fig. 3.
Secondary structure of parvovirus B19. (a) Ribbon diagram of VP2. The strands of the β-barrel (gray) are labeled sequentially A through I. The surface loops connecting the strands of the β-barrel are labeled by color: dark blue, BC loop; dark green, DE loop; light blue, EF loop; red, GH loop; light green, HI loop; yellow, C-terminal amino acids. The position of the disordered loop (amino acids 301–313) (dashed line) was modeled based on the corresponding AAV-2 loop. There is a RMSD of 0.3 Å between the 13 Cα atoms of AAV-2 and the modeled B19 structure, although the loop as a whole has been translated and rotated. (b) Surface topography of B19. The surface loops are color-coded according to the ribbon diagram. The disordered loop has been omitted.
Fig. 4.
Comparison of the secondary structure of parvovirus B19 (red) with AAV-2 (green) and FPV (blue). The structures in the stereodiagrams are plotted in the same orientation as in Fig. 3a.
The most obvious difference between the capsid surface of parvovirus B19 and that of FPV is the lack of prominent spikes at the icosahedral threefold axes. The protrusions adjacent to the depression at the threefold axis are smaller in B19 than in AAV-2, but their positions are the same (“threefold-proximal” peaks). The area around the threefold axes of the capsid is formed by the large loop between βG and βH (amino acids 235–454) and its symmetry-related equivalents (Fig. 3b). In AAV-2, a threefold-proximal peak is formed by a subloop of the GH loop (AAV-2 amino acids 348–379 and B19 amino acids 296–328) sandwiched by loop 3 (AAV-2 amino acids 293–341 and B19 amino acids 249–289) and loop 4 (AAV-2 amino acids 440–458 and B19 amino acids 388–403) of a symmetry-related molecule. The lack of prominent protrusions in B19 is mostly due to the shortening of loops 3 and 4 compared with AAV-2. The subloop of amino acids 296–328 in B19 and its equivalent in AAV-2 contains an insertion of 19 amino acids relative to FPV. The ends of this insertion are traceable in B19, but 13 amino acids at the tip of the subloop are disordered (Figs. 4 and 5a). In FPV, an insertion (amino acids 433–448 of FPV) in loop 4 forms the tip of the threefold spike. Because of this missing subloop in B19 and AAV-2, there is a small depression at the threefold axes. The base of the threefold structure is composed of the C-terminal portion of the GH loop (B19 amino acids 410–454), which is mostly conserved and structurally similar in all three viruses.
Fig. 5.
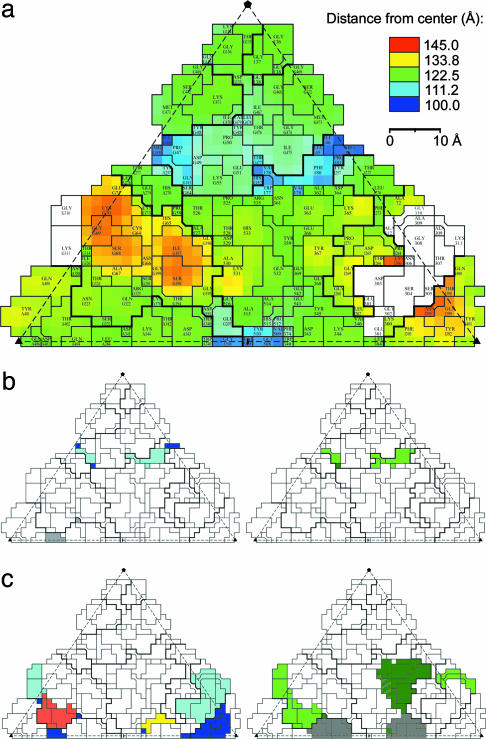
Road maps of the B19 capsid. (a) Surface colored according to distance from viral center. The disordered loop (amino acids 301–313) was modeled based on the corresponding AAV-2 loop and is colored white. (b) Overlap between regions corresponding to the epitope of the neutralizing anti-AAV-2 antibody A20 and conserved regions between B19 and AAV-2. (Left) B19 residues equivalent to the position of the conformational epitope of A20 in AAV-2 (41) are shown in color: dark blue, A20–1 (B19 amino acids 81–88); light blue, A20–2 (B19 amino acids 178–187); yellow, A20–4 (B19 amino acids 378–386). (Right) The conserved regions between B19 and AAV-2 are shown in color: dark green, amino acids 82–87; light green, amino acids 168–188. (c) Overlap between regions corresponding to AAV-2 heparan-binding sites and B19 neutralizing epitopes. (Left) The corresponding position of the major antigenic site of the neutralizing anti-AAV-2 antibody C37-B that prevents virus-cell binding (light blue, B19 amino acids 303–314) (41) and amino acids involved in heparan binding by AAV-2 (red, B19 amino acids 320–334; gray, B19 amino acid 345; and dark blue, B19 amino acids 373–399) (42, 44, 45). (Right) The epitopes of neutralizing anti-B19 antibodies S-309, S-340, and S-359 (28) that coincide entirely (light green, amino acids 309–330) or in parts (grey, amino acids 340–360; dark green, amino acids 359–382) with the heparan-binding sites of AAV-2.
A cylindrical structure at the fivefold icosahedral axis is formed by the DE loop (amino acids 129–148) and its symmetry-related equivalents. This fivefold structure in B19 is homologous to that of AAV-2 and FPV, but in B19 the tip of the loop is bent toward the central axis of the channel. Five symmetry-equivalent threonines (Thr-135) form an uncharged polar gate at the outside end of the fivefold channel, creating an opening of only ≈9 Å in diameter (distance of Cα atoms) in contrast to ≈18 Å (Asn-189) in AAV-2 and ≈14 Å (Thr-158) in FPV. Consistent with structures of other empty parvoviral capsids (24, 38), there is no electron density in the fivefold channel representing the glycine-rich N-terminal region of VP2. The HI loop (amino acids 460–480) extends the fivefold structure to a flower-like motif with the fivefold cylindrical structure at its center. This flower-like structure is surrounded by a canyon-like depression. The outer rim of the canyon in B19 is more prominent than that in AAV-2 and FPV and is composed of loop 1 (amino acids 59–77), loop 2 (amino acids 190–207), a subloop of the GH loop (amino acids 355–362), and an insertion of 10 aa (amino acids 525–536), unique to erythroviruses in the loop comprising amino acids 512–543 (Figs. 2 and 5a). The latter insertion forms the crest of the wall between the twofold depression and the canyon.
Discussion
Comparison of B19 with AAV-2. The recombinant B19 particles are structurally most similar to AAV-2, with which B19 shares a common host. However, the degree of similarity between B19 and AAV-2 (26% identity between major structural proteins) is far less than between B19 and other primate parvoviruses, such as LaLi (97%), V9 (98%), and SPV (67%). Therefore, assuming a similar rate of mutational acceptance, the divergence between B19 and AAV has probably occurred before the separate evolution of Homo sapiens and other primates; thus, their utilization of the same host may be coincidental. Phylogenetic analysis of parvoviruses shows that AAV clusters with avian parvoviruses (32), which might suggest a common avian origin for both B19 and AAV. Crossing of the human–avian host barrier has been observed for other viruses, such as for avian influenza virus A (39).
Heparan sulfate proteoglycan serves as a primary attachment receptor for AAV-2, whereas αVβ5 integrin has been implicated as a coreceptor for the AAV-2 internalization process (40). Similarly, α5β1 integrin is a cellular coreceptor of B19 that is required for viral entry into human cells (13). Therefore, structurally conserved surface loops might indicate the location of a homologous coreceptor binding site or host range determinants. Regions of greatest sequence similarity between B19 and AAV-2 that coincide with externalized amino acid side chains occur close to loop 1 (amino acids 82–87) and at the N terminus of loop 2 (amino acids 168–188) (Fig. 2). These sequences are located in the canyon around the fivefold axis (Fig. 5b). The regions of amino acids 79–88 and 178–187 in B19 correspond to parts of an epitope on AAV-2 recognized by an antibody that neutralizes AAV-2 infection after primary receptor attachment, possibly blocking the secondary receptor attachment site (41). This overlap suggests that the integrin coreceptors might bind to the canyon floor. Alternatively, conserved surface features in that region might constitute a binding site for the exposed unique region of VP1.
The biggest structural differences among parvoviruses occur in the architecture of the threefold spike formed by the large GH loop. Therefore, it is significant that the RMSD between Cα atoms of B19 and AAV-2 is only 2.0 Å for 80% of the residues in that region (Fig. 4). The surface features formed by the GH loop are a major immunodominant region in native B19 virions and in recombinant VP2 particles (28, 30). Regions known to affect the heparan sulfate binding of AAV-2 were located by peptide mapping and mutational analyses at or near the surface of the threefold-proximal peaks and the wall of the twofold depression (23, 41–44) (Fig. 5c). Epitopes that elicit neutralizing antibodies in B19 (28) correspond to regions in AAV-2 that are involved in heparan sulfate binding. Hence, this region might function as the primary receptor attachment site of B19 but does not correspond to the globoside binding site suggested by Chipman et al. (45). This discrepancy requires further research to clarify.
Erythrovirus. According to sequence homology and cell tropism, human parvovirus B19 is closely related to the nonhuman primate parvoviruses (9, 10, 32). Most of the highly conserved sequences among erythroviruses (amino acids 160–177, 182–194, 206–216, 246–257, 511–524 and 534–543) (Fig. 2) are located in the canyon and correspond in part to the region suggested to be involved in B19 and AAV-2 coreceptor binding (see above). The conservation of canyon residues has been suggested for picornaviruses as a means of avoiding host immune surveillance by sterically hindering the binding of neutralizing antibodies (46). However, the canyon of parvoviruses is far wider than in the case of picornaviruses, and, hence, the reason for the conservation of the canyon residues within Erythrovirus is probably a general property of concave surfaces. Nevertheless, because erythroviruses are linked by a common tropism, the greater conservation of the residues in the canyon might suggest the utilization of similar receptor molecules.
The conserved regions amino acids 511–524 and amino acids 534–543 flank an insertion unique to erythroviruses. The amino acid sequence of the insertion differs greatly between human parvovirus B19 and the nonhuman primate counterparts, suggesting that this external loop might play a role in the determination of host range. It may be significant that residues known to be involved in determination of the host range of FPV (amino acids 564 and 568) (47) are at about the same site. However, an antibody against the linear epitope comprising amino acids 520–544 of B19 VP2 did not neutralize the virus (28), suggesting that the loop is not directly involved in cell entry.
The Fivefold Structure. About one in five of the N-terminal regions of the structural proteins of parvoviruses are usually externalized by passing through a channel at the fivefold vertices of the capsid (18, 19). The channel, constructed of five copies of the DE loop, is occupied by a conserved glycine-rich region. The channel in B19 is 10 Å shorter than that in FPV, consistent with the difference in length of the glycine-rich sequences. The fivefold channel of the B19 VP2 capsids is closed at its outside end. However, the tip of the loop contains a Gly-Gly-Gly sequence (amino acids 136–138), which might provide flexibility to switch the channel from closed to open in the presence of the VP1 unique region. Within erythroviruses, the only significant amino acid change in the DE loop is that the gating residue 135 is a threonine in human viruses and a proline in nonhuman viruses.
A Nuclear Localization Signal. It has been shown that a nuclear localization signal (NLS) directs the transport of newly translated viral protein from the cytoplasm into the nucleus for the assembly of the viral particle (48, 49). A nonconventional NLS (amino acids 493–503) has been identified in the C-terminal region of B19 VP2 that is well conserved within erythroviruses (48). The sequence is located in about the same region as a nuclear localization motif found for minute virus of mice (49). The NLS of B19 is exposed on the surface of an isolated VP2 subunit and, thus, can be recognized by cellular nuclear import molecules. After assembly, the NLS is hidden because it is on the inner capsid surface.
Acknowledgments
We thank Susanne Modrow (Universität Regensburg, Regensburg, Germany) for the gift of recombinant baculovirus AcMNPV-VP2, the staff of BioCARS beamline BM14-ID-B at the Advanced Photon Source for support in collecting the x-ray diffraction data, Sharon Wilder and Cheryl Towell (Purdue University) for assistance in the preparation of this paper, and Chuan Xiao (Purdue University) for help with the roadmaps. The work was supported by National Institutes of Health Grant AI11219 (to M.G.R.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RMSD, rms deviation; SPV, simian parvovirus; PtPV, pig-tailed macaque parvovirus; RhPV, rhesus macaque parvovirus; AAV-2, adeno-associated virus 2; FPV, feline panleukopenia virus.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1S58).
References
- 1.Anderson, M. J., Jones, S. E., Fisher-Hoch, S. P., Lewis, E., Hall, S. M., Bartlett, C. L. R., Cohen, B. J., Mortimer, P. P. & Pereira, M. S. (1983) Lancet 1, 1378 (lett.). [DOI] [PubMed] [Google Scholar]
- 2.Naides, S. J. (1999) Curr. Infect. Dis. Rep. 1, 273–278. [DOI] [PubMed] [Google Scholar]
- 3.Brown, K. E. (2000) Baillieres Best. Pract. Res. Clin. Haematol. 13, 245–259. [DOI] [PubMed] [Google Scholar]
- 4.Katz, V. L., Chescheir, N. C. & Bethea, M. (1990) J. Perinatol. 10, 366–368. [PubMed] [Google Scholar]
- 5.Gilbert, G. L. (2000) Commun. Dis. Intell. 24, Suppl., 69–71. [DOI] [PubMed] [Google Scholar]
- 6.Orth, T., Herr, W., Spahn, T., Voigtlander, T., Michel, D., Mertens, T., Mayet, W. J., Dippold, W. & Meyer-zum, B. K. H. (1997) Eur. Heart J. 18, 524–525. [DOI] [PubMed] [Google Scholar]
- 7.van Regenmortel, M. H. V., Fauquet, C. M., Bishop, D. H. L., Carstens, E. B., Estes, M. K., Lemon, S. M., Maniloff, J., Mayo, M. A., McGeoch, D. J., Pringle, C. R. & Wickner, R. B. (2000) Virus Taxonomy: The Classification and Nomenclature of Viruses, The Seventh Report of the International Committee on Taxonomy of Viruses (Academic, San Diego).
- 8.Servant, A., Laperche, S., Lallemand, F., Marinho, V., De Saint, M. G., Meritet, J. F. & Garbarg-Chenon, A. (2002) J. Virol. 76, 9124–9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, K. E., Green, S. W., O'Sullivan, M. G. & Young, N. S. (1995) Virology 210, 314–322. [DOI] [PubMed] [Google Scholar]
- 10.Green, S. W., Malkovska, I., O'Sullivan, M. G. & Brown, K. E. (2000) Virology 269, 105–112. [DOI] [PubMed] [Google Scholar]
- 11.Brown, K. E., Anderson, S. M. & Young, N. S. (1993) Science 262, 114–117. [DOI] [PubMed] [Google Scholar]
- 12.Weigel-Kelley, K. A., Yoder, M. C. & Srivastava, A. (2001) J. Virol. 75, 4110–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weigel-Kelley, K. A., Yoder, M. C. & Srivastava, A. (2003) Blood 102, 3927–3933. [DOI] [PubMed] [Google Scholar]
- 14.Agbandje, M., Kajigaya, S., McKenna, R., Young, N. S. & Rossmann, M. G. (1994) Virology 203, 106–115. [DOI] [PubMed] [Google Scholar]
- 15.Ozawa, K. & Young, N. S. (1987) J. Virol. 61, 2627–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotmore, S. F., McKie, V. C., Anderson, L. J., Astell, C. R. & Tattersall, P. (1986) J. Virol. 60, 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawase, M., Momoeda, M., Young, N. S. & Kajigaya, S. (1995) Virology 211, 359–366. [DOI] [PubMed] [Google Scholar]
- 18.Tsao, J., Chapman, M. S., Agbandje, M., Keller, W., Smith, K., Wu, H., Luo, M., Smith, T. J., Rossmann, M. G., Compans, R. W. & Parrish, C. R. (1991) Science 251, 1456–1464. [DOI] [PubMed] [Google Scholar]
- 19.Xie, Q. & Chapman, M. S. (1996) J. Mol. Biol. 264, 497–520. [DOI] [PubMed] [Google Scholar]
- 20.Agbandje, M., McKenna, R., Rossmann, M. G., Strassheim, M. L. & Parrish, C. R. (1993) Proteins 16, 155–171. [DOI] [PubMed] [Google Scholar]
- 21.Llamas-Saiz, A. L., Agbandje-McKenna, M., Wikoff, W. R., Bratton, J., Tattersall, P. & Rossmann, M. G. (1997) Acta Crystallogr. D 53, 93–102. [DOI] [PubMed] [Google Scholar]
- 22.Simpson, A. A., Chipman, P. R., Baker, T. S., Tijssen, P. & Rossmann, M. G. (1998) Structure 6, 1355–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie, Q., Bu, W., Bhatia, S., Hare, J., Somasundaram, T., Azzi, A. & Chapman, M. S. (2002) Proc. Natl. Acad. Sci. USA 99, 10405–10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simpson, A. A., Hébert, B., Sullivan, G. M., Parrish, C. R., Zádori, Z., Tijssen, P. & Rossmann, M. G. (2002) J. Mol. Biol. 315, 1189–1198. [DOI] [PubMed] [Google Scholar]
- 25.Kajigaya, S., Fujii, H., Field, A., Anderson, S., Rosenfeld, S., Anderson, L. J., Shimada, T. & Young, N. S. (1991) Proc. Natl. Acad. Sci. USA 88, 4646–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenfeld, S. J., Young, N. S., Alling, D., Ayub, J. & Saxinger, C. (1994) Arch. Virol. 136, 9–18. [DOI] [PubMed] [Google Scholar]
- 27.Saikawa, T., Anderson, S., Momoeda, M., Kajigaya, S. & Young, N. S. (1993) J. Virol. 67, 3004–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato, H., Hirata, J., Kuroda, N., Shiraki, H., Maeda, Y. & Okochi, K. (1991) J. Virol. 65, 5485–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshimoto, K., Rosenfeld, S., Frickhofen, N., Kennedy, D., Hills, R., Kajigaya, S. & Young, N. S. (1991) J. Virol. 65, 7056–7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown, C. S., Jensen, T., Meloen, R. H., Puijk, W., Sugamura, K., Sato, H. & Spaan, W. J. M. (1992) J. Virol. 66, 6989–6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strassheim, M. L., Gruenberg, A., Veijalainen, P., Sgro, J. Y. & Parrish, C. R. (1994) Virology 198, 175–184. [DOI] [PubMed] [Google Scholar]
- 32.Lukashov, V. V. & Goudsmit, J. (2001) J. Virol. 75, 2729–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grenfell, B. T., Pybus, O. G., Gog, J. R., Wood, J. L., Daly, J. M., Mumford, J. A. & Holmes, E. C. (2004) Science 303, 327–332. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 35.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 37.Rossmann, M. G. & Johnson, J. E. (1989) Annu. Rev. Biochem. 58, 533–573. [DOI] [PubMed] [Google Scholar]
- 38.Wu, H., Keller, W. & Rossmann, M. G. (1993) Acta Crystallogr. D 49, 572–579. [DOI] [PubMed] [Google Scholar]
- 39.Fouchier, R. A., Schneeberger, P. M., Rozendaal, F. W., Broekman, J. M., Kemink, S. A., Munster, V., Kuiken, T., Rimmelzwaan, G. F., Schutten, M., Van Doornum, G. J., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 1356–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Summerford, C., Bartlett, J. S. & Samulski, R. J. (1999) Nat. Med. 5, 78–82. [DOI] [PubMed] [Google Scholar]
- 41.Wobus, C. E., Hugle-Dorr, B., Girod, A., Petersen, G., Hallek, M. & Kleinschmidt, J. A. (2000) J. Virol. 74, 9281–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, P., Xiao, W., Conlon, T., Hughes, J., Agbandje-McKenna, M., Ferkol, T., Flotte, T. & Muzyczka, N. (2000) J. Virol. 74, 8635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Opie, S. R., Warrington, K. H., Jr., Agbandje-McKenna, M., Zolotukhin, S. & Muzyczka, N. (2003) J. Virol. 77, 6995–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kern, A., Schmidt, K., Leder, C., Muller, O. J., Wobus, C. E., Bettinger, K., der Lieth, C. W., King, J. A. & Kleinschmidt, J. A. (2003) J. Virol. 77, 11072–11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chipman, P. R., Agbandje-McKenna, M., Kajigaya, S., Brown, K. E., Young, N. S., Baker, T. S. & Rossmann, M. G. (1996) Proc. Natl. Acad. Sci. USA 93, 7502–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rossmann, M. G., Arnold, E., Erickson, J. W., Frankenberger, E. A., Griffith, J. P., Hecht, H. J., Johnson, J. E., Kamer, G., Luo, M., Mosser, A. G., et al. (1985) Nature 317, 145–153. [DOI] [PubMed] [Google Scholar]
- 47.Truyen, U., Agbandje, M. & Parrish, C. R. (1994) Virology 200, 494–503. [DOI] [PubMed] [Google Scholar]
- 48.Pillet, S., Annan, Z., Fichelson, S. & Morinet, F. (2003) Virology 306, 25–32. [DOI] [PubMed] [Google Scholar]
- 49.Lombardo, E., Ramirez, J. C., Agbandje-McKenna, M. & Almendral, J. M. (2000) J. Virol. 74, 3804–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]