Abstract
A mutation in the canine multidrug resistance gene, MDR1, has previously been associated with drug sensitivities in two breeds from the collie lineage. We exploited breed phylogeny and reports of drug sensitivity to survey other purebred populations that might be genetically at risk. We found that the same allele, mdr1-1Δ, segregated in seven additional breeds, including two sighthounds that were not expected to share collie ancestry. A mutant haplotype that was conserved among affected breeds indicated that the allele was identical by descent. Based on breed histories and the extent of linkage disequilibrium, we conclude that all dogs carrying mdr1-1Δ are descendants of a dog that lived in Great Britain before the genetic isolation of breeds by registry (ca. 1873). The breed distribution and frequency of mdr1-1Δ have applications in veterinary medicine and selective breeding, whereas the allele's history recounts the emergence of formally recognized breeds from an admixed population of working sheepdogs.
Keywords: allele age, Canis familiaris, drug sensitivity, identity by descent, P-glycoprotein
The introduction of a new parasiticide in the 1980s (1) uncovered a preexisting mutation in dogs that predisposes animals to a potentially fatal neurotoxicosis (2, 3). The drug, ivermectin, exerts antiparasitic action by potentiating ligandgated chloride ion channels in the peripheral nervous system of several invertebrate phyla (4–7). The resulting influx of chloride ions silences synaptic transmissions, thereby causing lethal paralysis in nematode and arthropod parasites. Ivermectin is generally safe for use in domestic animals because the homologous mammalian targets are restricted to the CNS (8, 9) where they are shielded by the blood–brain barrier (reviewed in ref. 10). A principal component of this protective barrier is P-glycoprotein, an ATP-dependent drug transporter that moves a broad spectrum of substrates across several important tissue borders (11). P-glycoprotein is encoded by the multiple drug resistance gene, MDR1.
The earliest indication that ivermectin neurotoxicity was caused by a defect in the blood–brain barrier came from an observation that affected dogs had elevated concentrations of ivermectin in the CNS (12). Almost a decade later, a similar phenotype was observed in knockout mice lacking Abcb1a, the murine ortholog of MDR1 (13). Mealey et al. (14) investigated canine MDR1 as a candidate gene for ivermectin sensitivity and discovered that affected Collies were homozygous for a 4-bp deletion in the fourth exon. The mutation, mdr1-1Δ, causes a frameshift accompanied by multiple premature stop codons, presumably resulting in a severely truncated P-glycoprotein composed of <10% of the wild-type amino acid sequence. This allele probably results in a complete loss of P-glycoprotein function, although this fact has not yet been formally established (15). More than 20 therapeutic drugs are known substrates of P-glycoprotein. Recently, three of these drugs were found to interact with mdr1-1Δ and cause toxic reactions in dogs (16, 17); two were anticancer drugs that caused extreme cytotoxicity in heterozygous dogs. These results demonstrate that mdr1-1Δ defines a multidrug sensitivity that depends on many drug–genotype interactions.
The mdr1-1Δ allele that was discovered in the Collie breed has since been found in three Australian Shepherd dogs, one of which presented clinically with ivermectin neurotoxicity (18). Australian Shepherds are believed to share common ancestry with Collies, suggesting that mdr1-1Δ might stem from a single ancestral mutation that has been inherited identical by descent. Shetland Sheepdogs and Old English Sheepdogs, which also share the working collie lineage, have reportedly exhibited ivermectin sensitivity (19, 20), as have several nonherding breeds (20).
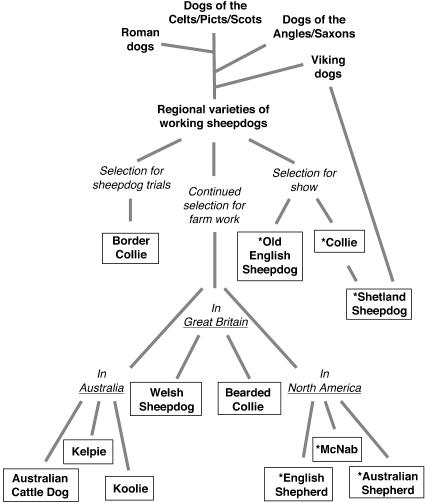
To determine whether mdr1-1Δ places additional breeds at risk for multidrug sensitivity, we selectively surveyed dog populations based on phylogeny (i.e., presumed relatedness to the Collie) and phenotype (i.e., reports of drug sensitivity). Four classes of dogs were tested: first, breeds from the collie lineage that were selected based on a composite of breed histories (Fig. 1); second, European herding breeds that were not thought to be closely related to the Collie; third, sighthounds and miscellaneous breeds that had exhibited drug sensitivities, often in response to ivermectin (ref. 21 and P. Ihrke, personal communication); and, fourth, a multibreed panel composed of over a thousand samples from >90 breeds. This panel was included to establish a general baseline of mdr1-1Δ frequency among purebred dogs.
Fig. 1.
The Collie Family Tree, a composite of anecdotal breed histories (adapted with permission from L. Rorem). The diagram depicts reported historical relationships among contemporary herding breeds that share the collie lineage out of Great Britain. The breeds shown were selectively surveyed for the presence of mdr1-1Δ. Breeds that segregated the mutation are shown with an asterisk.
We have found that two types of dog segregated the mutant allele: seven breeds from the collie lineage and two breeds of the sighthound class. The allele was identical by descent among both types as evidenced by a single ancestral haplotype. Strong linkage disequilibrium (LD) extending over several centimorgans implied a recent origin for mdr1-1Δ, whereas the distribution among multiple breeds suggested the mutation predates contemporary breeds.
Materials and Methods
Sample Ascertainment. DNA samples were obtained from a repository at the Veterinary Genetics Laboratory and through solicitation of volunteer breeders and owners. The breed designation and number of animals sampled are listed in Table 1 and Tables 5 and 6, which are published as supporting information on the PNAS web site. Samples were derived from buccal cells gathered with cytology brushes (Medical Packaging, Camarillo, CA). DNA was prepared from cheek swabs by using a described method (22).
Table 1. Observed frequencies of mdr1-1Δ in affected breeds.
| Allele, %
|
Genotype, %
|
||||
|---|---|---|---|---|---|
| Breed | No. of dogs | mdr1-1Δ | mdr1-1Δ/mdr1-1Δ | mdr1-1Δ/MDR1 | MDR1/MDR1 |
| Australian Shepherd | 178 | 16.6 | 1.7 | 29.8 | 68.5 |
| Australian Shepherd, Miniature | 56 | 25.9 | 3.6 | 44.6 | 51.8 |
| Collie | 263 | 54.6 | 31.2 | 46.8 | 22.0 |
| English Shepherd | 91 | 7.1 | 0 | 14.3 | 85.7 |
| Longhaired Whippet | 89 | 41.6 | 15.7 | 51.7 | 32.6 |
| McNab | 35 | 17.1 | 2.8 | 28.6 | 68.6 |
| Old English Sheepdog | 151 | 3.6 | 0 | 7.3 | 92.7 |
| Shetland Sheepdog | 190 | 8.4 | 1.1 | 14.7 | 84.2 |
| Silken Windhound | 84 | 17.9 | 1.2 | 33.3 | 65.5 |
Genotyping. Primers CfMDR1F or (5′-VIC-GGC TTG ATA GGT TGT ATA TGT TGG TG-3′) and CfMDR1Rev (5′-ATT ATA ACT GGA AAA GTT TTG TTT C-3′) were designed from the canine MDR1 sequence obtained from the GenBank database (accession no. AF045016). The primers bracketed the reported microdeletion in MDR1 (14). Both primers were based within the fourth exon, as inferred from the gene structures of annotated human (ENSG00000085563) and mouse (ENSMUSG00000040584) orthologs obtained from the respective ensembl genome databases (www.ensembl.org).
PCRs for genotyping MDR1 were performed in 20-μl reactions. Three microliters of buccal swab-derived DNA was added to each well along with primer. The samples were brought to 72°C before adding the reagent mix. Each 20-μl PCR contained 0.5 μM of each primer, 0.01 mM tetra-methyl-ammonium chloride (Sigma), 1.5 mM MgCl2, 1× buffer (PerkinElmer), 200 μM each of dNTP, and 1 unit of AmpliTaq DNA Polymerase (PerkinElmer). The thermocycling program for genotyping MDR1 consisted of an initial denaturation step of 93°C for 2 min; 35 cycles of 93°C for 20 sec, 55°C for 20 sec, and 72°C for 1 min; and a final extension step of 72°C for 7 min. After PCR, 0.4 μl of product was mixed with 2 μl of Promega 400 fluorescent ladder, denatured for 3 min at 95°C, and maintained at 5°C until loading. Aliquots of 1.5 μl each were loaded onto a 6% denaturing polyacrylamide gel and were analyzed on an ABI 377 instrument. Genotypes were scored with strand software (23).
Genome Mapping. The genome location of MDR1 was mapped by typing a described canine/hamster radiation hybrid panel (RHDF5000.2; ref. 24). Each of the 118 independent cell lines was assayed in duplicate for the presence of MDR1. When results for a cell line were discordant, the positive score was retained in the data set (4/118). The data were merged with published data (25) and were analyzed with carthagene to calculate interlocus distances and establish the order of loci (26).
Genetic linkage distances were measured with genotype data from several families. The Cornell Families is a described resource comprised of 18 sibships and 163 F2 progeny (27). Additional families included a Border Collie and Newfoundland intercross comprised of 20 F2 animals, a Border Collie family with 30 progeny, a Beagle family with 90 progeny, and an Australian Shepherd family with 60 progeny. The latter family segregated mdr1-1Δ, enabling linkage distances relative to this locus to be estimated. Linkage distances were computed from the genotype data with cri-map (28) by using the twopoint and chrompic options.
To estimate SE and confidence intervals for these genetic distances, we used a parametric bootstrap approach (29) with 10,000 replicates. New genetic mapping data were simulated based on the estimated linkage map and marker allele frequencies, while maintaining the same pattern of missing genotypes as in the observed data. We reestimated the map based on the simulated data, and estimated SE by the SD across simulations. We took the 95% confidence intervals to be from the 2.5–97.5 percentiles of the estimates from simulations.
Haplotype Analysis. Previously mapped microsatellite loci were selected for haplotyping from the canine radiation hybrid map (25) based on proximity to MDR1. For each primer pair, the forward oligonucleotide was labeled at the 5′ end with one of three fluorescent dyes: 6-FAM, VIC, or NED (Applied Biosystems). Reverse primers were synthesized by Operon Technologies (Alameda, CA). Primers for four markers and MDR1 were assembled into a panel suitable for multiplex PCR (Table 7, which is published as supporting information on the PNAS web site). The reaction conditions and thermocycling parameters for multiplex genotyping were the same as those used to amplify MDR1 individually.
Haplotypes were reconstructed with phase, a program that applies Bayesian methods to infer the phase of linked-marker alleles from population genotype data (30, 31). The program normally applies a stepwise mutation model that assumes that all haplotypes found in a population were generated in that population, which was not necessarily the case (e.g., introgression). For this reason, haplotypes with alleles not observed in the breed population were omitted in the estimation of that breed's haplotype frequencies.
Statistical Analyses. Deviation from Hardy–Weinberg equilibrium in each breed was assessed by a χ2 test. Allelic association was calculated according to a described measure (32, 33). Allele age was estimated from the extent of LD at mdr1-1Δ (33–35). The approach assumes that a mutation arose on a single chromosome carrying allele M at a closely linked marker locus. Recombination with nonmutant chromosomes, and/or mutation at the marker locus, erodes initial maximal LD. The number of generations since the founding mutation can thus be estimated by
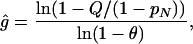 |
[1] |
where g is the age of the mutation, Q is the frequency of mutant chromosomes not carrying marker allele M, pN is the frequency of M on nonmutant chromosomes, and θ is the recombination fraction separating the mutation site and the marker locus. In estimating allele age, we generally ignored mutation given that microsatellite mutation rates in mammals are estimated to be 10–4 or lower (36). Because map distances between mdr1-1Δ and the marker loci used to estimate allele age were 1 cM or greater, mutation is expected to be a much weaker force than recombination in eroding LD. If mutation were an important force, then estimates of g obtained from Eq. 1 would tend to be too large. Although mutation is unlikely to affect the average decay of LD, mutation might be the basis for rare haplotypes found within a breed.
Results
Distribution Among Breeds. More than 4,000 samples from purebred dogs were surveyed for the presence of mdr1-1Δ. The allele was found in nine breeds (Table 1), including the Collie and Australian Shepherd, as reported (14, 18). A published Collie Family Tree, which was used to select collie-related breeds for testing, was generally predictive of the observed distribution of mdr1-1Δ (Fig. 1). Approximately half the breeds from this lineage segregated mdr1-1Δ. In contrast, the mutation was not found in any of the herding breeds whose origins traced back to continental Europe (Table 5). However, the allele was detected in two nonherding breeds, the Longhaired Whippet and the Silken Windhound, which were among the sighthound class of breeds tested because of anecdotal reports of ivermectin sensitivity.
Frequency Within Breeds. Allele and genotype frequencies were measured for each affected breed population (Table 1). The mutant allele frequency varied by breed, from <4% in the Old English Sheepdog to >50% in the Collie. A high frequency in the Collie and the Longhaired Whippet suggested a founder effect for each breed. The Collie data, when partitioned into geographic subpopulations, indicated a similarly high allele frequency in both British and American dogs (Table 8, which is published as supporting information on the PNAS web site), consistent with mdr1-1Δ having entered the Collie gene pool when the breed was formally established in Great Britain (37). Although fewer samples from the U.K. were available for the Shetland Sheepdog and Old English Sheepdog, the allele appeared to be more frequent in the British subpopulations (9 of 15 Shetland Sheepdogs, and 4 of 31 Old English Sheepdogs from the U.K. carried at least one copy of mdr1-1Δ). This result was also consistent with a geographic origin for mdr1-1Δ in Great Britain.
Identity by Descent. Identical alleles may arise by recurrent mutation, as with the S allele of the human β-globin gene (38) and the G380R allele of the human FGFR3 gene (39). Mealey et al. (14) noted that a palindrome is located 9 bp upstream of mdr1-1Δ and that such sequences sometimes serve as mutational hot spots (40). This finding allowed for the possibility that the identical alleles of herding breeds and sighthounds stemmed from independent mutation events. If mdr1-1Δ was identical by state, the marker alleles associated with mdr1-1Δ would likely differ between the two breed types.
To test allelic associations, MDR1 was mapped by radiation hybrid analysis (Fig. 2) and the four closest markers were selected to genotype individuals from affected breeds. Comparison of genotype data from the homozygote classes (where phase was known) revealed that the marker alleles most strongly associated with mdr1-1Δ were identical for three of the four loci in both herding breeds and sighthounds (Table 2). The conservation of these allelic associations indicated that mdr1-1Δ arose once, and that both herding breeds and sighthounds shared the allele identical by descent.
Fig. 2.
Genetic map of the MDR1 region of CFA14. The order of loci was established by radiation hybrid analysis. The confidence intervals (95%) for linkage distances are listed in parentheses. An asterisk denotes previously reported distances (25).
Table 2. Marker allele frequencies for mutant and wild-type chromosomes in herding breeds and sighthounds.
| C14.866
|
REN103E18
|
REN144I15
|
G01506
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Herding
|
Sighthound
|
Herding
|
Sighthound
|
Herding
|
Sighthound
|
Herding
|
Sighthound
|
||||||||||||
| Allele | Mut | Wt | Mut | Wt | Allele | Mut | Wt | Mut | Wt | Allele | Mut | Wt | Mut | Wt | Allele | Mut | Wt | Mut | Wt |
| 233 | 0.008 | 173 | 0.995 | 0.567 | 0.967 | 0.315 | 251 | 0.092 | 0.288 | 0.536 | 151 | 0.008 | |||||||
| 237 | 0.008 | 175 | 0.033 | 0.105 | 253 | 153 | 0.684 | 0.317 | 0.967 | 0.018 | |||||||||
| 239 | 0.670 | 0.267 | 177 | 0.203 | 0.136 | 255 | 0.500 | 0.464 | 155 | 0.230 | 0.056 | ||||||||
| 241 | 0.010 | 0.032 | 0.033 | 0.133 | 178 | 0.128 | 257 | 0.111 | 157 | 0.061 | 0.238 | 0.663 | |||||||
| 243 | 0.132 | 0.343 | 180 | 0.005 | 0.091 | 0.444 | 259 | 0.908 | 0.097 | 1.000 | 159 | 0.210 | 0.006 | ||||||
| 245 | 0.090 | 0.028 | 182 | 0.011 | 261 | 0.004 | 161 | 0.026 | 0.171 | 0.033 | 0.313 | ||||||||
| 247 | 0.363 | 0.247 | |||||||||||||||||
| 249 | 0.230 | 0.111 | 0.967 | ||||||||||||||||
| 251 | 0.052 | ||||||||||||||||||
| 253 | 0.277 | ||||||||||||||||||
Deduced from data of mdr1-1Δ and MDR1 homozygous classes where phase was known. MDR1 maps between C14.866 and REN103E18. Mut, mutant; Wt, wild-type. The highest allele frequency for each marker on the mutant chromosome is in bold.
Allele Origins. The strong LD exhibited in the sighthound breeds suggested that they acquired mdr1-1Δ recently and on a limited number of chromosomes (i.e., one that carried the “249” allele at the C14.866 locus). Full-haplotype analysis could address the origin of mdr1-1Δ in sighthounds, and possibly inform on the general processes that drove dispersal of mdr1-1Δ (e.g., founder effect, population admixture, focused introgression, etc.). Table 3 lists the frequency and distribution of mdr1-1Δ haplotypes for each breed. The diversity of haplotypes exhibited by the Collie, even when partitioned into geographic subpopulations, was consistent with prolonged segregation of mdr1-1Δ. In contrast, the Longhaired Whippet and the Silken Windhound segregated only a few haplotypes, which is consistent with recent introgression (possibly from a herding breed that favored haplotype II, such as the Australian Shepherd or Shetland Sheepdog). The diversity of haplotypes in the Old English Sheepdog (4 haplotypes from 10 mutant chromosomes) suggested that this breed had also segregated mdr1-1Δ for many generations. Given that three of four haplotypes were breed-specific, the Old English Sheepdog may have been one of the first breeds to diverge from the collie lineage.
Table 3. Frequencies of inferred four-locus haplotypes for mdr1-1Δ-bearing chromosomes among affected breeds.
| Haplotypes | Collie U.K. (n = 123) | Collie U.S. (n = 177) | Australian Shepherd (n = 48) | Miniature Australian Shepherd (n = 29) | English Shepherd (n = 13) | McNab (n = 12) | Old English Sheepdog (n = 10) | Shetland Sheepdog (n = 32) | Longhaired Whippet (n = 74) | Silken Windhound (n = 30) |
|---|---|---|---|---|---|---|---|---|---|---|
| I 239 mdr1 173 259 153 | 0.236 | 0.542 | 0.354 | 0.759 | 0.308 | 0.100 | 0.063 | |||
| 239 mdr1 173 259 157 | 0.041 | 0.692 | ||||||||
| 239 mdr1 173 259 155 | 0.577 | |||||||||
| 239 mdr1 173 259 159 | 0.600 | |||||||||
| 243 mdr1 173 255 153 | 0.200 | |||||||||
| 245 mdr1 173 251 153 | 0.147 | 0.125 | ||||||||
| 247 mdr1 173 255 159 | 0.100 | |||||||||
| II 249 mdr1 173 259 153 | 0.106 | 0.181 | 0.521 | 0.207 | 0.781 | 0.986 | 0.733 | |||
| 249 mdr1 173 259 157 | 0.017 | 0.042 | 1.000 | |||||||
| 249 mdr1 175 259 161 | 0.233 | |||||||||
| Other | 0.040 | 0.113 | 0.083 | 0.034 | 0.031 | 0.014 | 0.034 |
n, number of mutant chromosomes. Haplotypes I and II are bold, as are the most frequent haplotypes for each breed. The loci order is C14.866-mdr1-1Δ-REN103E18-REN144I15-GO1506. Table 9, which is published as supporting information on the PNAS web site, provides a full list of mutant haplotypes.
Allele Age. The broad distribution of haplotypes I and II (Table 3) suggested that both versions of the mutant chromosome existed in an ancestral population before the emergence of formal breeds (ca. 1873). This distribution afforded an opportunity to estimate the date of the founding mutation relative to the time of breed divergence, if several assumptions were made. First, it was assumed that only haplotypes I and II existed in this ancestral population, and that their frequencies at the time of divergence could be represented by their current relative frequencies (0.63 and 0.37, respectively). Similarly, the allele frequencies for C14.866 on wild-type chromosomes in the ancestral population were estimated from the present-day herding breed data. Linkage distances, shown in Fig. 2, were obtained by genotyping families composed of ≈600 meioses. For the interval between C14.866 and MDR1, θ ≈ 0.01–0.02. With this information, the number of generations needed to erode the initial maximal LD and arrive at the relative frequencies of haplotypes I and II could be estimated according to Eq. 1 (33–35). Given that either haplotype could represent the original mutant chromosome (haplotype I was more frequent, but the allelic association for 249 on haplotype II was stronger), separate allele ages were calculated based on the decay from I to II, and vice versa.
The resulting estimates suggested that the founding mutation predated the divergence of breeds by ≈40–120 generations. This range reflects the estimates obtained from Eq. 1, allowing for a factor of two uncertainty in the estimated recombination rates. With a generation time of ≈4 years (calculated from modern breeding data, Table 10, which is published as supporting information on the PNAS web site), the allele age estimates predicted that the mutation occurred sometime after the 14th century. Although the estimates were imprecise and based on several assumptions (which precluded calculating a confidence interval), the results suggested that mdr1-1Δ was not an ancient mutation (> 250 generations), but rather, was an allele that arose more recently within the collie lineage.
Discussion
We found that at least nine breeds of dog segregated an identical-by-descent allele of MDR1 that predisposes dogs to multidrug sensitivity (Figs. 3 and 4, which are published as supporting information on the PNAS web site). The present genetic survey was biased in favor of breed relatedness, so the clustering of mdr1-1Δ among herding breeds of the collie lineage was not unexpected. The allele was not found in the Border Collie, Bearded Collie, or Australian Cattle Dog, three collie-related breeds that have reportedly exhibited ivermectin sensitivity (21). The presence of drug sensitivity in these breeds implies that mdr1-1Δ may be present at a low frequency, or that another mutation is responsible. Negative results from a multibreed panel indicated that, in general, the baseline frequency among all purebred dogs is low. This panel consisted of as few as five dogs per breed, so additional breeds segregating mdr1-1Δ may yet be discovered.
Prevalence in Breeds. A principal aim of this study was to identify breeds that were at risk for multidrug sensitivity, and to characterize the degree of susceptibility for each breed population. Accurate measures of allele frequencies within breeds are difficult to obtain, and we caution against the straightforward clinical application of these data. Selection and nonrandom mating can rapidly change allele and genotype frequencies, and sample relatedness and geographic stratification can further confound measurements. Although American and British Collies had similarly high frequencies of mdr1-1Δ [which agreed with previous data for 40 Collies from the northwestern U.S (ref. 41 and Table 8)], the two geographic subpopulations favored different mutant haplotypes, presumably owing to genetic drift. Great Britain, until recently, required a 6-month quarantine of imported dogs, which may have isolated British subpopulations and aided drift. These results suggest that data gathered for one geographic subpopulation may not be generally applicable to other subpopulations.
Applications of mdr1-1Δ Genetics. An understanding of the genetic basis of differential drug response can be used to identify individuals that might be predisposed to adverse drug effects. Veterinarians can use DNA testing of mdr1-1Δ to screen animals before administering certain therapeutic drugs. Denying entire breeds an effective treatment, which is currently done with ivermectin and herding breeds, is now unnecessary; clinicians can treat animals based on genotype rather than breed affiliation. The pharmacogenetics approach can be extended to treatment with all drugs that are P-glycoprotein substrates. More than 20 drugs are known substrates (Table 4); each could exhibit a unique dose–toxicity curve among MDR1 genotypes. Genetic background differences among breeds may further modify mdr1-1Δ-based responses. Many potential drug–genotype interactions must therefore be examined to fully explore the phenotypic spectrum of mdr1-1Δ.
Table 4. P-glycoprotein substrate drugs.
Although pharmacogenetics affords a near-term solution to mdr1-1Δ, genotype-based selection could ultimately eliminate the mutation from breed gene pools. The allele and genotype frequency data reported here will aid in the design of breeding strategies. For the Shetland Sheepdog, where the allele frequency is low and the number of dogs is large, mdr1-1Δ could be removed in a few generations. For the Longhaired Whippet, the allele frequency is high and the number of dogs is small, hence a more gradual approach could be applied.
History of mdr1-1Δ. Dogs carrying mdr1-1Δ share a common ancestor that experienced remarkable evolutionary success, having contributed genetically to at least nine distinct breeds of dog. We propose that this animal lived in Great Britain in the 1800s, before the emergence of formal breeds. Before 1870, there were no established registries for sheepdogs, only regional varieties of working dogs that had been adapted to terrain, climate, breed of sheep, and working style. Industrialization in the 19th century brought changes in trade and transportation that may have facilitated admixture among these varieties. Socioeconomic changes almost certainly altered the role of working dogs because they were no longer needed to drive sheep over long distances to market. Although a few specialized strains rose in prominence, perhaps aided by success at field trial events (42), many strains such as the Galway Collie, the Dalesman, the Manx Sheepdog, and the Welsh Gray gradually began to disappear. The neglect of regional varieties may have contributed directly to the advent of dog shows, which aimed to preserve and restore strains by emphasizing form rather than function. The first bench show to admit herding dogs took place in Birmingham in 1860, with one class open to all “sheepdogs, colleys, yard, or keeper's dogs (42).” This show marked the beginning of an important transition in the history of sheepdogs, from regional variety to registered breed, and from anonymous working dog to pedigreed purebred (reviewed in ref. 43).
The first formal breeds to emerge from working sheepdog populations were the Collie, Old English Sheepdog, and Shetland Sheepdog. Several influential founders of the Collie breed, such as Old Cockie and Trefoil, were born in the 1860s (42). According to Baskerville, “Next to nothing was known of the pedigrees of the afore-mentioned dogs except that the majority of them came from a working strain of sheepdogs (37).” Working collies contributed genetically to the Shetland Sheepdog, which probably accounts for the presence of mdr1-1Δ in the latter breed. Thus, the allele may have already been prevalent among working collies by the 1890s.
The Old English Sheepdog was a founding member of the Kennel Club of England in 1873, and has probably been genetically isolated from other collie-related breeds since that time. Unlike the Shetland Sheepdog, the Old English Sheepdog is distinct from the Collie in size, shape, and behavior, so registered show Collies are unlikely to have been the source of mdr1-1Δ. Rather, admixture among the working progenitors of these two breeds is the more likely explanation. A collection of essays on pastoral life in 19th century Great Britain describes shepherds as using two complementary types of dog: the smaller collies that excelled at herding, and the larger, more versatile “old English type” that could drive, protect, and herd the flock (44). The use of both types by shepherds presumably afforded gene flow. Thus, the ancestral population that produced mdr1-1Δ was probably an admixed population of working sheepdogs. The ancestors of the Australian Shepherd, English Shepherd, and McNab also trace back to this ancestral population, roughly defined. Although these latter breeds were developed in North America in the 1900s, they were most likely derived from nondescript farm collies imported from Great Britain and Australia in the 1800s and early 1900s.
Several lines of evidence suggest that the mdr1-1Δ mutation event predated the formal establishment of British herding breeds, beginning in 1873. The high frequency of mdr1-1Δ in both subpopulations of Collies, the broad distribution of haplotypes I and II among multiple breeds, and the distinct haplotypes of the Old English Sheepdog together suggest that mdr1-1Δ was widely dispersed by the time breeds were being registered. Although based on several assumptions, the allele age estimates suggested that mdr1-1Δ was not an ancient allele (e.g., one introduced into the British Isles by Roman or Viking dogs; Fig. 1). Thus, the allele may not be broadly distributed beyond the collie lineage, except as a consequence of focused introgression.
The presence of the mutation in sighthounds may provide a more recent historical perspective on mdr1-1Δ. Reports of ivermectin sensitivity in these dogs were initially explained as having resulted from well known crosses between Queen Victoria's Collies and Borzois given to her by Czar Nicholas II (I. Combe, personal communication). However, the strong LD and the limited number of mutant haplotypes in sighthounds were consistent with a more recent event. The Longhaired Whippet is described as an ancient variety that was apparently restored in the 1950s by a single breeder who also bred Shetland Sheepdogs. It is interesting to speculate that mdr1-1Δ accompanied an allele for long hair during focused introgression, through either linkage or drift. Both the Longhaired Whippet and Shetland Sheepdog favor haplotype II (Table 3), which is consistent with this interpretation. The Silken Windhound was developed even more recently (in the 1980s) by crossing multiple sighthound breeds, including the Borzoi, Whippet, and Longhaired Whippet; the latter is the probable source of mdr1-1Δ (Fig. 4). This explanation is also consistent with a preference for haplotype II in both breeds (Table 3).
The Importance of Genetic Drift. Within each breed, the genotype frequencies were consistent with Hardy–Weinberg equilibrium. Thus, there was no evidence of selection or nonrandom mating with respect to mdr1-1Δ. Therefore, the allele frequency differences observed among breeds (4–50%) may be attributable to genetic drift, perhaps aided by the rapid expansion of formal breed populations since the late 1800s. The high frequency of mdr1-1Δ in the Longhaired Whippet (42%) is also likely to be a consequence of drift. This frequency is five-fold greater than that of the Shetland Sheepdog (8%), the suspected source of the allele. These results support the view that genetic drift, in addition to artificial selection, has played an important role in defining the genetic composition of breeds.
Implications for Complex Trait Mapping. The causative mutation for multidrug sensitivity was identified by the candidate gene approach (14). Had this strategy failed, a nonparametric mapping approach would have been necessary, given that drug response is multifactorial. Here, we used the presence of LD to establish that mdr1-1Δ was identical by descent, and to estimate the time since the founding mutation. This same LD would have signaled successful mapping of multidrug sensitivity. Allelic associations between mdr1-1Δ and the most distal marker, G01506 (≈7 cM interval), were sufficiently strong among breeds (from 0.47 to 1.00) to detect association with a case-control study. However, the appropriate genome-wide threshold for assessing the significance of such an LD signal remains to be determined. Combining the data for British and American Collies reduces the strength of allelic association from 0.60 and 0.72, respectively, to 0.23. Thus, the results reported here could be taken as proof of concept that complex traits are amenable to identity-by-descent mapping in dogs, but that the potential pitfalls encountered in human genetics (e.g., population stratification) also exist in canine genetics.
Deconstructing Breeds. Geneticists have previously attempted to describe canine breed ancestry by averaging the phylogenetic results of marker loci distributed across the genome, thereby characterizing breeds as monophyletic. We prefer to view dog breeds not as Linnaean subspecies, but rather as dynamic populations that have historically experienced admixture, introgression, and genetic isolation. The presence of mdr1-1Δ in the sighthound breeds supports this view, and confirms that different regions of the canine genome have distinct evolutionary histories (i.e., polyphyletic). Accordingly, breed phylogeny per se is perhaps less relevant than the phylogeny of individual traits. Each polymorphism, especially those of phenotypic effect, will recount an interesting story about the history of dogs and the origin of traits.
Acknowledgments
We thank T. Gilliland, K. Urish, M. Grady, S. Bentjen, L. Markegard, P. MacRoberts, and L. Millon for technical assistance; Drs. E. True, K. Mulvihill, and P. Ihrke for clinical expertise; M. Shipstone, A. Bell, C. Fychan, A. Ruhe, C. Gaiser, V. Geddes, C. Masterson, C. Mellersh, V. Magee, P. Hutchinson, and M. Henninger for facilitating sample collections; L. Rorem, C. A. Sharp, G. Kaye, J. Brackman, P. Hutchinson, and I. Combe for historical information; and all of the owners who contributed samples. This work was supported by proceeds from services offered at the Veterinary Genetics Laboratory at the University of California, Davis M.S. was supported in part by National Institutes of Health Grant R01-GM40282 and the Miller Institute of Basic Research in Science.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: LD, linkage disequilibrium.
References
- 1.Campbell, W. C., Fisher, M. H., Stapley, E. O., Albers-Schonberg, G. & Jacob, T. A. (1983) Science 221, 823–828. [DOI] [PubMed] [Google Scholar]
- 2.Preston, J. M. (1983) Vet. Rec. 112, 286 (lett.). [DOI] [PubMed] [Google Scholar]
- 3.Seward, R. L. (1983) J. Am. Vet. Med. Assoc. 183, 493 (lett.). [PubMed] [Google Scholar]
- 4.Cully, D. F., Vassilatis, D. K., Liu, K. K., Paress, P. S., Van der Ploeg, L. H., Schaeffer, J. M. & Arena, J. P. (1994) Nature 371, 707–711. [DOI] [PubMed] [Google Scholar]
- 5.Cully, D. F., Paress, P. S., Liu, K. K., Schaeffer, J. M. & Arena, J. P. (1996) J. Biol. Chem. 271, 20187–20191. [DOI] [PubMed] [Google Scholar]
- 6.Kane, N. S., Hirschberg, B., Qian, S., Hunt, D., Thomas, B., Brochu, R., Ludmerer, S. W., Zheng, Y., Smith, M., Arena, J. P., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 13949–13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ludmerer, S. W., Warren, V. A., Williams, B. S., Zheng, Y., Hunt, D. C., Ayer, M. B., Wallace, M. A., Chaudhary, A. G., Egan, M. A., Meinke, P. T., et al. (2002) Biochemistry 41, 6548–6560. [DOI] [PubMed] [Google Scholar]
- 8.Dawson, G. R., Wafford, K. A., Smith, A., Marshall, G. R., Bayley, P. J., Schaeffer, J. M., Meinke, P. T. & McKernan, R. M. (2000) J. Pharmacol. Exp. Ther. 295, 1051–1060. [PubMed] [Google Scholar]
- 9.Shan, Q., Haddrill, J. L. & Lynch, J. W. (2001) J. Biol. Chem. 276, 12556–12564. [DOI] [PubMed] [Google Scholar]
- 10.Fisher, M. H. & Mrozik, H. (1992) Annu. Rev. Pharmacol. Toxicol. 32, 537–553. [DOI] [PubMed] [Google Scholar]
- 11.Van Asperen, J., Mayer, U., Van Tellingen, O. & Beijnen, J. H. (1997) J. Pharm. Sci. 86, 881–884. [DOI] [PubMed] [Google Scholar]
- 12.Pulliam, J. D., Seward, R. L., Henry, R. T. & Steinberg, S. A. (1985) Vet. Med. 7, 33–40. [Google Scholar]
- 13.Schinkel, A. H., Smit, J. J., Van Tellingen, O., Beijnen, J. H., Wagenaar, E., Van Deemter, L., Mol, C. A., van der Valk, M. A., Robanus-Maandag, E. C., te Riele, H. P., et al. (1994) Cell 77, 491–502. [DOI] [PubMed] [Google Scholar]
- 14.Mealey, K. L., Bentjen, S. A., Gay, J. M. & Cantor, G. H. (2001) Pharmacogenetics 11, 727–733. [DOI] [PubMed] [Google Scholar]
- 15.Roulet, A., Puel, O., Gesta, S., Lepage, J. F., Drag, M., Soll, M., Alvinerie, M. & Pineau, T. (2003) Eur. J. Pharmacol. 460, 85–91. [DOI] [PubMed] [Google Scholar]
- 16.Mealey, K. L., Northrup, N. C. & Bentjen, S. A. (2003) J. Am. Vet. Med. Assoc. 223, 1453–1455. [DOI] [PubMed] [Google Scholar]
- 17.Sartor, L. L., Bentjen, S. A., Trepanier, L. & Mealey, K. L. (2004) J. Vet. Intern. Med. 18, 117–118. [DOI] [PubMed] [Google Scholar]
- 18.Nelson, O. L., Carsten, E., Bentjen, S. A. & Mealey, K. L. (2003) J. Vet. Intern. Med. 17, 354–356. [PubMed] [Google Scholar]
- 19.Houston, D. M., Parent, J. & Matushek, K. J. (1987) J. Am. Vet. Med. Assoc. 191, 78–80. [PubMed] [Google Scholar]
- 20.Scott, D. W., Miller, W. H. & Griffin, C. E. (1995) Muller and Kirk's Small Animal Dermatology (W.B. Saunders, Philadelphia).
- 21.Lovell, R. A. (1990) Vet. Clin. North. Am. Small Anim. Pract. 20, 453–468. [DOI] [PubMed] [Google Scholar]
- 22.Oberbauer, A. M., Grossman, D. I., Eggleston, M. L., Irion, D. N., Schaffer, A. L., Pedersen, N. C. & Belanger, J. M. (2003) Vet. Res. Commun. 27, 27–38. [DOI] [PubMed] [Google Scholar]
- 23.Toonen, R. J. & Hughes, S. S. (2001) BioTechniques 31, 1320–1324. [PubMed] [Google Scholar]
- 24.Vignaux, F., Hitte, C., Priat, C., Chuat, J. C., Andre, C. & Galibert, F. (1999) Mamm. Genome 10, 888–894. [DOI] [PubMed] [Google Scholar]
- 25.Breen, M., Jouquand, S., Renier, C., Mellersh, C. S., Hitte, C., Holmes, N. G., Cheron, A., Suter, N., Vignaux, F., Bristow, A. E., et al. (2001) Genome Res. 11, 1784–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schiex, T. & Gaspin, C. (1997) Proc. Int. Conf. Intell. Syst. Mol. Biol. 5, 258–267. [PubMed] [Google Scholar]
- 27.Neff, M. W., Broman, K. W., Mellersh, C. S., Ray, K., Acland, G. M., Aguirre, G. D., Ziegle, J. S., Ostrander, E. A. & Rine, J. (1999) Genetics 151, 803–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green, P., Fall, K. & Crooks, S. (1990) Documentation for CRI-MAP (Washington University, St. Louis), Version 2.4.
- 29.Efron, B. & Tibshirani, R. J. (1993) An Introduction to Bootstrap (Chapman & Hall/CRC, Boca Raton, FL).
- 30.Stephens, M., Smith, N. J. & Donnelly, P. (2001) Am. J. Hum. Genet. 68, 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens, M. & Donnelly, P. (2003) Am. J. Hum. Genet. 73, 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bengtsson, B. O. & Thomson, G. (1981) Tissue Antigens 18, 356–363. [DOI] [PubMed] [Google Scholar]
- 33.Risch, N., de Leon, D., Ozelius, L., Kramer, P., Almasy, L., Singer, B., Fahn, S., Breakefield, X. & Bressman, S. (1995) Nat. Genet. 9, 152–159. [DOI] [PubMed] [Google Scholar]
- 34.Serre, J. L., Simon-Bouy, B., Mornet, E., Jaume-Roig, B., Balassopoulou, A., Schwartz, M., Taillandier, A., Boue, J. & Boue, A. (1990) Hum. Genet. 84, 449–454. [DOI] [PubMed] [Google Scholar]
- 35.Slatkin, M. & Rannala, B. (2000) Annu. Rev. Genomics Hum. Genet. 1, 225–249. [DOI] [PubMed] [Google Scholar]
- 36.Chakraborty, R., Kimmel, M., Stivers, D. N., Davison, L. J. & Deka, R. (1997) Proc. Natl. Acad. Sci. USA 94, 1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baskerville, W. (1926) Show Collies (Rough and Smooth Coated) and Shetland Sheepdogs, with a Chapter on Working Sheepdogs. Origin, History, Show Points, and Working Qualities. How to Breed, Rear, and Prepare for Exhibition (Our Dogs Publishing Co., Ltd., Manchester, U.K.).
- 38.Pagnier, J., Mears, J. G., Dunda-Belkhodja, O., Schaefer-Rego, K. E., Beldjord, C., Nagel, R. L. & Labie, D. (1984) Proc. Natl. Acad. Sci. USA 81, 1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bellus, G. A., McIntosh, I., Smith, E. A., Aylsworth, A. S., Kaitila, I., Horton, W. A., Greenhaw, G. A., Hecht, J. T. & Francomano, C. A. (1995) Nat. Genet. 10, 357–359. [DOI] [PubMed] [Google Scholar]
- 40.Lewis, S., Akgun, E. & Jasin, M. (1999) Ann. N.Y. Acad. Sci. 870, 45–57. [DOI] [PubMed] [Google Scholar]
- 41.Mealey, K. L., Bentjen, S. A. & Waiting, D. K. (2002) Am. J. Vet. Res. 63, 479–481. [DOI] [PubMed] [Google Scholar]
- 42.Combe, I. (1987) Herding Dogs, Their Origins and Development in Britain (Faber & Faber Ltd., London).
- 43.Brackman, J. (2003) in AKC Gazette, ed. Paddock, A. (Berger, New York), pp. 32–35.
- 44.Gosset, A. L. J. (1911) Shepherds of Britain; Scenes From Shepherd Life Past and Present from the Best Authorities (Constable and Co., Ltd., London).
- 45.Sakaeda, T., Nakamura, T. & Okumura, K. (2002) Biol. Pharm. Bull. 25, 1391–1400. [DOI] [PubMed] [Google Scholar]
- 46.Schinkel, A. H., Wagenaar, E., van Deemter, L., Mol, C. A. & Borst, P. (1995) J. Clin. Invest. 96, 1698–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takano, M., Hasegawa, R., Fukuda, T., Yumoto, R., Nagai, J. & Murakami, T. (1998) Eur. J. Pharmacol. 358, 289–294. [DOI] [PubMed] [Google Scholar]
- 48.Schwab, M., Eichelbaum, M. & Fromm, M. F. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 285–307. [DOI] [PubMed] [Google Scholar]