ABSTRACT
Influenza virus neuraminidase (NA) drug resistance is one of the challenges to preparedness against epidemic and pandemic influenza virus infections. NA N1- and N2-containing influenza viruses are the primary cause of seasonal epidemics and past pandemics. The structural and functional basis underlying drug resistance of the influenza virus N1 NA is well characterized. Yet drug resistance of the N2 strain is not well understood. Here, we confirm that replacement of N2 E119 or I222 results in multidrug resistance, and when the replacements occur together, the sensitivity to NA inhibitors (NAI) is reduced severely. Using crystallographic studies, we showed that E119 replacement results in a loss of hydrogen bonding to oseltamivir and zanamivir, whereas I222 replacement results in a change in the hydrophobic environment that is critical for oseltamivir binding. Moreover, we found that MS-257, a zanamivir-oseltamivir hybrid inhibitor, is less susceptible to drug resistance. The binding mode of MS-257 shows that increased hydrogen bonding interactions between the inhibitor and NA active site anchor the inhibitor within the active site and allow adjustments in response to active-site modifications. Such stability is likely responsible for the observed reduced susceptibility to drug resistance. MS-257 serves as a next-generation anti-influenza virus drug candidate and serves also as a scaffold for further design of NAIs.
IMPORTANCE Oseltamivir and zanamivir are the two major antiviral drugs available for the treatment of influenza virus infections. However, multidrug-resistant viruses have emerged in clinical cases, which pose a challenge for the development of new drugs. N1 and N2 subtypes exist in the viruses which cause seasonal epidemics and past pandemics. Although N1 drug resistance is well characterized, the molecular mechanisms underlying N2 drug resistance are unknown. A previous report showed that an N2 E119V/I222L dual mutant conferred drug resistance to seasonal influenza virus. Here, we confirm that these substitutions result in multidrug resistance and dramatically reduced sensitivity to NAI. We further elucidate the molecular mechanism underlying N2 drug resistance by solving crystal structures of the N2 E119V and I222L mutants and the dual mutant. Most importantly, we found that a novel oseltamivir-zanamivir hybrid inhibitor, MS-257, remains more effective against drug-resistant N2 and is a promising candidate as a next-generation anti-influenza virus drug.
INTRODUCTION
Influenza virus is the causative agent of seasonal and pandemic flu infections and therefore poses a great threat to humanity. Currently, only M2 ion channel inhibitors and influenza virus neuraminidase (NA) inhibitors have been fully developed as specific anti-influenza virus drugs. However, M2 ion channel inhibitors are no longer recommended for use due to serious problems with drug resistance (1–4), leaving influenza virus NA as the most successful anti-influenza virus drug target. Yet influenza virus NA also develops various types of drug resistance (5–12). Therefore, understanding the molecular mechanisms of influenza virus NA drug resistance is critical for directing drug development and preparedness against potential future influenza epidemics and pandemics.
NA from influenza A virus, the causative agent of all known influenza pandemics, is classified into two phylogenetic groups: group 1, containing N1, N4, N5, and N8, and group 2, containing N2, N3, N6, N7, and N9 (13). So far, the structures of all subtypes have been solved (13–19). Influenza viruses containing N1 and N2 (i.e., H1N1, H3N2, and H2N2) are the most common types that infect humans and are therefore a great threat to public health (20–23). The structural and functional basis of N1 drug resistance has been thoroughly explored (24, 25). Yet comprehensive studies of N2 drug resistance or that of any group 2 NAs remain unreported.
Replacement of residue E119, yielding drug resistance, was commonly observed in N2 (26, 27). Five substitutions (Gly, Asp, Ala, Val, and Ile) have been reported to be associated with NA inhibitor resistance either in vivo or in vitro (26, 28–32). An E119V/I variant has been isolated from oseltamivir-treated and untreated patients. However, two clinical isolates containing the same E119V substitution exhibited different levels of oseltamivir resistance (32, 33), and the molecular mechanism remains unknown. E119G, E119A, and E119D mutations have also been selected in vitro by passaging influenza virus in the presence of zanamivir (28–30). Mutations at E119 have also been observed in influenza B viruses (34) but not in influenza A N1 viruses.
Multiple drug-resistant substitutions in N2 have been reported. Of particular interest is the dual replacements of residues E119 and I222, resulting in multidrug resistance with enhanced resistance to oseltamivir (27, 35). E119V has been found in naturally isolated influenza viruses, while the I222L and the E119V/I222L double substitutions have been studied using reverse genetics (35). Moreover, an E119V/I222V double substitution has also been identified from a natural influenza virus isolate which shows oseltamivir resistance (27). In this study, we determined the structural and functional basis of these substitutions using highly purified recombinant N2 proteins. Our results, at the soluble protein level, confirm that dual replacement of I222 and E119 results in severe oseltamivir resistance and a milder effect for zanamivir. Analysis of the crystal structures of the N2 I222L and E119V mutants and the N2-I222L/E119V dual mutant reveals that a loss of a salt bridge due to the E119V substitution and a change in the hydrophobic environment due to the I222L substitution likely confer multidrug resistance. However, we found that this drug resistance can be overcome by a novel hybrid inhibitor, MS-257, which has both the guanidino group of zanamivir and hydrophobic pentyloxy group of oseltamivir (36, 37). Previously, our group has reported the reduced susceptibility of MS-257 to a known oseltamivir resistance mutation, H274Y (H1N1) (38). The change in position of the double bond in comparison with oseltamivir and the H-bonding networks made by the guanidinium function were shown to be responsible for the reduced resistance susceptibility of MS-257 against the H274Y mutant in the N1 subtype. Moreover, MS-257 does not exhibit cross-reactivity with human neuraminidase isoforms, NEU3 and NEU4 (39), further supporting its role as a leading, next-generation influenza virus NA inhibitor.
MATERIALS AND METHODS
Cloning, expression, and purification of influenza virus NAs.
The ectodomain (residues 83 to 469 in N2 numbering) of N2 protein from A/RI/5+/1957 (H2N2) clone ×7FINA12 and the mutations were expressed in a baculovirus system for structural and functional analyses. The E119V, I222L, and E119V/I222L mutations were constructed based on p57N2. The cDNAs corresponding to the N2 ectodomain were inserted into pFastBac1 vector with an N-terminal gp67 signal peptide, a 6×His tag, a tetramer sequence, and a thrombin cleavage site. The constructed plasmids were used to transform DH10bac competent bacterial cells (Invitrogen), and the recombinant bacmid was identified by blue-white receptor selection. The recombinant baculovirus was obtained by following the manufacturer's protocol. Virus stocks were amplified with three sequential infections of sf9 cells. For N2 expression, Hi5 cells grown at 300 K were infected at a density of 2 × 106/ml with high-titer fourth-passage (P4) recombinant baculovirus stock. After culture in suspension for 60 h at 300 K, Hi5 cells were removed by centrifugation, and supernatants containing secreted, soluble NAs were applied to a HisTrap HP 5-ml column (GE Health) and then eluted using 20 mM Tris-HCl (pH 8.0), 50 mM NaCl with 20 mM imidazole, 50 mM imidazole, and 200 mM imidazole. The target proteins were digested with thrombin to remove the 6×His tag and the tetramer sequence. The cleaved N2 proteins were further purified by size exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare) with 20 mM Tris-HCl (pH 8.0) and 50 mM NaCl. High-purity NA fractions were pooled and concentrated using a 10-kDa-cutoff concentrator (Millipore).
Crystallization, drug soaking, data collection, and structure determination.
Crystals of NAs were obtained by hanging drop vapor diffusion at 291 K. Initial screening was performed using a commercial kit (Hampton Research). Diffraction quality crystals of p57N2 (wild type) were obtained as described previously (18) using 0.1 M bis-Tris propane (pH 9.0) and 10% (vol/vol) Jeffamine ED-2001 (pH 7.0). N2 E119V mutant crystals were obtained using 0.1 M bicine (pH 8.5) and 15% (wt/vol) polyethylene glycol 1500. Quality N2-I222L mutation at 7 mg/ml was crystallized in 0.1 M HEPES (pH 7.5) and 12% (wt/vol) polyethylene glycol 3350. The crystals of E119V/I222L protein were obtained using 5% (vol/vol) ±-2-methyl-2,4-pentanediol, 0.1 M HEPES (pH 7.5), and 10% (wt/vol) polyethylene glycol 10000. NA crystals were incubated in mother liquor containing 20 mM oseltamivir, zanamivir, or 10 mM MS-257 and then flash-cooled at 100 K. Diffraction data for the p57N2 wild type and E119V mutant were collected at KEK beamlines NE-3A and BL-5A, respectively, while I222L and E119V/I222L mutant data were collected at SSRF beamline BL-17U. Data for all crystals were indexed, integrated, and scaled with HKL2000 (40).
The structures of mutations were determined by molecular replacement using Molrep (41) from the CCP4 program with the structure of 1957 pandemic H2N2 neuraminidase (PDB code 4K1H) as the search model. Initial restrained rigid-body refinement was performed using REFMAC5 (42). Further rounds of refinement were performed using phenix (43), and model building was carried out with the program Coot (44). The stereochemical quality of the final model was assessed with the program PROCHECK (45) in CCP4. Final statistics for all the structures are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parameter | Value(s) for: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N2 wild type | E119V mutant | E119V mutant- oseltamivir | E119V mutant- zanamivir | I222L mutant | I222L mutant- oseltamivir | I222L mutant- zanamivir | Dual mutant | Dual mutant- oseltamivir | Dual mutant- zanamivir | N2 wild type-MS-257 | E119V mutant– MS-257 | I222L mutant– MS-257 | Dual mutant– MS-257 | |
| Data collection statistics | ||||||||||||||
| Space group | C2221 | C2221 | C2221 | C2221 | P21212 | P21212 | C2221 | C2221 | P21 | C2221 | C2221 | C2221 | C2221 | P21212 |
| Unit cell dimensions (a, b, c) | 114.8, 139.5, 140.1 | 114.5, 139.4, 139.9 | 113.7, 139.1, 140.1 | 114.8, 138,8, 139.6 | 131.0, 136.2, 135.7 | 131.0, 136.7, 135.5 | 114.4, 139.3, 139.8 | 114.1, 139.5, 140.0 | 90.4, 140.0, 90.4 | 114.5, 139.3, 139.8 | 115.2, 139.7, 140.2 | 114.2, 138.9, 140.0 | 114.0, 140.0, 140.0 | 131.4, 137.4, 135.4 |
| Unit cell dimensions (α, β, γ) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 100.63, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution range (Å) | 50.00–1.80 (1.86–1.80) | 50.00–2.40 (2.49–2.40) | 50.00–2.50 (2.59–2.50) | 50.00–2.10 (2.18–2.10) | 50.00–2.20 (2.28–2.20) | 50.00–1.90 (1.97–1.90) | 50.00–2.10 (2.18–2.10) | 50.00–1.70 (1.76–1.70) | 50.00–1.80 (1.86–1.80) | 50.00–2.10 (2.18–2.10) | 50.00–1.90 (1.97–1.90) | 50.00–2.20 (2.28–2.20) | 50.00–2.00 (2.07–2.00) | 50.00–1.90 (1.97–1.90) |
| Rmerge (%) | 10.2 (54.7) | 15.8 (59.4) | 17.1 (50.9) | 13.8 (47.5) | 16.7 (54.0) | 11.2 (53.1) | 13.8 (47.5) | 5.7 (20.7) | 9.2 (39.5) | 11.7 (3.7) | 12.8 (51.4) | 15.2 (51.2) | 12.7 (46.6) | 9.0 (47.8) |
| I/σ | 17.6 (2.5) | 11.3 (3.3) | 11.6 (3.9) | 11.7 (3.7) | 12.8 (3.8) | 19.4 (4.3) | 11.7 (3.7) | 46.8 (16.3) | 23.0 (6.4) | 13.8 (47.5) | 17.1 (5.0) | 14.9 (3.8) | 13.8 (3.7) | 19.3 (4.0) |
| Redundancy | 6.2 (5.9) | 5.2 (5.2) | 6.2 (6.0) | 5.2 (5.1) | 7.0 (6.9) | 7.7 (7.5) | 5.2 (5.1) | 14.8 (14.4) | 8.4 (8.1) | 5.2 (5.1) | 7.1 (7.3) | 7.4 (7.2) | 7.1 (6.9) | 6.0 (5.5) |
| Completeness (%) | 96.0 (98.1) | 99.7 (100) | 98.0 (99.8) | 99.7 (100) | 100 (100) | 99.9 (99.9) | 99.7 (100) | 99.0 (98.0) | 99.9 (99.8) | 99.7 (100) | 99.7 (100) | 100 (100) | 95.3 (92.6) | 99.0 (96.5) |
| Refinement | ||||||||||||||
| Resolution (Å) | 37.5–1.8 | 88.5–2.0 | 88.1–2.5 | 88.5–2.1 | 136.3–2.2 | 136.7–1.9 | 88.46–1.9 | 88.35–1.7 | 135.9–1.8 | 135.7–1.8 | 38.8–1.90 | 88.2–2.2 | 88.2–2.0 | 135.4–1.9 |
| Rwork/Rfree (%) | 16.0/17.8 | 24.5/26.3 | 20.1/24.2 | 21.3/24.5 | 22.2/25.7 | 22.9/24.9 | 18.7/21.3 | 20.5/21.9 | 19.5/21.2 | 20.8/21.8 | 15.5/17.8 | 21.5/24.2 | 19.5/21.8 | 20.6/22.0 |
| RMSDa | ||||||||||||||
| Bond length (Å) | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.006 | 0.005 | 0.005 | 0.005 | 0.005 | 0.007 | 0.006 | 0.006 | 0.005 |
| Bond angle (°) | 1.199 | 1.107 | 1.175 | 1.114 | 1.110 | 1.094 | 1.046 | 1.003 | 1.003 | 1.010 | 1.161 | 1.190 | 1.154 | 1.038 |
RMSD, root mean square deviation.
Sialidase inhibition assay.
Sialidase enzymatic activity was measured using the fluorogenic substrate 4-methylumbelliferyl-N-acetylneuraminic acid (4-MUNANA) according to previously reported methods (17, 18, 46, 47). Briefly, the appropriate protein concentrations were chosen after several rounds of preliminary tests. Ten microliters of purified protein was mixed with 10 μl of serial dilution inhibitors (at least five concentrations at an appropriate range) were incubated for 30 min at 308 K. Then 30 μl of 167 μM 4-MUNANA in 33 mM morpholineethanesulfonic acid (MES) and 4 mM CaCl2 (pH 6.0) was added to the solution to start the reaction at the same time. Both positive and negative controls were included in each assay. Fluorescence was read with a SpectraMax M5 (Molecular Devices); the excitation wavelength was 355 nm, and the emission wavelength was 460 nm. To measure the inhibitory effects of the compounds, proteins and inhibitors were preincubated for 30 min at 37°C, and the substrate was then added. All inhibition assays were done in triplicates and over three inhibitor concentrations and four concentrations of the substrate. The kinetic parameters were calculated by fitting the data to the appropriate Michaelis-Menten equations using GraphPad Prism.
RESULTS
The N2 E119V/I222L dual mutant displays multidrug resistance.
In this study, wild-type N2 and three recombinant mutant proteins were generated using a baculovirus expression system according to previously reported methods (14, 18, 48). All NA proteins displayed stable sialidase activity using a standard 4-MUNANA-based fluorescence assay originally developed by Portier et al. (47). The Michaelis-Menten constants (Km) of the N2 wild type and E119V, I222L, and E119V/I222L mutants are 11.2 μM, 10.8 μM, 95.1 μM, and 51.9 μM, respectively, which are consistent with values obtained in the virus-based assay using MUNANA (35).
The inhibitory constants (Ki) of MS-257, oseltamivir, and zanamivir were determined for the wild type and mutant N2s using the same fluorescence-based assay. The inhibitory constants are listed in Table 2. The hybrid inhibitor showed excellent inhibitory properties in neuraminidase inhibition assays. The data revealed that MS-257 has better inhibition against the N2 E119V mutant than oseltamivir. Among the three inhibitors tested, oseltamivir showed the highest inihibitory activity against the E119V mutant, a 50-fold increase in Ki value, whereas zanamivir and MS-257 showed only 3.8- and 4.8-fold increases in Ki values, respectively. A similar trend was observed in the case of the E119V/I222L double mutant as well. For oseltamivir, the Ki value against the E119V/I222L mutant is 232-fold higher than the value against wild-type N2, while the Ki values are only 37-fold and 36-fold higher in the cases of zanamivir and MS-257, respectively. It is interesting that the I222L mutation alone did not bring significant resistance to any of the three inhibitors tested (Table 2), but when present together with the E119V mutation (double mutant), it brings significant resistance to oseltamivir. The profiles of inhibition for zanamivir and MS-257 against these mutants are similar.
TABLE 2.
Enzyme kinetics and inhibitory activities of three NA inhibitors against N2 wild-type and mutant enzymes
| Enzyme | Km (μM) | Relative Kia |
||
|---|---|---|---|---|
| Oseltamivir | Zanamivir | MS-257 | ||
| Wild type | 11.2 (9.37–13.0) | 1.0 | 1.0 | 1.0 |
| E119V mutant | 10.8 (6.24–15.4) | 50.5 | 3.50 | 4.83 |
| I222L mutant | 95.1 (82.4–108) | 4.29 | 2.14 | 2.79 |
| E119V/I222L mutant | 51.9 (43.0–60.9) | 232 | 37.6 | 36.0 |
Relative Ki is Ki of the mutant/Ki of the wild type. The Kis of the wild type for oseltamivir, zanamivir, and MS-257 are 0.21 nM, 0.14 nM, and 0.18 nM, respectively.
Structures of the N2 E119V mutant in complex with oseltamivir and zanamivir.
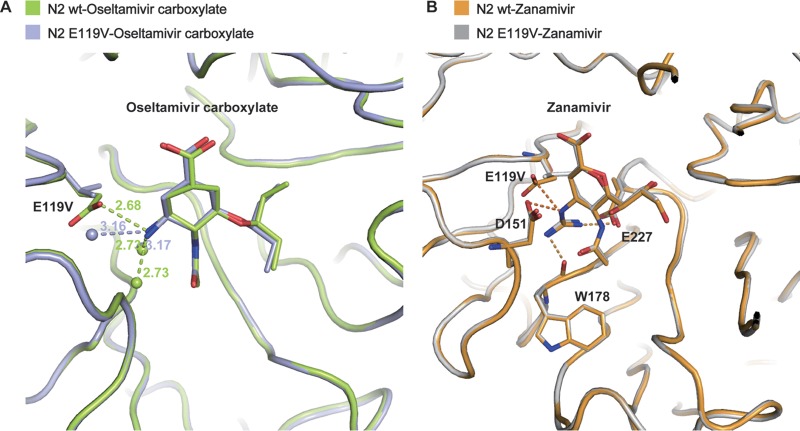
To understand the molecular basis of the resistance caused by E119V, crystal structures of the N2 E119V mutant were obtained in complex with oseltamivir and zanamivir. The N2 wild type-oseltamivir structure overlays almost exactly with the N2 E119V-oseltamivir structure, with a subtle repositioning of the cyclohexene ring and the hydrophobic group. The salt bridge between the oseltamivir amino group and the side chain of E119 is replaced by hydrogen bonds between the oseltamivir amino group and two water molecules because of the E119V substitution (Fig. 1).
FIG 1.
Antiviral resistance mechanism of the E119V mutant. (A) E119V causes oseltamivir resistance by losing the hydrogen bond between side chain of residue 119 and oseltamivir caboxylate amino group. The wild-type N2-oseltamivir and E119V mutant-oseltamivir are shown as limon and light blue sticks, respectively. For the water molecule distribution, the interactions between water molecules and oseltamivir amino group are stronger in the N2 wild type than in the E119V mutant. Water molecules are displayed as spheres. (B) The guanidino group of zanamivir forms more interactions with environmental residues (E227, W178, and D151), which compensate the loss of interaction due to E119V. The wild type-zanamivir and E119V mutant-zanamivir are shown as orange and silver sticks, respectively.
Comparison of the structures of the N2 wild type and N2 E119V mutant in complex with zanamivir shows that the inhibitors display nearly the same orientation, with a minor repositioning of the C-7 atom. N2-E119 forms a salt bridge with the zanamivir 4-guanidino group; however, the distance is further (3.33 Å) than that of the oseltamivir amino group (2.87 Å). Moreover, unlike the oseltamivir amino group, the zanamivir guanidino group displaces a water molecule beneath the 150 loop and forms additional interactions with the E227 carboxylate and the main-chain carbonyl of W178. After substitution of N2-E119 with valine, zanamivir also loses the salt bridge between residue 119 and the 4-guanidino group. However, the zanamivir 4-guanidino group still retains its typical interactions with E227, W178, and D151, which helps to explain why the E119V mutation results in only mild zanamivir resistance.
Weaker hydrophobic interactions in the I222L mutant result in oseltamivir resistance.
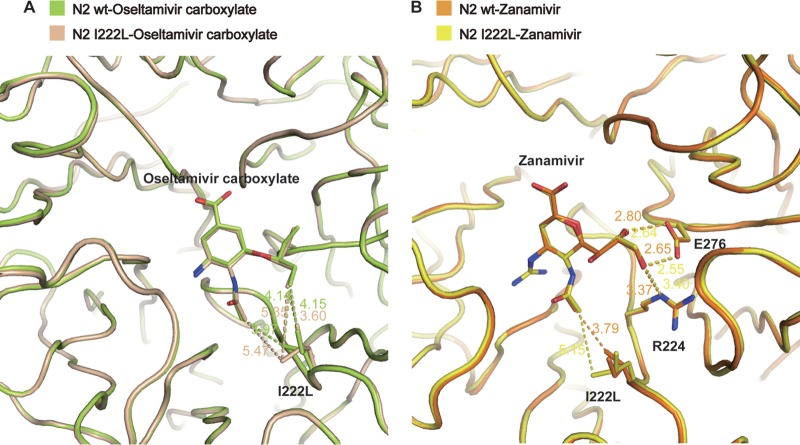
The oseltamivir 3-pentyloxy group binds to a hydrophobic pocket formed by I222, W178, and A246, as well as the carbon stems of the E276 and R224 side chains. These hydrophobic interactions are critical for oseltamivir potency, and the most common oseltamivir-resistant mutations interfere with these interactions (24, 49, 50). In wild-type N2, I222 is in an optimal position with its terminal C-δ 4.15 Å from the oseltamivir 3-pentyloxy group, and its C-γ2 carbon of 4.14 Å from the oseltamivir 3-pentyloxy and 3.97 Å from the oseltamivir N-acetyl carbon. However, in the I222L mutant, one of the terminal C-γ carbons is 3.60 Å from the 3-pentyloxy group, while the other terminal C-γ is 5.35 Å and 5.47 Å away from the 3-pentyloxy group and N-acetyl group, respectively (Fig. 2A).
FIG 2.
The polarity group of zanamivir enables inhibition of the I222L mutant. (A) Oseltamivir resistance caused by I222L mutant. In wild type-oseltamivir (limon), I222 is in an optimal position to form a hydrophobic interaction with the oseltamivir 3-pentyloxy group and N-acetyl carbon, while in I222L mutant-oseltamivir (wheat), the interactions between one of the L222 terminal C-γ atom and the oseltamivir carboxylate 3-pentyloxy group and N-acetyl group are weaker. (B) The zanamivir binding modes of the wild type (yellow) and I222L mutant (orange) are nearly the same. In both the wild-type and I222L mutant structures, the interactions among zanamivir glycerol moiety, E276, and R224 are nearly the same, which is the key factor to stable the zanamivir binding.
Although we did not observe any significant resistance to zanamivir, the binding mode was also slightly better in the wild-type N2 structure relative to the I222L mutant. In the wild-type structure, I222 is 3.79 Å (molecule B, 3.77 Å) away from the zanamivir N-acetyl carbon, whereas in the I222L mutant, L222 is 5.15 Å (molecule B, 5.10 Å) away. Yet in the I222L structure, the zanamivir glycerol moiety still retains its important interactions with E276 and R224 (Fig. 2B).
The E119V/I222L dual mutant results in severe resistance to oseltamivir but mild resistance to zanamivir.
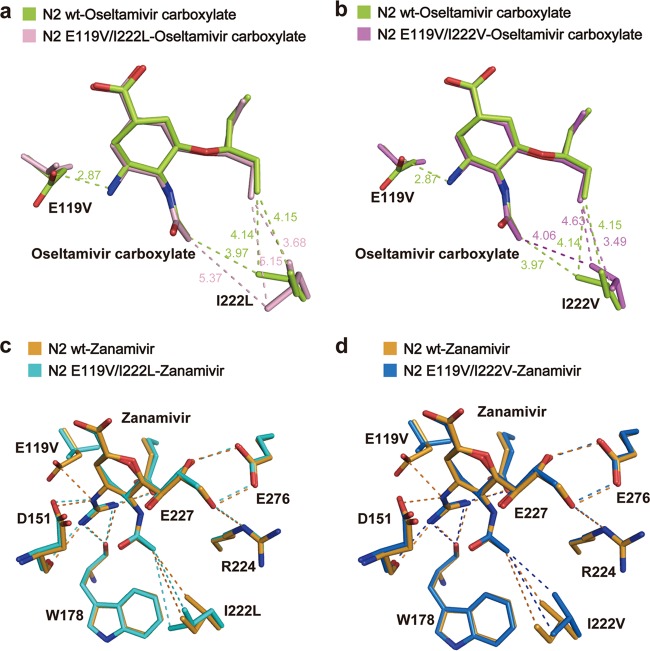
In order to determine the structural basis of E119/I222 group 2 NA resistance, we also solved crystal structures of the E119V/I222L dual mutant. Consistent with the single E119V and I222L substitutions, in the double mutant complex structure with oseltamivir, we observed the loss of the salt bridge with the amino group as well as weaker hydrophobic interactions between L222 and the 3-pentyloxy side chain (Fig. 3a). The presence of both substitutions therefore interferes with binding of all oseltamivir functional groups, with exception of the carboxylate group, and accordingly, the level of oseltamivir resistance is amplified significantly. In order to extrapolate these findings to the N2 E119V/I222V natural isolate, we replaced L222 with valine in our structure and observed a similar structural effect (Fig. 3b). In the N2 E119V/I222L-zanamivir complex structure we also observed a combination of the changes observed in the single mutant complex structures with no significant change in the positions of V119 and L222. Although the L222 single substitution resulted in a weakening of the hydrophobic interaction with the zanamivir N-acetyl group in the single mutant, it did not confer significant drug resistance in the single mutant. However, when the I222L mutation was present in combination with the E119V mutation (double bond), its effect seemed to be amplified in combination with the loss of the salt bridge between the residue 119 and the guanidino group.
FIG 3.
Structural basis of N2 dual resistance mutants. (a) The E119V/I222L dual mutant causes synergistic oseltamivir resistance. The wild type-oseltamivir and E119V/I222L mutant-oseltamivir are shown by limon and light pink sticks, respectively. (b) Model of the N2 E119V/I222V dual mutant-oseltamivir. The oseltamivir binding mode of the N2 E119V/I222V mutant (magenta) is similar to that of the E119V/I222L mutant. (c) The E119V/I222L dual mutant results in mild zanamivir resistance. The N2 wild type-zanamivir and E119V/I222L-zanamivir are colored orange and cyan, respectively. (d) Model of the N2 E119V/I222V dual mutant-zanamivir. The zanamivir binding mode of the E119V/I222V mutant (sky blue) is similar to that of the E119V/I222L mutant.
MS-257, a novel oseltamivir-zanamivir hybrid, is effective against multidrug-resistant N2.
Previously we found that oseltamivir can induce the opening of the N2 150 loop (16); however, this was never observed using zanamivir, which has a more positively charged guanidino group (51). Therefore, we hypothesized that the zanamivir guanidino group can help to overcome drug resistance through increased hydrogen bonding interactions with the NA active site. We recently reported a novel carbocycle, MS-257, which contains an oseltamivir 3-pentyloxy group in combination with a zanamivir 4-guanidino group (Fig. 4) (36–38). MS-257 is also similar to oseltamivir in that it is a carbocycle, yet it contains its double bond in the regioisomeric position corresponding to zanamivir. Inhibition assays indicate that MS-257 is both more potent than zanamivir and oseltamivir and resilient to drug-resistant mutations as predicted (Table 2).
FIG 4.
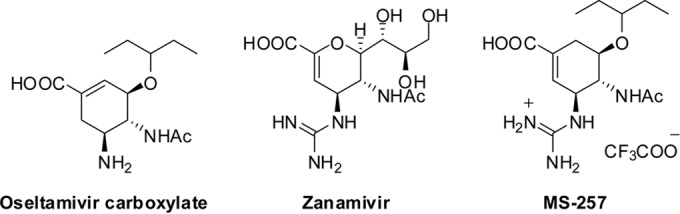
Chemical structures of influenza virus NA inhibitors.
In order to gain more insight into the structural basis, we solved the complex structures of MS-257 with wild-type N2 and our various E119V/I222L N2 mutants (Fig. 5). Despite its having its double bond in the zanamivir position, it is difficult to see any difference in the conformation of the MS-257 ring compared to our oseltamivir complex structures. As expected, in both the I222L and E119V/I222L complex structures, MS-257 also has weakened hydrophobic interactions in the same manner as oseltamivir. Furthermore, in the E119V and E119V/I222L complex structures, MS-257 still retains its salt bridges with E227 and D151, indicating that these additional interactions are critical for overcoming drug resistance. Interestingly, when bound to the E119V single mutant, the lipophilic pentyloxy group of MS-257 adopted a novel conformation.
FIG 5.
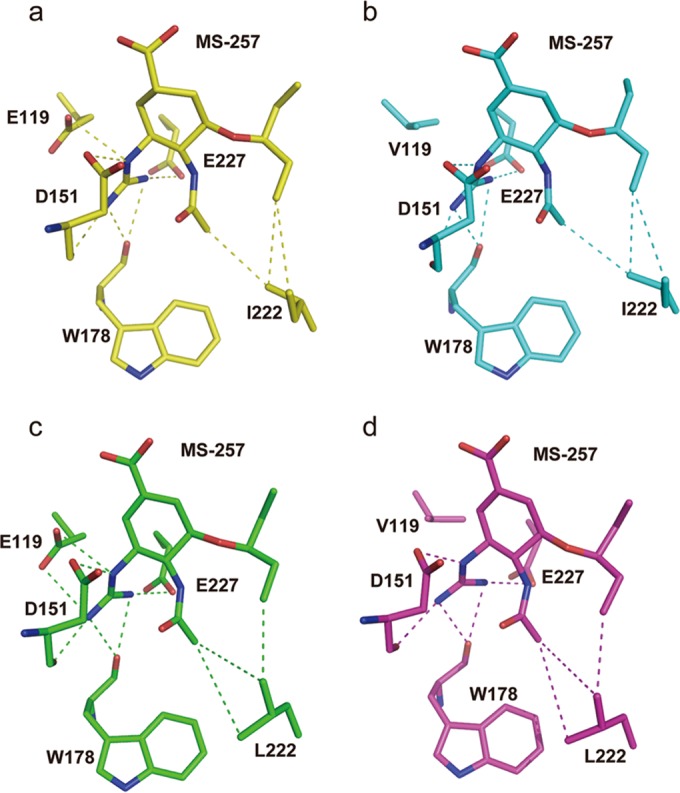
Effective mechanism of hybrid inhibitor MS-257. (a) Binding mode of MS-257 in wild-type N2 (yellow). (b) The binding mode of MS-257 in N2 E119V mutant (cyan) is similar to that of wild-type N2. (c) MS-257 also has weakened hydrophobic interactions in the I222L mutant N2 (green), which is similar to the case with oseltamivir. (d) Binding mode of MS-257 in the N2 E119V/I222L dual mutant (magenta).
DISCUSSION
In this study, we first confirmed, using highly purified recombinant NA, that replacement of E119 and I222 in N2 results in multidrug resistance. Previous studies have found different effects on NA activity and inhibitor binding when comparing virus and recombinant NAs. For example, the Wilson research group at Scripps Institute showed that recombinant N2 D151G mutants showed significantly decreased activity (52), which contrasted with the findings with complete influenza virus. Richard et al. concluded that passage of live influenza virus in the laboratory likely resulted in a reversion of some virus population to the wild type, leading to a rescue of NA activity (35). This illustrates that our results, which are consistent with previous results (35), are important for confirming that resistance due to substitution of E119 and I222 is a real phenomenon.
Interestingly, when N2 E119V and I222L substitutions occur together, the dual mutations result in both oseltamivir and zanamivir resistance, with oseltamivir resistance at a much higher level. Previous reports show that the N1 drug-resistant substitutions I223R (N1 numbering), S247N, and I117V/M also exhibit similar effects when combined with H275Y (N1 numbering) (53). However, zanamivir sensitivity is not significantly affected by the S247N and I117V/M N1 substitutions (5, 54). Similar to the case with N2 E119 and I222 substitution, zanamivir is also more effective than oseltamivir against the multidrug-resistant N1 with I223R and H275Y substitutions (25).
In all three N2 mutant complex structures, the oseltamivir amino group and zanamivir guanidino group both form salt bridges with D151. Yet the zanamivir guanidino group is larger and more positively charged and also engages in additional interactions with E227 and the main-chain carbonyl of W178 (16). Furthermore, we have previously observed that oseltamivir sometimes loses its interaction between its amino group and D151 (1), although in all of our complex structures and all other reported zanamivir structures, the 150 loop does remain closed (13, 17, 18). Therefore, we propose that these additional polar interactions with zanamivir are a major factor as to why the zanamivir resistance resulting from E119V and I222L in N2 is relatively mild.
In our previous study on the oseltamivir-induced opening of the N2 150 loop, we proposed that strengthening the interaction between the inhibitor and the 150 loop may help to overcome drug resistance (16). Therefore, we also included the structural and functional analyses of the novel inhibitor MS-257 (36, 37), which is essentially a hybrid between oseltamivir and zanamivir and contains a guanidino group with a binding site below the 150 loop. In accordance with our hypothesis, MS-257 remained very resilient toward the drug resistance substitutions relative to oseltamivir while retaining the same oseltamivir pentyloxy interactions and zanamivir guanidine interactions. Therefore, MS-257, which also shows no cross-reactivity with human NAs, serves as a lead scaffold for next-generation antiviral agents with reduced susceptibility to drug resistance.
ACKNOWLEDGMENT
We thank Xiaowu Chen for critical discussions.
Funding Statement
This work was supported by the National Natural Science Foundation of China (NSFC) (81330082 and 81301465), the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (XDB08020100), and the Natural Sciences and Engineering Research Council of Canada (NSERC). Yan Wu is supported by the Youth Innovation Promotion Association of the Chinese Academy of Sciences (Youth Innovation Promotion Association CAS) (2016086). George Fu Gao is a leading principal investigator of the NSFC Innovative Research Group (grant no. 81321063).
REFERENCES
- 1.Krumbholz A, Schmidtke M, Bergmann S, Motzke S, Bauer K, Stech J, Durrwald R, Wutzler P, Zell R. 2009. High prevalence of amantadine resistance among circulating European porcine influenza A viruses. J Gen Virol 90:900–908. doi: 10.1099/vir.2008.007260-0. [DOI] [PubMed] [Google Scholar]
- 2.Deyde V, Garten R, Sheu T, Smith C, Myrick A, Barnes J, Xu X, Shaw M, Klimov A, Gubareva L. 2009. Genomic events underlying the changes in adamantane resistance among influenza A(H3N2) viruses during 2006–2008. Influenza Other Respir Viruses 3:297–314. doi: 10.1111/j.1750-2659.2009.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deyde VM, Xu X, Bright RA, Shaw M, Smith CB, Zhang Y, Shu Y, Gubareva LV, Cox NJ, Klimov AI. 2007. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J Infect Dis 196:249–257. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- 4.Simonsen L, Viboud C, Grenfell BT, Dushoff J, Jennings L, Smit M, Macken C, Hata M, Gog J, Miller MA, Holmes EC. 2007. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol 24:1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- 5.Hurt AC LS, Speers DJ, Barr IG, Maurer-Stroh S. 2012. Mutations I117V and I117M and oseltamivir sensitivity of pandemic (H1N1) 2009 viruses. Emerg Infect Dis 18:109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seibert CW, Rahmat S, Krammer F, Palese P, Bouvier NM. 2012. Efficient transmission of pandemic H1N1 influenza viruses with high-level oseltamivir resistance. J Virol 86:5386–5389. doi: 10.1128/JVI.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stoner TD, Krauss S, DuBois RM, Negovetich NJ, Stallknecht DE, Senne DA, Gramer MR, Swafford S, DeLiberto T, Govorkova EA, Webster RG. 2010. Antiviral susceptibility of avian and swine influenza virus of the N1 neuraminidase subtype. J Virol 84:9800–9809. doi: 10.1128/JVI.00296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Cheung CL, Tai H, Zhao P, Chan JF, Cheng VC, Chan KH, Yuen KY. 2009. Oseltamivir-resistant influenza A pandemic (H1N1) 2009 virus, Hong Kong, China. Emerg Infect Dis 15:1970–1972. doi: 10.3201/eid1512.091057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le QM, Kiso M, Someya K, Sakai YT, Nguyen TH, Nguyen KH, Pham ND, Ngyen HH, Yamada S, Muramoto Y, Horimoto T, Takada A, Goto H, Suzuki T, Suzuki Y, Kawaoka Y. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437:1108. doi: 10.1038/4371108a. [DOI] [PubMed] [Google Scholar]
- 10.Monto AS, McKimm-Breschkin JL, Macken C, Hampson AW, Hay A, Klimov A, Tashiro M, Webster RG, Aymard M, Hayden FG, Zambon M. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother 50:2395–2402. doi: 10.1128/AAC.01339-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen HT, Fry AM, Gubareva LV. 2012. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir Ther 17:159–173. doi: 10.3851/IMP2067. [DOI] [PubMed] [Google Scholar]
- 12.Abed Y, Baz M, Boivin G. 2006. Impact of neuraminidase mutations conferring influenza resistance to neuraminidase inhibitors in the N1 and N2 genetic backgrounds. Antivir Ther 11:971–976. [PubMed] [Google Scholar]
- 13.Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, Hay AJ, Gamblin SJ, Skehel JJ. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 14.Li Q, Qi J, Zhang W, Vavricka CJ, Shi Y, Wei J, Feng E, Shen J, Chen J, Liu D, He J, Yan J, Liu H, Jiang H, Teng M, Li X, Gao GF. 2010. The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat Struct Mol Biol 17:1266–1268. doi: 10.1038/nsmb.1909. [DOI] [PubMed] [Google Scholar]
- 15.Sun X, Li Q, Wu Y, Wang M, Liu Y, Qi J, Vavricka CJ, Gao GF. 2014. Structure of influenza virus N7: the last piece of the neuraminidase “jigsaw” puzzle. J Virol 88:9197–9207. doi: 10.1128/JVI.00805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, Qin G, Gao F, Liu Y, Vavricka CJ, Qi J, Jiang H, Yu K, Gao GF. 2013. Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci Rep 3:1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Bi Y, Vavricka CJ, Sun X, Zhang Y, Gao F, Zhao M, Xiao H, Qin C, He J, Liu W, Yan J, Qi J, Gao GF. 2013. Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res doi: 10.1038/cr.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vavricka CJ, Li Q, Wu Y, Qi J, Wang M, Liu Y, Gao F, Liu J, Feng E, He J, Wang J, Liu H, Jiang H, Gao GF. 2011. Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS Pathog 7:e1002249. doi: 10.1371/journal.ppat.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Qi J, Liu Y, Vavricka CJ, Wu Y, Li Q, Gao GF. 2011. Influenza A virus N5 neuraminidase has an extended 150-cavity. J Virol 85:8431–8435. doi: 10.1128/JVI.00638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cox NJ, Subbarao K. 2000. Global epidemiology of influenza: past and present. Annu Rev Med 51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention. 2012. Update: influenza A (H3N2)v transmission and guidelines—five states, 2011. MMWR Morb Mortal Wkly Rep 60:1741–1744. [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention. 2011. Swine-origin influenza A (H3N2) virus infection in two children—Indiana and Pennsylvania, July–August 2011. MMWR Morb Mortal Wkly Rep 60:1213–1215. [PubMed] [Google Scholar]
- 23.Centers for Disease Control and Prevention. 2011. Limited human-to-human transmission of novel influenza A (H3N2) virus—Iowa, November 2011. MMWR Morb Mortal Wkly Rep 60:1615–1617. [PubMed] [Google Scholar]
- 24.Collins PJ, Haire LF, Lin YP, Liu J, Russell RJ, Walker PA, Skehel JJ, Martin SR, Hay AJ, Gamblin SJ. 2008. Crystal structures of oseltamivir-resistant influenza virus neuraminidase mutants. Nature 453:1258–1261. doi: 10.1038/nature06956. [DOI] [PubMed] [Google Scholar]
- 25.van der Vries E, Collins PJ, Vachieri SG, Xiong X, Liu J, Walker PA, Haire LF, Hay AJ, Schutten M, Osterhaus AD, Martin SR, Boucher CA, Skehel JJ, Gamblin SJ. 2012. H1N1 2009 Pandemic influenza virus: resistance of the I223R neuraminidase mutant explained by kinetic and structural analysis. PLoS Pathog 8:e1002914. doi: 10.1371/journal.ppat.1002914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okomo-Adhiambo M, Demmler-Harrison GJ, Deyde VM, Sheu TG, Xu X, Klimov AI, Gubareva LV. 2010. Detection of E119V and E119I mutations in influenza A (H3N2) viruses isolated from an immunocompromised patient: challenges in diagnosis of oseltamivir resistance. Antimicrob Agents Chemother 54:1834–1841. doi: 10.1128/AAC.01608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baz M, Abed Y, McDonald J, Boivin G. 2006. Characterization of multidrug-resistant influenza A/H3N2 viruses shed during 1 year by an immunocompromised child. Clin Infect Dis 43:1555–1561. doi: 10.1086/508777. [DOI] [PubMed] [Google Scholar]
- 28.Barnett JM, Cadman A, Burrell FM, Madar SH, Lewis AP, Tisdale M, Bethell R. 1999. In vitro selection and characterisation of influenza B/Beijing/1/87 isolates with altered susceptibility to zanamivir. Virology 265:286–295. doi: 10.1006/viro.1999.0058. [DOI] [PubMed] [Google Scholar]
- 29.Gubareva LV, Robinson MJ, Bethell RC, Webster RG. 1997. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J Virol 71:3385–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blick TJ, Tiong T, Sahasrabudhe A, Varghese JN, Colman PM, Hart GJ, Bethell RC, McKimm-Breschkin JL. 1995. Generation and characterization of an influenza virus neuraminidase variant with decreased sensitivity to the neuraminidase-specific inhibitor 4-guanidino-Neu5Ac2en. Virology 214:475–484. doi: 10.1006/viro.1995.0058. [DOI] [PubMed] [Google Scholar]
- 31.Whitley RJ, Hayden FG, Reisinger KS, Young N, Dutkowski R, Ipe D, Mills RG, Ward P. 2001. Oral oseltamivir treatment of influenza in children. Pediatr Infect Dis J 20:127–133. doi: 10.1097/00006454-200102000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Kiso M, Mitamura K, Sakai-Tagawa Y, Shiraishi K, Kawakami C, Kimura K, Hayden FG, Sugaya N, Kawaoka Y. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364:759–765. doi: 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- 33.Carr J, Ives J, Roberts NA, Tai CY, Mendel DB, Kelly L, Lambkin R, Oxford J. 2000. An oseltamivir treatment-selected influenza A/Wuhan/359/95 virus with an E119V mutation in the neuraminidase gene has reduced infectivity in vivo, p 23 In Second International Symposium on Influenza and other Respiratory Viruses. The Macrae Group, New York, NY. [Google Scholar]
- 34.Staschke KA, Colacino JM, Baxter AJ, Air GM, Bansal A, Hornback WJ, Munroe JE, Laver WG. 1995. Molecular basis for the resistance of influenza viruses to 4-guanidino-Neu5Ac2en. Virology 214:642–646. doi: 10.1006/viro.1995.0078. [DOI] [PubMed] [Google Scholar]
- 35.Richard M, Ferraris O, Erny A, Barthelemy M, Traversier A, Sabatier M, Hay A, Lin YP, Russell RJ, Lina B. 2011. Combinatorial effect of two framework mutations (E119V and I222L) in the neuraminidase active site of H3N2 influenza virus on resistance to oseltamivir. Antimicrob Agents Chemother 55:2942–2952. doi: 10.1128/AAC.01699-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohan S, McAtamney S, Haselhorst T, von Itzstein M, Pinto BM. 2010. Carbocycles related to oseltamivir as influenza virus group-1-specific neuraminidase inhibitors. Binding to N1 enzymes in the context of virus-like particles. J Med Chem 53:7377–7391. [DOI] [PubMed] [Google Scholar]
- 37.Niikura M, Bance N, Mohan S, Mario Pinto B. 2011. Replication inhibition activity of carbocycles related to oseltamivir on influenza A virus in vitro. Antiviral Res 90:160–163. doi: 10.1016/j.antiviral.2011.03.180. [DOI] [PubMed] [Google Scholar]
- 38.Kerry PS, Mohan S, Russell RJ, Bance N, Niikura M, Pinto BM. 2013. Structural basis for a class of nanomolar influenza A neuraminidase inhibitors. Sci Rep 3:2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albohy A, Mohan S, Zheng RB, Pinto BM, Cairo CW. 2011. Inhibitor selectivity of a new class of oseltamivir analogs against viral neuraminidase over human neuraminidase enzymes. Bioorg Med Chem 19:2817–2822. doi: 10.1016/j.bmc.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 40.Rossmann MG, van Beek CG. 1999. Data processing. Acta Crystallogr D Biol Crystallogr 55:1631–1640. doi: 10.1107/S0907444999008379. [DOI] [PubMed] [Google Scholar]
- 41.Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30:1022–1025. doi: 10.1107/S0021889897006766. [DOI] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RMM, Moss D, Thornton J. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 46.Gubareva LV, Webster RG, Hayden FG. 2001. Comparison of the activities of zanamivir, oseltamivir, and RWJ-270201 against clinical isolates of influenza virus and neuraminidase inhibitor-resistant variants. Antimicrob Agents Chemother 45:3403–3408. doi: 10.1128/AAC.45.12.3403-3408.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-d-N-acetylneuraminate) substrate. Anal Biochem 94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 48.Xu X, Zhu X, Dwek RA, Stevens J, Wilson IA. 2008. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J Virol 82:10493–10501. doi: 10.1128/JVI.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins PJ, Haire LF, Lin YP, Liu J, Russell RJ, Walker PA, Martin SR, Daniels RS, Gregory V, Skehel JJ, Gamblin SJ, Hay AJ. 2009. Structural basis for oseltamivir resistance of influenza viruses. Vaccine 27:6317–6323. doi: 10.1016/j.vaccine.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 50.Gubareva LV, Kaiser L, Matrosovich MN, Soo-Hoo Y, Hayden FG. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J Infect Dis 183:523–531. doi: 10.1086/318537. [DOI] [PubMed] [Google Scholar]
- 51.Dörwald FZ. 2012. Lead optimization for medicinal chemists. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 52.Zhu X, McBride R, Nycholat CM, Yu W, Paulson JC, Wilson IA. 2012. Influenza virus neuraminidases with reduced enzymatic activity that avidly bind sialic acid receptors. J Virol 86:13371–13383. doi: 10.1128/JVI.01426-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen HT, Fry AM, Loveless PA, Klimov AI, Gubareva LV. 2010. Recovery of a multidrug-resistant strain of pandemic influenza A 2009 (H1N1) virus carrying a dual H275Y/I223R mutation from a child after prolonged treatment with oseltamivir. Clin Infect Dis 51:983–984. doi: 10.1086/656439. [DOI] [PubMed] [Google Scholar]
- 54.Hurt AC, Lee RT, Leang SK, Cui L, Deng YM, Phuah SP, Caldwell N, Freeman K, Komadina N, Smith D, Speers D, Kelso A, Lin RT, Maurer-Stroh S, Barr IG. 2011. Increased detection in Australia and Singapore of a novel influenza A(H1N1)2009 variant with reduced oseltamivir and zanamivir sensitivity due to a S247N neuraminidase mutation. Euro Surveill 16(23):pii=19884. [PubMed] [Google Scholar]