Abstract
The transcriptional coactivators p300 and pCAF are necessary for the myogenic factor MyoD to initiate the expression of skeletal muscle genes. In addition to mediating histone acetylation, both of these factors can acetylate MyoD; however, the complexity of cellular systems used to study MyoD has impeded delineation of the specific roles of these two acetyltransferases. Therefore, we established a MyoD-dependent in vitro transcription system that permits us to determine the roles of p300 and pCAF during MyoD-dependent transcriptional activation. Consistent with results from cellular systems, we demonstrate that maximal levels of transactivation in vitro require both p300 and pCAF, as well as the cofactor acetyl CoA. Dissection of the steps leading to transcription initiation revealed that the activities of p300 and pCAF are not redundant. During the initial stages of transactivation, p300 acetylates histone H3 and H4 within the promoter region and then recruits pCAF to MyoD. Once tethered to the promoter, pCAF acetylates MyoD to facilitate the transactivation process. Thus, we have established that pCAF and p300 carry out sequential and functionally distinct events on a promoter leading to transcriptional activation. Further dissection of this in vitro transcription system should be highly useful toward elucidating the mechanism by which coactivators facilitate differential gene expression by MyoD.
MyoD and the related myogenic basic helix–loop–helix proteins (Myf5, myogenin, and MRF4) regulate skeletal myogenesis as heterodimers with E proteins (including E47, E12, E2-2, and HEB) (1), which bind to a consensus sequence of CANNTG (E-box) and activate muscle-specific gene expression (2). Studies in cultured cells demonstrated that exogenous expression of MyoD is sufficient to initiate the myogenic program (3, 4) and that MyoD orchestrates this process by establishing a temporally ordered differential regulation of gene expression (5). Numerous coactivators have been identified that interact with MyoD to regulate its activity in a cell (reviewed in ref. 6). However, the inherent complexity of cellular systems has limited our understanding of the functional interactions among cofactors.
Two coactivators required for transcriptional activation by MyoD are the acetyltransferase (AT) proteins p300 and pCAF (7, 8). Histone acetylation facilitates promoter recognition by proteins that possess a bromodomain (9) and promotes the transfer of histones H2A/H2B from nucleosomes to chaperone proteins during transactivation (10). Consistent with this role, MyoD binding to specific promoters has been shown to coincide with an increased acetylation of histones leading to gene activation (5). In addition, pCAF and p300 are both able to acetylate MyoD, but the functional significance of this modification is not clear (11, 12). Interestingly, p300 and pCAF are required for transactivation leading to muscle differentiation but the AT domain of p300 was dispensable for transcription from a MyoD-responsive reporter construct in cultured cells (7). In contrast, studies performed in Xenopus oocytes (using the p300-specific inhibitor Lys-CoA) demonstrated a critical role for this domain in MyoD-dependent transcriptional activation (13). Thus, the relative contribution of p300 and pCAF, alone or in combination, in the establishment of transcriptionally competent promoters at MyoD-responsive genes remains unclear.
To better understand the role of p300 and pCAF in mediating MyoD-dependent transactivation, we have established an in vitro transcription system using nucleosomal DNA templates. Using this system, we confirmed that both p300 and pCAF are required for maximal MyoD-dependent transactivation and demonstrate that these two ATs play distinct and sequential roles in the transactivation process: p300 acetylates histones H3 and H4 and recruits pCAF to the promoter whereas pCAF acetylates MyoD to enhance transcription initiation.
Materials and Methods
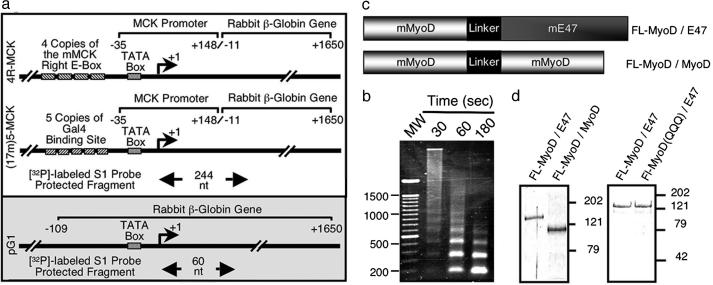
DNA Templates. Reporter DNA templates were generated by cloning a PCR-amplified fragment of the mouse muscle creatine kinase (MCK) promoter (–35 to +148) into the HindIII site of pAL3 (14). Either four copies of the MCK right E-box or five copies of the Gal4 DNA binding site were cloned into the SacI site immediately upstream of the MCK minimal promoter to generate the plasmids 4R-MCK, and (17m)5-MCK, respectively (see Fig. 1a). Nucleosomal arrays were generated by using recombinant Drosophila ACF complex and NAP-1 as described (15) and were confirmed by micrococcal nuclease analysis (16).
Fig. 1.
Components of a MyoD-dependent in vitro transcription system. (a) The structure of the 4R-MCK, 17m5-MCK, and pG1 templates are schematically represented. (b) Analysis of nucleosomal arrays on 4R-MCK DNA template. After chromatin assembly, 300 ng of 4R-MCK was digested by using 0.3 units of MNase (250 μl, 24°C) for varying amounts of time (16). DNA was separated on a 1.5% agarose gel and stained with ethidium bromide. (c) Schematic diagram of the structure of the tethered dimers FL-MyoD/MyoD and FL-MyoD/E47. (d) Coomassie blue staining of FL-MyoD/E47, FL-MyoD(QQQ)/E47, and FL-MyoD/MyoD purified from baculovirus-infected Sf-9 cells.
Protein Expression and Purification. The plasmid pFB-FL was generated by inserting an oligonucleotide encoding the Flag-epitope into the BamHI site of pFastBac (Invitrogen). The constructs pFB-FL-MyoD and pFB-FL-E47 were generated by cloning PCR-amplified cDNAs of mouse MyoD or E47 into BamHI-EcoRI-digested pFB-FL. To generate the FL-MyoD/E47 (pFB-FL-MyoD/E47) and FL-MyoD/MyoD (pFB-FL-MyoD/MyoD) tethered dimers (see Fig. 1c), the stop codon was removed from the N-terminal MyoD portion of the dimer, which was then fused to the C-terminal protein (either E47 or MyoD) by using a previously described linker peptide (17). FL-MyoD(QQQ)/E47, which contains the mutations K99Q, K102Q, and K104Q, was generated by using the QuikChange (Stratagene) protocol. The integrity of all constructs was confirmed by sequencing.
FL-MyoD, FL-E47, FL-MyoD/E47, FL-MyoD/MyoD, and FL-pCAF were expressed in baculovirus-infected Sf-9 cells and purified by using M2-Agarose resin (Sigma). Flag-tagged ACF complex was purified as described (15). Bacterially expressed His-Gal4-VP16 (18) and baculovirus-expressed His-p300(wt) (19) and His-p300(am) (20) proteins were purified by using Ni2+-NTA resin (Qiagen, Valencia, CA). The ability of the purified recombinant FL-MyoD, FL-MyoD/MyoD, FL-MyoD/E47 (see Fig. 5, which is published as supporting information on the PNAS web site), and FL-E47 (data not shown) to bind an E-Box was confirmed by using gel mobility shift analysis as described (21). The histone AT activity of purified FL-pCAF and His-p300(wt) was confirmed (see Fig. 6, which is published as supporting information on the PNAS web site) as described (22).
Transcription. Transcription reactions were carried out on 15 fmol of naked or nucleosomal 4R-MCK template, in the presence or absence of 1 pmol of transcriptional activator (FL-MyoD, FL-E47, FL-MyoD/E47, or FL-MyoD/MyoD), 25 fmol of FL-pCAF and/or His-p300(wt), HeLa nuclear extract (3 μg), and acetyl CoA (1 μM) at 27°C. After 30 min, 60 μg of HeLa nuclear extract (23) was added to reactions and left to incubate for 10 min at 30°C. Finally, dNTPs (0.5 mM) and the internal control template pG1 (6.5 fmol) were added to the reaction (total volume 50 μl) and allowed to proceed for 45 min. In experiments where single rounds of transcription were measured, 0.1% sarkosyl was added to reactions 30 s after the dNTPs and then allowed to continue for 45 min. After deproteination, RNA was recovered and subjected to S1 nuclease analysis as described (18). Transcripts originating from the 4R-MCK template gave rise to a protected fragment of 244 nt whereas transcripts originating from pG1 generated a protected fragment of 60 nt (see Fig. 1a).
Acetylation of MyoD. FL-MyoD/E47 (5 pmol) was incubated with the nucleosomal 4R-MCK template (75 fmol) and acetyl CoA (1 μM) in the presence or absence of 125 fmol of either FL-pCAF, His-p300(wt), or His-p300(am) for 30 min at 27°C in a total volume of 250 μl. Proteins were then precipitated with 4 vol of cold (–20°C) acetone, redissolved in 1× gel loading dye, separated on an 8% SDS/PAGE gel, and analyzed by Western blot. Acetylation of FL-MyoD/E47 was detected by using an antibody recognizing acetylated proteins (AbCAM) whereas total FL-MyoD-E47 was detected by using the Flag-M2 antibody (Sigma).
Chromatin Immunoprecipitation (ChIP). ChIP reactions were carried out as described (24). Reactions were set up as described for transcription experiments, except that, instead of adding rNTPs and pG1 template to initiate transcription, the DNA template was digested with the restriction endonuclease HaeIII for 1 h. Fragmented nucleosomes were subjected to immunoprecipitation by using either an anti-acetyl-Histone H3 or an anti-acetyl-Histone H4 antibody (Upstate Biotechnology, Lake Placid, NY). Antibody-bound nucleosomes were recovered by using protein A-coated Dynabeads (Dynal, Great Neck, NY) applied to a nylon membrane by using a Slot Blotter (Bio-Rad), and subjected to Southern blot analysis by using 32P-labeled template-specific probes. The promoter proximal and distal probe recognized an HaeIII fragment of –193 to +317, and +2258 to +2692, respectively.
Results
Establishing an in Vitro Transcription System. To establish a MyoD-dependent in vitro transcription system, we used the 4R-MCK reporter [four copies of the right E-box from the mouse MCK promoter (25) immediately upstream of the mouse MCK minimal promoter, which was then fused to the rabbit β-globin gene (see Fig. 1a)]. The 4R-MCK template was incorporated into nucleosomal arrays (Fig. 1b) by using the previously described ACF complex chromatin assembly system (15).
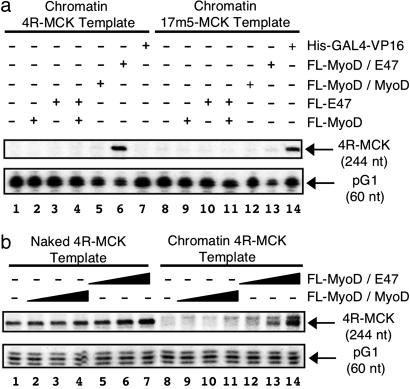
To compare MyoD and E47 as homo- and heterodimers, we expressed and purified MyoD and E47 in several different forms (Fig. 1 c and d). In addition to the Flag-tagged monomer versions of MyoD (FL-MyoD) and E47 (FL-E47), we generated tethered versions of these proteins to create either a forced MyoD homodimer (FL-MyoD/MyoD) or a forced MyoD/E47 heterodimer (FL-MyoD/E47) because these fusion proteins will allow us to study heterodimer binding without competition from homodimers. The purified basic helix–loop–helix proteins were able to bind an E-box-containing probe in gel mobility shift assays, and, similar to the progesterone receptor (26), the recombinant proteins required a small amount of nuclear extract for efficient binding (data not shown). Next, we examined the ability of these different forms of basic helix–loop–helix proteins to activate transcription on the nucleosomal 4R-MCK template. His-p300(wt) and FL-pCAF were included in all initial experiments because these proteins have been suggested to be necessary for MyoD-dependent transcription (7). Under these conditions, we found that the forced MyoD/E47 heterodimers (FL-MyoD/E47) were efficient inducers of transactivation from the 4R-MCK promoter (Fig. 2a, lane 6). In contrast, FL-MyoD and FL-E47 homodimers were unable to activate transcription on this template (Fig. 2a, lanes 1–3 and 5). Surprisingly, the combination of MyoD and E47 monomers also failed to activate transcription (Fig. 2a, lane 4), possibly due to competition between the mixed population of homo- and heterodimers or altered stability of dimer subpopulations in the in vitro transcription reaction.
Fig. 2.
MyoD/E47 forced-heterodimers activate transcription on 4R-MCK template. (a) Transactivation by MyoD/E47 requires the presence of E-boxes within the promoter region. Transactivation by FL-MyoD, FL-E47, FL-MyoD/MyoD, FL-MyoD/E47 (1 pmol), or His-Gal4/VP16 (50 fmol) was examined on 15 fmol of nucleosomal 4R-MCK or 17m5-MCK DNA template in the presence of FL-pCAF, His-p300(wt), acetyl CoA, and HeLa nuclear extract as described in Materials and Methods. (b) MyoD/E47 heterodimers, and not MyoD/MyoD homodimers, activate transcription on 4R-MCK templates. Varying amounts (0.1–1 pmol) of either FL-MyoD/MyoD or FL-MyoD/E47 were incubated with naked (lanes 1–7) or nucleosomal (lanes 8–14) 4R-MCK templates (15 fmol) as described in Materials and Methods. Transcripts generated in a and b were analyzed by S1 nuclease analysis where RNA originating from the 4R-MCK and pG1 (control) template generated 244- and 60-nt fragments, respectively (see Fig. 1a). Please note that the band at 249 nt in b, lanes 9–14 represents a MyoD-inducible alternative transcription start site observed from nucleosomal templates in the presence of some (but not all) HeLa nuclear extracts (F.J.D. and S.J.T., unpublished observations).
The activation of transcription by FL-MyoD/E47 was specific for a promoter containing an E-box because no transactivation was observed in the presence of FL-MyoD/E47 on a control template where the four E-boxes of the 4R-MCK template were replaced by five copies of a Gal4-binding site [(17m)5-MCK; Fig. 2a, lane 13]. Analogously, Gal4-VP16 was an efficient activator of transcription on the (17m)5-MCK template (Fig. 2a, lane 14), but not the 4R-MCK template (Fig. 2a, lane 7). Similar to our results on the nucleosomal DNA, FL-MyoD/E47 is significantly more efficient than FL-MyoD/MyoD at activating transcription from the naked 4R-MCK (Fig. 2b, lanes 1–7). Therefore, the E47 moiety of the MyoD/E47 heterodimer is required for maximal levels of transactivation on both naked and nucleosomal templates.
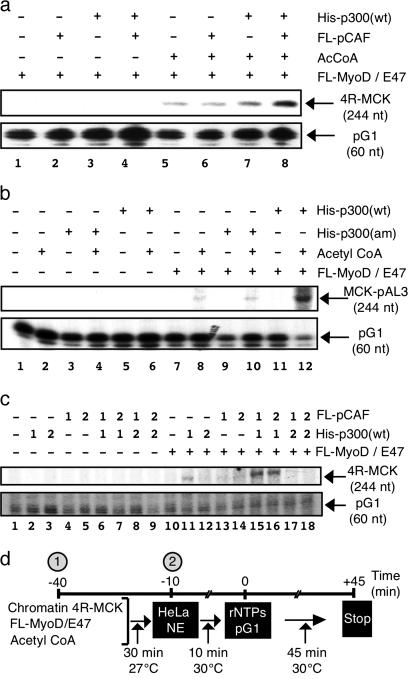
Investigating the Role of p300 and pCAF in MyoD-Dependent Transactivation. Having established a MyoD-dependent in vitro transcription system, we next set out to examine the role of the transcriptional coactivators p300 and pCAF in MyoD-dependent transactivation. When transcription reactions were performed in the absence of the cofactor acetyl CoA, neither His-p300(wt) nor FL-pCAF (alone or in combination) could stimulate transactivation by FL-MyoD/E47 (Fig. 3a). Furthermore, the addition of acetyl CoA was not sufficient to stimulate transcription by FL-MyoD/E47 when FL-pCAF alone was added to the reaction (Fig. 3a, lane 6). Modest levels of transcription were reproducibly observed when both His-p300(wt) and acetyl CoA were present in the transcription reaction (Fig. 3a, lane 7; see also Fig. 3c, lane 11). However, the presence of both His-p300(wt) and FL-pCAF (along with acetyl CoA) resulted in a synergistic activation of transcription (Fig. 3 a, lane 8, and c, lanes 15 and 16) that depended on the presence of FL-MyoD/E47 (data not shown; see also Fig. 3 b and c). This synergistic activation of transcription is critically dependent on the AT domain of p300 because no synergy with FL-pCAF is observed when the p300 AT domain mutant [His-p300(am)] is substituted into the reaction (Fig. 3b, lane 10). This finding suggests that p300 and pCAF play complementary roles in the acetyl CoA-dependent process of MyoD-mediated transactivation.
Fig. 3.
The ATs p300 and pCAF act synergistically to activate transcription on nucleosomal 4R-MCK template. (a) Maximal transactivation by FL-MyoD/E47 requires pCAF, p300, and acetyl CoA. FL-MyoD/E47 transcription from the nucleosomal 4R-MCK template was examined in the presence or absence of FL-pCAF (25 fmol), His-p300(wt) (25 fmol), and/or acetyl CoA (1 μM). (b) The AT activity of p300 is required for transactivation by MyoD/E47. Transcription reactions were performed as in Fig. 3a, except that pCAF was present in all reactions whereas acetyl CoA and/or wild-type [His-p300(wt)] or AT mutant [His-p300(am)] forms of p300 were included as indicated. (c) p300 is acting before pCAF to mediate transactivation by MyoD/E47. Transcription was performed in the presence or absence of FL-MyoD/E47 on the nucleosomal 4R-MCK. Where indicated, FL-pCAF and/or His-p300(wt) was added to the reactions either at the beginning of the transcription reaction (time –40 min)(marked by a 1), at the same time as the HeLa nuclear extract (time –10) (marked by a 2), or not at all (marked by a minus sign). (d) Timeline of transcription reaction showing when different components are added to the system.
To better understand the relationship between His-p300(wt) and FL-pCAF during the transactivation process, the ATs were added to the reaction at different times. In all experiments, His-p300(wt) had to be added at the earliest time point to observe transactivation whether FL-pCAF was present (Fig. 3c, compare lanes 15 and 16 with lanes 17 and 18) or not (Fig. 3c, compare lane 11 with lane 12). In contrast, FL-pCAF could be added to transcription reactions at the same time as the nuclear extract (–10 min) without affecting its ability to act synergistically with His-p300(wt) to stimulate transactivation (Fig. 3c, compare lanes 15 and 16). When transcription reactions were limited to the initiation step by the addition of sarkosyl (to block reinitiation), His-p300(wt) and FL-pCAF continued to act synergistically in the activation of transcription (data not shown), demonstrating that both pCAF and p300 are functioning during transcription initiation. Together, these experiments suggest that pCAF requires prior p300 activity to potentiate maximal transactivation by MyoD/E47.
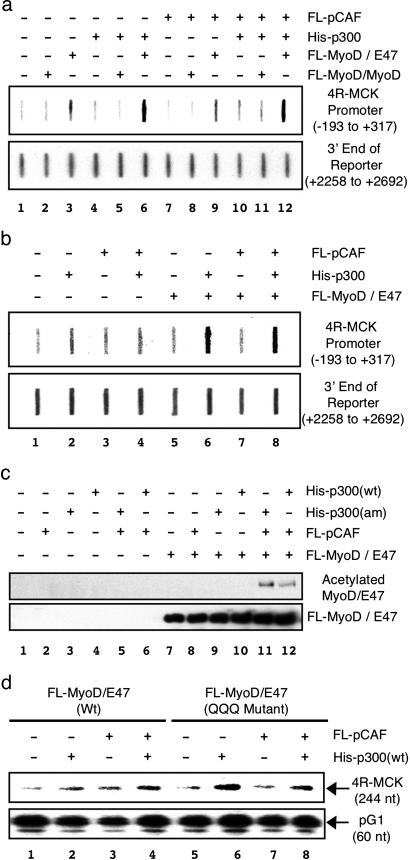
To determine the targets of the two different ATs during MyoD-dependent transcription, we first examined the ability of His-p300(wt) and/or FL-pCAF to carry out histone H3- and/or H4-specific acetylation of nucleosomes on the 4R-MCK template using ChIP analysis. In the presence of FL-MyoD/E47, His-p300(wt) significantly increased acetylation of both histone H3 (Fig. 4a, lane 6) and H4 (Fig. 4b lane 6) within the 4R-MCK promoter, but not at the distal region of the reporter gene. In contrast, no increase in acetylation was observed in either region of the template in the presence of FL-pCAF alone (Fig. 4a, compare lanes 3 and 9). Furthermore, no increase in acetylation was observed when both His-p300(wt) and FL-pCAF were added to the reaction compared with His-p300(wt) alone (Fig. 4a, compare lanes 6 and 12; Fig. 4b, compare lanes 6 and 8), suggesting that pCAF is not facilitating acetylation of histones. Consistent with its inability to activate transcription, FL-MyoD/MyoD was unable to recruit histone H3 AT activity to the 4R-MCK promoter (Fig. 4a, lanes 2, 5, 8, and 11). Interestingly, FL-MyoD/E47 was able to recruit a histone H3 AT activity to the 4R-MCK promoter region in the absence of His-p300(wt) and FL-pCAF (Fig. 4a, lane 3). The presence of a small amount of functional histone AT activity in the nuclear extract explains why a low level of MyoD-dependent transactivation was observed in the absence of exogenous coactivators (Fig. 3a, lane 5). These results suggests that p300 (and not pCAF) is acting to acetylate the promoter and that the transcriptional synergy between the two ATs is not due to increased histone acetylation.
Fig. 4.
p300 acetylates histones H3 and H4, whereas pCAF acetylates MyoD. (a) FL-MyoD/E47 recruits His-p300(wt) to the 4R-MCK promoter to mediate histone H3 acetylation. ChIP experiments were performed on the nucleosomal 4R-MCK template in the presence or absence of FL-MyoD/MyoD or FL-MyoD/E47. Where indicated, FL-pCAF and/or His-p300(wt) were included in the reaction. Nucleosomal 4R-MCK templates immunoprecipitated by using an anti-acetylated histone H3-specific antibody were analyzed by Southern (slot) blotting by using a promoter (–193 to +317) or distal (+2268 to +2652) region-specific probe. (b) FL-MyoD/E47 recruits His-p300(wt) to the 4R-MCK promoter to mediate histone H4 acetylation. Experiments were performed as described above, except that an anti-acetylated histone H4 antibody was used. (c) pCAF mediates acetylation of MyoD/E47. FL-MyoD/E47 was incubated with nucleosomal 4R-MCK template in the presence or absence of FL-pCAF, His-p300(wt), or His-p300(am) for 30 min at 27°C as indicated. Western blot was used to detect acetylated FL-MyoD/E47 (anti-acetylated protein antibody) and total FL-MyoD/E47 (anti-Flag antibody). (d) His-pCAF is not required for maximal transactivation by FL-MyoD(QQQ)/E47. Transcription reactions were performed by using either FL-MyoD/E47 or FL-MyoD(QQQ)/E47 on a nucleosomal 4R-MCK template in the presence or absence of His-p300(wt) and/or FL-pCAF as indicated.
To further address the functional role of His-p300(wt) and FL-pCAF, we explored the possibility that one or both of these proteins may be acetylating MyoD, leading to increased transactivation. On their own, neither FL-pCAF nor His-p300(wt) produced detectable levels of MyoD acetylation, whereas FL-MyoD/E47 was acetylated when both coactivators were included in the reaction (Fig. 4c). Importantly, MyoD was also acetylated in the presence of pCAF and the AT mutant form of p300 [His-p300(am); Fig. 4c, lane 11]. This finding suggests that pCAF, and not p300, is acetylating the FL-MyoD/E47 heterodimer. To examine whether “preacetylation” of MyoD would eliminate the need for either FL-pCAF or His-p300(wt) in our transcription system, we used a MyoD/E47 heterodimer (FL-MyoD(QQQ)/E47) that mimics acetylation by mutations K99Q, K102Q, and K104Q (Fig. 1d). When transcription was performed in the presence of FL-MyoD(QQQ)/E47, FL-pCAF alone did not stimulate transactivation on the nucleosomal 4R-MCK template (Fig. 4d, lane 7). In contrast, His-p300(wt) alone activated transcription much better in the presence of FL-MyoD(QQQ)/E47 compared with FL-MyoD/E47 (Fig. 4d, compare lanes 2 and 6). The level of transactivation observed in the presence of His-p300(wt) was not further increased by the addition of FL-pCAF (Fig. 4d, compare lanes 6 and 8). Interestingly, the combination of FL-pCAF and His-p300(wt) were unable to acetylate FL-MyoD(QQQ)/E47 (data not shown), suggesting that K99, K102, and/or K104 of MyoD are the only sites acetylated by the coactivators. Taken together, these results strongly suggest that pCAF is facilitating MyoD-dependent transactivation through its ability to acetylate MyoD, whereas p300 is acting to acetylate histones H3 and H4 within the promoter region.
Discussion
In this article, we describe the establishment of, to our knowledge, the first in vitro transcription system where a member of the basic helix–loop–helix family of transcriptional activators overcomes the repressive effect of nucleosomes to activate transcription. By using this system, we have been able to delineate distinct and sequential roles for the coactivators p300 and pCAF in the process of MyoD-dependent transactivation. Finally, we demonstrate that MyoD/E47 heterodimers (and not MyoD or E47 homodimers) drive transactivation from the 4R-MCK promoter.
Consistent with previous studies demonstrating that p300 and pCAF were coactivators of MyoD (7, 8, 11, 12, 27), we observed that both of these ATs are necessary for maximal levels of transcriptional activation. Although pCAF was not sufficient for MyoD-dependent transcription, when p300 was also present, the two coactivators synergize, leading to a strong activation of transcription. The synergy between pCAF and p300 observed in this study, along with transfection and microinjection studies (7), demonstrates that p300 and pCAF perform complementary functions during MyoD-dependent transcriptional activation. The events mediated by both of these ATs occur during transcription initiation because this synergy continues to be observed when sarkosyl is used to prevent transcription reinitiation. Chronologically, p300 is functioning to acetylate both histone H3 and H4 within the promoter region in an event before the need for pCAF. Use of the preacetylated heterodimer demonstrated that, after nucleosome modification, pCAF facilitates transactivation through direct acetylation of MyoD. Thus, the sequential actions of p300 and pCAF are necessary to establish a stable activator/coactivator-containing complex within the promoter region leading to transactivation.
A direct interaction between p300 and MyoD (by means of its acidic activation domain) has been described (8), suggesting a mechanism by which p300 is recruited to the promoter. This recruitment likely leads to the acetylation of nucleosomes within the promoter through the strong histone AT activity of p300 (22). Although pCAF can also interact directly with MyoD in vitro, transfection studies have suggested that p300 mediates the interaction between these two proteins in cells (7). Thus, pCAF is probably recruited to the MyoD-bound promoter by means of p300, and this recruitment might be facilitated and/or stabilized by the interaction between the bromodomain of pCAF and the acetylated histone H3 tail within the nucleosomes (9). Once stably associated with the promoter, pCAF then acetylates MyoD (11), which in turn could stabilize the interaction between MyoD and p300 (27) or aid in the recruitment of other bromodomain containing coactivators (28). Stable interaction between MyoD and p300 would facilitate the recruitment of the RNA Pol II transcriptional machinery by means of its ability to interact with the RNA Helicase A (29). This sequence of events is highly similar with elaborate ChIP studies examining recruitment of coactivators to the pS2 promoter by the liganded estrogen receptor (ER) (30). There, it was shown that, in an initial unproductive cycle of coactivator recruitment, p300 was associated with the ER-bound pS2 promoter in the absence of other known AT proteins, resulting in acetylation of histone H3 within this region. Then, in a second wave of coactivator recruitment, pCAF and the p300-related protein CBP arrived on the ER-bound promoter, leading to productive transcription initiation. Thus, it seems that sequential coactivator recruitment to promoters is a common mechanism by which several different families of activators establish a transcriptionally competent promoter (31).
Consistent with previous work performed in transfected cell systems (1), the use of tethered dimers in our transcription system clearly establishes MyoD/E47 heterodimers as the functional transcriptional activator at the 4R-MCK promoter. The role of E47 is not limited to providing a dimerization partner to facilitate DNA binding because homodimers of MyoD are able to bind an E-box in the context of nucleosomes (F.J.D. and S.J.T., unpublished observations). A role for E47 in facilitating recruitment of coactivators to the promoter would not be surprising because homodimers of this protein mediate expression of specific genes in B cells (32). Thus, both MyoD and E47 possess the ability to interact directly with chromatin remodeling factors and the basal transcriptional machinery (32–34). Although this redundancy in activities may help establish a more stable interaction between MyoD/E47 and the transcriptional machinery, we propose that these heterodimers may provide a scaffolding that is conformationally preferable for establishing a coactivator bridge between the proximal promoter elements (E-boxes) and the minimal promoter. Consistent with this finding, FL-MyoD/MyoD homodimers were unable to recruit p300 histone AT activity to the promoter region (Fig. 3d) although a direct interaction between the coactivator and MyoD has been described in vitro (8). In the scaffolding model, this result would suggest that the second MyoD molecule in the homodimer causes steric hindrance to prevent a strong interaction between the dimer and p300. Furthermore, FL-pCAF was unable to acetylate histones within the FL-MyoD/E47-bound promoter although the AT efficiently acetylates histone H3 in conditions where neither the enzyme nor substrate is limiting (see Fig. 6). This result suggests that, under conditions where pCAF concentrations are limiting, the AT is recruited to the promoter in a manner that favors its interaction with MyoD rather than histone H3. Based on these results, we suggest that the promoter of the 4R-MCK template likely creates an architecture that is favorable to the synergistic interaction between MyoD/E47 and the coactivators (including p300 and pCAF) necessary to activate transcription (35). The importance of promoter architecture on coactivator recruitment has been established for the IFN-β promoter, which requires all transcriptional activators present on the promoter to be properly aligned to ensure efficient transcriptional activation (36). Although MyoD/E47 heterodimers activate transcription from the 4R-MCK promoter, it remains to be determined whether this heterodimer pair is active on all MyoD-responsive promoters. The use of this in vitro transcription system to study natural MyoD-responsive promoters will certainly be highly useful for refining our understanding of the role of different dimer pairs in the context of promoter architectures, as well as for unraveling the mechanism by which MyoD orchestrates differential gene expression during myogenesis.
Acknowledgments
We thank J. T. Kadonaga (University of California at San Diego, La Jolla) for providing baculovirus encoding ACF-1, ISWI, and NAP-1; W. L. Kraus (Cornell University, Ithaca, NY) for baculovirus encoding p300 (both wild-type and mutant); and Y. Nakatani (Dana–Farber Cancer Institute, Boston) for baculovirus encoding pCAF; the National Cell Culture Center for preparing HeLa cell pellets; and E. Bengal and M. Brand for helpful insight. This work was supported by National Institutes of Health Grant AR45113 (to S.J.T.) and a Muscular Dystrophy Association Development Grant (to F.J.D.). F.J.D. is a Senior Research Fellow of the Canadian Institutes of Health Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MCK, muscle creatine kinase; AT, acetyltransferase; ChIP, chromatin immunoprecipitation.
References
- 1.Lassar, A. B., Davis, R. L., Wright, W. E., Kadesch, T., Murre, C., Voronova, A., Baltimore, D. & Weintraub, H. (1991) Cell 66, 305–315. [DOI] [PubMed] [Google Scholar]
- 2.Murre, C., McCaw, P. S., Vaessin, H., Caudy, M., Jan, L. Y., Jan, Y. N., Cabrera, C. V., Buskin, J. N., Hauschka, S. D., Lassar, A. B., et al. (1989) Cell 58, 537–544. [DOI] [PubMed] [Google Scholar]
- 3.Davis, R. L., Weintraub, H. & Lassar, A. B. (1987) Cell 51, 987–1000. [DOI] [PubMed] [Google Scholar]
- 4.Weintraub, H., Tapscott, S. J., Davis, R. L., Thayer, M. J., Adam, M. A., Lassar, A. B. & Miller, A. D. (1989) Proc. Natl. Acad. Sci. USA 86, 5434–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergstrom, D. A., Penn, B. H., Strand, A., Perry, R. L., Rudnicki, M. A. & Tapscott, S. J. (2002) Mol. Cell 9, 587–600. [DOI] [PubMed] [Google Scholar]
- 6.McKinsey, T. A., Zhang, C. L. & Olson, E. N. (2002) Curr. Opin. Cell Biol. 14, 763–772. [DOI] [PubMed] [Google Scholar]
- 7.Puri, P. L., Sartorelli, V., Yang, X. J., Hamamori, Y., Ogryzko, V. V., Howard, B. H., Kedes, L., Wang, J. Y., Graessmann, A., Nakatani, Y. & Levrero, M. (1997) Mol. Cell 1, 35–45. [DOI] [PubMed] [Google Scholar]
- 8.Sartorelli, V., Huang, J., Hamamori, Y. & Kedes, L. (1997) Mol. Cell. Biol. 17, 1010–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhalluin, C., Carlson, J. E., Zeng, L., He, C., Aggarwal, A. K. & Zhou, M. M. (1999) Nature 399, 491–496. [DOI] [PubMed] [Google Scholar]
- 10.Ito, T., Ikehara, T., Nakagawa, T., Kraus, W. L. & Muramatsu, M. (2000) Genes Dev. 14, 1899–1907. [PMC free article] [PubMed] [Google Scholar]
- 11.Sartorelli, V., Puri, P. L., Hamamori, Y., Ogryzko, V., Chung, G., Nakatani, Y., Wang, J. Y. & Kedes, L. (1999) Mol. Cell 4, 725–734. [DOI] [PubMed] [Google Scholar]
- 12.Polesskaya, A., Duquet, A., Naguibneva, I., Weise, C., Vervisch, A., Bengal, E., Hucho, F., Robin, P. & Harel-Bellan, A. (2000) J. Biol. Chem. 275, 34359–34364. [DOI] [PubMed] [Google Scholar]
- 13.Lau, O. D., Kundu, T. K., Soccio, R. E., Ait-Si-Ali, S., Khalil, E. M., Vassilev, A., Wolffe, A. P., Nakatani, Y., Roeder, R. G. & Cole, P. A. (2000) Mol. Cell 5, 589–595. [DOI] [PubMed] [Google Scholar]
- 14.Sassone-Corsi, P., Duboule, D. & Chambon, P. (1985) Cold Spring Harbor Symp. Quant. Biol. 50, 747–752. [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., Levenstein, M. E., Fyodorov, D. V., Kutach, A. K., Kobayashi, R. & Kadonaga, J. T. (1999) Genes Dev. 13, 1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pazin, M. J. & Kadonaga, J. T. (1998) in Chromatin: A Practical Approach, ed. Gould, H. (Oxford Univ. Press, Oxford), pp. 173–194.
- 17.Neuhold, L. A. & Wold, B. (1993) Cell 74, 1033–1042. [DOI] [PubMed] [Google Scholar]
- 18.Dilworth, F. J., Fromental-Ramain, C., Remboutsika, E., Benecke, A. & Chambon, P. (1999) Proc. Natl. Acad. Sci. USA 96, 1995–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraus, W. L. & Kadonaga, J. T. (1998) Genes Dev. 12, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraus, W. L., Manning, E. T. & Kadonaga, J. T. (1999) Mol. Cell. Biol. 19, 8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berkes, C. A., Bergstrom, D. A., Penn, B. H., Seaver, K. J., Knoepfler, P. S. & Tapscott, S. J. (2004) Mol. Cell 14, 465–477. [DOI] [PubMed] [Google Scholar]
- 22.Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H. & Nakatani, Y. (1996) Cell 87, 953–959. [DOI] [PubMed] [Google Scholar]
- 23.Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. (1983) Nucleic Acids Res. 11, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dilworth, F. J., Ramain-Fromental, C., Yamamoto, K. & Chambon, P. (2000) Mol. Cell 6, 1049–1058. [DOI] [PubMed] [Google Scholar]
- 25.Davis, R. L. & Weintraub, H. (1992) Science 256, 1027–1030. [DOI] [PubMed] [Google Scholar]
- 26.Thackray, V. G., Toft, D. O. & Nordeen, S. K. (2003) Mol. Endocrinol. 17, 2543–2553. [DOI] [PubMed] [Google Scholar]
- 27.Polesskaya, A., Naguibneva, I., Duquet, A., Bengal, E., Robin, P. & Harel-Bellan, A. (2001) Mol. Cell. Biol. 21, 5312–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng, L. & Zhou, M. M. (2002) FEBS Lett. 513, 124–128. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima, T., Uchida, C., Anderson, S. F., Lee, C.-G., Hurwitz, J., Parvin, J. D. & Montminy, M. (1997) Cell 90, 1107–1112. [DOI] [PubMed] [Google Scholar]
- 30.Metivier, R., Penot, G., Hubner, M. R., Reid, G., Brand, H., Kos, M. & Gannon, F. (2003) Cell 115, 751–763. [DOI] [PubMed] [Google Scholar]
- 31.Dilworth, F. J. & Chambon, P. (2001) Oncogene 20, 3047–3054. [DOI] [PubMed] [Google Scholar]
- 32.Bain, G., Gruenwald, S. & Murre, C. (1993) Mol. Cell. Biol. 13, 3522–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heller, H. & Bengal, E. (1998) Nucleic Acids Res. 26, 2112–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massari, M. E., Grant, P. A., Pray-Grant, M. G., Berger, S. L., Workman, J. L. & Murre, C. (1999) Mol. Cell 4, 63–73. [DOI] [PubMed] [Google Scholar]
- 35.Bengal, E., Flores, O., Rangarajan, P. N., Chen, A., Weintraub, H. & Verma, I. M. (1994) Proc. Natl. Acad. Sci. USA 91, 6221–6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merika, M., Williams, A. J., Chen, G., Collins, T. & Thanos, D. (1998) Mol. Cell 1, 277–287. [DOI] [PubMed] [Google Scholar]