Abstract
Little is known about the expression levels or function of micro-RNAs (miRNAs) in normal and neoplastic cells, although it is becoming clear that miRNAs play important roles in the regulation of gene expression during development [Ambros, V. (2003) Cell 113, 673–676; McManus, M. T. (2003) Semin. Cancer Biol. 13, 253–258]. We now report the genomewide expression profiling of miRNAs in human B cell chronic lymphocytic leukemia (CLL) by using a microarray containing hundreds of human precursor and mature miRNA oligonucleotide probes. This approach allowed us to identify significant differences in miRNome expression between CLL samples and normal CD5+ B cells; data were confirmed by Northern blot analyses and real-time RT-PCR. At least two distinct clusters of CLL samples can be identified that were associated with the presence or absence of Zap-70 expression, a predictor of early disease progression. Two miRNA signatures were associated with the presence or absence of mutations in the expressed Ig variableregion genes or with deletions at 13q14, respectively. These data suggest that miRNA expression patterns have relevance to the biological and clinical behavior of this leukemia.
MicroRNAs (miRNAs) represent a class of small, functional, noncoding RNAs of 19–23 nt cleaved from ≈60- to 110-nt hairpin precursors (1, 2). Hundreds of miRNAs have been identified in plants and animals. The miRNAs are involved in various biological processes, including cell proliferation and cell death during development, stress resistance, and fat metabolism, through the regulation of gene expression (3). Some miRNAs, such as miR-15a or miR-16–1 (4, 5), are widely expressed, whereas others, such as miR-1 in mammalian heart (6, 7) or miR-223 in granulocytes and macrophages (5), are expressed in a tissue-specific manner. Little else is known about miRNA expression patterns or function in normal or neoplastic cells.
Understanding of the molecular pathogenesis of B cell chronic lymphocytic leukemia (CLL), the most common adult leukemia in the Western world, is incomplete. We have shown previously that miR-15a and miR-16-1 are located at chromosome 13q14.3 within a 30-kb region of loss in CLL cells and that both genes are deleted and/or down-regulated in the majority of the analyzed CLL cell samples (4). These results provided the indication that deletion of miRNAs might be associated with a human malignancy. We also reported that 98 of the identified 186 miRNAs are located at fragile sites, minimal loss of heterozygosity regions, minimal regions of amplification, or common break-point regions in human cancers (8), suggesting that miRNAs might play a large and unanticipated role in the pathogenesis of human cancer.
Methods
Tissue Samples and CLL Samples. Forty-seven samples were used for this study, including 41 samples from 38 patients with CLL and 6 normal samples, including one lymph node, tonsillar CD5+ B cells from two normal donors, and blood mononuclear cells (MNC) from three normal donors. For three cases, two independent samples were collected and processed. CLL samples were obtained after informed consent from patients diagnosed with CLL at the CLL Research Consortium institutions. Briefly, blood was obtained from CLL patients, and MNC were isolated through Ficoll/Hypaque gradient centrifugation (Amersham Pharmacia Biotech) and processed for RNA extraction according to described protocols (9). For the majority of samples, clinical and biological information, such as age at diagnosis, sex, Rai stage, presence/absence of treatment, ZAP-70 expression, and IgVH gene mutation status were available (see Table 4, which is published as supporting information on the PNAS web site).
Cell Preparation. MNC from peripheral blood of normal donors were separated by Ficoll-Hypaque density gradients. T cells were purified from these MNC by rosetting with neuraminidase-treated sheep erythrocyte and depletion of contaminant monocytes (Cd11b+); natural killer cells (CD16+) and B lymphocytes (CD19+) were purified by using magnetic beads (Dynabeads, Unipath, Milan) and specific mAbs (Becton Dickinson). Total B cells and CD5+ B cells were prepared from tonsillar lymphocytes as described (10). Briefly, tonsils were obtained from patients in the pediatric age group undergoing routine tonsillectomies, after informed consent. Purified B cells were prepared by rosetting T cells from MNC with neuraminidase-treated sheep erythrocyte. To obtain CD5+ B cells, purified B cells were incubated with anti-CD5 mab followed by goat anti-mouse Ig conjugated with magnetic microbeads. CD5+ B cells were positively selected by collecting the cells retained on the magnetic column MS by Mini MACS system (Miltenyi Biotec, Auburn, CA). The degree of purification of the cell preparations was >95%, as assessed by flow cytometry.
RNA Extraction and Northern Blots. Total RNA isolation and blots were performed as described (4). After RNA isolation, the washing step with ethanol was not performed or if performed, the tube walls were rinsed with 75% ethanol without perturbing the RNA pellet (9). For reuse, blots were stripped by boiling in 0.1% aqueous SDS/0.1× SSC for 10 min and reprobed. 5S rRNA stained with ethidium bromide served as a sample loading control.
Microarray Experiments. RNA blot analysis was performed as described (11). Briefly, labeled targets from 5 μg of total RNA was used for hybridization on each KCC/TJU miRNA microarray chip containing 368 probes in triplicate, corresponding to 245 human and mouse miRNA genes. All probes on these microarrays are 40-mer oligonucleotides spotted by contacting technologies and covalently attached to a polymeric matrix. The microarrays were hybridized in 6× SSPE (0.9 M NaCl/60 mM NaH2PO4·H2O/8 mM EDTA, pH 7.4)/30% formamide at 25°C for 18 h, washed in 0.75× TNT (Tris·HCl/NaCl/Tween 20) at 37°C for 40 min, and processed by using a method of direct detection of the biotin-containing transcripts by streptavidin-Alexa647 conjugate. Processed slides were scanned by using a PerkinElmer ScanArray XL5K Scanner, with the laser set to 635 nm, at Power 80 and PMT 70 setting, and a scan resolution of 10 μm.
Data Analysis. Expression profiles were analyzed in duplicate independent experiments starting for three different patients. Raw data were normalized and analyzed in genespring software version 6.1.1 (Silicon Genetics, Redwood City, CA). genespring generated an average value of the three spot replicates of each miRNA. After data transformation (to convert any negative value to 0.01), normalization was performed by using a per-chip on median normalization method and a normalization to specific samples, expressly to the two CD5+ B cell samples, used as common reference for miRNA expression. Hierarchical clustering was generated, for both genes and conditions, by using standard correlation as a measure of similarity. To identify genes with statistically significant differences between sample groups (i.e., CLL cells and CD5+ B cells, CLL and MNC, CLL samples with or without IgVH mutations, or CLL cases with or without 13q14.3 deletion), a Welch's approximate t test for two groups (variances not assumed equal) with a P value cutoff of 0.05 and Benjamini and Hochberg false discovery rate as multiple testing correction were performed.
Real-Time PCR. Quantitative real-time PCR was performed as described (12). Briefly, RNA was reverse-transcribed to cDNA with gene-specific primers and Thermoscript, and the relative amount of each miRNA to tRNA for initiator methionine was described, using the equation 2-dCT, where dCT = (CTmiRNA – CTU6 or HUMTMI RNA). The set of analyzed miRNAs included miR-15a, miR-16-1, miR-18, miR-20, and miR-21. The primers used were as published (12).
Western Blotting. Protein lysates were prepared from the leukemia cells of seven CLL patients and isolated tonsillar CD5+ B cells. Western blot was performed with a polyclonal Pten antibody (Cell Signaling Technology, Beverly, MA) and was normalized by using an anti-actin antibody (Sigma).
Microarray Data Submission. All data were submitted by using miamexpress to the Array Express database, and each of the 38 CLL samples described here received an ID number ranging from SAMPLE169194SUB621 to SAMPLE169234SIUB621.
Results and Discussion
Distinct miRNome Expression Signatures in Various Human Tissues Revealed with an Oligonucleotide Microchip. To investigate expression of each member of this class of genes in normal and neoplastic cells, we developed a microarray chip that can examine global expression levels for each known member of the miRNome (defined as the full cellular complement of miRNA) (11). This chip contains gene-specific 40-mer oligonucleotide probes generated from 161 human and 84 mouse precursor miRNA collected from the Sanger database (July 2003 release) or from published papers (9, 13, 14). In some instances, two different oligonucleotide probes were designed, one containing the active sequence and the other specific for the precursor, enabling us to analyze expression of the mature miRNA and its precursor at the same time. We tested this platform by using a panel of 20 human normal tissues and identified specific miRNA expression signatures for each tissue type (11). Based on these signatures, hematopoietic tissues cluster together in a group distinct from that of nonhematopoietic tissues, showing that miRNA expression profiles differ with cell/tissue type, suggesting that abnormal cell/tissues will also have distinctive miRNA expression profiles.
B-CLL Cells Present Distinct miRNome Signatures in Respect with CD5+ Cells. To further investigate the involvement of miRNA in CLL, we analyzed the miRNome expression in 38 individual CLL cell samples. One normal lymph node sample and five samples from healthy donors, including two tonsillar CD5+ B lymphocyte samples and three blood MNC samples, were included for comparison. Unsupervised hierarchical clustering revealed homogenous but distinctive expression profiles for each sample type, with all CLL samples falling into a cluster that was distinct from that of the lymph node or normal lymphoid cell samples (Fig. 1). First, we compared miRNA expression in CLL cells vs. normal CD5+ B cells, considered as a normal cell counterpart to the CLL B cells, or CLL cells vs. normal blood MNC. As expected, two groups of differentially expressed miRNAs, the first composed of 55 genes and the second of 29 genes, had statistically significant differences in expression levels between the various groups (P < 0.05 using the Welch t test as described in Methods) (Table 1 and Table 5, which is published as supporting information on the PNAS web site). Of note, only six miRNAs are shared between the two lists, a result confirming our previous data showing distinct miRNome signatures in CD5+ B cells and leucocytes (11). When both pre-miRNA and mature miRNA were observed to be dysregulated (such as for miR-123, miR-132, or miR-136), the same type of variation in CLL samples with respect to CD5 or MNC was noted in every case. Also, for some miRNA genomic clusters all members were aberrantly regulated (such as the up-regulated 7q32 group encompassing miR-96–miR182–miR183), whereas for others only some members were abnormally expressed (such as the 13q31 genomic cluster where two of six members, miR-19 and miR-92-1, were strongly up-regulated and two, miR-17 and miR-20, were moderately down-regulated) (Tables 1 and 5). The results illustrate the complexity of the patterns of miRNA expression in CLL and argue for the existence of mechanisms regulating individual miRNA genes that map in the same chromosome region. In confirmation of the data accuracy, miR-223, reported to be expressed at high levels in granulocytes (5), was expressed at significantly lower levels in the CLL samples than in the MNC, but at about the same level as that noted for CD5+ B cells (which generally constitute less than a few percent of blood MNC).
Fig. 1.
Two distinct miRNA signatures characterize the CLL samples. Thirty-eight CLL patients were analyzed; from three of them two different cell samples were processed and analyzed. In all instances, the different samples from the same patients clustered together. The main miRNA-associated CLL clusters are presented. The control samples are: MNC; Ly, lymph node; CD5+, selected CD5+ B lymphocytes.
Table 1. Top 25 miRNAs differentially expressed in CLL cells versus CD5+ cells.
| miRNA | Chromosome location | FRA associated | P value | Type |
|---|---|---|---|---|
| mir-213 | 1q31.3-q32.1 | 1.47E-33 | Down | |
| mir-183-prec | 7q32 | FRA7H | 1.26E-23 | Up |
| mir-190 | 15q21 | FRA15A | 1.48E-20 | Up |
| mir-24-1-prec | 9q22.1 | FRA9D | 7.35E-20 | Up |
| mir-33 | 22q13.2 | 1.56E-18 | Up | |
| mir-19a | 13q31 | 5.16E-17 | Up | |
| mir-140 | 16q22.1 | 2.41E-16 | Up | |
| mir-123 | 9q34 | 2.80E-16 | Up | |
| mir-10b | 2q31 | 1.10E-15 | Up | |
| mir-15b-prec | 3q26.1 | 5.79E-14 | Up | |
| mir-92-1 | 13q31 | 1.70E-12 | Up | |
| mir-188 | Xp11.23-p11.2 | 6.08E-11 | Up | |
| mir-154 | 14q32 | 1.14E-10 | Up | |
| mir-220 | Xq25 | 2.14E-09 | Down | |
| mir-217 | 2p16 | 3.85E-09 | Up | |
| mir-101 | 1p31.3 | FRA1C | 1.26E-08 | Up |
| mir-141-prec | 12p13 | 1.39E-08 | Up | |
| mir-153-prec | 2q36 | 1.48E-08 | Up | |
| mir-196-2 | 12q13 | FRA12A | 4.94E-08 | Up |
| mir-134 | 14q32 | 6.01E-08 | Up | |
| mir-141 | 12p13 | 7.91E-08 | Up | |
| mir-132 | 11q12 | 1.68E-07 | Up | |
| mir-192 | 11q13 | 2.00E-07 | Down | |
| mir-181b-prec | 1q31.2-q32.1 | 3.26E-06 | Up |
For a complete list see Table 5. The name of each miRNA is as in the miRNA Registry, and the disregulation of either active molecule or precursor is specified in the name. The type of altered expression is presented in the last column.
At the top of the CLL vs. CD5+ B cell list are several miRNAs located exactly inside fragile sites (miR-183 at FRA7H, miR-190 at FRA12A, and miR-24-1 at FRA9D) and miR-213. The mature miR-213 molecule is expressed at lower levels in all of the CLL samples, and the precursor miR-213 is reduced in expression in 62.5% of the samples. miR-16-1, at 13q14.3, which we previously reported to be down-regulated in the majority of CLL cases by Northern analysis (4), was expressed at low levels in 45% of CLL samples. An identical mature miR-16 exists on chromosome 3; because the 40-mer oligonucleotide for both miR-16 sequences from chromosome 13 (miR-16-1) and chromosome 3 (miR-16-2) exhibit the same 23-mer mature sequence, very similar profiles were observed. However, because we observed very low levels of miR-16-2 expression in CLL samples by Northern blot, the expression observed is contributed mainly by miR-16-1. The other miRNA of 13q14.3, miR-15a, was expressed at low levels in ≈25% of CLL cases. Overall, these data demonstrate that CLL is a malignancy with extensive alterations of miRNA expression and suggest a role for distinct miRNAs in the pathogenesis of human B cell malignancies (Fig. 1 and Fig. 3, which is published as supporting information on the PNAS web site).
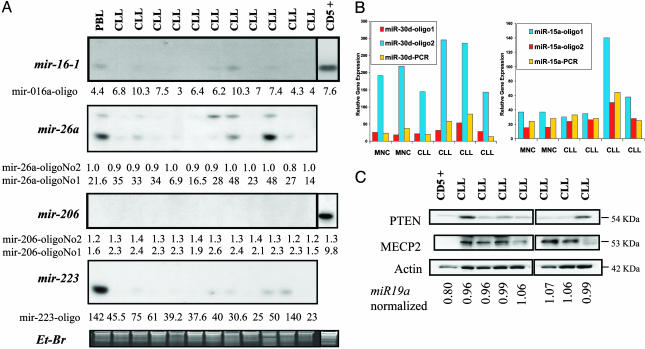
Validation of the microarray data was supplied for four miRNAs by Northern blot analyses: miR-16-1, located within the region of deletion at 13q14.3, miR-26a, on chromosome 3 in a region not involved in the pathogeneses of CLL, and miR-206 and miR-223 that are down-regulated (see above) in the majority of samples. For all four miRNAs, the Northern blot analyses confirmed the data obtained by using the microarray (Fig. 2A). We also performed real-time RT-PCR to measure expression levels of precursor molecules for five genes (miR-15a, miR-16–1, miR-18, miR-21, and miR-30d), and we found results concordant with the chip data (examples for miR-15a and miR-30d are presented in Fig. 2B).
Fig. 2.
Microarray data confirmation. (A) Northern blots of CLL RNAs showing concordance with the microarray data. Names of miRNA and specific oligonucleotides spotted on the array are presented. The numbers correspond to absolute expression value of each miRNA (determined by a per-chip on median normalization) and, therefore can be compared with the band intensity on Northern blots. 5S RNA stained with ethydium bromide (Et-Br) was used as loading control. The significance of the two oligos designed for some miRNAs is described in Tables 4–6. Generally, the oligoNo1 corresponds to the active part, whereas the oligoNo2 corresponds to the precursor miR molecule. PBL, peripheral blood leukocytes. (B) Real-time PCR quantification of miRNA expression in MNC and CLL samples. Expression of miR-15a (Left) and miR-30d (Right) by microarray (yellow bar) is compared with the levels detected with the two oligo specific for the active part (red bar) or precursor miRNA (blue bar). Correlations only with the expression levels of pre-miRNA (blue bar) but not active miRNA (red bar) were found. The data represent the mean of triplicate real-time PCRs from a single DNA sample. (C) Variations in the expression of Pten protein, a target of miR-19a are paralleled, in six of seven CLLs, by miR-19a microarray expression. The numbers represent MeCP2, the methyl-CpG-binding protein excluded as a putative target of miR-19a was used as the negative control. The numbers correspond to absolute expression value of miR-19a (determined by a per-chip on median normalization). Actin was used as loading control.
miRNA Signatures in B-CLL Cells Are Associated with the Levels of ZAP-70. Second, we asked whether CLL cells from different patients shared the same miRNA expression patterns. Unsupervised hierarchical clustering generated two clearly distinguishable miRNA signatures within the set of CLL samples, one closer to the miRNA expression profile observed in human MNC and the other clearly different (Fig. 1). The top 25 differentially expressed miRNA in these two signatures (at P < 0.001) include genes known or suggested to be involved in cancer (Table 2 and Table 6, which is published as supporting information on the PNAS web site). The precursor of miR-155 is overexpressed in the majority of childhood Burkitt's lymphoma (15), miR-21 is located at the fragile site FRA17B (8), and miR-26a is at 3p21.3, a region frequently deleted region in epithelial cancers, whereas miR-92-1 and miR-17 are at 13q32, a region amplified in malignant lymphomas (16). Furthermore, we asked whether there are some clinico-biological factors that can help to discriminate between the two clusters. We found a high difference in the levels of ZAP-70 between the two groups: 66% (6/9) patients from the first cluster vs. 25% (4/16) patients from the second one (Fig. 1) have low levels of ZAP-70 (<20%) (P = 0.04 at χ test). The mean value of ZAP-70 was 19% (±31% SD) vs. 35% (±30% SD), respectively; otherwise the two clusters can discriminate between patients who express and who do not express this protein (at levels <20% ZAP-70 is considered as nonexpressed) (Table 4). ZAP-70 is a tyrosine kinase, which is a strong predictor of early disease progression, and low levels of expression are proved to be a finding associated with good prognosis (17). To validate these important differences further studies with larger numbers of patients are required.
Table 2. miRNAs differentially expressed between the two main CLL clusters.
| miRNA | Chromosome location | P value | Cancer-associated genomic regions |
|---|---|---|---|
| miR-17-prec | 13q31 | 0.00000000 | Amp-folicular Ly/Del-HCC |
| miR-20 | 13q31 | 0.00000000 | Amp-folicular Ly/Del-HCC |
| miR-103-2 | 20p13 | 0.00000001 | |
| miR-30d-prec | 8q24.2 | 0.00000002 | |
| miR-106 | Xq26.2 | 0.00000006 | Del-advanced ovarian cancer |
| miR-26b | 2q35 | 0.00000006 | |
| miR-103-1 | 5q35.1 | 0.00000006 | |
| miR-25 | 7q22 | 0.00000007 | FRA7F |
| miR-30a | 6q12-13 | 0.00000008 | |
| miR-21 | 17q23.2 | 0.00000008 | Amp-neuroblastoma; FRA17B |
| miR-107 | 10q23.31 | 0.00000008 | |
| miR-92-1-prec | 13q31 | 0.00000024 | Amp-folicular Ly/Del-HCC |
| miR-27a | 19p13.2 | 0.00000024 | |
| miR-23a | 19p13.2 | 0.00000032 | |
| miR-92-2 | Xq26.2 | 0.00000040 | Del-advanced ovarian cancer |
| miR-30b | 8q24.2 | 0.000004 | |
| miR-26a | 3p21 | 0.000009 | Del-epithelial malignancies |
| miR-93-1 | 7q22 | 0.000009 | Amp-folicular Ly/Del-HCC; FRA7F |
| miR-194 | 1q41 | 0.000015 | FRA1H |
| miR-155 | 21q21 | 0.000028 | Amp-colon cancer; childhood Burkit Lymphoma |
| miR-153-2 | 7q36 | 0.000028 | t(7;12)(q36;p13)-AML |
| miR-193-prec | 17q11.2 | 0.000044 | Del-Muci/Sero ovarian cancer |
| miR-130a | 11q12 | 0.0001 | |
| miR-23b | 9q22.1 | 0.0001 | Del-urothelial cancer; FRA9D |
| miR-30c | 6q13 | 0.0001 | |
| miR-139 | 11q13 | 0.0001 |
For a complete list see Table 6. The name of each miRNA is as in the miRNA Registry, and the disregulation of either active molecule or precursor is specified in the name. The location in minimally deleted or minimally amplified or breakpoint regions or in fragile sites is presented. HCC, hepatocellular carcinoma; AML, acute myeloid leukemia; FRA, fragile site.
Chromosome 13q14 Deletions and the Expression of Mutated IgVH Correlates with miRNAs Expression. Third, we asked whether the microarray data revealed specific molecular signatures predictive for subsets of CLL that differ in clinical behavior. CLL cases harbor deletions at chromosome 13q14.3 in ≈50% of cases (18). As a single cytogenetic defect, these CLL patients have a relatively good prognosis, compared with patients with leukemia cells harboring complex cytogenetic changes (19). It was also shown that deletion at 13q14.3 was associated with the presence of mutated IgVH genes (20), another good prognostic factor. By comparing expression data of CLL samples with or without deletions at 13q14, we found that miR-16-1 was expressed at low levels in leukemias harboring deletions at 13q14 (P = 0.03, ANOVA). We also found that miR-24-2, miR-195, miR-203, miR-220, and miR-221 are expressed at significantly reduced levels, whereas miR-7-1, miR-19a, miR-136, miR-154, miR-217, and the precursor of miR-218-2 are expressed at significantly higher levels in the samples with 13q14.3 deletions, respectively (Table 3). All of these genes are located in different regions of the genome and differ in their nucleotide sequences, excluding the possibility of cross-hybridization, suggesting the existence of functional miRNA networks in which hierarchical regulation may be present, with some miRNA (such as miR-16-1) controlling or influencing the expression of other miRNA.
Table 3. miRNA signatures associated with prognosis in B-CLL.
| miRNA | Chromosome location | P value | Association | Observation |
|---|---|---|---|---|
| miR-7-1 | 9q21.33 | 0.030 | 13q14 normal | |
| miR-16-1 | 13q14.3 | 0.030 | IGVH mutations negative | |
| 0.023 | 13q14 deleted | |||
| miR-19a | 13q31 | 0.024 | 13q14 normal | |
| miR-24-2 | 19p13.2 | 0.033 | 13q14 deleted | |
| miR-29c | 1q32.2-32.3 | 0.018 | IGVH mutations positive | Cluster miR-29c-miR 102 |
| miR-102 | 1q32.2-32.3 | 0.023 | IGVH mutations positive | Cluster miR-29c-miR 102 |
| miR-132 | 17p13.3 | 0.033 | IGVH mutations negative | |
| miR-136 | 14q32 | 0.045 | 13q14 normal | |
| miR-154 | 14q32 | 0.020 | 13q14 normal | |
| miR-186 | 1p31 | 0.038 | IGVH mutations negative | |
| miR-195 | 17p13 | 0.036 | 13q14 deleted | |
| miR-203 | 14q32.33 | 0.026 | 13q14 deleted | |
| miR-217-prec | 2p16 | 0.005 | 13q14 normal | |
| miR-218-2 | 5q35.1 | 0.019 | 13q14 normal | |
| miR-220 | Xq25 | 0.026 | 13q14 deleted | |
| miR-221 | Xp11.3 | 0.021 | 13q14 deleted |
The name of each miRNA is as in the miRNA Registry, and the disregulation of either active molecule or precursor is specified in the name. IGVH, IgVH genes.
We also looked for a specific signature profile associated with the expression of mutated IgVH, a favorable prognostic marker (20). Indeed, a distinct miRNA signature composed of five differentially expressed genes (miR-186, miR-132, miR-16-1, miR-102, and miR-29c) distinguished CLL samples that expressed mutated IgVH gene from those that expressed unmutated IgVH genes (Table 3), suggesting that miRNA expression profiles have prognostic significance in CLL. A confirmation of our results is the observation that the common element between the del 13q14.3-related and the IgVH-related signatures is miR-16-1. This gene is located in the common deleted region 13q14.3 and the presence of this particular deletion is associated with good prognosis. Therefore, miRNAs may expand the spectrum of adverse prognostic markers in CLL, such as expression of ZAP-70, unmutated IgVH, CD38, deletion at chromosome 11q23, or loss or mutation of TP53.
PTEN Protein Expression Inversely Correlates with the Levels of miR19 Expression on the Microchip. A present limitation in our understanding of the function of miRNA is our lack of knowledge about specific targets. However, two recent publications have presented algorithms with which to identify putative targets for miRNA in human (21) or Drosophila (22). Also, it might be possible to relate genomewide miRNA expression with data obtained from analyses by using large EST microarrays to identify miRNA gene targets, thereby revealing the functional consequences of altered miRNA gene expression in physiological or pathological states. We selected miR-19a, because its targets were experimentally determined (21), and we found it as a significantly up-regulated miRNA in CLL cells in respect to CD5+ B cells (Tables 3 and 5). One of its targets is PTEN (21), a tumor suppressor gene expressed in both CD5+ and CLL cells. It was reported that Pten protein is absent in CLL with a normal PTEN genotype, suggesting down-regulation of protein synthesis, only partly explained by promoter methylation (23). Analyzing a limited set of CLL cases, we have found an inverse correlation between the levels of expression of miR-19a and Pten protein in six of seven samples (86%), a result not observed for Mecp2 protein (methyl-CpG-binding protein), a predicted target but not experimentally confirmed (24) (Fig. 2C). Thus, using the combination of Western blot and our microarray platform, it is possible to analyze the relation between expression of miRNA and putative targets in large sets of clinical samples.
Genomic expression profiling using microarrays has proven to be a powerful approach for relating gene expression profiles with cell signaling in normal and disease states (25). We have developed a microarray chip for the analysis of expression of hundreds of miRNAs. Despite the continuous expansion of the number of known miRNA in various organisms, the miRNA microchip appears to be a powerful tool for analyzing miRNA expression patterns and quantitative expression level assays. This is important in light of the reported variations in expression levels of several miRNAs: reduced expression of miR-15a and miR-16-1 in CLL (4), miR-143 and miR-145 in colon cancers (26), and let-7 in lung cancers (27), or amplification of the precursor of miR-155 in Burkitt lymphomas (15), and the 13q31-q32 miRNA cluster (miR-17, miR-91, miR-18, miR-19a, miR-20, miR-19b, and miR-92) in malignant lymphomas (16). By applying the miRNA microarray to the study of human CLL, we demonstrated that <200 miRNA genes could reveal distinct miRNA signatures associated with specific bio-pathological features, including factors related to prognosis. The results suggest that miRNA plays important roles in the pathogenesis of human CLL.
Acknowledgments
This work was supported by National Institutes of Health Grant P01-CA81534 (to the CLL Research Consortium), National Cancer Institute Grant CA076259, the Italian Ministero dell'Istruzione, dell'Universitae della Ricerca, and the Italian Ministero della Salute.
Abbreviations: miRNA, microRNA; CLL, chronic lymphocytic leukemia; MNC, mononuclear cells.
References
- 1.Moss, E. G. (2003) in MicroRNAs in Noncoding RNAs: Molecular Biology and Molecular Medicine), eds. Barciszewski, J. & Erdmann, V. (Landes Bioscience, Georgetown, TX), pp. 98–114.
- 2.Bartel, D. P. (2004) Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 3.Ambros, V. (2003) Cell 113, 673–676. [DOI] [PubMed] [Google Scholar]
- 4.Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., Aldler, H., Rattan, S., Keating, M., Rai, K., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, C. Z., Li, L., Lodish, H. F. & Bartel, D. P. (2004) Science 303, 83–86. [DOI] [PubMed] [Google Scholar]
- 6.Lee, R. C. & Ambros, V. (2001) Science 294, 862–864. [DOI] [PubMed] [Google Scholar]
- 7.Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W. & Tuschl, T. (2002) Curr. Biol. 12, 735–739. [DOI] [PubMed] [Google Scholar]
- 8.Calin, G. A., Sevignani, C., Dumitru, C. D., Hyslop, T., Noch, E., Yendamuri, S., Shimizu, M., Rattan, S., Bullrich, F., Negrini, M. & Croce, C. M. (2004) Proc. Natl. Acad. Sci. USA 101, 2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. (2001) Science 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 10.Dono, M., Zupo, S., Leanza, N., Melioli, G., Fogli, M., Melagrana, A., Chiorazzi, N. & Ferrarini, M. (2000) J. Immunol. 164, 5596–5604. [DOI] [PubMed] [Google Scholar]
- 11.Liu, C.-G., Calin, G. A., Meloon, B., Gamliel, N., Sevignani, C., Ferracin, M., Dumitru, D. C., Shimizu, M., Zupo, S., Dono, M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmittgen, T. D., Jiang, J., Liu, Q. & Yang, L. (2004) Nucleic Acid Res. 32, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M. & Dreyfuss, G. (2002) Genes Dev. 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim, L. P., Glasner, M. E., Yekta, S., Burge, C. B. & Bartel, D. P. (2003) Science 299, 1540. [DOI] [PubMed] [Google Scholar]
- 15.Metzler, M., Wilda, M., Busch, K., Viehmann, S. & Borkhardt, A. (2004) Genes Chromosomes Cancer 39, 167–169. [DOI] [PubMed] [Google Scholar]
- 16.Ota, A., Tagawa, H., Karnan, S., Tsuzuki, S., Karpas, A., Kira, S., Yoshida, Y. & Seto, M. (2004) Cancer Res. 64, 3087–3095. [DOI] [PubMed] [Google Scholar]
- 17.Orchard, J. A., Ibbotson, R. E., Davis, Z. A., Wiestner, A., Rosenwald, A., Thomas, P. W., Hamblin, T. J., Staudt, L. M. & Oscier, D. G. (2004) Lancet 363, 105–111. [DOI] [PubMed] [Google Scholar]
- 18.Bullrich, F. & Croce, C. M. (2001) Chronic Lymphoid Leukemia (Dekker, New York).
- 19.Dohner, H., Stilgenbaue, S., Benner, A., Leupolt, E., Krober, A., Bullinger, L., Dohner, K., Bentz, M. & Lichter, P. (2000) N. Engl. J. Med. 343, 1910–1916. [DOI] [PubMed] [Google Scholar]
- 20.Oscier, D. G., Gardiner, A., Mould, S. J., Glide, S., Davis, Z. A., Ibbotson, R. E., Corcoran, M. M., Chapman, R. M., Thomas, P. W., Copplestone, J. A., et al. (2002) Blood 100, 1177–1184. [PubMed] [Google Scholar]
- 21.Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P. & Burge, C. B. (2003) Cell 115, 787–798. [DOI] [PubMed] [Google Scholar]
- 22.Rajewsky, N. & Socci, N. D. (2004) Dev. Biol. 267, 529–535. [DOI] [PubMed] [Google Scholar]
- 23.Soria, J. C., Lee, H. Y., Lee, J. I., Wang, L., Issa, J. P., Kemp, B. L., Liu, D. D., Kurie, J. M., Mao, L. & Khuri, F. R. (2002) Clin. Cancer Res. 8, 1178–1184. [PubMed] [Google Scholar]
- 24.Muller, H. M., Fiegl, H., Goebel, G., Hubalek, M. M., Widschwendter, A., Muller-Holzner, E., Marth, C. & Widschwendter, M. (2003) Br. J. Cancer. 89, 1934–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butte, A. (2002) Nat. Rev. 1, 951–960. [DOI] [PubMed] [Google Scholar]
- 26.Michael, M. Z., O'Connor, S. M., van Holst Pellekaan, N. G., Young, G. P. & James, R. J. (2003) Mol. Cancer Res. 1, 882–891. [PubMed] [Google Scholar]
- 27.Takamizawa, J., Konishi, H., Yanagisawa, K., Tomida, S., Osada, H., Endoh, H., Harano, T., Yatabe, Y., Nagino, M., Nimura, Y., et al. (2004) Cancer Res. 64, 3753–3756. [DOI] [PubMed] [Google Scholar]