Abstract
We report herein a dual-purpose role for polyacidic domains in an aqueous-phase polymer amphiphile assembly. In addition to their typical role as ionized water-solubilizing and self-repulsive motifs, we find that polycarboxylic acid domains uniquely enable high levels of hydrophobic drug encapsulation. By attenuated total reflectance infrared spectroscopy, we find significant differences in the carbonyl stretching region of the nanoparticles formed by polyacidic amphiphiles relative to those in soluble, single-domain poly(acrylic acid), suggesting that stabilization may be derived from limited ionization of the carboxylate groups upon assembly. Acidic-hydrophobic diblock polyacrylates were prepared and coassembled with up to 60 wt % camptothecin (CPT) into nanoparticles, the highest loading reported to date. Controlled release of bioactive CPT from polymer nanoparticles is observed as well as protection from human serum albumin-induced hydrolysis. Surface protection with PEG limits uptake of the CPT-loaded nanoparticles by MCF-7 breast cancer cells, as expected. Acidic-hydrophobic polymer amphiphiles thus have the hallmarks of a useful and general drug delivery platform and are readily accessible from living radical polymerization of cheap, commercially available monomers. We highlight here the potential utility of this common polymer design in high-capacity, controlled-release polymer nanoparticle systems.
Graphical Abstract
INTRODUCTION
Polymer nanoparticle delivery vehicles typically incorporate drug cargo into the hydrophobic core of the particle.1 Disruption of core packing by the drug creates an instability that limits loading levels and renders the coassembly susceptible to “burst” release kinetics in which the majority of the encapsulated drug is released in a few hours. This problem has been addressed by design of favorable noncovalent polymer–drug interactions in the core2 as well as covalent drug modification3–7 and cross-linking of the loaded particle.8 Noncovalent approaches to encapsulation and platform stabilization are attractive in that they present potentially general packaging solutions with minimal processing. Nitrogen heterocycles have been used as hydrogen bonding elements to direct both the assembly of polymer nanostructures9,10 as well as drug loading within polymer nanoparticles.11 Although the carboxylate group has been previously used as a site for polymer nanoparticle stabilization12–14 via cross-linking acylation of diamines, it is typically considered as a solubilizing domain15–17 due to its ionized nature at physiological pH. However, the neutral acid group is considerably more hydrophobic18 than that of the carboxylate and can direct assembly19 through formation of hydrogen-bonded dimers (Figure 1). In a macromolecular context, hydrogen bonding interactions with poly(acrylic acid) have been reported to guide layer-by-layer assembly in water20,21 as well as mucoadhesion of hydrogels.22 Colombani and others have shown that pKa can be significantly elevated in polymer acids,23 similar to pKa shifts found in proteins and nucleic acids.24,25 We hypothesized that limited or altered ionization of acidic-hydrophobic diblock polymers could result in assembly stabilization via interchain hydrogen bonding and increased chain hydrophobicity. Although we able to interrogate polymer ionization state in situ using attenuated total reflectance infrared spectroscopy (ATR-IR), the high interfacial tension26 between water and hydrophobic cores such as polystyrene27–29 and poly-n-butyl acrylate (PnBA)26,30,31 dominates the assembly process, obfuscating the contribution of the polyacid domain to stability. We instead used drug encapsulation and release as a functional assay for stability; notably, these polyacidic amphiphiles have not been previously studied as carriers. We found that a minimum degree of polymerization (DP) of 50 was required for drug loading, suggesting that drug encapsulation occurs in the hydrophobic core; however, the polyacidic domain lends considerable stabilization to drug loading. To test the extent to which polyacidic domains stabilize drug encapsulation, we prepared block amphiphiles (Figure 1) comprised of a PnBA hydrophobic block (degree of polymerization (DP) = 50) and polar block (DP = 30–100) by RAFT polymerization.32 Polyacidic amphiphiles could be coassembled into nano-particles with camptothecin (CPT), a hydrophobic natural product inhibitor of topoisomerase I.3 The union of potent cytoxicity with poor pharmacokinetics has made CPT the targeted cargo of many carrier platforms with varying degrees of sophistication.3 Generally, carriers that noncovalently encapsulate CPT exhibit low weight percent loading (<8%);3,7 covalent drug modification can render higher loadings, though this is not a completely general strategy for increasing drug carrier capacity.4,6,33,34 Surprisingly, the simple polyacrylate amphiphiles studied herein could be packaged with up to 60 wt % of CPT at up to 90% efficiency, the highest noncovalent3,33,35,36 CPT loading within a polymer compartment to date (Table 1).
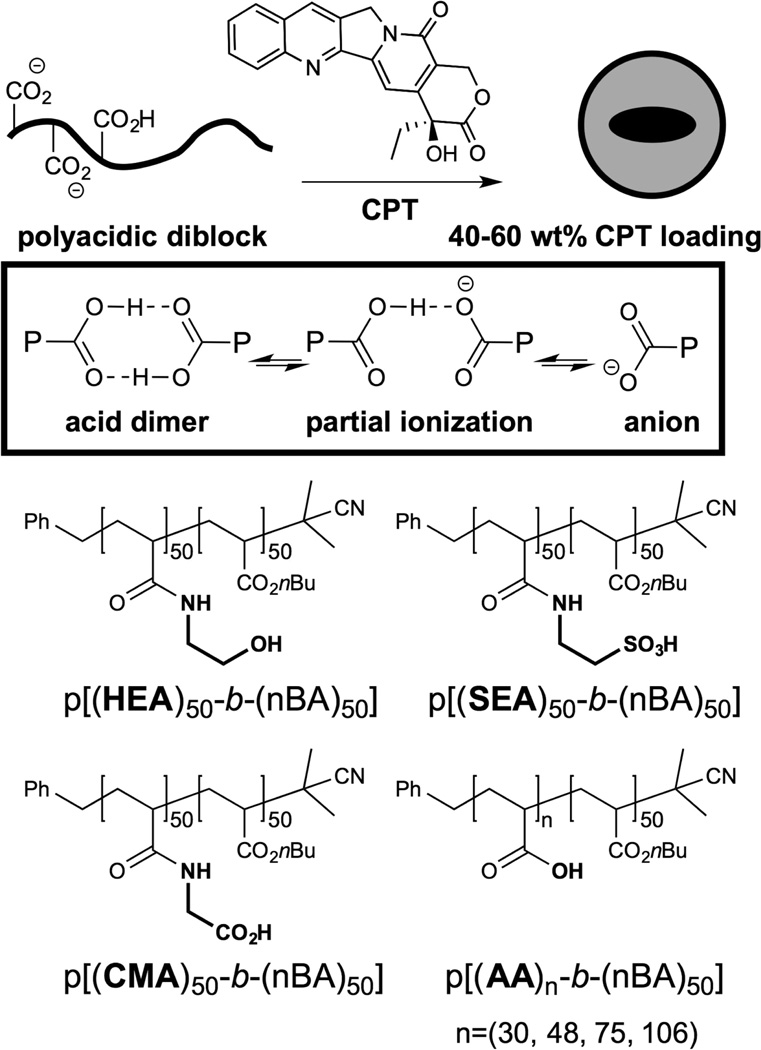
Figure 1.
Structures of diblock polymer amphiphiles. Potential carboxylic acid interactions are illustrated at top. Polymers are differentiated as indicated by side chains in bold. Macro-CTA polymers from RAFT polymerization were capped with AIBN.
Table 1.
CPT-Loaded Polymer Nanoparticles
| polymer | wt % CPTa | loading efficiency | NP diam (nm)b |
|---|---|---|---|
| p[(AA)30-b-(nBA)50] | >1000 | ||
| 42% | 90% | 180 | |
| p[(AA)75-b-(nBA)50] | 51% | 69% | 170 |
| p[(AA)106-b-(nBA)50] | 60% | 75% | 170 |
| p[(CMA)50-b-(nBA)50] | 40% | 67% | 200 |
| p[(SEA)50-b-(nBA)50] | 28% | 38% | 210 |
| p[(HEA)50-b-(nBA)50] | >1000 |
CPT in polymer nanoparticles (NP).
Average diameter of CPT-loaded particles; PDI was <0.2 for all nanoparticle samples except p[(AA)30-b-(nBA)50] and p[(HEA)50-b-(nBA)50] (>0.5). Empty NPs all had diameters of 70 nm by DLS and PDIs < 0.2.
EXPERIMENTAL SECTION
Materials and Instrumentation
Poly(ethylene glycol) methyl ether-2-(dodecylthiocarbono-thioylthio)-2-methylpropionate (PEG-5000 Macro-CTA) and butyl acrylate monomers were purchased from Sigma-Aldrich and used as provided. Polyacrylic acid standard (5.6 kDa) was purchased from American Polymer Standards Corporation. Azobis(isobutyronitrile) (AIBN) was purchased from Sigma-Aldrich and recrystallized from methanol prior to use. Chain transfer agent (CTA) 3-(((benzylthio)carbonothioyl)thio)propanoic acid37 (1) and N-hydroxysuccinimidyl (NHS) acrylate38 were prepared according to literature procedures. 1HNMR spectra were recorded on a Bruker Avance 400 MHz spectrometer. Dynamic light scattering (DLS) and ambient transmission electron microscopy (TEM) measurements were performed using a Malvern Nano series Zetasizer and an FEI 120 kV Tecnai G2 spirit TEM, respectively.
General Procedures
All polymerizations were run at 75 °C under N2 atmosphere in degassed dimethylformamide (DMF). Polymerization was initiated at 75 °C by addition of AIBN, monitored by 1H NMR, and quenched after 50% conversion by cooling to RT and venting to air. Solvent and unreacted monomer were removed under vacuum. All polymers were characterized by 1H NMR and GPC using poly(methyl methacrylate) (PMMA) as a standard.
p(nBA)50 Macro-CTA
n-Butyl acrylate (5.12 g, 40 mmol) was reacted in DMF (40 mL) with CTA 1 (110 mg, 0.4 mmol) and AIBN (6.5 mg, 0.040 mmol), yielding p(nBA)50 macro-CTA as a yellow oil.
p[(tBA)m-b-(nBA)50] (m = 30, 48, 75, 106)
Macro-CTA p(nBA)50 (0.256 g, 40 µmol) and 2 m equivalents of t-butyl acrylate were reacted with AIBN (0.65 mg, 4 µmol) as described above. Progress of the reaction was followed by 1HNMR spectroscopy and halted at 50% conversion to obtain the desired monomer ratio.
p[(AA)m-b-(nBA)50]
First, 0.5 g of p[(tBA)m-b-(nBA)50] was dissolved in 9 mL of 1:1 CH2Cl2:trifluoroacetic acid (TFA) and stirred at RT for 18 h and then concentrated to dryness under vacuum. Residual TFA was removed by dialysis (Spectra/Por 1, MWCO 6–8 kDa, Spectrum Laboratories) of the polymers in 5 mL of 1:1 H2O:THF against 1 L of ddH2O for 72 h. The white cloudy suspension remaining in the dialysis bag was freeze-dried to yield a white powder as the final product.
p[(NHS)50-b-(nBA)50]
MacroCTA p(nBA)50 (0.256 g, 0.04 mmol) was reacted with NHS acrylate (0.68 g, 4.02 mmol), and AIBN (0.65 mg, 4 µmole) was reacted in 2 mL of DMF to 50% conversion. The cooled and vented reaction solution was added dropwise to diethyl ether to precipitate p[(NHS)50-b-(nBA)50] as a faint-yellow solid (0.60 g, 50% conversion).
AIBN End-Capping
Removal of the trithiocarbonate end group follows a known procedure39 with slight modifications. Briefly, macro-CTA polymers were dissolved in DMF at ~40 mM concentration. The solution was sparged with N2 for 30 min, treated with AIBN (30 equiv), then heated to 85 °C for 3 h under N2 atmosphere. For p[(NHS)50-b-(nBA)50], the polymer solution was precipitated and washed with cold diethyl ether (3 × 50 mL) to yield a white solid (>95% mass recovery). The p[(tBA)m-b-(nBA)50] polymers have similar solubility to that of the trithiocarbonate side product formed with AIBN and required dialysis purification (Spectra/Por 1, MWCO 6–8 kDa, Spectrum Laboratories) against 200 mL of THF solution changed every 24 h for 72 h. The polymer solution in the dialysis bag was collected and concentrated by vacuum to yield a colorless, transparent oil (>90% mass recovery).
p[(SEA)50-b-(nBA)50]
p[(NHS)50-b-(nBA)50] (0.092 g) was dissolved in DMSO (3 mL) with 2-aminoethanesulfonic acid (0.1128 g, 0.9 mmol) and diisopropylethylamine (DIEA) (0.1165 g, 0.9 mmol) and heated to 70 °C overnight. DIEA was removed under vacuum, and polymer product was precipitated with diethyl ether and collected by centrifuge (4000 g, 5 min). The pellet was dissolved in 2 mL of distilled water, dialyzed against 0.1 M NaCl (24 h) and ddH2O (48 h), and then freeze-dried to obtain p[(SEA)50-b-(nBA)50] as a hygroscopic white solid.
p[(CMA)50-b-(nBA)50]
p[(NHS)50-b-(nBA)50] (0.100 g) was dissolved in DMSO (3 mL) with glycine (75 mg, 1 mmol) and diisopropylethylamine (DIEA) (0.126 g, 1 mmol) and heated to 70 °C overnight. The reaction was worked up and polymer purified as above to yield p[(CMA)50-b-(nBA)50] as a white solid.
p[(HEA)50-b-(nBA)50]
p[(NHS)50-b-(nBA)50] (0.100 g) was dissolved in DMSO (3 mL) with 2-aminoethanol (61 mg, 1 mmol) and diisopropylethylamine (DIEA) (0.126 g, 1 mmol) and stirred at room temperature overnight. The reaction was worked up and polymer purified as above to yield p[(HEA)50-b-(nBA)50] as a translucent hygroscopic solid.
TEM
Polymer-CPT nanoparticle sample, or polymer alone in Dulbecco’s phosphate buffered saline (DPBS) at 0.10 mg/mL of polymer equivalents, was mounted onto a 200 mesh Cu grid with a Formvar-carbon film. Sample treatment time was 60 s. The sample solution was then blotted dry by a small piece of filter paper and air-dried for 3 min. The sample grid was then negatively stained with uranyl acetate and air-dried prior to TEM.
Gel Permeation Chromatography (GPC)
GPC was performed in THF (flow rate of 1 mL/min) using four Styragel HR4 (Waters) columns (7.8 × 300 mm) connected in series. GPC traces were collected and analyzed by ChromPerfect software. Number-average molecular weights were determined with respect to PMMA standards.
Attenuated Total Reflectance Infrared Spectroscopy (ATR-IR)
FT-IR spectra were recorded on a Thermo Scientific Nicolet iS5 spectrometer with iD7 diamond ATR accessory and are uncorrected. Spectra were collected and processed using ThermoFisher OMNIC version 9. Samples (5–10 µL) were applied directly to the diamond crystal plate with background spectra recorded prior to polymer analysis. Polymer suspensions or solutions (50–100 mg/mL) were prepared in D2O with 500 mM NaCl and adjusted to the desired pD using DCl in D2O or NaOD in D2O. Commercially available poly(acrylic acid) (PAA standard, 78mer, 5.6 kDa) and diblock amphiphile, p[(AA)48-b-(nBA)50] (10 kDa), were studied. Polymers were dissolved in D2O, rotovapped to dryness 3–4 times, and then lyophilized from D2O to prepare samples for IR analysis in deuterated buffer.
Preparation of CPT-Loaded Nanoparticles
A solution of camptothecin and polymer in 95:5 methanol:TFA was dried under vacuum to yield a colorless, transparent thin film. The film was hydrated with pH 7.4 DPBS to yield a suspension that was disrupted by ultrasonication at 2% power amplitude. Sample was sonicated for 5–15 min (25 s on, 40 s off). Nonencapsulated camptothecin was removed by centrifuge (6000g, 2 min). The polymer/CPT weight ratio was varied to optimize drug loading to obtain maximum wt % CPT, 200 nm particle size, and PDI < 0.2 (Supporting Information).
In Vitro CPT (Dialysis) Release Assay
First, 500 µL of polymer-CPT suspension (0.3 mg/mL CPT) was dialyzed (Spectra/Por 1, MWCO 6–8 kDa, Spectrum Laboratories) against 1 L of DPBS at 37 °C with 400 rpm stirring. The reservoir buffer was changed every 12 h. At each time point, the total volume of polymer/CPT suspension inside the dialysis bag was measured using a pipetman to account for any possible volume change during the dialysis process. Five microliter aliquots were withdrawn from the dialysis bag and diluted into 95 µL of methanol, and the absorbance (370 nm) of the solution was measured. Remaining CPT was thus calculated using the CPT extinction coefficient in methanol (ε370 = 27692 cm−1 M−1).
CPT Sedimentation
Polymer-CPT nanoparticle suspensions (1 mL, 0.1 mg/mL CPT) were prepared in pH 7.4 DPBS or pH 5.5 (10 mM NaOAc, 160 mM NaCl) and incubated at 37 °C with 400 rpm stirring. At each time point, 100 µL of nanoparticle suspension was withdrawn into a 1.5 mL eppendorf tube and centrifuged for 2 min at 6000g. The aggregates precipitated at the bottom of the eppendorf tube; 5 µL of supernatant was carefully removed and diluted into 95 µl of methanol. UV absorbance (370 nm) of the sample was used to calculate the amount of remaining suspended CPT.
Acid-Induced Aggregation of CPT-Loaded Particles
Freshly prepared 1 mg/mL camptothecin encapsulated in 42% loaded p(AA)48-b-(nBA)50–CPT nanoparticles was diluted into 0.2 mg/mL of 1 mL of either pH 7.4 10 mM DPBS buffer or pH 5.5 10 mM NaOAc, 150 mM NaCl buffer. The diluted samples at both pH 7.4 and 5.5 were incubated at 37 °C with 400 rpm stirring. At the appropriate time points, 50 µL of each sample was withdrawn and analyzed by dynamic light scattering.
HSA Hydrolysis of CPT
Polymer-CPT nanoparticles (0.1 mg/mL CPT) were incubated at 37 °C with 400 rpm stirring in pH 7.0 DPBS supplemented with 3.5% human serum albumin (HSA). At each time point, 10 µL aliquots of the nanoparticle suspension were withdrawn and diluted into 190 µL of MeOH to precipitate HSA. Following sedimentation of HSA (10000g, 2 min), the supernatant was carefully removed and dried under nitrogen flow. The residue was redissolved into 100 µL of buffer (pH 6.5, 50 mM NaOAc) and analyzed by reverse-phase HPLC. Gradient program with A–B solvent system: 5– 40% B, 0–5 min; 40–80% B, 5–20 min; 80–100% B, 20–25 min. Elution solvents: A = 0.05 M NaOAc, pH 6.5; B = 99% acetonitrile, 1% H2O. Ring-opened and intact CPT quantities were determined by integration of HPLC peaks. Free camptothecin at 0.1 mg/mL was analyzed as a precipitate using the same method. For a more precise hydrolysis rate to be measured, free camptothecin was serially diluted in 3.5% HSA. At 0.025 mg/mL, negligible light scattering was observed; thus, this concentration was used to establish the HSA hydrolysis rate.
Cell Viability (MTT) Assay
MCF-7 cells were seeded in a 96-well plate (Corning, opaque bottom) at a density of 2 × 104 per well, with 90 µL of cell growth medium, and allowed to adhere overnight to reach approximately 60% confluency. Drug stock solutions were prepared at 300 µM CPT concentration and incubated in pH 6.8 DPBS (3.5% HSA) for 18 h at 37 °C or freshly prepared in DPBS without HSA treatment. These 300 µM stock solutions were serially diluted to 100, 30, 10, 3, 1, 0.3, 0.1, and 0.01 µM, and then 10 µL of each solution (including the 300 µM solution) was added to each well with 90 µL of medium, resulting in final CPT concentrations of 30, 10, 3, 1, 0.3, 0.1, 0.03, 0.01, and 0.001 µM. Cells were incubated for an additional 48 h and then assayed for viability using methylthiazolyl-diphenyltetrazolium bromide (MTT). Briefly, 20 µL of 5 mg/mL MTT solution in DPBS was added to each well, followed by incubation at 37 °C for 4 h after which the medium containing MTT reagent was carefully removed. Then, 100 µL of DMSO was added to each well, and the 96-well plate was shaken at 150 rpm for 30 min until no purple crystals were visible at the bottom of each well. The absorbance was then recorded at 570 nm using a SpectraMax Plus 384 microplate reader (Molecular Devices), and the absorbance at 650 nm was subtracted as background. The normalized viability of each cell sample is calculated using the equation
where A is the absorbance of the cell sample at 570 nm, and Actrl is the absorbance of cells treated only with DPBS.
Fluorescence-Activated Cell Sorting (FACS) Assay of CPT Cellular Uptake
Human breast cancer (MCF-7) cells were maintained in minimum essential media (MEM) supplemented with 10% FBS and 100 units/ml of penicillin and streptomycin. Twenty-four hours before cellular uptake experiments, cells were seeded at a density of 2 × 105 cells/ml. CPT-loaded particles from p[(AA)100-b-(nBA)50] (42 wt % CPT) and PEG-b-p[(AA)50-b-(nBA)50] (33 wt % CPT) were prepared and diluted 10× in DPBS. Equivolume samples were applied to cells to ensure parallel cultural conditions. After 6 h of incubation, cells were washed twice with DPBS, trypsinized, and fixed with 4% paraformaldehyde. All FACS samples were analyzed with a BD FACSAria III, with excitation at 355 nm, and fluorescence was detected at 420–470 nm. All cytometer settings were kept unchanged for samples of the same cell line. Fluorescence median value from three individual experiments were normalized, averaged, and then plotted against CPT concentration.
RESULTS AND DISCUSSION
Diblock polyacrylate amphiphiles were prepared using standard RAFT polymerization conditions and known chain transfer agents.32 The general amphiphile design features a hydrophobic poly-n-butyl acrylate block (DP = 50) and a polar acrylate block (DP ≈ 50–100). Notably, the critical micelle concentrations (CMCs) for all polymers were similar (7–9 µg/mL, Supporting Information) regardless of the polar block, suggesting that the lower limit of assembly remains driven by the high interfacial tension between the hydrophobic PnBA domain and water. Consistent with this notion, the CMC value was unchanged over pH 1–9 for all polymers, including those ionized in this pH range. However, significant differences in camptothecin encapsulation properties were observed as a function of the polar domain. Drug-loading in the poly(acrylic acid) (PAA) series was correlated with the length of the PAA: no loading was observed with a 30mer, but 42 wt % loading was observed with a 48mer PAA and a 9 wt % increase was seen for each ~29mer PAA extension (Table 1). Replacement of acrylic acid (AA) in p[(AA)48-b-(nBA)50] with carboxymethyl acrylamide yields p[(CMA)50-b-(nBA)50], which provides similar CPT loading (40 wt %). However, replacement of AA with sulfonic acid ethylacrylamide (SEA) or hydroxyethylacrylamide (HEA) diminishes or abolishes CPT loading, respectively. These findings suggest that the carboxylic acid domain plays a unique role in drug loading beyond its function as an anionic polyelectrolyte40 for particle suspension.
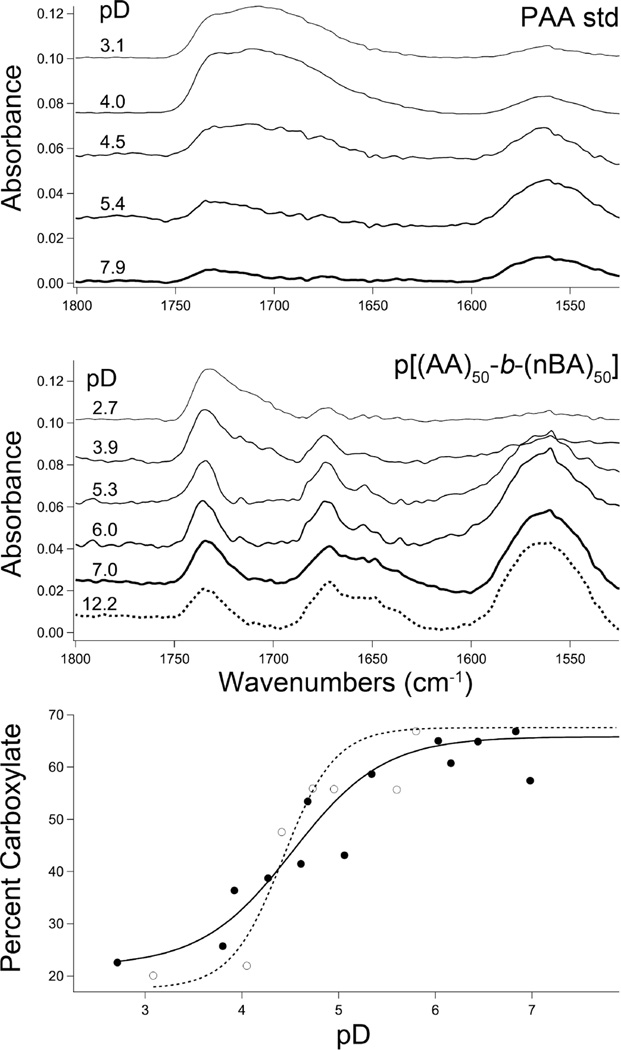
We speculated that partial chain neutralization imparts the carboxylic acid polymers with superior encapsulation properties over those of the sulfonic acid polymer. Indeed, PAA domains can have pKa’s in the range of 5–6.5, significantly shifted toward neutral by the local macromolecular environment relative to small molecule carboxylic acids.23 Global ionization state of poly(acrylic acid) (5.6 kDa standard) and p[(AA)48-b-(nBA)50] was interrogated using ATR-IR spectroscopy in deuterated saline water, adjusted to acidic, neutral, and basic pD at constant ionic strength (Figure 2). The PAA standard was chosen to benchmark expectations for known polyacids outside the context of diblock amphiphile assembly. The diblock p[(AA)48-b-(nBA)50] was readily assembled as nanoparticles under basic pD conditions, whereas the poly(acrylic acid) (PAA) standard did not exhibit detectable assembly (DLS); under acidic conditions (pD < 4.5), sonication was required to suspend both polymers. Analysis via ATR-IR indicated the presence of hydrogen-bonded carboxylic acid (1705 cm−1) and carboxylate (1560 cm−1) CO stretches in each macromolecular sample;41,42 as pD was increased, the carboxylate IR absorbance increased at the expense of the carboxylic acid IR band for both samples, providing a readout of polymer ionization state (Figure 2). This titration data indicates 50% ionization at pD’s of approximately 4.5 and 4.8 for the PAA standard and diblock, respectively, reflective of apparent pKa. The diblock exhibits a broadened transition and a moderately elevated pKa. However, the diblock spectra reveal an additional, unassigned species in the carbonyl stretching region that increases with ionization, which is absent in the PAA standard spectra (Figure 2). This new broad band centered at ~1672 cm−1 is indicative of a carbonyl species with CO bond strength intermediate between those of a typical carboxylate (1560 cm−1) and carboxylic acid (1710 cm−1), which appears as a shoulder on the ester band at 1732 cm−1; the invariant n-butyl ester band of the diblock was used to normalize the spectra. The 1672 cm−1 region is typically indicative of amide carbonyls, which are absent in the polymer sample. We speculate that the 1672 cm−1 band could derive from the carbonyls in a carboxylate-carboxylic acid hydrogen bond, in which a proton (or deuteron) is shared nearly equally between two carboxylates (Figure 1). This bonding configuration would be consistent with a carbonyl bond strength between that of a carboxylate, which has a strong O– donor, and an acid with a weaker OH donor. Further, the band is a broad multiplet and could contain energetically similar signatures from both hydrogen bond donor and acceptor carbonyl species. Although carboxylic acid dimerization via hydrogen bonding is more common, the sharing of a proton between chemically similar donors and acceptors, such as a carboxylic acid and a carboxylate, is known to be exceptionally strong, and this could be the origin of overall assembly stabilization and high drug loading capacity in the polyacidic diblocks.43 The 1672 cm−1 band intensity increases as the solution transitions from acidic to neutral, representing a significant fraction of the carbonyl population, but the spectra remain constant from neutral to strongly basic (Figure 2). Over this pH range, from neutral to basic, the drug-loaded polyacidic diblock amphiphiles are stable suspensions, which correlates with the formation of a stable, partially ionized state. Although the PAA standard and diblock have similar fractional ionization profiles (Figure 2), acid in PAA standard transforms to carboxylate whereas acid in the diblock transforms into both carboxylate and the unassigned 1672 cm −1 band. Further studies are needed to make unequivocal peak assignment of the 1672 cm −1 band, but it is clear that the carboxylic acid groups behave differently in the assembled diblock system relative to that in the PAA standard upon ionization. We hypothesize that this spectroscopic signature is correlated with the remarkable encapsulation and release properties of the polyacidic diblock amphiphiles.
Figure 2.
(Top) ATR-IR of the carbonyl region of (top) PAA standard and (middle) diblock p[(AA)48-b-(nBA)50] in D2O with 500 mM NaCl at the pD’s indicated. (bottom) Plot of pD vs % carboxylate from ATR-IR data of PAA standard (○) and diblock p[(AA)48-b-(nBA)50] (●). Carboxylate percentage was calculated from total carbonyl peak intensities of acid, carboxylate, and new peak at 1710, 1560, and 1672 cm−1, respectively.
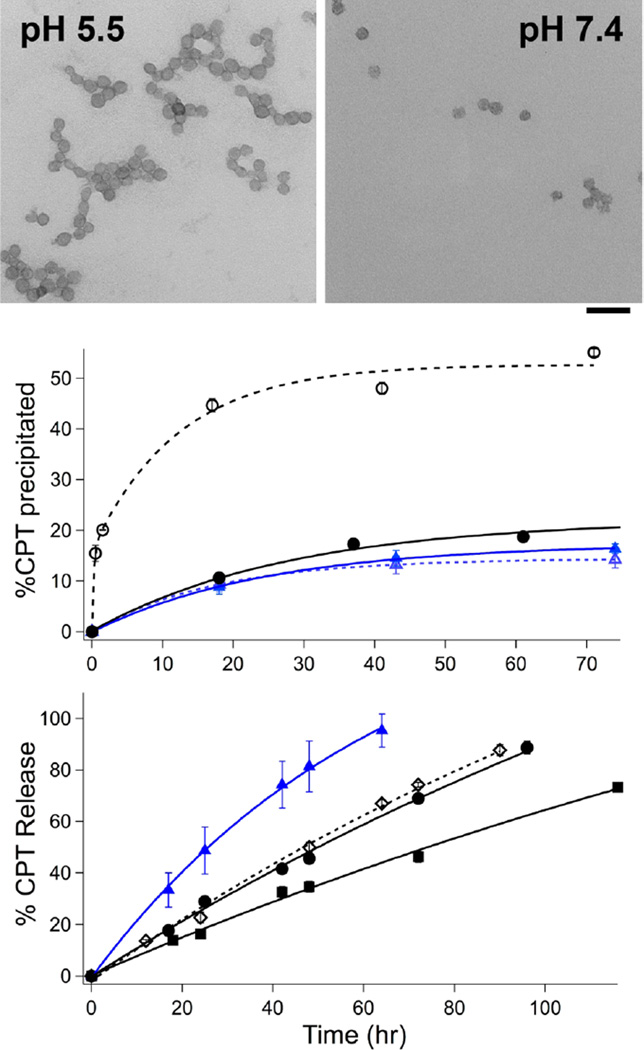
As drug-loaded particles were not stably suspended under the high polymer concentrations required for reliable ATR-IR analysis, we limited studies on the assembly properties of drug-loaded amphiphiles to dynamic light scattering (DLS) and transmission electron microscopy (TEM). The apparent particle size as judged by DLS increased to ~150 from ~70 nm upon CPT loading (Table 1). TEM analysis revealed that 70 and 150 nm nanoparticles observed by DLS were in fact clusters of 30 nm p[(AA)48-b-(nBA)50] particles with diameters roughly corresponding to size observed by scattering (Figure 3). These data suggest that the nanoparticles exist as small clusters that further agglomerate upon CPT loading due to increased hydrophobicity. The CPT-loaded p[(AA)48-b-(nBA)50] particles are sensitive to pH-induced assembly: both DLS and TEM indicated particle aggregation upon acidification to pH 5.5 from 7.4, consistent with neutralization of carboxylate groups (Figure 3). In contrast, sulfonic acid nanoparticles did not change in size from pH 7.4–1.0, reflective of constant ionization state. Sulfonic acid is ~106× more acidic than carboxylic acid and would be expected to be more fully ionized between pH 5.5–7.4. Thus, the diblock p[(SEA)50-b-(nBA)50] may represent the inherent drug capacity of an anionic hydrophobic skeleton without the stabilization afforded by acid neutralization. Indeed, together with the IR results, these data suggest that partial chain ionization could correlate with increased drug-loading stability by PAA diblock polymers relative to that of other related polyacid diblocks with lower pKa, such as the sulfonic acid (SEA) diblock p[(SEA)50-b-(nBA)50].
Figure 3.
(top) Uranyl acetate-stained TEM images of CPT-loaded p[(AA)48-b-(nBA)50] (43 wt %) in DPBS. Scale bar = 100 nm. (middle) Passive CPT sedimentation (RT, DPBS) in p[(AA)48-b-(nBA)50] at pH 5.5 (○) and pH 7.4 (●) and in p[(SEA)50-b-(nBA)50] at pH 5.5 (△) and pH 7.4 (▲). (bottom) CPT release assay via dialysis in DPBS, at pH 7.4 at 37 °C, from p[(SEA)50-b-(nBA)50] (▲), p[(CMA)50-b-(nBA)50] (◊), p[(AA)48-b-(nBA)50] (●), and p-[(AA)106-b-(nBA)50] (■). Loading levels were 28, 45, 40, and 60 wt %, respectively. Initial CPT encapsulation was identical for all samples.
Drug loading in the SEA polymer assembly is not only lower and less efficient than in the PAA polymers, it is less stable (Figure 3). Nanoparticles of p[(SEA)50-b-(nBA)50] released 50% of its CPT cargo after 24 h, whereas p[(AA)48-b-(nBA)50] released only ~22% of its drug cargo during the same time period. Notably, p[(CMA)50-b-(nBA)50 has a release profile nearly identical to that of p[(AA)48-b-(nBA)50], ruling out the possibility that the amide side chain linkage diminishes drug loading. The slowest release rates were observed with p[(AA)106-b-(nBA)50], underscoring the correlation of stable drug encapsulation with the length of the polyacidic domain.
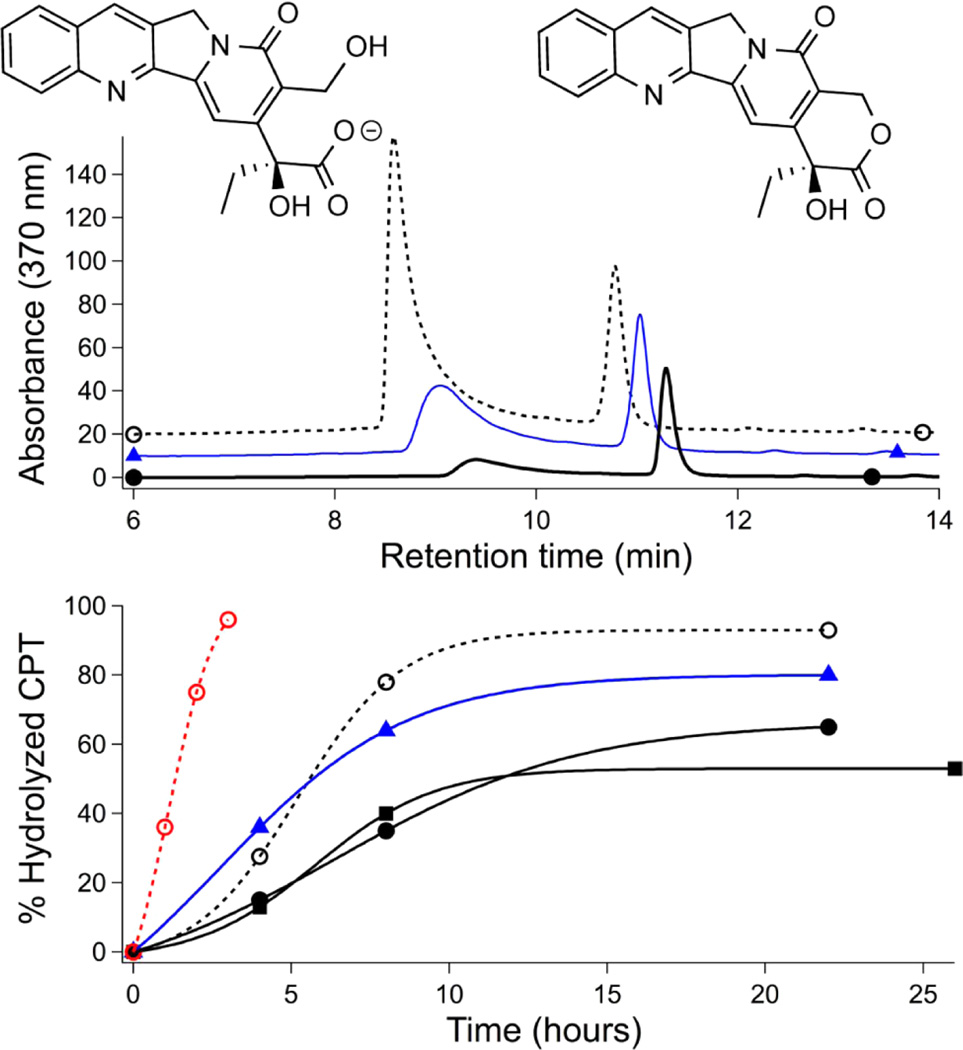
We next tested the degree to which polymer encapsulation could protect CPT from lactone hydrolysis, which ablates cytotoxicity. Intact CPT lactone could be recovered from nanoparticle suspensions after 3 weeks on the benchtop (RT, DPBS, pH 7.4); release was unaffected by the addition of bovine serum albumin.44 However, CPT hydrolysis is greatly accelerated in human serum albumin (HSA) relative to that in nonhuman albumins due to the preferential binding of HSA to the ring-opened CPT carboxylate.45–47 We thus incubated CPT-loaded particles under more biomedically relevant conditions (37 °C, 3.5% HSA) and monitored hydrolysis by extraction and HPLC (Figure 4). Whereas polymer encapsulation easily enables CPT suspension at 0.1 mg/mL, naked CPT precipitates at this concentration. HSA hydrolysis of CPT precipitate required 22 h, whereas soluble CPT (0.025 mg/mL) was completely ring-opened in just ~2 h. Over 24 h exposure, 20% cargo protection was afforded by polysulfonate p[(SEA)50-b-(nBA)50], whereas 50 and 60% of CPT cargo could be protected from HSA hydrolysis by p[(AA)48-b-(nBA)50] and p[(AA)106-b-(nBA)50], respectively. Overall, in the context of drug delivery, the several hours of protection provided by the poly(carboxylic acid) carriers is sufficient time for circulating nanoparticles to accumulate in solid tumors through enhanced permeation retention effects.48
Figure 4.
Hydrolysis of CPT alone (○, dashed), loaded in p[(SEA)50-b-(nBA)50] (▲, blue), p[(AA)48-b-(nBA)50] (●, black), and p-[(AA)106-b-(nBA)50] (■) nanoparticles after incubation in DPBS at pH 7.4 at 37 °C with 3.5% human serum albumin. (top) Representative HPLC traces of hydrolyzed (~9.2 min) and intact (11.3 min) CPT after 18 h. (bottom) CPT hydrolysis over time. CPT concentration was 0.1 mg/mL for all samples except for a free CPT sample at 0.025 mg/mL (red dashed line).
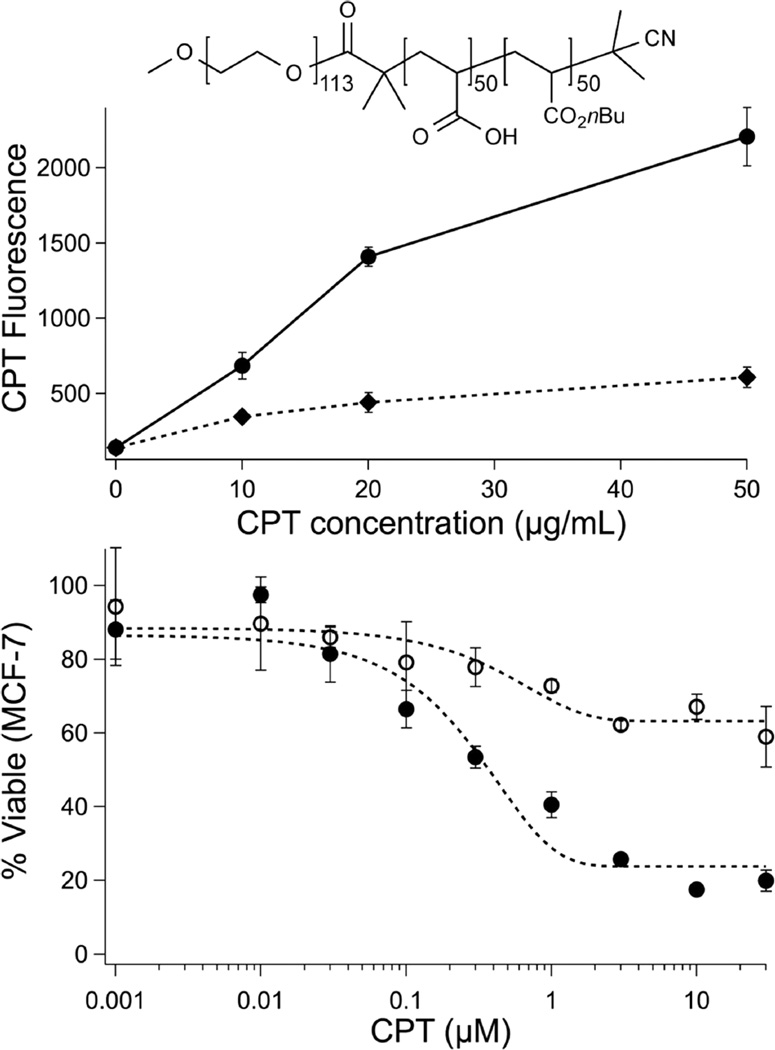
The efficacy of polymer-packaged CPT was tested in MCF-7 breast cancer cells using a cell death assay (MTT) following 18 h pretreatment with HSA, as described above (Figure 5). Whereas free CPT cytotoxicity was greatly diminished by HSA-driven hydrolysis, polymer-encapsulated CPT retained close to full activity for the entire poly(acrylic acid) series, p[(AA)n-b-(nBA)50]. Closer inspection indicates that cell death is commensurate with an ~50% reduction in effective CPT concentration, as predicted by the HSA hydrolysis experiment. Efficient cellular uptake of anionic nanoparticles was expected during this time frame,49 and indeed, FACS analysis confirmed uptake of CPT-loaded p[(AA)48-b-(nBA)50] nanoparticles into MCF-7 breast cancer cells after a 6 h incubation. Together, the MTT and FACS data suggest that cell death arises through intracellular release of drug rather than extracellular CPT release. Importantly, it was possible to block cell uptake by surface PEGylation of the loaded particles (Figure 5). This was accomplished with a moderate decrease in drug capacity using a PEG-functionalized RAFT chain transfer agent.50 Surface-protected51 p[PEG5kD-(AA)50-b-(nBA)50] particles could be loaded with similarly high levels of CPT (33–54 wt %), indicating the compatibility of this polymer platform with conventional methods51 used to prepare carriers for systemic delivery. Notably, although it was possible to form drug-loaded particles composed entirely of p[PEG5kD-(AA)50-b-(nBA)50], the high degree of unimer PEGylation (100%) results in destabilization of the assembly and increased release rates similar to those the sulfonic acid diblock (Supporting Information). Immunoevasive protection of liposomes can be achieved with a much lower degree of surface PEG (~8 mol %);51 thus, we anticipate that it will be possible to balance drug loading, release, and immunoevasive properties by decreasing the mole percentage of PEG polymer within the polyacidic assembly.
Figure 5.
(top) Fluorescence of MCF-7 cells gated on CPT emission via FACS analysis after treatment with CPT-loaded p[(AA)48-b-(nBA)50] at 42 wt % (●) and p[PEG5kD-(AA)50-b-(nBA)50] at 33 wt % (⧫, structure inset) at indicated concentrations. (below) MCF-7 cell viability (MTT) as a function of initial CPT concentration after 18 h treatment with HSA. CPT was delivered as free CPT (○) and at 42 wt % in p[(AA)48-b-(nBA)50] particles (●).
CONCLUSIONS
These findings demonstrate the utility of acidic polymer amphiphiles as high-capacity drug carriers. This function is likely a consequence of altered carboxylic acid ionization behavior in the polyacid diblock amphiphiles.24,25 Partial carboxylate neutralization may stabilize high drug loading assemblies by hydrogen bonding43 and/or increased hydrophobicity18 in the same way that synthetic polymer acids52,53 are subject to proton-triggered assembly54 and membrane activity.55,56 We speculate that hydrogen bonding side chains with pKa’s near neutral pH may be generally useful in preparation of high-capacity macromolecular drug carriers. Indeed, polyacidic domains may have played an unappreciated role in previously studied drug carrier systems.16,57 Additionally, this carrier function is found in 10–14 kD polymers, which are expected to be cleared in vivo without toxic accumulation by renal excretion.58,59 It is particularly striking that the desirable features of high drug capacity and controlled release can be obtained from facile polymerization of commercially available monomers. We anticipate that the well-studied class of acidic-hydrophobic diblock polymer amphiphiles will find expanded use as carriers for sensitive hydrophobic cargo.
Supplementary Material
Acknowledgments
Research was supported in part by NSF-DMR and NIH.
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Synthetic schemes, polymer characterization, additional sedimentation, and DLS and release data (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Chu DSH, Schellinger JG, Shi J, Convertine AJ, Stayton PS, Pun SH. Acc. Chem. Res. 2012;45(7):1089–1099. doi: 10.1021/ar200242z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim S, Park K. CRC Press; 2010. pp. 513–551. [Google Scholar]
- 3.Venditto VJ, Simanek EE. Mol. Pharmaceutics. 2010;7(2):307–349. doi: 10.1021/mp900243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheetham AG, Zhang P, Lin Y-A, Lock LL, Cui H. J. Am. Chem. Soc. 2013;135(8):2907–2910. doi: 10.1021/ja3115983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai K, He X, Song Z, Yin Q, Zhang Y, Uckun FM, Jiang C, Cheng J. J. Am. Chem. Soc. 2015;137(10):3458–3461. doi: 10.1021/ja513034e. [DOI] [PubMed] [Google Scholar]
- 6.Kim SH, Kaplan JA, Sun Y, Shieh A, Sun H-L, Croce CM, Grinstaff MW, Parquette JR. Chem. - Eur. J. 2015;21(1):101–105. doi: 10.1002/chem.201404520. [DOI] [PubMed] [Google Scholar]
- 7.Hatefi A, Amsden B. Pharm. Res. 2002;19(10):1389–1399. doi: 10.1023/a:1020427227285. [DOI] [PubMed] [Google Scholar]
- 8.O’Reilly RK, Hawker CJ, Wooley KL. Chem. Soc. Rev. 2006;35(11):1068–1083. doi: 10.1039/b514858h. [DOI] [PubMed] [Google Scholar]
- 9.De Greef TFA, Smulders MMJ, Wolffs M, Schenning A, Sijbesma RP, Meijer EW. Chem. Rev. 2009;109:5687–5754. doi: 10.1021/cr900181u. [DOI] [PubMed] [Google Scholar]
- 10.Aida T, Meijer EW, Stupp SI. Science. 2012;335:813–817. doi: 10.1126/science.1205962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Z, Bong D. Langmuir. 2013;29:144–150. doi: 10.1021/la304457y. [DOI] [PubMed] [Google Scholar]
- 12.Lee S-M, Chen H, O’Halloran TV, Nguyen ST. J. Am. Chem. Soc. 2009;131:9311–9320. doi: 10.1021/ja9017336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JO, Sahay G, Kabanov AV, Bronich TK. Biomacromolecules. 2010;11:919–926. doi: 10.1021/bm9013364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bronich TK, Keifer PA, Shlyakhtenko LS, Kabanov AV. J. Am. Chem. Soc. 2005;127:8236–8237. doi: 10.1021/ja043042m. [DOI] [PubMed] [Google Scholar]
- 15.Rösler A, Vandermeulen GWM, Klok H-A. Adv. Drug Delivery Rev. 2012;64(Supplement (0)):270–279. [Google Scholar]
- 16.Inoue T, Chen G, Nakamae K, Hoffman AS. J. Controlled Release. 1998;51(2–3):221–229. doi: 10.1016/s0168-3659(97)00172-7. [DOI] [PubMed] [Google Scholar]
- 17.Greene AC, Zhu J, Pochan DJ, Jia X, Kiick KL. Macromolecules. 2011;44(7):1942–1951. doi: 10.1021/ma102869y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wimley WC, White SH. Nat. Struct. Biol. 1996;3(10):842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 19.Etter MC. Acc. Chem. Res. 1990;23(4):120–126. [Google Scholar]
- 20.Sukhishvili SA, Granick S. Macromolecules. 2002;35(1):301–310. [Google Scholar]
- 21.Kim B-S, Park SW, Hammond PT. ACS Nano. 2008;2(2):386–392. doi: 10.1021/nn700408z. [DOI] [PubMed] [Google Scholar]
- 22.Park H, Robinson JR. Pharm. Res. 1987;4(6):457–464. doi: 10.1023/a:1016467219657. [DOI] [PubMed] [Google Scholar]
- 23.Colombani O, Lejeune E, Charbonneau C, Chassenieux C, Nicolai T. J. Phys. Chem. B. 2012;116(25):7560–7565. doi: 10.1021/jp3012377. [DOI] [PubMed] [Google Scholar]
- 24.Wilcox JL, Bevilacqua PC. J. Am. Chem. Soc. 2013;135(20):7390–7393. doi: 10.1021/ja3125299. [DOI] [PubMed] [Google Scholar]
- 25.Harris TK, Turner GJ. IUBMB Life. 2002;53(2):85–98. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 26.Jacquin M, Muller P, Cottet H, Crooks R, Theodoly O. Langmuir. 2007;23:9939–9948. doi: 10.1021/la700370f. [DOI] [PubMed] [Google Scholar]
- 27.Mai Y, Eisenberg A. Chem. Soc. Rev. 2012;41:5969–5985. doi: 10.1039/c2cs35115c. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Eisenberg A. J. Am. Chem. Soc. 1996;118:3168–3181. [Google Scholar]
- 29.Zhang L, Eisenberg A. Science. 1995;268:1728–1731. doi: 10.1126/science.268.5218.1728. [DOI] [PubMed] [Google Scholar]
- 30.Colombani O, Ruppel M, Burkhardt M, Drechsler M, Schumacher M, Gradzielski M, Schweins R, Müller AHE. Macromolecules. 2007;40(12):4351–4362. [Google Scholar]
- 31.Nicolai T, Colombani O, Chassenieux C. Soft Matter. 2010;6:3111–3118. [Google Scholar]
- 32.Keddie DJ, Moad G, Rizzardo E, Thang SH. Macromolecules. 2012;45(13):5321–5342. [Google Scholar]
- 33.Soukasene S, Toft DJ, Moyer TJ, Lu H, Lee H-K, Standley SM, Cryns VL, Stupp SI. ACS Nano. 2011;5(11):9113–9121. doi: 10.1021/nn203343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y, Jin E, Zhang B, Murphy CJ, Sui M, Zhao J, Wang J, Tang J, Fan M, Van Kirk E, Murdoch WJ. J. Am. Chem. Soc. 2010;132:4259. doi: 10.1021/ja909475m. [DOI] [PubMed] [Google Scholar]
- 35.Chiang Y-T, Yen Y-W, Lo C-L. Biomaterials. 2015;61:150–161. doi: 10.1016/j.biomaterials.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe M, Kawano K, Yokoyama M, Opanasopit P, Okano T, Maitani Y. Int. J. Pharm. 2006;308:183–189. doi: 10.1016/j.ijpharm.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 37.Thang SH, Chong YK, Mayadunne RTA, Moad G, Rizzardo E. Tetrahedron Lett. 1999;40(12):2435–2438. [Google Scholar]
- 38.Mammen M, Dahmann G, Whitesides GM. J. Med. Chem. 1995;38(21):4179–4190. doi: 10.1021/jm00021a007. [DOI] [PubMed] [Google Scholar]
- 39.Perrier S, Takolpuckdee P, Mars CA. Macromolecules. 2005;38(6):2033–2036. [Google Scholar]
- 40.Förster S, Hermsdorf N, Böttcher C, Lindner P. Macromolecules. 2002;35(10):4096–4105. [Google Scholar]
- 41.Max J-J, Chapados C. J. Phys. Chem. A. 2004;108(16):3324–3337. [Google Scholar]
- 42.Dong J, Ozaki Y, Nakashima K. Macromolecules. 1997;30(4):1111–1117. [Google Scholar]
- 43.Hibbert F, Emsley J. In: Adv. Phys. Org. Chem. Bethell D, editor. Vol. 26. Academic Press; 1991. pp. 255–379. [Google Scholar]
- 44.Lu J, Owen SC, Shoichet MS. Macromolecules. 2011;44:6002–6008. doi: 10.1021/ma200675w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mi Z, Burke TG. Biochemistry. 1994;33(42):12540–12545. doi: 10.1021/bi00208a002. [DOI] [PubMed] [Google Scholar]
- 46.Burke TG, Mi Z. J. Med. Chem. 1994;37(1):40–46. doi: 10.1021/jm00027a005. [DOI] [PubMed] [Google Scholar]
- 47.Opanasopit P, Yokoyama M, Watanabe M, Kawano K, Maitani Y, Okano T. J. Controlled Release. 2005;104(2):313–321. doi: 10.1016/j.jconrel.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 48.Maeda H, Nakamura H, Fang J. Adv. Drug Delivery Rev. 2013;65(1):71–79. doi: 10.1016/j.addr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 49.Bandyopadhyay S, Xia X, Maiseiyeu A, Mihai G, Rajagopalan S, Bong D. Macromolecules. 2012;45:6766–6773. doi: 10.1021/ma301536f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartels JW, Cauet SI, Billings PL, Lin LY, Zhu J, Fidge C, Pochan DJ, Wooley KL. Macromolecules. 2010;43:7128–7138. doi: 10.1021/ma1002112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Immordino ML, Dosio F, Cattel L. Int. J. Nanomedicine. 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- 52.Yin X, Hoffman AS, Stayton PS. Biomacromolecules. 2006;7:1381–1385. doi: 10.1021/bm0507812. [DOI] [PubMed] [Google Scholar]
- 53.Murthy N, Robichaud JR, Tirrell DA, Stayton PS, Hoffman AS. J. Controlled Release. 1999;61:137–143. doi: 10.1016/s0168-3659(99)00114-5. [DOI] [PubMed] [Google Scholar]
- 54.Ghadiri MR, Granja JR, Milligan RA, McRee DE, Khazanovich N. Nature. 1993;366:324–327. doi: 10.1038/366324a0. [DOI] [PubMed] [Google Scholar]
- 55.Hunt JF, Rath P, Rothschild KJ, Engelman DM. Biochemistry. 1997;36(49):15177–15192. doi: 10.1021/bi970147b. [DOI] [PubMed] [Google Scholar]
- 56.Ghadiri MR, Granja JR, Buehler LK. Nature. 1994;369:301–304. doi: 10.1038/369301a0. [DOI] [PubMed] [Google Scholar]
- 57.Hu Y, Ding Y, Ding D, Sun M, Zhang L, Jiang X, Yang C. Biomacromolecules. 2007;8(4):1069–1076. doi: 10.1021/bm0608176. [DOI] [PubMed] [Google Scholar]
- 58.Maeda H, Kabanov A. Polymer drugs in the clinical stage: advantages and prospects. Vol. 519. Springer Science & Business Media; 2003. [Google Scholar]
- 59.Fraser JR, Laurent TC, Pertoft H, Baxter E. Biochem. J. 1981;200(2):415–424. doi: 10.1042/bj2000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.