Abstract
The growing number of people suffering from obesity and type 2 diabetes mellitus (T2DM) is a global health problem that results in increased mortality from their complications, mainly cardiovascular diseases. Although the relationship between obesity and T2DM is well established, the common molecular pathomechanisms are still under investigation. Recently, it has been suggested that epigenetic modifications may be involved in both obesity and T2DM development. Epigenetics plays a pivotal role in the regulation of gene expression by the reversible modifications of chromatin structure without any changes in DNA sequence. Epigenetic modifications include DNA methylation, posttranslational histone modifications and miRNA interference. Therefore, the aim of this article is to discuss the current knowledge on epigenetic modifications in adipose tissue and their association with obesity and T2DM.
Keywords: epigenetic modifications, adipose tissue, obesity, type 2 diabetes mellitus
Introduction
According to the World Health Organization, obesity and type 2 diabetes mellitus (T2DM), which frequently occur together (diabesity), have become a global 21st century epidemic. Both obesity and T2DM lead to serious life-threatening complications, especially cardiovascular diseases. Epidemiological data show that between 60% and 90% of T2DM patients are overweight or obese [1]. Moreover, Ford et al. suggest that one kilogram of weight gain increases the risk of T2DM development by 4.5% to 9% [2]. However, it is ambiguous whether obesity is a risk factor or a cause of T2DM development. It was found that the development of obesity and/or T2DM may be associated with changes in gene expression evoked by genetic and environmental factors [3]. Moreover, it was demonstrated that gene expression is also under the control of epigenetic mechanisms responsible for switching between active and inactive states of chromatin [4].
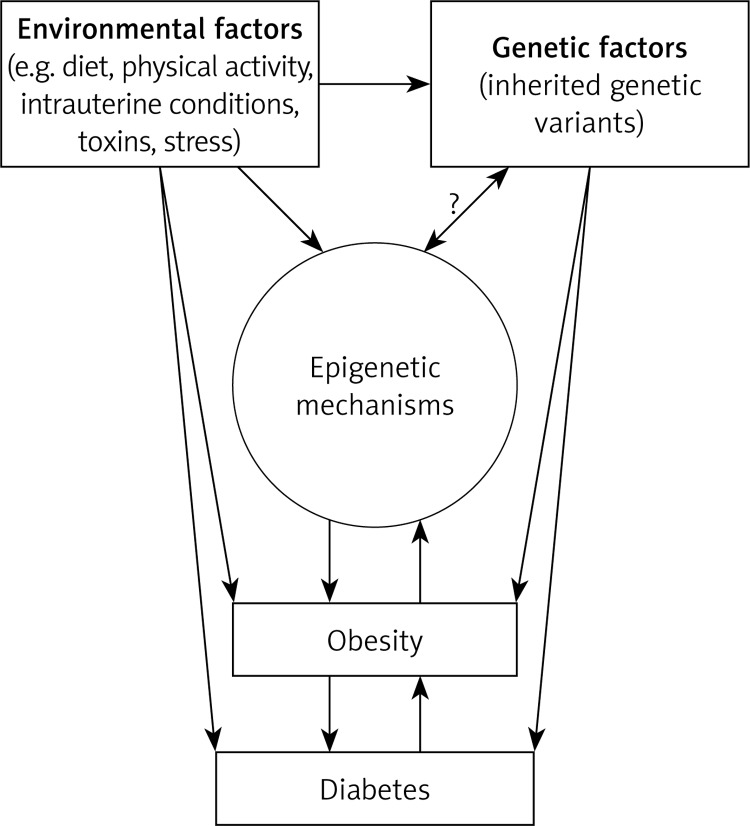
Notable evidence demonstrating the involvement of epigenetic mechanisms in the development of metabolic disorders comes from the “Dutch famine birth cohort” study. In this study, children of Western Netherlands’ women, affected by the Nazi embargo, born between November 1943 and February 1947, have been followed up since 1996. Children conceived by malnourished women (caloric intake of 400–800 per day) were smaller and more prone to develop several chronic diseases, including cardiovascular disease, obesity, diabetes, elevated blood pressure, and impaired glucose tolerance in later life [5]. Noteworthy, other observational studies showed that children conceived by mothers with gestational diabetes were also more susceptible to obesity, T2DM or metabolic syndrome [6]. Taken together these observations suggest that the fetal developmental environment may evoke significant epigenetic modifications that predispose the offspring to metabolic diseases (Figure 1).
Figure 1.
Proposed interplay between environment, genetic factors, epigenetic mechanisms, obesity and diabetes
Twin studies are considered as a crucial approach to unravel the interplay between heritable and environmental factors in the etiology of various diseases, including metabolic disorders [7]. These studies provide an opportunity to separate the influence of common environmental from genetic factors on disease phenotype by comparing monozygotic twins to dizygotic twins. Monozygotic twin studies are considered as unbiased due to the fact that they are genetically identical (age, sex) and share congruent environmental influences (cohort effects, maternal influences and common environment), whereas dizygotic twins share on average 50% of genetic variants [8]. The analysis of cardiometabolic risk factors using a structural equation model performed on 63 monozygotic and 38 dizygotic adult twins revealed that some risk factors were strongly associated with heritability (weight, waist circumference), whereas others (fasting insulin, fasting blood glucose and total cholesterol) were significantly influenced by the environment [9]. Therefore, it seems that twin studies give an excellent opportunity to assess the influence of environment and heritability on epigenetic pattern and related disease phenotype.
The main goal of this manuscript is to present contemporary findings related to epigenetic modifications in adipose tissue and their association with obesity and type 2 diabetes.
Epigenetics – definition and background
The term “epigenetics” was introduced in the 1940s by Waddington, as an attempt to define the role of genetics in developmental processes [10]. Nowadays, epigenetics is a discipline exploring gene expression involving functional changes in chromatin structure and non-coding RNAs without direct change in nucleotide sequences of DNA. Epigenetic regulation includes modifications of chromatin proteins, methylation of DNA and miRNA interference. The local packing of chromatin, loose (euchromatin) or tight (heterochromatin), determines the access of proteins responsible for gene expression. Euchromatin, a transcriptionally active form of chromatin, consists of nucleosomes (DNA double-helix DNA wrapped around the octamer of core histone proteins: dimers of H2A, H2B, H3 and H4 histones) connected via internucleosomal DNA, which is known as ‘beads on a string’. Heterochromatin, an inactive form of chromatin, arises as a result of H1 histone interaction with nucleosomes. Epigenetic modifications of DNA and core histone proteins may weaken or strengthen the interaction between nucleosomes and H1 histones, which results in loosening or tightening of chromatin structure, altering the access of proteins to genes [4]. Epigenetic changes were found to be involved in developmental and differentiation processes, i.e. repetitive elements silencing, inactivation of chromosome X during sex determination, and genomic imprinting during gametogenesis. A growing body of evidence suggests that changes in the epigenome induced by the environment may alter gene expression and lead to the development of various diseases, including cancers, obesity and diabetes [11].
DNA methylation is a reversible process performed by DNA methyltransferases (DNMTs) and DNA demethylases (DNAdeMeths). Methylation of DNA involves the addition of a methyl group to cytosine in specific regions, called CpG islands. Areas of lower transcriptional activity were detected as highly methylated, while hypomethylation of cytosine was associated with increased gene expression [12].
Posttranslational modifications of core histones include acetylation, methylation, phosphorylation, ubiquitination, and sumoylation [13]. Currently, much attention is focused on core histones’ methylation and acetylation of lysine and arginine. Key enzymes responsible for modifications of N-terminal tails of H2A, H2B, H3 and H4 are histone acetyltransferases (HATs), histone deacetyltransferases (HDACs), histone methyltransferases (HMTs) and histone demethylases (HDMs) [11]. The effect of acetylation and methylation on the regulation of gene expression was found to be dependent on the site of modification. It was reported that acetylation of H3 histone at lysine-9/14 sites (H3KAc) and methylation of H3 histone at lysine-4 (H3K4me) were associated with activation of gene expression. Contrariwise, methylation of H3 histone at lysine-9 (H3K9me) or methylation of H3 histone at lysine-27 (H3K27me) was demonstrated to inhibit gene expression. HMTs were shown to mediate the addition of two or three methyl groups to a specific lysine residue [14].
Interestingly, enzymes involved in histone epigenetic modifications were also demonstrated to modify non-histone proteins, such as nuclear factor-κB (NF-κB) and p53 [15]. Both NF-κB and p53 are transcription factors, engaged in the control of cell survival and the inflammatory process.
In addition to chromatin structure modification, another mechanism of epigenetic regulation is gene silencing through single-stranded non-coding RNAs [16]. miRNAs are small molecules, approximately 19 to 33 nucleotides in length, derived from endogenous transcripts generating a hairpin structure, that interact with specific regions of mRNA. Pairing between miRNA and mRNA results in translation inhibition, as a result of degradation of mRNA and/or destabilization of mRNA structure [17, 18]. Recent evidence identified involvement of various miRNAs in the pathogenesis of several diseases, including obesity and diabetes.
Adipose tissue as a site of initiation of metabolic disorders
Adipose tissue contains different types of cells, including mainly adipocytes and a stromal vascular fraction composed of fibroblasts, preadipocytes, vascular endothelial cells and immune cells (macrophages, lymphocytes) [19]. Apart from energy storage, cushioning and insulating the body, adipose tissue was established as an important endocrine organ, secreting various hormones, cytokines (adipocytokines) and free fatty acids (FFAs) [20]. Adipocytes and adipose tissue macrophages express various receptors including peroxisome proliferator-activated receptors γ (PPARγ), fundamentally important for adipose tissue function. The PPARγ are crucial for differentiation and apoptosis of adipocytes, as well as the regulation of lipogenesis-related genes. Additionally, activation of PPARγ is a part of the regulation of adipocytokine expression, including leptin and tumor necrosis factor α (TNF-α) and interleukin-6 (IL6) [21].
Adipose tissue is situated in various body departments: under the skin (subcutaneous fat), around internal organs (visceral fat: mesenteric, epididymal white adipose tissue, and perirenal depots), bone marrow (yellow fat marrow) and in breasts. Excess of visceral adipose tissue (VAT), also called central obesity, is considered an essential factor for the development of glucose intolerance and T2DM associated with insulin resistance (tissues’ impaired ability to respond properly to insulin action) [22, 23]. Adipose tissue constantly deals with FFA turnover. Whether FFAs will enter the adipocyte, being reassembled in the form of triglycerides, or leave the cell (lipolysis) is controlled by the relation between insulin and leptin level [24]. Leptin is an adipocytokine that stimulates the inflammatory response and prevents lipid accumulation in adipose tissue, while high insulin levels exhibit an antilipolytic effect, promoting the storage of triglycerides in adipocytes [25]. In obese individuals, hypertrophic visceral adipocyte cells have a lower density of insulin receptors and become resistant to insulin, exhibiting a hyperlipolytic state. As a result, FFAs are fluxed into the liver, which in turn impairs liver metabolism, leading to glucose overproduction [24]. Chronically elevated levels of glucose and lipids exert toxic effect (gluco- and lipotoxicity) on various organs, resulting in serious metabolic disorders [26]. Additionally, hypertrophic adipocytes facilitate the migration of macrophages to the stromal fraction, initiating and promoting the inflammatory process [27]. Moreover, the enhancement of obesity-related chronic low grade inflammation is promoted via FFA oxidation in the liver, which results in reactive oxygen species (ROS) generation [28]. Reactive oxygen species production by macrophages and monocytes is also stimulated by pro-inflammatory cytokines. It was recognized that the secretory profile of adipose tissue in overweight/obese people is altered [29]. The expression of pro-inflammatory cytokines (TNF-α, IL-6) increases, whereas expression of cytokines improving insulin sensitivity (e.g. adiponectin) is reduced. It is well known that both oxidative stress and chronic low-grade inflammation having an impact on insulin secretion and insulin sensitivity may exaggerate the development of T2DM and atherosclerosis-associated diseases.
Epigenetic modifications in adipose tissue
DNA methylation pattern
Studies indicate the involvement of DNA methylation in differentiation of adipocytes and its effect on expression of genes related to obesity and diabetes.
One major study exploring the role of DNA methylation in adipogenesis was conducted by Chapman et al. They found that treatment of mouse embryo fibroblast (10T1/2) with an inhibitor of mammalian DNA methylation, 5-azacytidine, gave origin to new cell types, including preadipocytes (TA1, TA2) featuring the accumulation of lipid droplets. Additionally, insulin- and dexamethasone-induced differentiated TA1 cells showed changes typical for mature adipocytes. A significant increase in glycerol phosphate dehydrogenase activity and the level of proteins involved in fatty acid and triglyceride synthesis, as well as changes in total mRNA expression, was observed [30]. Wabitsch et al. reported that using 5-aza-deoxycytidine, a DNA hypomethylation agent, during differentiation of a cell line derived from liposarcoma (LiSa-2), resulted in a significant increase in leptin mRNA expression [31].
Recently published results of the study performed by Malodobra-Mazur et al. on differentiated 3T3-L1 adipocytes suggested that the level of proinflammatory cytokine expression might be regulated by stearoyl-CoA desaturase 1 (SCD1) through DNA methylation [32]. SCD1, an enzyme highly expressed in differentiated adipocytes, is responsible for the biosynthesis of monounsaturated fatty acids from saturated fatty acids. It is suggested that SCD1 may be involved in obesity-related inflammation [33]. Findings indicate that over-expression of SCD1 is associated with global DNA hypomethylation, whereas silencing or inhibition of SCD1 evokes global hypermethylation. Additionally, it was found that the promoter methylation pattern in CpG islands of interleukin-10 receptor a (IL10ra), interleukin-4 receptor a (IL4ra), interleukin-6 signal transducer (IL6st), and transforming growth factor β1 (TGFβ1) genes, was changed by SCD1 overexpression and correlated with changes in expression of the IL10ra, IL4ra, TGFβ1, and IL6st genes [32].
Bouchard et al. investigated the DNA methylation pattern and changes in gene expression in subcutaneous adipose tissue of obese/overweight postmenopausal women before and after a 6-month caloric restriction diet [34]. Participants were grouped into high and low diet responders according to changes of body fat percentage after completion of the experiment. Differences in DNA methylation between high and low responders were observed at 35 loci before and at 3 loci after dietary intervention. Several loci comprised genes involved in the control of weight (potassium voltage-gated channel shaker-related subfamily member 3 [KCNA3], nuclear factor I/X [NFIX]), insulin secretion and diabetes (insulinoma-associated 1 [INSM1], GLIS family zinc finger 3 [GLIS3], cholecystokinin B receptor [CCKBR]). It was found that gene expression changes appeared only after dietary intervention, and included 644 genes engaged in metabolic pathways (327 genes up-regulated and 317 down-regulated). Comparing high diet responders to low diet responders, the highest overexpression was observed for the gene encoding cholesteryl ester transfer protein (CETP) and the highest downexpression was found for phosphoserine phosphatase (PSPH) mRNA. The CETP is involved in cholesteryl ester and triglyceride transfer among plasma lipoproteins, whereas PSPH is responsible for L-serine formation. Recently, CETP was also proposed to play a role in metabolism and storage of lipids in adipose tissue [35].
The relationship between 8-week low-caloric diet and methylation level in the promoter regions as well as the expression level of leptin and TNF-β determined in human subcutaneous adipose tissue was assessed by Cordero et al. [36]. Subjects were divided into responder and non-responder groups on the basis of changes in weight, lipid profile, fat mass percentage, and systolic and diastolic blood pressure after diet completion. A lower methylation pattern in the promoters of leptin and TNF-α detected before starting the diet was found in responders as compared to non-responders, but there were no differences after diet completion. The level of mRNAs for leptin and TNF-α did not differ significantly between responders and non-responders before implementation of the diet. However, after dietary intervention leptin expression level was significantly decreased in the responder group, but no changes in TNF-α expression level were observed. These observations suggested that methylation level of leptin and TNF-α promoters may be an epigenetic predictor of responsiveness to a caloric restriction diet.
Rönn et al. [37] examined the effect of 6-month exercise intervention on DNA methylation pattern at 476,753 CpG sites in adipose tissue of healthy men previously representing low physical activity. They found that the global DNA methylation pattern was altered at 17,975 CpG sites in response to exercise. The authors also observed that after exercise adipose tissue displayed differential methylation at 236 CpG sites of 197 gene regions along with a distinct alteration in mRNA expression of these genes. Moreover, the relation between DNA methylation and expression of obesity and T2DM candidate genes was explored. Among 476,753 CpG sites after exercise, 24 CpG sites with a difference in DNA methylation were located in 18 candidate genes for obesity, whereas 45 differentially methylated CpG sites were observed in 21 T2DM candidate genes. However, changes in gene expression in parallel with differential DNA methylation were found only for 2 obesity candidate genes (cytoplasmic polyadenylation element binding protein 4 [CPEB4], serologically defined colon cancer antigen 8 [SDCCAG8]) and 4 T2DM-related genes (hematopoietically expressed homeobox [HHEX], insulin-like growth factor 2 mRNA binding protein [2IGF2BP2], juxtaposed with another zinc finger gene 1 [JAZF1], and transcription factor 7-like 2 [TCF7L2]). Of note, 2 genes (histone deacetylase 4 [HDAC4], nuclear receptor co-repressor 2 [NCOR2]) exhibiting increased DNA methylation accompanied by decreased mRNA expression in adipose tissue after exercise were selected to assess their effect on adipocyte metabolism. HDAC4 plays a significant role in the repression of glucose transporter 4 (GLUT4) transcription in adipocytes, whereas NCOR2 regulates the expression of genes involved in lipid metabolism and adipogenesis, as well as recruitment of various histone deacetylases, including HDAC4. As expected, siRNA-induced silencing of HDAC4 and NCOR2 resulted in increased lipogenesis in 3T3-L1 adipocytes.
Posttranslational core histone modifications
Among investigated posttranslational core histone modifications in adipose tissue, the largest body of evidence has accumulated for H3 histone.
Steger et al. found that the process of preadipocyte differentiation was associated with transient epigenetic modification of H3 histone [38]. 3T3-L1 preadipocytes were induced to differentiate into mature adipocytes by the treatment with insulin, glucocorticoids and cAMP signaling inducers. H3 histone modifications (H3K4 trimethylation – H3K4me3, H3K4 dimethylation – H3K4me2, and H3K9 acetylation – H3K9ac) overlapping 74 gene regions were investigated at days 0, 1, and 10 of 3T3-L1 differentiation. Among these genes’ regions, 20 exhibited increased levels of H3K4me3, H3K4me2 and H3K9ac at the end of differentiation (day 10), as compared to undifferentiated cells (day 0). Of note, only PPARγ regions showed increased levels for all three modifications on the first day of differentiation. The induction of PPARγ expression was detected ~24 h after the initiation of differentiation and was sustained in mature 3T3-L1 adipocytes. Steger et al. found that PPARγ transactivation was a consequence of a transient assemble of glucocorticoid receptor (GR) and CCAAT/enhancer-binding protein β (CEBPβ) with p300 and mediator subunit 1 (MED1), stimulating histone H3 acetylation at several enhancers, including enhancers for PPARγ. Importantly, this assemblage occurred only at the start of adipogenesis (day 1).
Tateishi et al. demonstrated that genetic knockout of specific H3K9 demethylase, jumonji C-domain containing protein (Jhdm2a), increased mice susceptibility to obesity, as a result of a decrease in PPARα expression in skeletal muscles and brown fat [39]. Jhdm2a deficiency led to abnormal fat distribution and hyperlipidemia. It was also found that other genes involved in the PPAR pathway were down-regulated, including those participating in fatty acid metabolism (uncoupling protein 2 [Ucp2], medium-chain acyl-CoA dehydrogenase [MCAD], light-chain acyl-CoA dehydrogenase [LCAD], very long chain acyl-CoA dehydrogenase [VLCAD]) and glycerol release (aquaporin 7, [Aqp7]).
Okuno et al. investigated the role of another jumonji C-domain containing protein responsible for demethylation of H3K9me2 (plant homeodomain finger 2 [PHF2]; also called JHDM1E) in the process of adipogenesis [40]. They generated transgenic PHF2-knockout mice that exhibited a partial neonatal death, growth delay, and reduction of body weight. Moreover, transgenic mice showed lipoatrophic changes, mainly in epididymal and subcutaneous white adipose tissue and slight lipoatrophy in mesenteric adipose tissue. In vitro studies on mice 3T3-L1 fibroblasts and vascular stromal cells with knockout of PHF2 induced to differentiate into mature adipocytes showed impairment of adipogenesis. It was reported that PHF2 was a coactivator of metabolism-related transcription factors through interaction with CEBPα and demethylation of H3K9me2 in the promoters of CEBPα-regulated adipogenic genes, including PPARγ, CEBPα, and fatty acid binding protein 4 (FABP4) [40].
Hino et al. demonstrated that inhibition of lysine-specific demethylase-1 (LSD1) – via siRNA and selective inhibitors – increased expression of metabolism-related genes (PPARγ coactivator-1α [PGC-1α], pyruvate dehydrogenase kinase 4 [PDK4], protein kinase A regulatory subunit 2α [RIIα], adipose triglyceride lipase [ATGL], fatty acid transporter protein 1 [FATP1]) in 3T3-L1 differentiating adipocytes [41]. Transcription repression by LSD1 is associated with removal of a methyl group from mono- and dimethylated K4 of H3. They also reported that the inhibition of LSD1 activity in differentiating 3T3-L1 cells led to a significant increase in mono-methylated K4 of H3 (H3K4me) and enhanced mitochondrial metabolism (detected by increased mitochondrial membrane potential and increased mitochondrial oxidative phosphorylation). Results from experiments performed on mice demonstrated that consumption of a high fat diet for 6 weeks induced an obese phenotype with the increase of LSD1 expression and decreased expression of PGC-1α, PDK4, FATP1 in adipose tissue.
Enhanced H3K4me2 in promoters/enhancers, transcribed regions and increased expression of genes encoding proteins participating in the immune response and cellular remodeling (C-type lectin domain family 4 member D [Clec4d], toll-like receptor 8 [Tlr8], ATPase H+ transporting lysosomal V0 subunit D2 [Atp6v0d2], procollagen type VIII α1 [Col81a], fibulin-2 [Fbln2] and fibulin-5 [Fbln5], actin α1 [Acta1], matrix metallopeptidase 12 [MMP12]) in mesenteric adipose tissue of insulin-resistant diabetic (db/db) mice in comparison to non-diabetic mice were detected by Fujimoto et al. [42]. Clec4d and Tlr8 play an important role in macrophage infiltration in response to pathogens. However, macrophage infiltration was found to be involved in pathogenesis of insulin resistance and diabetes mediated by chronic low grade inflammation [43]. Up-regulated Atp6v0d2, encoding an enzyme activating mitochondrial electron transport chain, was shown to be connected with increased reactive oxygen species’ generation and reduced insulin sensitivity in adipose tissue [44]. The authors suggest that observed over-expression of Col81a, Fbln2, Fbln5, Acta1 and MMP12 in db/db mice might be associated with change in adipocytes’ shape during transition from non-insulin resistant to insulin-resistant phenotype and related conversion from non-obese to obese phenotype.
Abu-Farha et al. performed an analysis of proteome in peripheral blood mononuclear cells (PBMCs) and subcutaneous adipose tissue of obese and lean individuals before and after 3 months of exercise [45]. Significant differences in the level of 47 proteins in PBMCs were found in obese compared to lean individuals. Among them distinct up-regulation of thrombospondin 1 (TSP1) and down-regulation of histone deacetylase 4 (HDAC4) in obese individuals as compared to lean participants were noted. Exercise intervention affected the level of 38 proteins in PBMCs in obese subjects (17 proteins increased, 21 proteins decreased). Additionally, they observed that the level of TSP1 and HDAC4 changed after exercise both in PBMCs and subcutaneous adipose tissue to the level observed in lean subjects. The functional assays revealed that the expression of HDAC4 significantly disturbed TNF-α-dependent activation of NF-κB. The authors suggested that HDAC4 might protect against obesity, as the expression pattern of HDAC4 in obese subjects before and after physical exercise correlated with physical, clinical and metabolic parameters.
miRNA
A growing amount of evidence indicates that miRNAs play a crucial role in the regulation of cellular metabolism in adipose tissue, and in the pathogenesis of obesity and diabetes. Ferland-McCollough et al. performed a study on adipose tissue from low-birth-weight (LBW) adult humans, prediabetic adult rats and on 3T3-L1 mice adipocytes that identified the role of programmed changes of miR-483-3p expression in the relationship between early-life nutrition and metabolic disorders in later life [46]. Microarray analysis of miRNAs in epididymal adipose tissue taken from offspring of rats fed with a low-protein diet showed significant up-regulation of miR-483-3p. Moreover, the level of miR-483-3p expression was also increased in subcutaneous adipose tissue of young adult LBW men in comparison to healthy control subjects. The molecular target of miR-483-3p is growth differentiation factor-3 (GDF3), a member of the bone morphogenetic protein (BMP) family and the TGF-β superfamily, which participates in cell growth and differentiation of embryonic and adult tissue. GDF3 was also found to be involved in the regulation of adipocyte development, adiposity and energy expenditure in 3T3-L1 cells and primary cultures of human adipocytes [47]. Both LBW human and prediabetic rat adipose tissues exhibiting an increased level of miR-483-3p had a decreased level of GDF3 expression. Using 3T3-L1 mice adipocytes the authors demonstrated that during the differentiation process (0, 4, 7, 9 days) a gradual decrease of miR-483-3p expression and increase in GDF3, PPARγand FABP4 expression occurred. In contrast, miR-483-3p inhibition evoked a decrease of GDF3, PPARγ and FABP4 expression during 3T3-L1 adipocyte differentiation.
Esau et al. performed an analysis of expression and functional assays of various miRNAs on human white differentiating adipocytes [48]. They found that the levels of miR-143 increased during differentiation (days 0, 1, 4, 7, 10) and antisense oligonucleotides (ASO) complementary to miR-143 inhibited adipocyte differentiation. Additionally, they demonstrated that miR-143 inhibition increased expression of extracellular-signal-regulated kinase 5 (ERK5), a member of mitogen-activated protein kinase responsible for the regulation of cell proliferation and differentiation.
A summary of epigenetic changes found in adipose tissue is presented in Table I.
Table I.
Epigenetic modifications in adipose tissue
| Intervention | Detected epigenetic modification | Expression | Process/function in adipose tissue | Reference |
|---|---|---|---|---|
| Treatment with DNA hypomethylation agent – 5-aza-deoxycytidine | DNA hypomethylation | ↑ Leptin | Regulation of fat storage | [31] |
| Overexpression of SCD1 | ↓ Methylation in promoter of IL10ra | ↑ IL10ra | Inflammation | [32] |
| ↑ Methylation in promoter of IL4ra | ↑ IL4ra | |||
| ↑ Methylation in promoter of TGFβ | ↓ TGFβ | |||
| ↑ Methylation of IL6st | ↓ IL6st | |||
| 6 months of exercise | ↑ Global DNA methylation | Changes in expression of CPEB4, SDCCAG8, 2IGF2BP2, JAZF1, HHEX, TCF7L2, HDAC4, NCOR | Metabolism | [37] |
| Induction of differentiation | H3K9me3, H3K9me2, H3K9ac | ↑ PPARγ | Adipogenesis | [38] |
| Knockout of Jhdm2a | Demethylation of H3K9me | ↑ PPARα, ↓ Ucp2, ↓ MCAD, ↓ LCAD, ↓ VLCAD, ↓ Aqp7 | Abnormal fat distribution, hyperlipidemia and susceptibility to obesity in mice | [39] |
| Knockout of LSD1 | Inhibition of H3K4me, H3K4me2 demethylation | ↑ PGC-1α, PDK4, RIIα, ATGL, FATP1 | Increased fat metabolism | [41] |
| H3K4me2 in db/db obese mice vs. non-obese mice | ↑ Clec4d, ↑ TLR8, ↑ Atp6v0d2, ↑ Col81a, ↑ Fbln5, ↑ Fbln2, ↑ Acta1, ↑ MMP12 | Immune response, cellular remodeling associated with conformation from non-obese insulin-sensitive to obese insulin-resistant phenotype | [42] | |
| Downexpression of miR-483-3p | ↑ GDF3, PPARγ, FABP4 | Development, differentiation, fat metabolism | [46] | |
| Downexpression of miR-143 | ↑ ERK5 | Proliferation, differentiation | [48] |
Conclusions and future implications
Evidence from in vitro, animal and human studies strongly suggests that epigenetic mechanisms in adipose tissue may be involved in the pathogenesis of metabolic disorders, as they affect the expression of many genes participating in metabolism, fat storage and cellular remodeling, the immune response and adipogenesis.
Due to the incompletely understood pathogenesis of obesity and T2DM, it is necessary to search for genetic and environmental factors that may have an impact on the development and progression of these metabolic disorders. Findings from this research may indicate new targets for the prevention and treatment of these conditions. A growing number of both experimental and clinical studies suggest that epigenetic modifications may become such targets. This hypothesis is at least partially supported by the results of randomized clinical trials showing that early intensive glycemic control in diabetic patients might delay the development of diabetes and its complications. Early normalization of glycemia may induce epigenetic changes affecting gene expression and in consequence reduce hyperglycemia-associated oxidative stress and advanced glycation of protein and lipids. Therefore, further research to explore epigenetic mechanisms may reveal important information for the pathogenesis, treatment and prevention of diabesity.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Stumvoll M, Goldstein BJ, van Haeften TW. Type 2 diabetes: principles of pathogenesis and therapy. Lancet. 2005;365:1333–46. doi: 10.1016/S0140-6736(05)61032-X. [DOI] [PubMed] [Google Scholar]
- 2.Ford ES, Williamson DF, Liu S. Weight change and diabetes incidence: findings from a national cohort of US adults. Am J Epidemiol. 1997;146:214–22. doi: 10.1093/oxfordjournals.aje.a009256. [DOI] [PubMed] [Google Scholar]
- 3.van Vliet-Ostaptchouk JV, Snieder H, Lagou V. Gene-lifestyle interactions in obesity. Curr Nutr Rep. 2012;1:184–96. doi: 10.1007/s13668-012-0022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 5.Roseboom TJ, van der Meulen JH, Ravelli AC, Osmond C, Barker DJ, Bleker OP. Effects of prenatal exposure to the Dutch famine on adult disease in later life: an overview. Twin Res. 2001;4:293–8. doi: 10.1375/1369052012605. [DOI] [PubMed] [Google Scholar]
- 6.Dunn GA, Bale TL. Epigenetic programming of obesity and diabetes by in utero exposure to gestational diabetes mellitus. Endocrinology. 2011;152:2228–36. [Google Scholar]
- 7.Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends Genet. 2011;27:116–25. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell JT, Saffery R. The value of twins in epigenetic epidemiology. Int J Epidemiol. 2012;41:140–50. doi: 10.1093/ije/dyr179. [DOI] [PubMed] [Google Scholar]
- 9.Jermendy G, Horváth T, Littvay L, et al. Effect of genetic and environmental influences on cardiometabolic risk factors: a twin study. Cardiovasc Diabetol. 2011;10:96–104. doi: 10.1186/1475-2840-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- 11.Reddy MA, Natarajan R. Epigenetic mechanisms in diabetic vascular complications. Cardiovasc Res. 2011;90:421–9. doi: 10.1093/cvr/cvr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–90. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 13.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Pradhan S, Chin HG, Estève PO, Jacobsen SE. SET7/9 mediated methylation of nonhistone proteins in mammalian cells. Epigenetics. 2009;4:383–7. doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammond SM. Dicing and slicing. The core machiner of the RNA interference pathway. FEBS Lett. 2005;579:5822–9. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 18.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961–6. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schipper HS, Prakken B, Kalkhoven E, Boes M. Adipose tissue-resident immune cells: key players in immunometabolism. Trends Endocrinol Metab. 2012;23:407–15. doi: 10.1016/j.tem.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Coelho M, Oliveira T, Fernandes R. Biochemistry of adipose tissue: an endocrine organ. Arch Med Sci. 2013;9:191–200. doi: 10.5114/aoms.2013.33181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma AM, Staels B. Peroxisome proliferator-activated receptor gamma and adipose tissue: understanding obesity-related changes in regulation of lipid and glucose metabolism. J Clin Endocrinol Metab. 2007;92:386–95. doi: 10.1210/jc.2006-1268. [DOI] [PubMed] [Google Scholar]
- 22.Montague CT, O’Rahilly S. The perils of portliness: causes and consequences of visceral adiposity. Diabetes. 2000;49:883–8. doi: 10.2337/diabetes.49.6.883. [DOI] [PubMed] [Google Scholar]
- 23.Kern PA, Ranganathan S, Li C, Wood L, Ranganathan G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am J Physiol Endocrinol Metab. 2001;280:745–51. doi: 10.1152/ajpendo.2001.280.5.E745. [DOI] [PubMed] [Google Scholar]
- 24.Després JP, Lemieux I. Abdominal obesity and metabolic syndrome. Nature. 2006;444:881–7. doi: 10.1038/nature05488. [DOI] [PubMed] [Google Scholar]
- 25.Muoio DM, Dohm GL, Fiedorek FT, Jr, Tapscott EB, Coleman RA. Leptin directly alters lipid partitioning in skeletal muscle. Diabetes. 1997;46:1360–3. doi: 10.2337/diab.46.8.1360. [DOI] [PubMed] [Google Scholar]
- 26.Poitout V, Robertson RP. Minireview: secondary beta-cell failure in type 2 diabetes: a convergence of glucotoxicity and lipotoxicity. Endocrinology. 2002;143:339–42. doi: 10.1210/endo.143.2.8623. [DOI] [PubMed] [Google Scholar]
- 27.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW., Jr Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernández-Sánchez A, Madrigal-Santillán E, Bautista M, et al. Inflammation, oxidative stress, and obesity. Int J Mol Sci. 2011;12:3117–32. doi: 10.3390/ijms12053117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skurk T, Alberti-Huber C, Herder C, Hauner H. Relationship between adipocyte size and adipokine expression and secretion. J Clin Endocrinol Metab. 2007;92:1023–33. doi: 10.1210/jc.2006-1055. [DOI] [PubMed] [Google Scholar]
- 30.Chapman AB, Knight DM, Dieckmann BS, Ringold GM. Analysis of gene expression during differentiation of adipogenic cells in culture and hormonal control of the developmental program. J Biol Chem. 1984;259:15548–55. [PubMed] [Google Scholar]
- 31.Wabitsch M, Brüderlein S, Melzner I, Braun M, Mechtersheimer G, Möller P. LiSa-2, a novel human liposarcoma cell line with a high capacity for terminal adipose differentiation. Int J Cancer. 2000;88:889–94. doi: 10.1002/1097-0215(20001215)88:6<889::aid-ijc8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 32.Malodobra-Mazur M, Dziewulska A, Kozinski K, et al. Stearoyl-CoA desaturase regulates inflammatory gene expression by changing DNA methylation level in 3T3 adipocytes. Int J Biochem Cell Biol. 2014;55:40–50. doi: 10.1016/j.biocel.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Hulver MW, Berggren JR, Carper MJ, et al. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab. 2005;2:251–61. doi: 10.1016/j.cmet.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouchard L, Rabasa-Lhoret R, Faraj M, et al. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr. 2010;91:309–20. doi: 10.3945/ajcn.2009.28085. [DOI] [PubMed] [Google Scholar]
- 35.Izem L, Morton RE. Possible role for intracellular cholesteryl ester transfer protein in adipocyte lipid metabolism and storage. J Biol Chem. 2007;282:21856–65. doi: 10.1074/jbc.M701075200. [DOI] [PubMed] [Google Scholar]
- 36.Cordero P, Campion J, Milagro FI, et al. Leptin and TNF-alpha promoter methylation levels measured by MSP could predict the response to a low-calorie diet. J Physiol Biochem. 2011;67:463–70. doi: 10.1007/s13105-011-0084-4. [DOI] [PubMed] [Google Scholar]
- 37.Rönn T, Volkov P, Davegårdh C, et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose Tissue. PLoS Genet. 2013;9:e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steger DJ, Grant GR, Schupp M, et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24:1035–44. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–61. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okuno Y, Ohtake F, Igarashi K, et al. Epigenetic regulation of adipogenesis by PHF2 histone demethylase. Diabetes. 2013;62:1426–34. doi: 10.2337/db12-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hino S, Sakamoto A, Nagaoka K, et al. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat Commun. 2012;3:758–70. doi: 10.1038/ncomms1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimoto S, Goda T, Mochizuki K. In vivo evidence of enhanced di-methylation of histone H3 K4 on upregulated genes in adipose tissue of diabetic db/db mice. Biochem Biophys Res Commun. 2011;404:223–37. doi: 10.1016/j.bbrc.2010.11.097. [DOI] [PubMed] [Google Scholar]
- 43.Arce I, Martínez-Muñoz L, Roda-Navarro P, Fernández-Ruiz E. The human C-type lectin CLECSF8 is a novel monocyte/macrophage endocytic receptor. Eur J Immunol. 2004;34:210–20. doi: 10.1002/eji.200324230. [DOI] [PubMed] [Google Scholar]
- 44.Pietrement C, Sun-Wada GH, Silva ND, et al. Distinct expression patterns of different subunit isoforms of the V-ATPase in the rat epididymis. Biol Reprod. 2006;74:185–94. doi: 10.1095/biolreprod.105.043752. [DOI] [PubMed] [Google Scholar]
- 45.Abu-Farha M, Tiss A, Abubaker J, et al. Proteomics analysis of human obesity reveals the epigenetic factor HDAC4 as a potential target for obesity. PLoS One. 2013;8:e75342. doi: 10.1371/journal.pone.0075342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferland-McCollough D, Fernandez-Twinn DS, Cannell IG, et al. Programming of adipose tissue miR-483-3p and GDF-3 expression by maternal diet in type 2 diabetes. Cell Death Differ. 2012;19:1003–12. doi: 10.1038/cdd.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W, Yang Y, Meng Y, Shi Y. GDF-3 is an adipogenic cytokine under high fat dietary condition. Biochem Biophys Res Commun. 2004;321:1024–31. doi: 10.1016/j.bbrc.2004.07.058. [DOI] [PubMed] [Google Scholar]
- 48.Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–5. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]