Abstract
Low-molecular weight heparins (LMWH) prepared by partial depolymerization of unfractionated heparin are used globally to treat coagulation disorders on an outpatient basis. Patent protection for several LMWH has expired and abbreviated new drug applications have been approved by the Food and Drug Administration. As a result, reverse engineering of LMWH for biosimilar LMWH has become an active global endeavor. Traditionally, the molecular weight distributions of LMWH preparations have been determined using size exclusion chromatography (SEC) with optical detection. Recent advances in liquid chromatography–mass spectrometry methods have enabled exact mass measurements of heparin saccharides roughly up to degree-of-polymerization 20, leaving the high molecular weight half of the LMWH preparation unassigned. We demonstrate a new LC–MS system capable of determining the exact masses of complete LMWH preparations, up to dp30. This system employed an ion suppressor cell to desalt the chromatographic effluent online prior to the electrospray mass spectrometry source. We expect this new capability will impact the ability to define LMWH mixtures favorably.
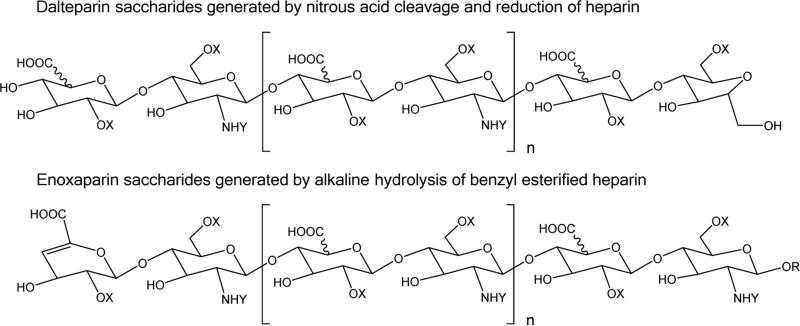
Pharmaceutical heparin is a sulfated, linear polysaccharide prepared from porcine intestinal mucosa. Consisting of repeating uronic acid-glucosamine disaccharide units modified with an average of 2.3 sulfate groups, unfractionated heparin has an average molecular weight of 17 800 Da with a high degree of polydispersity.1 Pharmaceutical heparin, used as an anticoagulant in surgical suites, is neutralizable with protamine and requires constant patient monitoring. Heparin-induced thrombocytopenia, a serious risk of heparin treatment, results from an immunological reaction against a complex between heparin and platelet factor 4.2,3 Low molecular weight heparins (LMWH) were developed in order to minimize such risks to allow administration as an outpatient anticoagulant. LMWH are produced from unfractionated heparin by limited chemical or enzymatic cleavage4 to produce a biopolymer with average molecular weight of 4000–6000 Da, depending on the preparation.5 The chemical structure and pharmacological activities of LMWH depend on the method used for partial depolymerization4 (see Figure 1).
Figure 1.
General structures for dalteparin and enoxaparin saccharides. X = H or SO3H, Y = Ac or SO3H.
Since their introduction, LMWH have captured a large share of the U.S. heparin market. This has driven effort to develop biosimilar heparins, the first of which for the enoxaparin LMWH was approved by the Food and Drug Administration in 2010.6 Because the complexity is vast, it is not possible to characterize every molecular species in LMWH preparations; thus, the FDA approval was based on the ability to reverse engineer LMWH using analytical and pharmacologic assay data representative of the preparation. Such biosimilars development depends on the demonstration of acceptably low risk for use of the biosimilar heparin. The ability to demonstrate acceptable risk improves with increases in analytical capability.
Molecular weight distributions for unfractionated heparin and LMWH have been measured traditionally using size exclusion chromatography with refractive index detection.1,7,8 In order to increase the detail of the chromatographic LMWH separations, we and others have analyzed LMWH fractions using hydrophilic interaction chromatography (HILIC)-MS.9,10 While the chromatographic resolution of HILIC is higher than that of SEC, the ability to interpret the mass spectral data is limited by the presence of abundant ammonium adducted forms of the LMWH oligosaccharide ions. This adduction results from pairing of ammonium cations with deprotonated oligosaccharide anions, resulting in proton transfer reactions that decrease the magnitude of ion charge. Thus, abundant ions with low degree of charge and high degree of ammonia adduction are observed. As a result it is not possible to extract neutral masses from LMWH saccharides larger than degree-of-polymerization (dp) 20 using LC–MS. Similar problems limit the use of MS to detect LMWH eluting from a SEC column since ammonium ions must be included in the mobile phase.
Tinzaparin LMWH was analyzed using 6 μm particle size SEC columns.11 These investigators used a self-regenerating cation exchange column to remove ammonium salts prior to the MS source. Chromatographic resolution was modest but they reported ability to identify ions corresponding to Tinzaparin oligomers up to dp18. The availability of ultra-performance SEC columns with 1.7 μm particles greatly improved the chromatographic resolution for LC–MS separation of LMWH.12 Again, the LMWH extracted mass spectra suffered from ammonium adduction, limiting the ability to interpret data for >dp18.
In this work, we solved this ammonium adduction problem for SEC-MS of LMWH by using an online ion suppressor to exchange remove ammonium ions for protons, dramatically reducing the abundances of ammonium adducted LMWH ions. This allowed us to measure mass profiles for LMWH up to dp30, essentially the complete LMWH biopolymeric range. We developed a specialized bioinformatics approach to interpret the data.
EXPERIMENTAL SECTION
Additional experimental details are provided in the Supporting Information Methods section.
Materials
The United States Pharmacopeia standard for enoxaparin sodium was purchased from Sigma-Aldrich (St. Louis, MO). Dalteparin sodium as Fragmin was obtained from Pharmacia Corporation (Kalamazoo, MI).
Stock solutions of enoxaparin sodium and dalteparin sodium were prepared at 10 nmol/μL in water. They were then desalted by dialysis against 50 mM ammonium formate (pH 6.8) in methanol/water (20:80) using 500 Da molecular weight membranes (The Nest Group, Southborough, MA).
SEC LC–MS Analysis with Ion Suppressor
Tandem Acquity UPLC BEH columns (4.6 mm × 150 mm and 4.6 mm × 300 mm, Waters Corp., Milford, MA) were used to separate the LMWH preparations using conditions similar to a published method.12 The mobile phase contained 50 mM ammonium formate (pH 6.8) in methanol/water (80:20). The constituents were eluted in 90 min at a flow rate of 75 μL/min.
The LC column was connected to a chemically regenerated ion suppressor (ACRS 500, 2 mm, Thermo Fisher Scientific/Dionex, San Jose, CA) for online desalting. The ion suppressor exchanged ammonium salts to their corresponding organic acids, thereby eliminating the problem of ammonium adduction of the saccharide ions. A solution of 100 mM sulfuric acid was used to regenerate the suppressor. The suppressor effluent was connected to the standard ESI source of a Q-Executive Plus mass spectrometer (Thermo Fisher Scientific, San Jose, CA). The mass spectra were acquired using the negative ionization mode at resolution 75 000 and m/z range 200–2000 Th with three microscans. The source parameters for the analyses were optimized to minimize sulfate loss and included a spray voltage of 3.8 kV, a capillary temperature of 280 °C, a sheath gas flow of 10, and an S-lens rf level of 40.
Data Deconvolution
The acquired spectra were manually examined and averaged over retention time ranges using the Xcalibur software (version 3.0.63, ThermoFisher Scientific, San Jose, CA). The combined spectra were autodeconvoluted by an in-house software, GAGdecon (https://github.com/BostonUniversityCBMS/gagdecon). GAGdecon uses a database search algorithm for structural assignment at the MS level. It maps the isotopic distribution of theoretical species using a python implementation of the BRAIN (Baffling Recursive Algorithm for Isotopic distribution calculations)13,14 available at (https://github.com/BostonUniversityCBMS/brainpy) to the experimental data.
GAGdecon works by constructing a combinatorial database of GAG compositions, computing their theoretical monoisotopic mass at a range of user specified charge states. The composition includes the number of monosaccharide species, HexN, HexA, HexNAc, dHexA, ManN, as well as substituents and adducts, Ac, Na, K, Ca, and Li. The disaccharide pattern was extrapolated from the user input GAG species, and the counts of each monosaccharide scale with chain length (degree of polymerization, dp), which may either be a range or a fixed value. GAGdecon can process either centroid or profile data, using a Lorentzian peak shape fit to calculate the centroids for profile peaks. Once the input spectra have been preprocessed, the database search is carried out by searching each theoretical GAG composition and charge state, generating a theoretical isotopic cluster based on the elemental composition of the theoretical molecular composition, including the first 95% of the isotopic pattern. Each theoretical isotopic pattern was fitted to the peak list, producing a score of the form:
Oi is the abundance of the ith observed peak and Ti is the abundance of the ith theoretical peak. Optimal fits minimize G. Each fit with a score below a parametrized threshold is reported to the user.
RESULTS AND DISCUSSION
We analyzed the two most widely used LMWH in the U.S., dalteparin and enoxaparin, the general structures for which are shown in Figure 1.
SEC-MS of Dalteparin
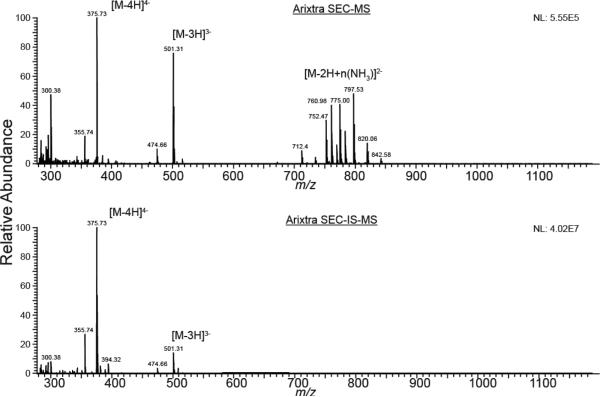
Dalteparin is produced by limited nitrous acid depolymerization of unfractionated heparin, resulting in chains that range from 5600 to 6400 Da (European Pharmacopeia range)5 that terminate with an anhydromannitol residue at the reducing end.15 Notably, a side reaction occurs in that corresponds to GlcN ring contraction without chain cleavage (Figure S1).16 SEC-MS of a synthetic heparin saccharide (Arixtra) in the absence of an ion suppressor results in abundant ammonium adducted ions that complicate the assignment of exact masses and saccharide compositions. Use of a postcolumn ion suppressor results in observation of deprotonated ions with an absence of ammonium adducts (Figure 2).
Figure 2.
SEC-MS extracted mass spectra for Arixtra. Top: mass spectrum with no ion suppression, showing abundant ammonium adducts for the 2– charge state. Bottom: use of an online ion suppressor results in deprotonated ions with no ammonium adducts.
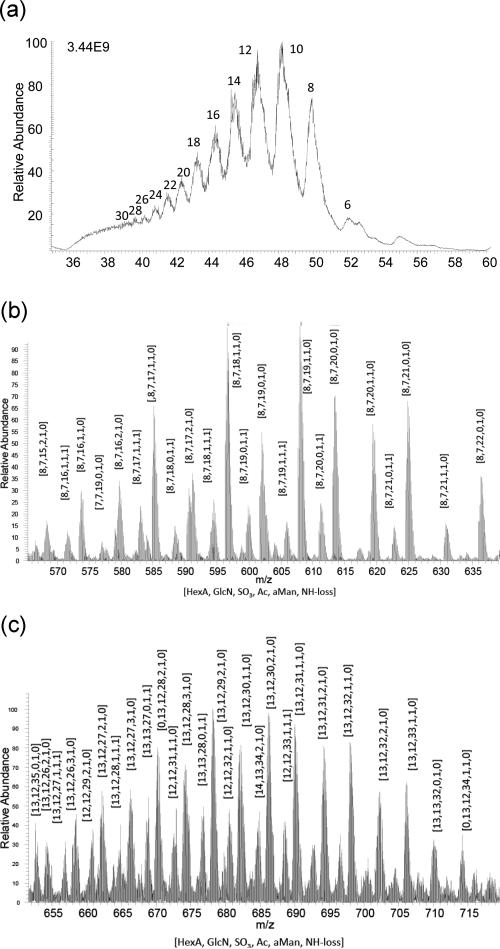
Figure 3a shows the total ion chromatogram (TIC) for SEC-ion suppressor (IS)-MS of dalteparin. The total ion chromatograms were reproducible as shown by triplicate analyses (Figure S2a). Distinct chromatographic peaks are observed corresponding to saccharides ranging from dp6–30 as labeled in the figure. The extracted mass spectrum for dp16 saccharides obtained using the ion suppressor is shown in Figure 3b in which deprotonated ions are observed without ammonium adducts. This absence of adduction greatly facilitates interpretation of the mass spectral data; ammonium adducts increase the incidence of overlapping isotopic clusters and limit the size of heparin saccharides that yield interpretable mass spectra. We noticed several pairs of isotope clusters separated by 15.0 Da in the data. We reasoned that these resulted from deamination of heparin chains without chain cleavage, resulting from the nitrous acid depolymerization reaction used in the formulation of dalteparin.16 This loss corresponds to the substitution of an amino group with a proton for a net loss of NH (15.01 Da) (Figure S1). This pattern of paired isotopic clusters differing by NH units was observed throughout the SEC-IS-MS data set on dalteparin. The extracted mass spectrum of dp26 saccharides (Figure 3c) show absence of ammonium adducts that allow unambiguous assignment of compositions. Saccharides resulting from NH loss were too low in abundances to be assigned.
Figure 3.
(a) SEC-IS-MS total ion chromatogram for Dalteparin. The oligomer size (degree-of-polymerization) is shown for each chromatographic peak, (b) extracted mass spectrum for 43.6–44.7 min, and (c) extracted mass spectrum for 39.9–40.4 min.
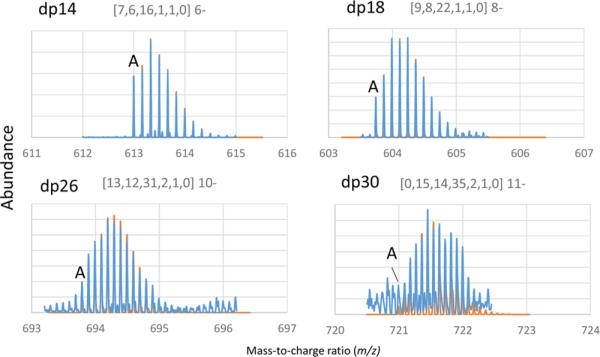
In previous work, we used hydrophilic interaction chromatography (HILIC) MS for analysis of heparins.9,10,17 In these studies we used Horn deconvolution18–20 to convert mass spectra data to neutral masses. As shown in Figure 4, the decrease in abundances of the monoisotopic ions for LMWH saccharides that occur as molecular weight increases poses a serious problem for identification of monoisotopic ions for dp >18. For the dp14 and dp18 saccharides, the monoisotopic peak is readily identifiable. This identification becomes more difficult for larger saccharides as shown by the dp26 and dp30 examples because of the lower abundance of the monoisotopic peak. By overlaying the calculated isotopic patterns (shown in orange), we found that it was possible to assign dalteparin saccharides accurately through dp30.
Figure 4.
Expanded m/z ranges for (a) dp 8 [7,6,16,1,1,0] 6–, (b) dp 18 [9,8,22,1,1,0] 8–, (c) dp 26 [13,12, 31, 2, 1, 0] 10–, and (d) dp30 [15,14,35,2,1,0] 11–. Compositions are given as [HexA, HexN, SO3, Ac, aMan, NH loss]. Experimental data (blue) overlay theoretical isotope patterns (orange). Theoretical isotopic distributions were calculated using IsoPro 3.1 at 40 000 resolution.
In order to determine the LMWH compositions, it is essential for the algorithm to assign charge states correctly. Horn deconvolution seeks to identify the monoisotopic peak using a molecular averagine. This task becomes increasingly difficult as the LMWH oligomer size increases, to the point that it cannot easily be determined even by manual inspection (see examples for dp26 and dp30 in Figure 4). We therefore developed a bioinformatics algorithm (GAGdecon) to accomplish the task of isotopic pattern matching and scoring for LMWH data sets. For this, the algorithm generated theoretical dalteparin compositions and exact isotope patterns, see Supporting Information File Set 1.
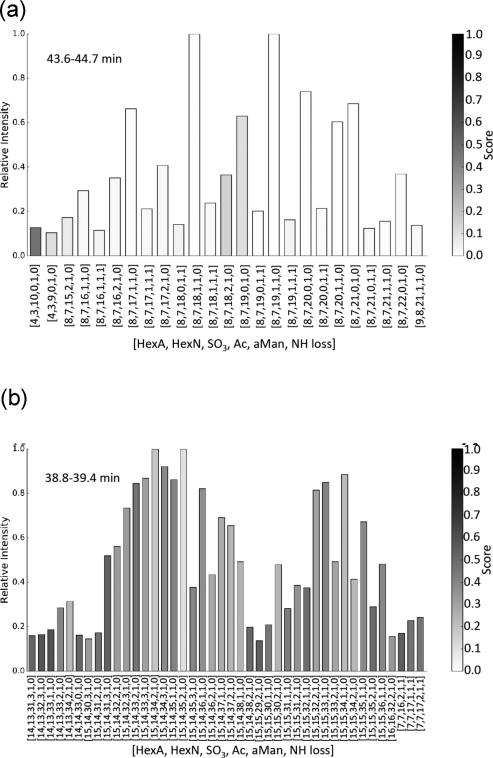
As described in the methods section, exact masses for LMWH saccharides were assigned by fitting calculated isotope patterns. The complete set of scored isotope cluster matches for dalteparin are shown in Supporting Information File Set 1. The compositional profile aggregated for all observed charge states for dalteparin dp16 saccharides is shown in Figure 5a. Scores were higher (poorer) for saccharides with NH losses, owing to the lower abundances and correspondingly greater difficulty in matching isotope clusters. For dp30 saccharides (Figure 5b) it was possible to assign only the compositions without NH losses with acceptable confidence. The complete set of compositional profiles are shown in Figure S3. These compositional profiles show a small degree of incorrect compositions for low abundance saccharides due to incorrect charge state assignments at half the correct value. This is due to multiple well-scoring interpretations of the signal which share peaks. These cases are uncommon, requiring that the isotopic pattern of one composition be similar on a relative scale to the overlapped components of another composition. Often, a difference in scores can be used to discriminate the correct fit.
Figure 5.
Compositions, scores, and abundances of dalteparin saccharides: (a) retention time 43.6–44.6 min, corresponding to dp16 and (b) retention time 38.8–39.4 min, corresponding to dp30. Ring contraction is given as NH loss in the compositions. The lowest score values reflect the greatest confidence.
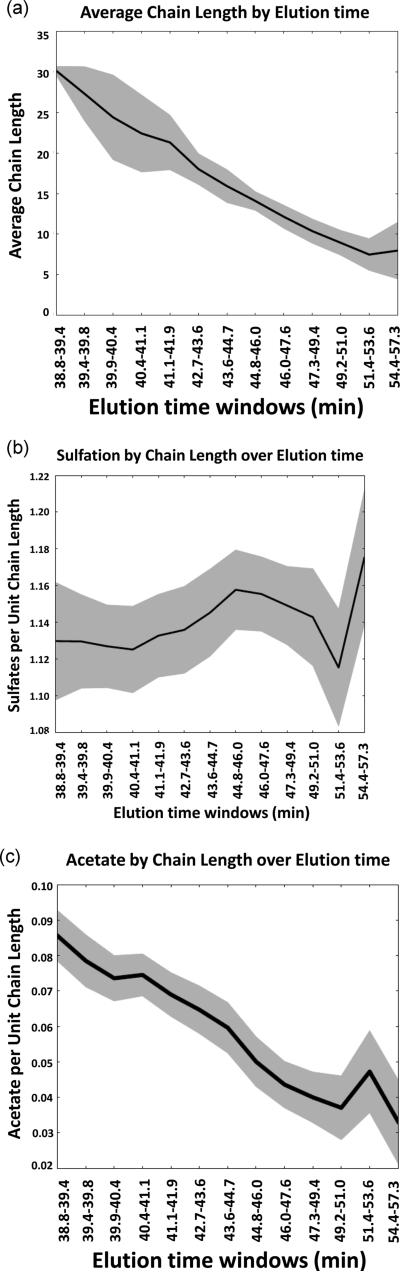
In order to validate that the assignment of saccharide compositions was correct, we plotted the average saccharide chain length versus elution time. The resulting plot (Figure 6a) shows that average chain length diminishes with elution time as expected. This is consistent with overall correctness of assignment of saccharide compositions. Instances of incorrect assignments would skew this trend. This plot also demonstrates that charge state assignments were generally correct. We next calculated the number of sulfation per saccharide chain length across the SEC elution profile (Figure 6b). This showed that the number of sulfate groups ranged from 1.10 to 1.14, indicating a relatively narrow distribution of sulfation per saccharide across the elution profile. This corresponds to an average of 2.3 sulfate groups per disaccharide unit for dalteparin, in close agreement with the expected value.1 We also plotted the number of acetate groups per dalteparin disaccharide unit as shown in Figure 6c. This plot showed that the number acetate groups per disaccharide unit diminished as the dalteparin chain length diminished. This is expected since nitrous acid cleavage occurs at N-sulfated glucosamine residues to produce anhydromannitol, requiring the absence of acetate from the reducing terminal residue.
Figure 6.
(a) Plot of dalteparin chain length as a function of SEC retention time. The shaded regions surrounding the trend lines indicate standard deviation of the mean at each point. (b) Plot of number of sulfate groups per unit chain length. The shaded region for the relationships indicate the confidence interval of the coefficient for the sample. (c) Plot of number of acetate groups per unit chain length. The shaded regions surrounding the trend lines indicate standard deviation of the mean at each point.
SEC-IS-MS of Enoxaparin
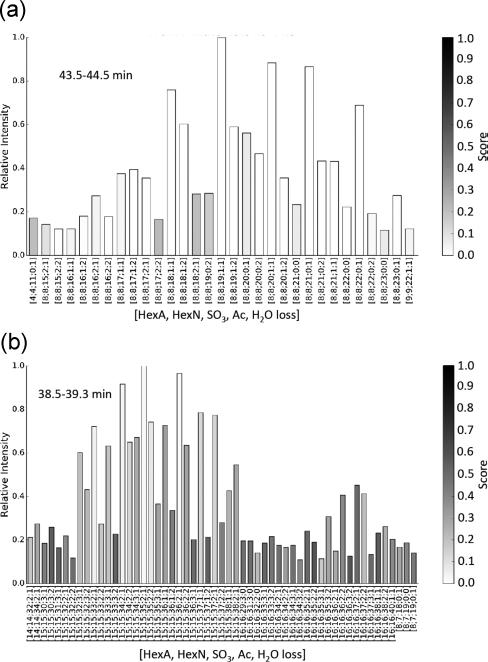
Enoxaparin is produced by alkaline hydrolysis of heparin benzyl esters.4 The package insert for the commercially available Lovenox formulation states that 20% of the molecular weight distribution is below 2000 Da, 68% between 2000 and 8000 Da, and 15% above 8000 Da.21 The European Pharmakopeia range is listed as 3800–5000 Da.5 As with dalteparin, SEC-IC-MS of enoxaparin showed high reproducibility (Figure S2b). In comparison with that of dalteparin (Figure 3a), the SEC-IS-MS total ion chromatogram (TIC) of enoxaparin (Figure 7a) shows more abundant dp4 saccharides. The 43.7–44.7 min retention time window (Figure 7b) shows a range of enoxaparin dp16 saccharides in three compositional forms. The fully saturated saccharides correspond to enoxaparin containing the original saturated nonreducing HexA residue of the parent heparin chains. The saccharides showing one water loss correspond to chains cleaved by alkaline hydrolysis in which a Δ4,5-unsaturated uronic acid (ΔHexA) is present on the nonreducing chain ends. Saccharides of the same mass would be produced by cyclic ester formation at the reducing end. The saccharides showing two water losses correspond to saccharides with Δ-unsaturated nonreducing ends and a cyclic ester formed from the reducing end glucosamine (also known as a cyclic acetal). These compositions are given as [HexA, GlcN, SO3, Ac, H2O-loss] in the figure. Fully saturated enoxaparin dp16 saccharides (0 water losses) were low in abundances (Figure 7b). The assignments for a summed mass spectrum for the 39.9–40.4 min retention time range, corresponding to dp30 are shown in Figure 7c. The complete compositional profiles for enoxaparin are shown in Figure S4. As shown by the data included in Supporting Information File Set 2, GAGdecon assigns the enoxaparin oligomer isotopic clusters with acceptable confidence. As shown in Figure 8a for dp16, abundances for enoxaparin saccharides fully saturated, with one water loss and with two water losses, respectively, fall in the order 1 > 2 > 0. The same order is observed for dp30 saccharides (Figure 8b). As shown by the shading (darker shade indicating lower confidence), the confidence of assignments for dp30 saccharides of low relative abundances is low due to the difficulty of assigning isotopic patterns for isotope clusters with low signal-to-noise ratios.
Figure 7.
SEC-IS-MS of enoxaparin (a) total ion chromatogram, (b) summed mass spectrum for the 43.7–44.7 min retention time window, corresponding to dp16, (c) summed mass spectrum for the 39.9–40.4 min retention time window, corresponding to dp26–28. Compositions are given as [ΔHexA, HexA, GlcN, SO3, Ac, H2O-loss].
Figure 8.
Compositions, scores and abundances of enoxaparin saccharides. (a) Retention time 43.5–44.5 min, corresponding to dp16. (b) Retention time 38.5–39.3 min, corresponding to dp30. The lowest score values reflect the greatest confidence.
As with daltepaprin, we plotted the average chain length versus SEC elution time (Figure 9a). This plot showed that as expected the average chain length for enoxaparin diminished with time indicating that GAGdecon identified enoxaparin compositions correctly. We plotted the sulfation divided by saccharide chain length to determine the density of sulfation of enoxaparin per unit chain length (Figure 9b). We found that sulfation of enoxaparin saccharides diminished with decreasing chain length. This trend is consistent with less sulfated regions of the precursor chains being of increased likelihood to undergo alkaline hydrolysis. We next plotted the number of acetate groups per enoxaparin saccharide chain length (Figure 9c). This showed that the number of sulfate groups per unit chain length varied to a small degree for the range of saccharides detected in the SEC-IS-MS runs. This is consistent with acetylated glucosamine playing little influence on the alkaline hydrolysis reaction.
Figure 9.
(a) Plot of enoxaparin chain length as a function of SEC retention time. The shaded regions surrounding the trend lines indicate standard deviation of the mean at each point. (b) Plot of number of sulfate groups per unit chain length. The shaded region for the relationships indicate the confidence interval of the coefficient for the sample. (c) Plot of number of acetate groups per unit chain length. The shaded regions surrounding the trend lines indicate standard deviation of the mean at each point.
CONCLUSIONS
The United States Food and Drug Administration has defined five criteria for demonstrating sameness for LMWH abbreviated new drug applications.6 Among these, improvements in physical and chemical methods that occur over time were recognized to increase the ability to characterize sameness of LMWH preparations. While the establishment of sameness impacts both the development of abbreviated new drugs for LMWH and LMWH batch release criteria, the identification of contaminants remains a critical need. This was shown by the 2007–2008 heparin contamination crisis in which criminal adulteration of the heparin severely impacted the global heparin supply.22–25
We demonstrate the ability to produce exact mass values on the entire polymeric distribution of LMWH preparations using SEC-IS-MS. This capability adds a dimension to traditional SEC analysis of LMWH distributions by enabling calculation of elemental compositions for each detected LMWH species. We assert that this new capability will impact the ability to characterize the compositions of LMWH preparations in the global market favorably.
The components used in this first time SEC-IS-MS method for LMWH profiling are commercially available. It was necessary, however, to develop specialized bioinformatics to assign the compositions of the LMWH preparation. This involved assigning elemental compositions from theoretically calculated values. Using this approach, we defined the distribution of saccharides present in the LMWH preparations. We suggest that this approach will be useful for analyzing LMWH as part of drug development and quality control efforts.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes for Health Grants P41GM104603 and R21HL131554. Thermo-Fisher Scientific provided access to the Q-Exactive plus mass spectrometer and the ACRS 500 ion suppressor used in this work.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.6b03081.
Methods and additional figures (PDF)
File Set 1: dalteparin scores for isotope compositions for designated retention time windows (ZIP)
File Set 2: enoxaparin scores for isotope compositions for designated retention time windows (ZIP)
The authors declare no competing financial interest.
REFERENCES
- 1.Mulloy B, Heath A, Shriver Z, Jameison F, Al Hakim A, Morris TS, Szajek AY. Anal. Bioanal. Chem. 2014;406:4815–4823. doi: 10.1007/s00216-014-7940-3. [DOI] [PubMed] [Google Scholar]
- 2.Benton RE, Gersh BJ. South. Med. J. 1998;91:133–137. doi: 10.1097/00007611-199802000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Mikhailov D, Young HC, Linhardt RJ, Mayo KH. J. Biol. Chem. 1999;274:25317–25329. doi: 10.1074/jbc.274.36.25317. [DOI] [PubMed] [Google Scholar]
- 4.Linhardt RJ, Gunay NS. Semin. Thromb. Hemost. 1999;25(Suppl 3):5–16. [PubMed] [Google Scholar]
- 5.Gray E, Mulloy B. J. Thromb. Haemostasis. 2009;7:1218–1221. doi: 10.1111/j.1538-7836.2009.03461.x. [DOI] [PubMed] [Google Scholar]
- 6.Docket No. FDA-2003-P-0273, 2010.
- 7.Mulloy B. Handb. Exp. Pharmacol. 2012;207:77–98. doi: 10.1007/978-3-642-23056-1_5. [DOI] [PubMed] [Google Scholar]
- 8.Mulloy B, Gray E, Barrowcliffe TW. Thromb. Haemost. 2000;84:1052–1056. [PubMed] [Google Scholar]
- 9.Staples GO, Naimy H, Yin H, Kileen K, Kraiczek K, Costello CE, Zaia J. Anal. Chem. 2010;82:516–522. doi: 10.1021/ac901706f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Zhang F, Zaia J, Linhardt RJ. Anal. Chem. 2012;84:8822–8829. doi: 10.1021/ac302232c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henriksen J, Ringborg LH, Roepstorrf P. J. Mass Spectrom. 2004;39:1305–1312. doi: 10.1002/jms.723. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, Chen X, Zhu Z, Zhan X, Wu Y, Song L, Kang J. Anal. Chem. 2013;85:1819–1827. doi: 10.1021/ac303185w. [DOI] [PubMed] [Google Scholar]
- 13.Dittwald P, Claesen J, Burzykowski T, Valkenborg D, Gambin A. Anal. Chem. 2013;85:1991–1994. doi: 10.1021/ac303439m. [DOI] [PubMed] [Google Scholar]
- 14.Hu H, Dittwald P, Zaia J, Valkenborg D. Anal. Chem. 2013;85:12189–12192. doi: 10.1021/ac402731h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shively JE, Conrad HE. Biochemistry. 1976;15:3932–3942. doi: 10.1021/bi00663a005. [DOI] [PubMed] [Google Scholar]
- 16.Guo YC, Conrad HE. Anal. Biochem. 1989;176:96–104. doi: 10.1016/0003-2697(89)90278-9. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y, Shi X, Yu X, Leymarie N, Staples GO, Yin H, Killeen K, Zaia J. Anal. Chem. 2011;83:8222–8229. doi: 10.1021/ac201964n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horn DM, Zubarev RA, McLafferty FW. J. Am. Soc. Mass Spectrom. 2000;11:320–332. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 19.Jaitly N, Mayampurath A, Littlefield K, Adkins JN, Anderson GA, Smith RD. BMC Bioinf. 2009;10:87. doi: 10.1186/1471-2105-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maxwell E, Tan Y, Tan Y, Hu H, Benson G, Aizikov K, Conley S, Staples GO, Slysz GW, Smith RD, Zaia J. PLoS One. 2012;7:e45474. doi: 10.1371/journal.pone.0045474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovenox (enoxaparin sodium) package insert, 1998.
- 22.Sasisekharan R, Shriver Z. Thromb. Haemost. 2009;102:854–858. doi: 10.1160/TH09-02-0083. [DOI] [PubMed] [Google Scholar]
- 23.Guerrini M, Shriver Z, Bisio A, Naggi A, Casu B, Sasisekharan R, Torri G. Thromb. Haemost. 2009;102:907–911. doi: 10.1160/TH09-02-0079. [DOI] [PubMed] [Google Scholar]
- 24.Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, Capila I, Lansing JC, Guglieri S, Fraser B, Al-Hakim A, Gunay NS, Zhang Z, Robinson L, Buhse L, Nasr M, Woodcock J, Langer R, Venkataraman G, Linhardt RJ, Casu B, Torri G, Sasisekharan R. Nat. Biotechnol. 2008;26:669–675. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Zhang Z, Linhardt RJ. Nat. Prod. Rep. 2009;26:313–321. doi: 10.1039/b819896a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.