SUMMARY
GGGGCC repeat expansions in C9ORF72 are the most common genetic cause of both ALS and FTD. To uncover underlying pathogenic mechanisms, we found DNA damage was greater, in an age-dependent manner, in motor neurons differentiated from iPSCs of multiple C9ORF72 patients than control neurons. Ectopic expression of the dipeptide repeat (DPR) protein (GR)80 in iPSC-derived control neurons increased DNA damage, suggesting poly(GR) contributes to DNA damage in aged C9ORF72 neurons. Oxidative stress was also increased in C9ORF72 neurons in an age-dependent manner. Pharmacological or genetic reduction of oxidative stress partially rescued DNA damage in C9ORF72 neurons and control neurons expressing (GR)80 or (GR)80-induced cellular toxicity in flies. Moreover, interactome analysis revealed that (GR)80 preferentially bound to mitochondrial ribosomal proteins and caused mitochondrial dysfunction. Thus, poly(GR) in C9ORF72 neurons compromises mitochondrial function and causes DNA damage in part by increasing oxidative stress, revealing another pathogenic mechanism in C9ORF72-related ALS and FTD.
Keywords: ALS, C9ORF72, DNA damage, DPR, FTD, Oxidative stress, RAN translation, Repeats
INTRODUCTION
GGGGCC (G4C2) repeat expansions in C9ORF72 are the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Rapid progress has been made recently in unraveling the pathogenic mechanisms elicited by expanded G4C2 repeats. For instance, knockdown or knockout of C9orf72 expression in mice does not cause any obvious neurodegeneration, suggesting that partial loss of C9ORF72 is not a major initiating factor in ALS and FTD (Lagier-Tourenne et al., 2013; Koppers et al., 2015; Atanasio et al., 2016; O’Rourke et al., 2016; Jiang et al., 2016). On the other hand, ectopic expression of expanded G4C2 repeats flanked by human introns results in the formation of nuclear RNA foci and production of dipeptide repeat (DPR) proteins through a process called repeat-associated non-AUG (RAN) translation without causing gross neurodegeneration in flies or some mouse models (Tran et al., 2015; O’Rourke et al., 2015; Peters et al., 2015; Jiang et al., 2016), although some behavioral phenotypes were observed when expanded G4C2 repeats were expressed at higher levels or in a different genetic background (Jiang et al., 2016; Liu et al., 2016).
Many proteins bind to G4C2 repeat RNA in cell lysates, and some have been reported to colocalize with RNA foci in patient cells; yet, none of them has been shown to play a key role in disease in a manner like that demonstrated for Muscleblind in myotonic dystrophy (Mohan et al., 2014). On the other hand, in flies and mice, overexpressing noninterrupted, expanded G4C2 repeats in the molecular context of poly(A)+ RNA has detrimental effects that are likely due to the production of toxic DPR proteins (Mizielinska et al., 2014; Chew et al., 2015; Freibaum et al., 2015; Tran et al., 2015). In cellular and animal models, expanded G4C2 repeats and some DPR proteins compromise nucleocytoplasmic transport (Freibaum et al., 2015; Jovičić et al., 2015; Zhang et al., 2015; Boeynaems et al., 2016; Zhang et al., 2016) and cause nucleolar stress (Kwon et al., 2014; Wen et al., 2014; Yang et al., 2015). However, it is not known whether other cellular pathways are dysregulated in an age-dependent manner by endogenously expressed expanded G4C2 repeats or DPR proteins. To address this important issue, here we examined ChAT+ motor neurons differentiated from multiple existing and newly generated iPSC lines from C9ORF72 patients and age-matched control subjects.
RESULTS
Generation and Characterization of New Control and C9ORF72 iPSC Lines and Their Differentiation into Motor Neurons
To better understand ALS/FTD pathogenesis in iPSC models, it is critical to study neurons derived from multiple control and patient subjects. To this end, in addition to iPSC lines we generated previously (Almeida et al., 2012, 2013; Zhang et al., 2013), we generated multiple new integration-free iPSCs lines from two control subjects and three C9ORF72 patients. Control line 35L11 and C9ORF72 lines 42L12, 40L3, and 40L8 were used recently in our collaborative study (Freibaum et al., 2015); the other lines are described here (Table S1). Briefly, human dermal fibroblasts were transfected with episomal plasmids expressing OCT3/4, SOX2, KLF4, c-MYC, LIN28, and NANOG. New lines from independent reprogramming events were characterized by expression of pluripotency markers (Figure S1A, B), normal karyotype (Figure S1C) and their ability to spontaneously differentiate into cells of the three germ layers (Figure S1D). In the present study, we also used an iPSC line (30iC9-ALS) generated from a C9ORF72 ALS patient by Sareen et al. (2013).
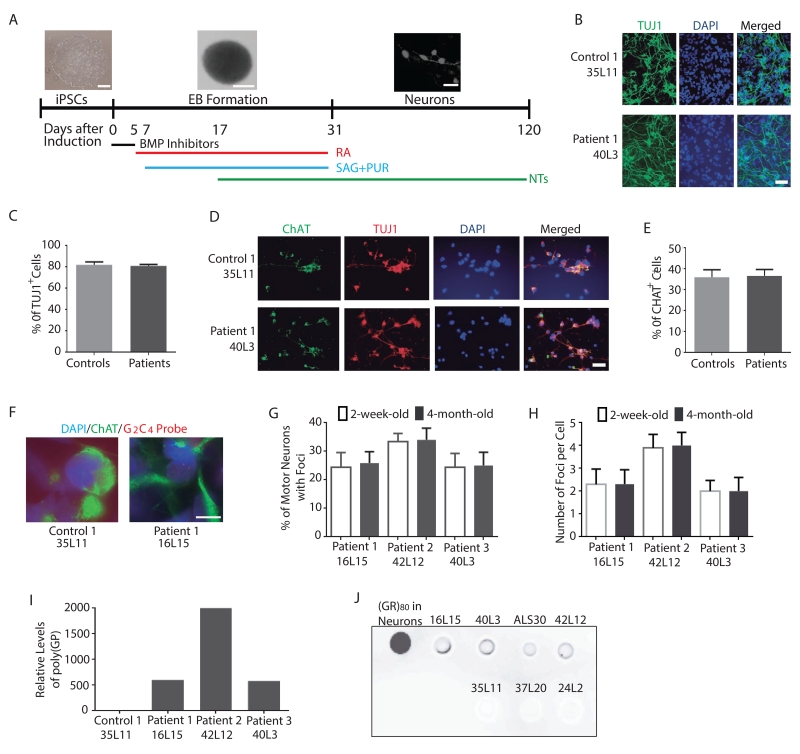
We differentiated multiple iPSC lines into spinal cord motor neurons as described by Amoroso et al. (2013) (Figure 1A). More than 80% of cells in these cultures were TUJ1+ neurons (Figure 1B, C), and about 35% of neurons were ChAT+ motor neurons (Figure 1D, E). Based on the percentages of TUJ1+ and CHAT+ motor neurons in the culture, the C9ORF72 repeat expansions did not seem to affect neuronal differentiation per se (Figure 1E). Further characterization revealed that nuclear RNA foci were present in 2-week-old iPSC-derived ChAT+ motor neurons of three C9ORF72 patients but not in control neurons (Figure 1F). The percentage of motor neurons with foci (Figure 1G) and the average number of foci per cell (Figure 1H) did not change from 2-week-old to 4-month old cultures. We also detected the production of RAN translation products, such as poly(GP) by ELISA assay (Figure 1I) and poly(GR) by dot blot assay (Figure 1J). Thus, this cellular model recapitulates key pathogenic hallmarks of C9ORF72 ALS/FTD.
Figure 1. Characterization of Motor Neuron Cultures Differentiated from Control and C9ORF72 iPSC Lines.
(A) Schematic of the motor neuron differentiation protocol.
(B) Representative images of TUJ1+ neurons from one control and one C9ORF72 iPSC line. Scale bar: 50 μm.
(C) Average percentage of TUJ1+ neurons in cultures of three control and three C9ORF72 patients. No statistically significant difference was found.
(D) Immunostaining of TUJ1+ and ChAT+ motor neurons in control and C9ORF72 neuron cultures. Scale bar: 50 μm.
(E) Average percentage of ChAT+ motor neurons among all neurons of three control and three C9ORF72 patients.
(F) RNA foci are present in iPSC-derived ChAT+ motor neurons of three C9ORF72 patients but not control subjects. Representative images of neurons from one control (35L11) and one patient (16L15) are shown. DAPI: blue; ChAT: green; RNA foci: red. Scale bar: 20 μm.
(G) Percentage of motor neurons with foci. Data are from two independent cultures.
(H) Average number of foci per ChAT+ motor neuron. Data are from two independent cultures. All the values in panels C, E, G and H are mean ± S.E.M.
(I) Poly(GP) is present in 2-week-old C9ORF72 but not control iPSC-derived motor neuron cultures.
(J) Dot blot analysis of Poly(GR) in 2-month-old high-yield motor neuron cultures.
Increased DNA Damage and P53 Activation in iPSC-Derived C9ORF72 Motor Neurons
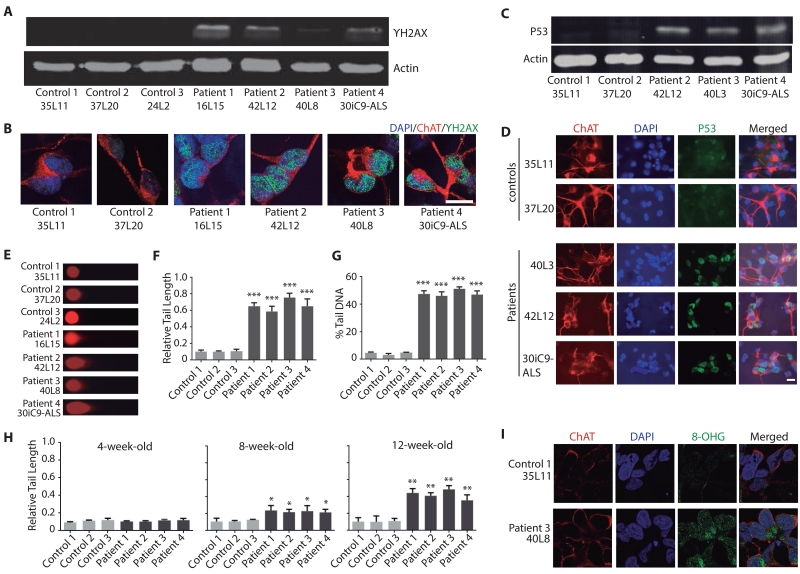
One challenge of using iPSC models is to reveal molecular and cellular phenotypes relevant to age-dependent neurodegenerative diseases. Previously, we demonstrated age-dependent phenotypes (e.g., decreased miR-124 levels) in aged iPSC-derived patient neurons (Gascon et al., 2014). Thus, human motor neurons differentiated from multiple iPSC lines were first cultured for 4 months. Western blot analysis revealed an increased expression of γH2AX, a marker of DNA double-strand breaks, in iPSC-derived motor neuron cultures of all four C9ORF72 patients (Figure 2A). Because other neuronal types are present in these cultures, we performed double-immunostaining analysis to examine DNA damage specifically in motor neurons. We found that the level of γH2AX+ was greater in the nucleus of 4-month-old ChAT+ C9ORF72 motor neurons than in control neurons (Figure 2B).
Figure 2. iPSC-Derived C9ORF72 Motor Neurons Show Increased DNA Damage.
(A) γH2AX expression is increased in 4-month-old C9ORF72 motor neuron cultures.
(B) Immunostaining for γH2AX in 4-month-old ChAT+ C9ORF72 motor neurons. Scale bar: 20 μm.
(C) Total p53 expression is elevated in 4-month-old C9ORF72 motor neuron cultures.
(D) P53 expression is increased in 4-month-old iPSC-derived ChAT+ C9ORF72 motor neurons. Scale bar: 20 μm.
(E) DNA strand breaks are more abundant in 4-month-old C9ORF72 motor neuron cultures than in control motor neurons, as measured by comet assay. Representative images are shown.
(F) Relative tail length from 50 4-month-old cells of each subject.
(G) Percentage of DNA in the tail in 50 cells per subject from 4-month-old motor neuron cultures.
(H) Age-dependent increase in DNA damage in C9ORF72 motor neuron cultures. Values in panels F–H are mean ± S.E.M. *: p < 0.05; **: p < 0.01; ***: p < 0.0001 by one-way ANOVA and Tukey’s multiple-comparison test.
(I) Increased RNA damage in 3-month-old C9ORF72 motor neurons as shown by anti-8-OHG antibody staining. Scale bar: 10 μm.
See also Figure S2.
Increased DNA damage activates the p53 pathway, an important component of the response to DNA damage and other cellular stresses (Lakin and Jackson, 1999). Indeed, western blot analysis showed higher levels of p53 in 4-month-old C9ORF72 motor neuron cultures than in control cultures (Figure 2C). C9ORF72 motor neuron cultures also had increased expression of other markers of DNA damage, such as ATR, which is activated by DNA strand breaks, and GADD45, which is regulated by p53 (Figure S2A, B). The p53 mRNA levels were the same in both cultures (Figure S2C), suggesting that the increase in p53 protein reflects posttranslational regulation, as in other cell types (Lakin and Jackson, 1999). To further demonstrate that p53 protein is increased in C9ORF72 motor neurons in these cultures, we found elevated p53 levels in 4-month-old iPSC-derived ChAT+ motor neurons (Figure 2D). DNA damage induces p53 phosphorylation, such as at serine 15, to stabilize the protein (Lakin and Jackson, 1999). Consistent with this notion, p53 phosphorylated at serine 15 (p-p53) was more abundant in ChAT+ motor neurons than in control neurons (Figure S2D).
To determine if the observed increase in DNA damage was age-dependent, we first examined p53 and found that it was not elevated in 2-week-old iPSC-derived C9ORF72 motor neurons (Figure S2E, F). Then we also analyzed the extent of DNA strand breaks by the comet assay (Figure 2E), and quantified the relative tail length (Figure 2F) and the percentage of DNA in the tail (Figure 2G). This analysis further confirmed a significant increase in DNA damage in 4-month-old C9ORF72 motor neuron cultures, compared with control motor neuron cultures. Then we examined earlier time points using a recently published iPSC differentiation protocol (Du et al., 2015), which generates cultures in which more than 80% of cells are ChAT+ motor neurons (Figure S3A–D). DNA damage became detectable at around 8-week-old cultures, and these changes became gradually more pronounced in 3- and 4-month-old cultures (Figures 2H, S2G). It seems that poly(GR) expression is higher in 4-month-old cultures than that in 2-month-old cultures (Figure S2H), although the dot blot analysis is not highly quantitative. DNA damage was not detected in astrocytes (Figure S2I) even though these cells show RNA foci (Figure S2J) and presumably express DPR proteins as well. Thus, C9ORF72 motor neurons show a gradual accumulation of DNA damage during aging process in vitro. Finally, RNA damage was also increased in 3-month-old C9ORF72 motor neurons (Figure 2I).
(GR)80 Expression Increases DNA Damage in Control iPSC-Derived Human Motor Neurons
The iPSC-derived C9ORF72 motor neurons produce both nuclear RNA foci and DPR proteins, such as poly(GR) (Figure 1J), but the cause of the DNA damage in these cells is unclear. In a Drosophila model expressing 160 copies of G4C2 repeats in the molecular context of human intronic and adjacent exonic sequences, modest toxicity at higher temperature correlates with increased DPR protein production but not with the number of nuclear G4C2 sense RNA foci (Tran et al., 2015). Moreover, poly(GR) is toxic in a number of cellular and Drosophila models (Kwon et al., 2014; Mizielinska et al., 2014; Wen et al., 2014; Freibaum et al., 2015; Jovičić et al., 2015; Yang et al., 2015).
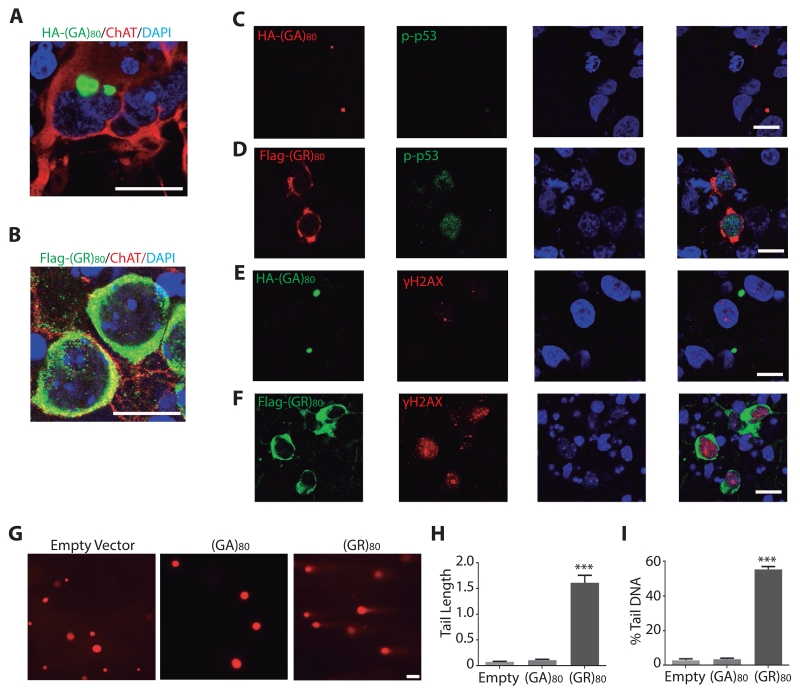
To examine the effect of (GR)80 in human neurons, we differentiated one control iPSC line into 2-week-old ChAT+ motor neurons that were transduced with lentiviral vectors expressing DPR proteins. (GA)80 formed perinuclear inclusions (Figure 3A), but (GR)80 was mostly localized in the cytoplasm (Figure 3B), consistent with earlier observations in Drosophila and patient brains (Yang et al., 2015; Schludi et al., 2015). Expression of (GR)80 in control motor neurons activated the p53 pathway (Figure 3D), increased the level of γH2AX (Figure 3F) and DNA strand breaks (Figure 3G–I). In contrast, expression of (GA)80 for 48 h did not induce these molecular changes (Figure 3). Thus, (GR)80 but not (GA)80 was sufficient to induce DNA damage when transiently expressed in human motor neurons. Moreover, (GR)80 but not (GA)80 expression compromised the cell survival in iPSC derived motor neuron cultures (Figure S3E). Furthermore, using our previously established Drosophila model of (GR)80 toxicity (Yang et al., 2015), we found that partial inhibition of the p53 pathway—either through heterozygosity of p53 loss-of-function allele or p53 RNAi knockdown or expression of a dominant-negative form of p53—partially suppressed (GR)80-induced toxicity (Figure S3F). Thus, DNA damage–induced activation of the p53 pathway contributes to poly(GR) toxicity.
Figure 3. (GR)80 Expression Induces DNA Damage in Control iPSC-Derived Human Motor Neurons.
(A,B) Subcellular localization of HA-tagged (GA)80 (A) and Flag-tagged (GR)80 (B) in control iPSC-derived motor neurons. DAPI: blue; ChAT: red; Flag: green. Scale bar: 20 μm.
(C) (GA)80 expression has little effect on the level of p-p53 in control iPSC-derived human motor neurons.
(D) The level of p-p53 is increased in (GR)80-transduced ChAT+ human motor neurons.
(E) (GA)80 expression does not affect γH2AX level.
(F) (GR)80 expression increases the level of γH2AX in control iPSC-derived motor neurons. Scale bar in C–F: 20 μm.
(G) (GR)80 but not (GA)80 expression increases DNA fragmentation in control motor neurons as measured by the comet assay. Scale bar: 20 μm.
(H, I) Tail length (H) and percentage of DNA in the tail (I) are increased in (GR)80- but not (GA)80-expressing human motor neurons. 50 cells from each condition were analyzed. Values are mean ± S.E.M. ***: p < 0.0001, by one-way ANOVA and Tukey’s multiple-comparison test.
See also Figure S3.
(GR)80 Compromises Mitochondrial Function and Increased Oxidative Stress Contributes to DNA Damage and Toxicity Induced by (GR)80
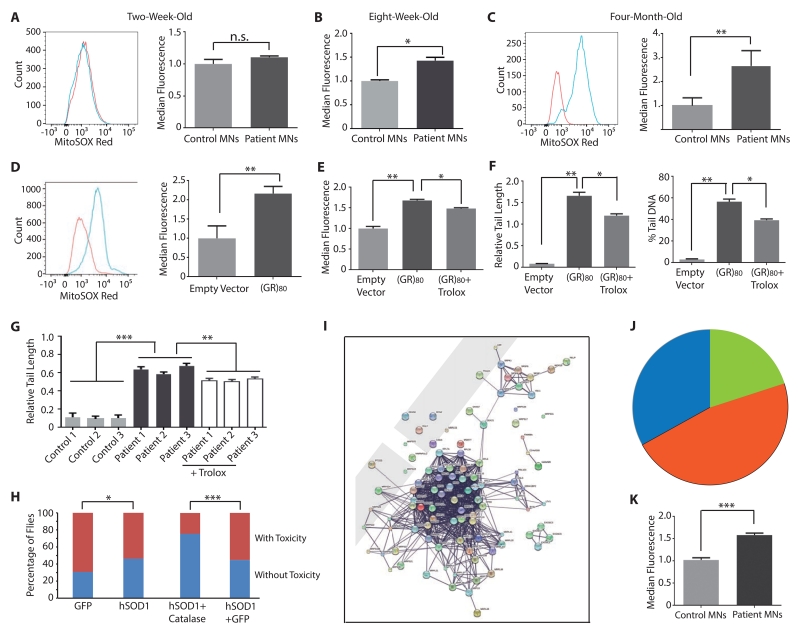
To understand the mechanism underlying (GR)80 toxicity, we assessed oxidative stress, a common cause of DNA damage (Madabhushi et al., 2014). First, we used MitoSOX Red, a mitochondrial dye that produces red fluorescence upon oxidation by superoxide, to measure the relative production of reactive oxygen species (ROS) in high-yield control and C9ORF72 motor neuron cultures. ROS production was not elevated in 2- (Figure 4A) or 4-week-old (not shown) iPSC-derived C9ORF72 motor neuron cultures. However, it started to increase in 8-week-old C9ORF72 neuron cultures (Figure 4B) and continued to increase in 12-week-old (not shown) and 4-month-old C9ORF72 neuron cultures (Figure 4C), which correlated well with the observed age-dependent increase in DNA damage in these cells (Figure 2F-G). Similar results were obtained with dihydrorhodamine 123, another ROS indicator (Figure S4A, B). ROS production measured by both indicators was also increased in 3-month-old iPSC-derived C9ORF72 cortical neurons (Figure S4C, D).
Figure 4. Increased ROS Contributes to the DNA Damage Phenotype in 4-Month-Old iPSC-Derived C9ORF72 Motor Neuron Cultures.
(A–C) Mitochondrial ROS levels in 2-week-old (A), 2-month-old (B) and 4-month-old (C) iPSC-derived high-yield motor neuron cultures of three controls (red) and three C9ORF72 patients (blue). **: p < 0.01 by Student’s t test.
(D) (GR)80 expression (blue) increases mitochondrial ROS in 2-week-old iPSC-derived control motor neuron cultures. **: p < 0.01 by Student’s t test.
(E) (GR)80-induced increase in mitochondrial ROS is partially rescued by Trolox treatment. *: p < 0.05; **: p < 0.01 by one-way ANOVA and Tukey’s multiple-comparison test.
(F) Trolox partially rescues the DNA damage phenotype caused by (GR)80 in 2-week-old control iPSC-derived motor neuron cultures. *: p < 0.05; **: p < 0.01 by one-way ANOVA and Tukey’s multiple-comparison test.
(G) DNA damage was partially reduced when Trolox was added to 3-month old C9ORF72 motor neuron cultures for additional 3 weeks. **: p < 0.01; ***: p < 0.001 by one-way ANOVA and Tukey’s multiple-comparison test.
(H) Ectopic expression of both hSOD1 and catalase suppresses (GR)80 toxicity in the fly wing. *: p < 0.05; **: p < 0.01; ***: p < 10−12 by chi-square test for categorical data.
(I) The interactome of top 100 proteins that bind to (GR)80 after RNase A treatment. The protein names are visible after zoom-in.
(J) The majority of top 100 (GR)80-interacting proteins are ribosomal proteins.
(K) Mitochondrial membrane potential is increased in 4-month-old C9ORF72 motor neurons. Values are mean ± S.E.M. ***: p < 0.001.
In control iPSC-derived motor neurons expressing (GA)80, ROS production, as measured by MitoSOX Red (Figure S4E) or dihydrorhodamine 123 (Figure S4F), was not affected. However, ROS levels in control iPSC-derived motor neuron cultures were significantly increased by (GR)80 expression (Figures 4D, S4G). This increase was partially prevented by Trolox, a water-soluble antioxidant and vitamin E analog (Le Gal et al., 2015) (Figures 4E and S4H). Importantly, the extent of DNA strand breaks was reduced in these cells (Figure 4F), indicating that increased DNA damage was caused in part by poly(GR)-induced oxidative stress. We also found that reduction of oxidative stress by Trolox partially reduced DNA damage in 3-month old C9ORF72 neuron cultures (Figures S4I and 4G). Furthermore, expression of catalase (Figure S4J) or hSOD1 (Figure 4H) or both (Figure 4H), enzymes that function in the same molecular pathway to suppress oxidative stress in flies (Milton et al., 2011), partially suppressed (GR)80-induced cell toxicity in flies. Thus, increased oxidative stress in part contributes to DNA damage and cellular toxicity in C9ORF72-related ALS/FTD.
To further understand the molecular mechanisms underlying (GR)80 toxicity, we expressed (GR)80-GFP or GFP alone in HEK293 cells and performed interactome analysis. We identified 403 proteins that interact with (GR)80-GFP but not GFP without RNase A treatment. Remarkably, about half of these proteins are ribosomal and other RNA binding proteins (Table S2). Then we treated cell lysates with RNase A to destroy RNA-mediated interactions and repeated the same analysis. We identified 237 (GR)80-specific interacting proteins (Table S2), including Lamin B receptor and several subunits of the exosome complex, consistent with our earlier findings that they are genetic modifiers of C9ORF72 repeat toxicity in Drosophila (Freibaum et al., 2015). Interactome and protein functional classification analyses reveal several major groups of proteins whose significance in disease remains to be further investigated (Figure S4K, L). However, among the top 100 (GR)80-interacting proteins, 67 are ribosomal proteins, and among them, two-thirds are mitochondrial ribosomal proteins that are required for translation of 13 subunits of mitochondrial complexes (Figure 4I, J). Indeed, we found that mitochondrial membrane potential was increased in iPSC-derived high-yield C9ORF72 motor neurons (Figure 4K) and in control neurons that expressed (GR)80 (Figure S4M). Thus, poly(GR) preferentially binds to mitochondrial ribosomal proteins and compromises mitochondrial function.
DISCUSSION
This study shows that oxidative stress and DNA damage are increased in iPSC-derived C9ORF72 motor neurons in an age-dependent manner. Poly(GR) preferentially binds to mitochondrial ribosomal proteins and compromises mitochondrial function, which likely leads to increased oxidative stress in human neurons. More importantly, pharmacological or genetic reduction of oxidative stress partially suppressed these detrimental effects in human motor neurons and flies. These results suggest another pathogenic mechanism in C9ORF72-related ALS/FTD.
To understand how cytoplasmic poly(GR) causes DNA damage, we assessed oxidative stress. We found that oxidative stress is elevated in control human neurons expressing (GR)80 as well as in C9ORF72 motor neurons in an age-dependent manner, which is in part responsible for increased DNA damage. More prolonged antioxidant treatment would likely have resulted in stronger inhibition of DNA damage. Interestingly, interactome analysis revealed that (GR)80 preferentially binds to mitochondrial ribosomal proteins (Figure 4), suggesting mitochondria is a major target of poly(GR) toxicity and compromised mitochondrial function may lead to increased oxidative stress. (GR)80 also binds to Lamin B receptor and many non-ribosomal RNA binding proteins such as Matrin 3. The functional significance of these biochemical interactions remains to be further investigated in human neurons. Unlike short poly(GR) proteins such as synthetic (GR)20, which is highly localized to the nucleolus when applied to cultured cells (Kwon et al., 2014), (GR)80 is mostly localized to the cytosol in both mammalian and Drosophila cells (Yang et al., 2015). Similarly, poly(GR) in C9ORF72 patient brains are mostly cytoplasmic and largely absent from the nucleolus (Schludi et al., 2015). Thus, DPR proteins of different length may interact with different sets of proteins.
The increase in oxidative stress and DNA damage in iPSC-derived C9ORF72 motor neurons is age-dependent. Deterioration of nuclear pore integrity and nucleocytoplasmic transport with age (D’Angelo et al., 2009; Mertens et al., 2015) might result in additional leakage of unspliced C9ORF72 pre-mRNA or G4C2 repeat-containing intron 1 transcripts, leading to increased production of toxic DPR proteins. In addition, DPR accumulation could be facilitated by compromised proteasome and autophagy activities during aging as well. Pharmacological or genetic suppression of oxidative stress decreases poly(GR)-induced DNA damage, suggesting that oxidative stress is one of the key causes of the DNA damage in iPSC-derived C9ORF72 neurons. Thus, reducing oxidative stress is a potential therapeutic strategy for C9ORF72-related ALS/FTD. Moreover, these age-dependent phenotypes can be used as quantifiable assays for testing effectiveness of potential therapeutic drugs on C9ORF72 human neurons.
EXPERIMENTAL PROCEDURES
For details on all methods, please see Supplemental Information.
Supplementary Material
Highlights.
C9ORF72 motor neurons show age-dependent increase in oxidative stress and DNA damage.
(GR)80 expression in control motor neurons increases oxidative stress and DNA damage.
(GR)80 binds mitochondrial ribosomal proteins and caused mitochondrial dysfunction.
Reducing oxidative stress partially rescues DNA damage in C9ORF72 motor neurons.
ACKNOWLEDGMENTS
We thank Dr. L. Ranum for poly(GR) antibody, the Cedars-Sinai Medical Center iPSC Core for an ALS iPSC line, the Bloomington Drosophila Stock Center for some fly lines, and the UMMS Confocal Core, Flow Cytometry Core, and Proteomics and Mass Spectrometry Facility for help. This work was supported by grants from the ALS Association and ALS Therapy Alliance (F.-B.G.), the Packard Center for ALS Research and Target ALS (F.-B.G., T.F.G., and L.P.), Association for FTD and the Angel Fund (S.A.), and grants from the National Institutes of Health: R01NS057553, R01NS079725, and R21NS086318 (F.-B.G.), P01AG19724 (B.L.M.), P01NS084974 (L.P.), and R21NS089979 (T.F.G.). We apologize some papers are not cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures, two tables, and Experimental Procedures that can be found with this article online.
AUTHOR CONTRIBUTIONS
R.L.-G. performed all the iPSC experiments with some help from S.A. and H.T. Y.L. identified (GR)80-interascting proteins. Y.L. and D.Y. studied Drosophila models. T.F.G. and L.P. measured poly(GP) levels. A.K. and B.L.M. provided patient skin biopsies. R.L.-G., S.A. and F.-B.G. analyzed data and wrote the manuscript. F.-B.G. supervised the project.
REFERENCES
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso MW, Croft GF, Williams DJ, O’Keeffe S, Carrasco MA, Davis AR, Roybon L, Oakley DH, Maniatis T, Henderson CE, Wichterle H. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 2013;33:574–586. doi: 10.1523/JNEUROSCI.0906-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin W-L, DeJesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasio A, Decman V, White D, Ramos M, Ikiz B, Lee HC, Siao CJ, Brydges S, LaRosa E, Bai Y, et al. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep. 2016;6:23204. doi: 10.1038/srep23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Michiels E, Gijselinck I, Sieben A, Jovičić A, De Baets G, Scheveneels W, Steyaert J, Cuijt I, et al. Drosophila screen connects nuclear transport genes to DPR pathology in c9ALS/FTD. Sci. Rep. 2016;6:20877. doi: 10.1038/srep20877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang Y-J, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME, et al. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348:1151–1154. doi: 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–295. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang P-W, Pham JT, Haeusler AR, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZW, Chen H, Liu H, Lu J, Qian K, Huang CL, Zhong X, Fan F, Zhang SC. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 2015;6:6626. doi: 10.1038/ncomms7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovičić A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW, 3rd, Sun S, Herdy JR, Bieri G, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 2015;18:1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers M, Blokhuis AM, Westeneng HJ, Terpstra ML, Zundel CA, Vieira de Sá R, Schellevis RD, Waite AJ, Blake DJ, Veldink JH, et al. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann. Neurol. 2015;78:426–438. doi: 10.1002/ana.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, Stauffer JE, Jafar-Nejad P, Drenner K, Schulte D, et al. Gain of Toxicity from ALS/FTD-Linked Repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron. 2016;90:535–550. doi: 10.1016/j.neuron.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li H-R, Jiang J, Watt AT, Chun S, Katz M, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. USA. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakin ND, Jackson SP. Rwegulation of DNA damage in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- Le Gal K, Ibrahim MX, Wiel C, Sayin VI, Akula MK, Karlsson C, Dalin MG, Akyürek LM, Lindahl P, Nilsson J, et al. Antioxidants can increase melanoma metastasis in mice. Sci. Transl. Med. 2015;7:308re8. doi: 10.1126/scitranslmed.aad3740. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, Yachnis AT, Ranum LP. C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron. 2016;90:521–534. doi: 10.1016/j.neuron.2016.04.005. [DOI] [PubMed] [Google Scholar]
- Madabhushi R, Pan L, Tsai LH. DNA damage and its links to neurodegeneration. Neuron. 2014;83:266–282. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Paquola AC, Ku M, Hatch E, Böhnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, et al. Directly reprogrammed human neurons retain aging-associated transcriptomic signatures and reveal age-related nucleocytoplasmic defects. Cell Stem Cell. 2015;17:705–718. doi: 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton VJ, Jarrett HE, Gowers K, Chalak S, Briggs L, Robinson IM, Sweeney ST. Oxidative stress induces overgrowth of the Drosophila neuromuscular junction. Proc. Natl. Acad. Sci. USA. 2011;108:17521–17526. doi: 10.1073/pnas.1014511108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Grönke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IOC, Pietrzyk J, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan A, Goodwin M, Swanson MS. RNA-protein interactions in unstable microsatellite diseases. Brain Res. 2014;1584:3–14. doi: 10.1016/j.brainres.2014.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng S-M, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- O’Rourke JG, Bogdanik L, Yáñez A, Lall D, Wolf AJ, Muhammad AK, Ho R, Carmona S, Vit JP, Zarrow J, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–9. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JG, Bogdanik L, Muhammad AK, Gendron TF, Kim KJ, Austin A, Cady J, Liu EY, Zarrow J, Grant S, et al. C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron. 2015;88:892–901. doi: 10.1016/j.neuron.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters OM, Cabrera GT, Tran H, Gendron TF, McKeon JE, Metterville J, Weiss A, Wightman N, Salameh J, Kim J, et al. Human C9ORF72 hexanucleotide expansion reproduces RNA foci and dipeptide repeat proteins but not neurodegeneration in BAC transgenic mice. Neuron. 2015;88:902–909. doi: 10.1016/j.neuron.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, O’Rourke JG, Meera P, Muhammad AKMG, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 2013;5:208ra149–208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schludi MH, May S, Grässer FA, Rentzsch K, Kremmer E, Küpper C, Klopstock T, German Consortium for Frontotemporal Lobar Degeneration, Bavarian Brain Banking Alliance. Arzberger T, Edbauer D. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. 2015;130:537–555. doi: 10.1007/s00401-015-1450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Almeida S, Moore J, Gendron TF, Chalasani U, Lu Y, Du X, Nickerson JA, Petrucelli L, Weng Z, Gao F-B. Differential toxicity of nuclear RNA foci versus dipeptide repeat proteins in a Drosophila model of C9ORF72 FTD/ALS. Neuron. 2015;87:1207–1214. doi: 10.1016/j.neuron.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Abdallah A, Li Z, Lu Y, Almeida S, Gao F-B. FTD/ALS associated poly(GR) protein impairs the Notch pathway and is recruited by poly(GA) into cytoplasmic inclusions. Acta Neuropathol. 2015;130:525–535. doi: 10.1007/s00401-015-1448-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci. 2016;19:668–677. doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.