ABSTRACT
Nonhealing chronic wounds are all unique in origin and circumstance, and attempting to isolate a single etiology for the failure of a wound to heal is daunting. Wounds represent complex systems of multispecies fungal and bacterial biofilms. The survival strategies of interactive microbial communities have led to cooperative evolutionary strategies that culminate in biofilm formation. In microbial dysbiosis, biofilms are beneficial to both bacterial and fungal communities but detrimental to the host. Fungi benefit by a surge in their virulence factors, while bacteria become tolerant to antibacterials as a consequence of living under the protective umbrella of the biofilm matrix. This interkingdom cooperation negatively impacts the host, as the fungi and bacteria produce extracellular enzymes that inflict tissue damage, leading to an increase in proinflammatory cytokines, which results in oxidative damage and apoptotic cell death.
COMMENTARY
Chronic wounds are the most financially burdensome skin disease, affecting nearly 6.5 million patients in the United States, with an annual expenditure of nearly $25 billion for treatment alone (1). Additionally, the prevalence of chronic wounds is on the rise, driven by an aging population and growing incidence of diabetes mellitus and obesity worldwide (2). Furthermore, infections of combat wounds are also on the increase (3, 4). Microbial infections are recognized as one of the many destructive processes that delay wound healing. Conventional diagnostic cultures of wounds are largely biased toward microbes that are able to grow rapidly in standard culture media and are presumed to be significant. Lack of reliable diagnostic measures for wound infections leads to nontargeted use of antimicrobials, promoting development of resistant microbial strains and/or killing of potentially beneficial commensal bacteria. Therefore, further understanding of the complex relationship between microbes and delayed healing is critical for development of more evidence-based treatment strategies.
Recent studies investigating the contribution of the microbial communities to health and disease are starting to show that bacteria, fungi, and viruses contribute to our health status (5). Early glimpses at ways the mycobiome might play a positive role in our bodies have been achieved despite the many technical challenges (5). More exciting and as shown in the paper by Kalan et al. (6) and work by our group (7) is the realization that not only do bacteria and fungi coexist in different body sites, but they also interact and have evolved to cooperate in a way that is beneficial to their existence and detrimental, in some cases, to the host. This cooperation represents evolutionary strategies aimed at protecting themselves from the host and antimicrobial insults.
In their article, Kalan et al., unlike earlier studies which focused on bacteria (8, 9), longitudinally profiled 100 nonhealing diabetic foot ulcers using high-throughput sequencing and showed that up to 80% of wounds contain fungi. In contrast, cultures performed in parallel captured only 5% of colonized wounds. The findings that fungi exist in the vast majority of wounds are important since wound infections are currently considered to be bacterial in nature. This perception could be due to the fact that culture-based diagnostic approaches are largely biased toward microbes that are able to grow rapidly in standard culture media and are presumed to be significant.
In total, 17 fungal phylotypes were identified (with a relative abundance of >1%) belonging to the phylum Ascomycota or Basidiomycota, with Cladosporium herbarum, a known environmental fungus associated with allergy (present in 41% of the samples and 56% of subjects), followed by the pathogen Candida albicans (22% of samples and 47% of subjects) as the two most abundant species. Analysis of the effect of antibiotic use on microbial diversity showed that subjects who received antibiotics had significantly higher Shannon diversity indices (for all visits combined) than those subjects who did not receive an antibiotic (P = 0.029). However, diversity over time did not significantly fluctuate before, during, or after antibiotic administration.
A very significant finding of the study by Kalan et al. (6) is the discovery that the mycobiome was associated with clinical outcomes. Specifically, mean proportions of fungal pathogens (and not fungi associated with allergens) were higher in nonhealing wounds and those that ultimately resulted in amputation. This association extended to wound necrosis, which was distinctly associated with pathogenic fungal species and not allergenic molds. Analysis of the fungal community at baseline visit (where specimens were collected over viable wound tissue, not necrotic tissue at the initial clinical presentation and before the wound was surgically debrided of dead tissue and/or biofilms) and subsequent visits showed that the fungal distribution was only significant at the initial presentation. This suggests that the mycobiome at the presentation visit may have utility as a diagnostic marker of time to heal, as well as act as an indicator of poor prognosis (necrosis and amputation). If confirmed in future studies, this discovery may address a glaring gap in wound management, namely, the lack of reliable diagnostic markers. Therefore, further understanding of the complex relationship between microbes and delayed healing is critical for development of more evidence-based treatment strategies.
Chronic microbial infections in the form of biofilms are increasingly recognized as a common cause of delayed healing through various mechanisms (10). Biofilms are microbes embedded in a polymeric matrix that protects them from antimicrobials and resists host defenses. The study of biofilms has introduced a new paradigm of chronic microbial infections: instead of free-floating (planktonic) microbes causing disease patterns that can be reproduced following Koch’s postulates, biofilms are attached polymicrobial communities in which relationships between microbes can alter disease outcome. The use of molecular diagnostic techniques applied to biofilms in chronic wounds has shown many microbes to be in a “viable but nonculturable” state, highlighting the limitations of conventional culture techniques for understanding the composition of biofilms (11).
In the study by Kalan et al. (6), mixed-species biofilms (C. albicans and Citrobacter freundii or Trichosporon asahii and Staphylococcus simulans) formed rapidly in vitro and revealed close interactions between bacterial and fungal cells, with yeast cells forming the biofilm “core” and bacteria associating with the biofilm periphery, coating yeast cells and hyphae as they rapidly grew out of the agar surface (within 48 h). This observation of cooperative mixed biofilms agrees with the findings described in our recent study (7) where we discovered significant intra- as well as interkingdom associations in the bacteriome and mycobiome of Crohn’s disease (CD) patients. In our study, C. tropicalis exhibited significant positive association with Serratia marcescens and Escherichia coli. Similar interkingdom associations between bacterial and fungal communities in CD were recently reported by Sokol et al. (12), who showed that fungal genera (mostly Saccharomyces and Malassezia) were positively correlated with several bacterial taxa in CD.
Furthermore, like Kalan et al. (6), our in vitro studies showed that C. tropicalis, E. coli, and S. marcescens cooperate to form robust biofilms comprising fungal hyphae (7). Biofilms render the organisms resistant to antimicrobial agents and protect them from immune cells (13–15). Moreover, fungal filamentation is a known Candida virulence factor that damages host tissues and triggers specific host immune responses (16–19). Distinct interspecies interactions in this biofilm environment were clearly evident, where E. coli tended to be closely associated with the fungal cell walls, while S. marcescens used its fimbriae to form a “bridge” between C. tropicalis and E. coli that stabilized the bacterium-fungus biofilm structure. Interestingly, Castro et al. (20) described analogous interactions between S. marcescens and Trypanosoma cruzi cells that were mediated by d-mannose-recognizing pili in insect guts. These reports show that interkingdom interactions have evolved in mixed biofilms in many varied niches.
One can speculate why the microbial communities have developed cooperative evolutionary strategies culminating in the development of robust thick biofilms (Fig. 1). In other words, why have fungi and bacteria evolved to adopt a biofilm lifestyle? Why would an individual species contribute to the group at the expense of its own interests? Evolutionary biologists conducted studies to understand the mechanisms sustaining the persistence of cooperation and suggested that spatial community structure (as exists in biofilms) provides a solution as to how cooperation might develop and remain stable (21). These studies provide evidence to show that within a biofilm, cooperation simultaneously results in strategies that ensure the stability of cooperative traits by directly or indirectly reducing the presence of cheaters (22). Because of its role in biofilm structure, the biofilm matrix (consisting primarily of extracellular polysaccharides [EPSs]) plays an important role in maintaining stability and cooperation within biofilms. In this regard, EPS producers play a dual role—they outcompete nonproducers in the presence of solute gradient (including the oxygen and resource gradient) and altruistically push their partners into a more nutrient-rich environment. In thick biofilms, cells far removed from the nutrient source or outside the biofilm milieu experience low levels of available nutrients, while those microbial cells living closer to the periphery will benefit from the available resources.
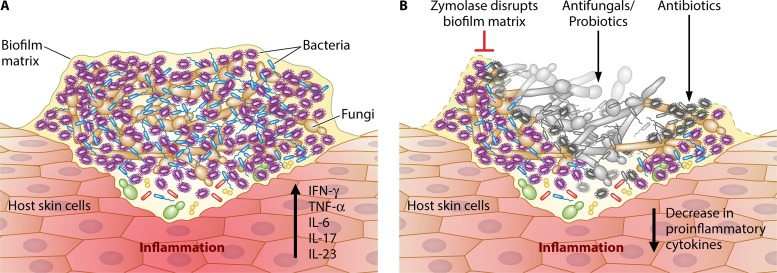
FIG 1 .
Interkingdom cooperation between fungi and bacteria. Chronic wounds are complex systems of multispecies fungal and bacterial biofilms. These biofilms provide a protected milieu for microbes living in close proximity. Fungal cells form the biofilm core while bacteria associate around the periphery of the cells. The fungal hyphae and microbial-secreted enzymes/metabolites facilitate invasion of the skin epidermis/dermis leading to host tissue damage and inflammatory response manifested by an increase in proinflammatory cytokine production (panel A). Panel B shows disruption of the biofilm matrix by zymolase thereby unmasking the microbes. Consequently, treatment with antifungal agents (e.g., echinocandins) and antibiotics leads to microbial cell death (gray/black color) and a decrease in the production of proinflammatory cytokines. IFN-γ, gamma interferon; TNF-α, tumor necrosis factor alpha; IL-6, -17, and -23, interleukins 6, 17, and 23, respectively.
Such strategic benefits apply to fungal and bacterial mixed-species biofilm, as reported by Kalan et al. and Hoarau et al. (6, 7). The fungi benefit by gaining virulence factors (e.g., increased ability to form hyphae and to secrete extracellular enzymes, such as aspartic proteinase [23] relative to a monoculture biofilm), thereby enhancing their ability to invade the host. Bacteria, on the other hand, develop antibacterial tolerance afforded by living under the protective fungal matrix umbrella. Evidence to support this concept has been shown recently with E. coli or Staphylococcus aureus and C. albicans (24, 25). This inter- and intrakingdom cooperation impacts the host immune system, where levels of proinflammatory cytokines (e.g., Th17 cytokines) may increase under the influence of enteric pathogens and immunomodulatory components of fungal biofilms (e.g., fungal β-d-glucans and bacterial lipopolysaccharides), causing increased oxidative damage and apoptotic cell death. Additionally, microbe-induced production of mucolytic enzymes may lead to barrier dysfunction, resulting in tissue damage and lesion formation. In this regard, separate studies have shown that the bacterium Ruminococcus gnavus and the fungal pathogen C. albicans produce mucolytic enzymes that can degrade the protective mucin layer of the gut epithelium, contributing to lesion formation (26, 27).
In conclusion, observation of diverse fungal communities in chronic nonhealing wounds and their ability to form interkingdom biofilms with bacterial species emphasizes not only their paramount importance, but also the complexity of studying whole microbial communities, their interspecies interactions, and implications in chronic disease. The finding that the mycobiome is associated with clinical outcomes raises the possibility that this community may have potential clinical utility as a diagnostic and prognostic biomarker.
ACKNOWLEDGMENTS
Funding support is acknowledged from National Institutes of Health grants R01DE024228 and RO1DE17846, the Oral HIV AIDS Research Alliance (OHARA; BRSACURE-S-11-000049-110229), and a Cleveland Digestive Diseases Research Core Center (DDRCC) Pilot and Feasibility project (supported by NIH/NIDDK P30 DK097948).
The views expressed in this Commentary do not necessarily reflect the views of this journal or of ASM.
Funding Statement
Funding support is acknowledged from the National Institutes of Health grants number R01DE024228 and RO1DE17846, the Oral HIV AIDS Research Alliance (OHARA, BRSACURE-S-11-000049-110229), and a Cleveland Digestive Diseases Research Core Center (DDRCC) Pilot and Feasibility project (supported by NIH/NIDDK P30 DK097948).
Footnotes
For the article discussed, see http://dx.doi.org/10.1128/mBio.01058-16.
Citation Ghannoum M. 2016. Cooperative evolutionary strategy between the bacteriome and mycobiome. mBio 7(6):mBio.01951-16. doi:10.1128/mBio.01951-16.
REFERENCES
- 1.Bickers DR, Lim HW, Margolis D, Weinstock MA, Goodman C, Faulkner E, Gould C, Gemmen E, Dall T, American Academy of Dermatology Association, Society for Investigative Dermatology . 2006. The burden of skin diseases: 2004 a joint project of the American Academy of Dermatology association and the Society for Investigative Dermatology. J Am Acad Dermatol 55:490–500. doi: 10.1016/j.jaad.2006.05.048. [DOI] [PubMed] [Google Scholar]
- 2.Sen CK, Gordillo GM, Roy S, Kirsner R, Lambert L, Hunt TK, Gottrup F, Gurtner GC, Longaker MT. 2009. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen 17:763–771. doi: 10.1111/j.1524-475X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warkentien T, Rodriguez C, Lloyd B, Wells J, Weintrob A, Dunne JR, Ganesan A, Li P, Bradley W, Gaskins LJ, Seillier-Moiseiwitsch F, Murray CK, Millar EV, Keenan B, Paolino K, Fleming M, Hospenthal DR, Wortmann GW, Landrum ML, Kortepeter MG, Tribble DR, Infectious Disease Clinical Research Program Trauma Infectious Disease Outcomes Study Group . 2012. Invasive mold infections following combat-related injuries. Clin Infect Dis 55:1441–1449. doi: 10.1093/cid/cis749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church D, Elsayed S, Reid O, Winston B, Lindsay R. 2006. Burn wound infections. Clin Microbiol Rev 19:403–434. doi: 10.1128/CMR.19.2.403-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghannoum M. 1 February 2016. The mycobiome. Scientist http://www.the-scientist.com/?articles.view/articleNo/45153/title/The-Mycobiome/.
- 6.Kalan L, Loesche M, Hodkinson BP, Heilmann K, Ruthel G, Gardner SE, Grice EA. 2016. Redefining the chronic-wound microbiome: fungal communities are prevalent, dynamic, and associated with delayed healing. mBio 7:e01058-16. doi: 10.1128/mBio.01058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA. 2016. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio 7:e01250-16. doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gontcharova V, Youn E, Sun Y, Wolcott RD, Dowd SE. 2010. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol J 4:8–19. doi: 10.2174/1874285801004010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuttle MS, Mostow E, Mukherjee P, Hu FZ, Melton-Kreft R, Ehrlich GD, Dowd SE, Ghannoum MA. 2011. Characterization of bacterial communities in venous insufficiency wounds using conventional culture and molecular diagnostic methods. J Clin Microbiol 49:3812–3819. doi: 10.1128/JCM.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjarnsholt T, Kirketerp-Møller K, Jensen PØ, Madsen KG, Phipps R, Krogfelt K, Høiby N, Givskov M. 2008. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen 16:2–10. doi: 10.1111/j.1524-475X.2007.00283.x. [DOI] [PubMed] [Google Scholar]
- 11.Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L. 3 February 2016. Fungal microbiota dysbiosis in IBD. Gut doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandra J, Zhou G, Ghannoum MA. 2005. Fungal biofilms and antimycotics. Curr Drug Targets 6:887–894. doi: 10.2174/138945005774912762. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn DM, Ghannoum MA. 2004. Candida biofilms: antifungal resistance and emerging therapeutic options. Curr Opin Invest Drugs 5:186–197. [PubMed] [Google Scholar]
- 15.Polke M, Hube B, Jacobsen ID. 2015. Candida survival strategies. Adv Appl Microbiol 91:139–235. doi: 10.1016/bs.aambs.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Cavalcanti YW, Morse DJ, da Silva WJ, Del-Bel-Cury AA, Wei X, Wilson M, Milward P, Lewis M, Bradshaw D, Williams DW. 2015. Virulence and pathogenicity of Candida albicans is enhanced in biofilms containing oral bacteria. Biofouling 31:27–38. doi: 10.1080/08927014.2014.996143. [DOI] [PubMed] [Google Scholar]
- 17.Gil ML, Gozalbo D. 2006. TLR2, but not TLR4, triggers cytokine production by murine cells in response to Candida albicans yeasts and hyphae. Microbes Infect 8:2299–2304. doi: 10.1016/j.micinf.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Kumamoto CA, Vinces MD. 2005. Contributions of hyphae and hyphaco-regulated genes to Candida albicans virulence. Cell Microbiol 7:1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- 19.Van der Graaf CA, Netea MG, Verschueren I, van der Meer JW, Kullberg BJ. 2005. Differential cytokine production and Toll-like receptor signaling pathways by Candida albicans blastoconidia and hyphae. Infect Immun 73:7458–7464. doi: 10.1128/IAI.73.11.7458-7464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro DP, Seabra SH, Garcia ES, de Souza W, Azambuja P. 2007. Trypanosoma cruzi: ultrastructural studies of adhesion, lysis and biofilm formation by Serratia marcescens. Exp Parasitol 117:201–207. doi: 10.1016/j.exppara.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Kovacs A. 24 November 2014. Impact of spatial distribution on the development of mutualism in microbes. Front Microbiol doi: 10.3389/fmicb.2014.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Travisano M, Velicer GJ. 2004. Strategies of microbial cheater control. Trends Microbiol 12:72–78. doi: 10.1016/j.tim.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Mendes A, Mores AU, Carvalho AP, Rosa RT, Samaranayake LP, Rosa EA. 2007. Candida albicans biofilms produce more secreted aspartyl protease than the planktonic cells. Biol Pharm Bull 30:1813–1815. doi: 10.1248/bpb.30.1813. [DOI] [PubMed] [Google Scholar]
- 24.Kong EF, Tsui C, Kucharikova S, Andes D, Van Dijck P, Jabra-Rizk MA. 2016. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. mBio 7:e01365-16. doi: 10.1128/mBio.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Brucker K, Tan Y, Vints K, De Cremer K, Braem A, Verstraeten N, Michiels J, Vleugels J, Cammue B, Thevissen K. 2015. Fungal β-1,3-glucan increases ofloxacin tolerance of Escherichia coli in a polymicrobial E. coli/Candida albicans biofilm. Antimicrob Agents Chemother 59:3052–3058. doi: 10.1128/AAC.04650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. 2010. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 27.Colina AR, Aumont F, Deslauriers N, Belhumeur P, De Repentigny L. 1996. Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infect Immun 64:4514–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]