Abstract
Hypoxia and inflammatory cytokines like interleukin-6 (IL6) are strongly linked to cancer progression, and signal in part through the transcription factor Ccaat/enhancer binding protein δ (C/EBPδ, CEBPD), which has been shown to promote mesenchymal features and malignant progression of glioblastoma. Here we report a different role for C/EBPδ in breast cancer. We found that the C/EBPδ protein is expressed in normal breast epithelial cells and in low-grade cancers. C/EBPδ protein (but not mRNA) expression correlates with estrogen receptor (ER+) and progesterone receptor (PGR) expression and longer progression-free survival of breast cancer patients. Specifically in ER+ breast cancers, CEBPD–but not the related CEBPB–mRNA in combination with IL6 correlated with lower risk of progression. Functional studies in cell lines showed that ERα promotes C/EBPδ expression at the level of protein stability by inhibition of the FBXW7 pathway. Furthermore, we found that C/EBPδ attenuates cell growth, motility and invasiveness by inhibiting expression of the SNAI2 (Slug) transcriptional repressor, which leads to expression of the cyclin dependent kinase inhibitor CDKN1A (p21CIP1/WAF1). These findings identify a molecular mechanism by which ERα signaling reduces the aggressiveness of cancer cells, and demonstrate that C/EBPδ can have different functions in different types of cancer. Furthermore, our results support a potentially beneficial role for the IL-6 pathway specifically in ER+ breast cancer and call for further evaluation of the role of intra-tumoral IL-6 expression and of which cancers might benefit from current attempts to target the IL-6 pathway as a therapeutic strategy.
Keywords: breast cancer, estrogen receptor, transcription, C/EBP, prognosis, SNAI2, FBXW7
Introduction
The inflammatory responses are essential defense mechanisms for the organism, and are also engaged in wound healing and tissue regeneration. However, inflammatory signals including the cytokine interleukin-6 can also contribute to many diseases including cancer49. For instance, the involution of the mammary gland subsequent to lactation involves inflammatory processes that facilitate breast cancer progression33. Paradoxically, molecules that promote mammary epithelial cell (MEC) death during involution, such as the transcription factor STAT3, can contribute to breast cancer progression and metastasis42. The transcription factor Ccaat/enhancer binding protein δ (C/EBPδ, CEBPD), which is a target of STAT3 and a mediator of inflammatory cytokine signaling5, 39 also promotes MEC death during mammary gland involution50. In contrast to IL-6 and STAT3, which are strongly linked to progression and metastasis of many cancer types including breast cancer49, the role of C/EBPδ in cancer is less clear. By crossing a Cebpd null mutation into MMTV-Neu transgenic mice expressing the Neu (Erbb2) proto-oncogene in mammary epithelial cells, we found that C/EBPδ acts as a tumor suppressor by attenuating mammary tumor multiplicity, while also acting as a tumor promoter by increasing the incidence of metastasis to the lungs3.
In support of a role in tumor progression, C/EBPδ promotes inflammatory signaling and cell survival under hypoxia by inhibiting the expression of FBXW73, 4, a tumor suppressor whose expression is frequently lost in glioblastoma22. In fact, C/EBPδ is overexpressed in glioblastoma and is a driver of glioblastoma progression5, 12. Also in pancreatic cancer - along with IL-6- and in urothelial carcinoma CEBPD is overexpressed and is a marker of poor prognosis30, 52. Furthermore, Cebpd mRNA expression correlates with STAT3 activity and metastasis in the MMTV-Neu mouse mammary tumor model40. In contrast, CEBPD is downregulated at the mRNA level in several cancer types, including cervical, liver, and breast cancer; and CEBPD mRNA expression is part of one signature predicting better survival for breast cancer patients5, 35, 38. Cell culture models mostly support the tumor suppressor-like functions for C/EBPδ. In myeloid and prostate cancer cell lines C/EBPδ promotes differentiation and inhibits growth5. C/EBPδ downregulates expression of cyclin D in cells in culture36, but in a small cohort of breast cancer tissues C/EBPδ correlated positively with cyclins D1 and E as well as RB1 and p16/CDKN2A34. In basal-type breast epithelial cell lines, C/EBPδ inhibits migration and growth in soft agar, and ectopic C/EBPδ inhibits clonal outgrowth of MCF-7 cells25, 36, 47. In light of these disparate findings on C/EBPδ’s role in different cancers and breast cancer model systems, we investigated C/EBPδ expression in human breast cancer tissues and analyzed endogenous C/EBPδ functions with relevant subtype-specific cancer cell lines.
Our study shows that in contrast to C/EBPδ’s role in inflammation and as a driver of glioblastoma progression, abundant expression of C/EBPδ is a good prognostic marker in estrogen receptor alpha positive (ER+) breast cancer, which accounts for approximately three quarters of all breast cancers. These findings suggest that the role of these “inflammatory molecules” may be subtype-specific and call for further investigation, especially in light of ongoing efforts to develop inhibitors of the IL-6 and STAT3 pathway for breast cancer.
Results
C/EBPδ protein is expressed in normal human and mouse breast epithelial cells
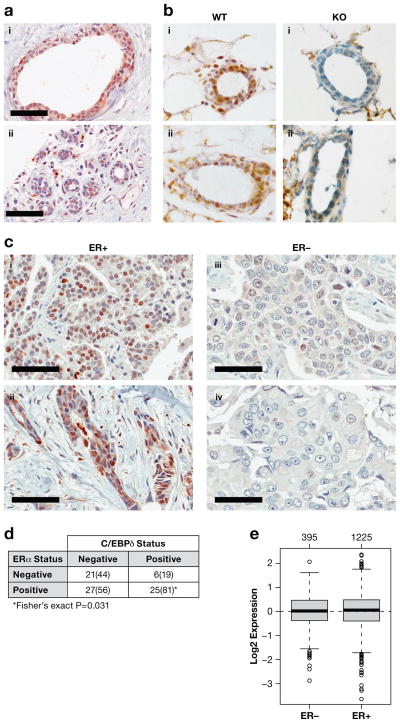
Genomic approaches showed that CEBPD mRNA levels are highest in normal breast, decrease with cancer progression and are inversely correlated with tumor grade38. Similar data can be retrieved with the on-line tool “Gene-expression based outcome for breast cancer online” (Supplementary Figure S1). Because C/EBPδ expression can be regulated at the level of protein stability4, 12, 47 its mRNA levels are not always predictive of protein expression. Therefore, we developed an antibody suitable for detection of C/EBPδ by IHC (Supplementary Figure S2). In normal breast, C/EBPδ protein was detected in luminal epithelial cells, with expression also in some basal and stromal cells (Figure 1a). This result contrasted our previous observation in mice, where C/EBPδ was not detectable in formalin-fixed mammary glands of nulliparous animals50. However, upon assessment of staining in frozen sections we indeed confirmed C/EBPδ protein expression in mammary epithelial cells of the adult mouse mammary gland (Figure 1b). These results demonstrate that C/EBPδ protein is a component of normal mammary epithelial cells in the human and mouse mammary gland.
Figure 1. C/EBPδ is expressed in normal breast epithelial cells and ER+ breast cancer.
A) Immunohistochemistry of C/EBPδ of two independent human breast tissue specimen (scale bar = 60 μm). B) Immunohistochemistry of C/EBPδ on frozen sections of abdominal mammary glands from 4–month old Cebpd wild-type (WT) and null mutant (KO) mice at diestrous, two independent specimen each (i–ii). C) Immunohistochemistry of C/EBPδ in human breast cancer tissues with ER status as indicated. i: ductal papillary adenocarcinoma; ii–iv: invasive ductal carcinoma (scale bar = 60 μm). D) Correlation analysis of ERα and nuclear C/EBPδ staining in carcinoma cells of breast cancer tissue microarray 1 (TMA-1). Number of specimen (percentages for C/EBPδ in parenthesis) scored as positive or negative for C/EBPδ and ERα are shown. See Supplementary Figure S3A for distribution of C/EBPδ staining frequency and intensity. E) CEBPD mRNA expression in breast cancer tissues according to ER status as analyzed by GOBO (http://co.bmc.lu.se/gobo)43. The number of samples per group is shown above the plots.
C/EBPδ protein but not mRNA is enriched in hormone receptor positive breast cancer and correlates with markers of good prognosis
To address C/EBPδ protein expression in breast cancer we first chose a tissue microarray (TMA-1) that included tumors from a dataset in which CEBPD mRNA was part of a gene expression signature that identified patients with longer survival35. This TMA revealed that C/EBPδ protein was also present in the carcinoma cell nuclei of a subset of cancer tissues and significantly enriched in ER+ tumors (Figure 1c–d). Positive correlations were also found with lower tumor grade (Spearman correlation coefficient: C.C. = −0.344; P = 0.0019) and two markers of the luminal subtype: cytokeratin 19 (C.C. = 0.30; P = 0.0092) and progesterone receptor (PGR; C.C. = 0.38; P = 0.0007). There were no significant correlations with EGFR, CK14, or p53. Interestingly, across breast cancer subtypes, there was no correlation of CEBPD mRNA levels with ERα status in this cohort (data not shown) or three other larger datasets (Figure 1e and Supplementary Figure S3b). This result demonstrates preferential expression of C/EBPδ protein, but not mRNA, in ER+ tumors.
Next, we analyzed an independent cohort with 292 breast cancer cases (TMA-2), which identified 30% of the tumors as C/EBPδ-positive (>50% staining), 75% of which were ER+ (>10% staining). Again, C/EBPδ expression correlated positively with ERα and PGR, and inversely with tumor grade (Table 1). Within ER+ cancers C/EBPδ also correlated positively with phosphorylated ERK (pERK), which can be an independent indicator of good prognosis14, citations within), and inversely with the hypoxia indicators carbonic anhydrase IX and VEGF, which are poor prognostic markers11. No significant correlations were found with (a) the clinical parameters of tumor size, age, or lymph node status; (b) the proliferation indicators Ki67 and cyclin D1; or (c) the proteins p27 (CDKN1B) or HER2. Taken together, these analyses revealed that the C/EBPδ protein is preferentially expressed in hormone receptor positive breast cancers and correlates with pathological indicators of a more benign tumor phenotype.
Table 1.
Correlation analysis of nuclear C/EBPδ staining in breast carcinoma cells (TMA-2)
| n | C.C.1 | P-value | |
|---|---|---|---|
| All tumors | |||
| ER | 292 | 0.121 | 0.039 |
| PGR | 286 | 0.255 | <0.001 |
| Grade | 290 | −0.215 | <0.001 |
| ER+ tumors | |||
| PR | 187 | 0.315 | <0.001 |
| Grade | 192 | −0.282 | <0.001 |
| pERK | 179 | 0.249 | 0.001 |
| CA IX | 164 | −0.196 | 0.012 |
| VEGF | 185 | −0.175 | 0.017 |
C.C.: Spearman’s rank-order correlation coefficient.
ERα promotes C/EBPδ protein stability through inhibition of the FBXW7 pathway
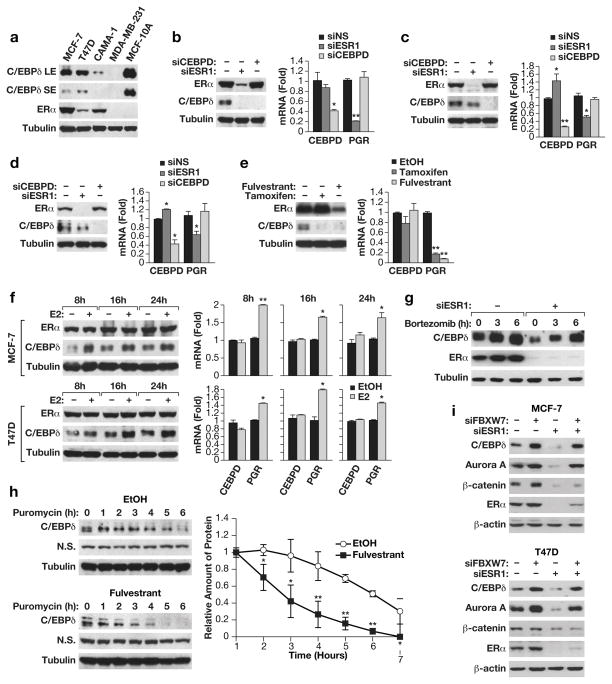
To study the potential mutual regulation of C/EBPδ and hormone receptors, we used RNA interference in cell culture models. While C/EBPδ levels are significantly lower in breast cancer cell lines compared to the non-tumorigenic MCF-10A and MCF-12A cells47, it was detectable in the three ER+ lines MCF-7, T47D and CAMA-1 (Figure 2a). In these cell lines, silencing of CEBPD did not change the level of ERα protein or ERα activity (Figures 2b–d), which was inferred from the mRNA levels of the progesterone receptor (PGR), a well-established target gene of ERα. On the other hand, silencing of the ERα gene (ESR1) did reduce C/EBPδ protein levels although CEBPD mRNA levels remained unchanged or increased modestly (Figures 2b–d). Similar results were obtained in MCF-7 cells when ERα activity was inhibited with tamoxifen or ERα expression was downregulated by fulvestrant (Figure 2e). In contrast, addition of estradiol increased C/EBPδ protein levels, again without affecting CEBPD mRNA expression (Figure 2f and Supplementary Figure S4a). Taken together these data show that ERα promotes C/EBPδ expression at the level of the protein.
Figure 2. ERα supports C/EBPδ protein stability through inhibition of the FBXW7 pathway.
A) Western blot analysis of C/EBPδ and ERα protein expression in ER+ breast cancer cell lines MCF-7, T47D and Cama-1; ER-, basal MDA-MB-231 cells; and non-tumorigenic, basal MCF-10A cells. SE, LE: short and long exposure, respectively. Tubulin was used as loading control. B) Western analysis (left) of C/EBPδ and ERα protein levels and Quantitative PCR (QPCR) analysis (right) of CEBPD and PGR mRNA levels in MCF-7 cells after nucleofection with siRNAs against non-specific control (-, siNS), C/EBPδ (siCEBPD) or ERα (siESR1) (n=3,* P<0.05 and ** P<0.01 when compared to siNS). C) Western and QPCR analysis of T47D cells as in panel B. D) Western and QPCR analysis of Cama-1 cells as in panel B. E) Western and QPCR analysis of MCF-7 cells treated for 48 h with 1 μM of fulvestrant, tamoxifen, or ethanol (EtOH) as solvent (**P<0.01 when compared to EtOH, n=3). F) Western and QPCR analysis of MCF-7 and T47D cells treated with β-estradiol (E2, 1nM) for the indicated times (*P<0.05, ** P<0.01; n=3). G) Western analysis of C/EBPδ and ERα protein in MCF-7 cells transfected with siRNA against ERα (siESR1) or non-specific control (-) and treated 48 h later with (bortezomib (5 μM) for the indicated times. DMSO was used as solvent control. H) Representative Western analysis (top panels) of MCF-7 cells treated with Fulvestrant (1 μM) or solvent control (EtOH) and 48 h later with puromycin (15 μg/ml) for the indicated times. Tubulin and a non-specific (N.S.) band are shown as loading controls. The bottom panel shows quantification of C/EBPδ protein expression normalized to the non-specific band from three independent experiments (* P<0.05 and ** P<0.01). I) Western analysis of the indicated proteins in whole cell extracts from MCF-7 and T47D cells transfected with siRNA against FBXW7 (siFBXW7), ERα (siESR1), or non-specific control (-).
To address the mechanism by which ERα promotes C/EBPδ protein expression, we assessed C/EBPδ protein stability. We had previously shown that the C/EBPδ protein is unstable in breast cancer cell lines due to the SIAH2 E3 ubiquitin ligase47. Therefore, the proteasome inhibitors bortezomib or MG132 alone increased C/EBPδ protein levels (Figure 2g and Supplementary Figure S4b). However, these drugs could also at least partially recover C/EBPδ protein expression when ERα expression was depleted. To more directly assess C/EBPδ protein stability, cells were treated with fulvestrant (Figure 2h) or tamoxifen (Supplementary Figure S5a) plus the protein synthesis inhibitor puromycin. Under these conditions, compared to vehicle control, C/EBPδ protein levels decreased more quickly when ERα was inhibited. These data show that ERα promotes C/EBPδ protein stability.
Next we asked whether ERα was attenuating C/EBPδ degradation by the SIAH2 E3 ligase47. However, silencing of SIAH2 could not rescue C/EBPδ protein when ERα was depleted (Supplementary Figure S5b). To confirm SIAH2 silencing we assessed its mRNA expression levels, which revealed that ERα supports SIAH2 expression at least at the level of the mRNA (Supplementary Figure S5c). Taken together, these results rule out the SIAH2 pathway as the target for ERα-mediated CEBPδ protein stabilization.
The F-box protein FBXW7, a substrate binding domain of SKP1-Cullin 1-F box protein (SCF)-type E3 ubiquitin ligases, can also target C/EBPδ for proteasomal degradation 4. FBXW7 interacts with most substrates – including C/EBPδ – only after phosphorylation of their degron sequence by GSK-3β4, 16. GSK-3β is constitutively active unless it is phosphorylated by kinases such as Akt on serine 9 58. ERα can inhibit GSK-3β activity either through activation of Akt kinase and/or by direct interaction with GSK-3β 6, 9, 55. Accordingly, we also observed that inhibition of ERα in MCF-7 cells by either siRNA or fulvestrant reduced the inhibitory phosphorylation of GSK-3β on Serine 9 (Supplementary Figure S5d). In agreement with activation of the GSK-3β kinase58, targets of GSK-3β mediated protein degradation such as Aurora kinase A53 and β-catenin48 were also reduced when ERα was inhibited. Silencing of FBXW7 in MCF-7 (Figure 2i and Supplementary Figures S5e–f) and T47D (Figure 2i) cells not only increased basal levels of C/EBPδ, as expected4, but completely recovered C/EBPδ protein expression when ERα was inhibited by either fulvestrant (Supplementary Figures S5e-f) or RNAi (Figure 2i). Similar results were obtained for Aurora A kinase, which is also a target of the SCFFBXW7 degradation pathway 53. FBXW7-silencing also led to a subtle but reproducible increase in ERα protein. However, the expression level of β-catenin protein, which is degraded by the FBXW7-independent β-TrCP complex48, was not rescued by FBXW7-silencing. Consistent with regulation of expression through protein stability, the mRNA levels of Aurora A (AURKA), β-catenin (CTNNB1) and CEBPD were not affected by silencing of ERα and/or FBXW7 (Supplementary Figure S5g). Taken together, our data indicate that ERα supports C/EBPδ protein stability by inhibiting the GSK-3β-FBXW7 pathway and thereby preventing proteasomal degradation of FBXW7 substrates.
Identification of C/EBPδ regulated genes/pathways
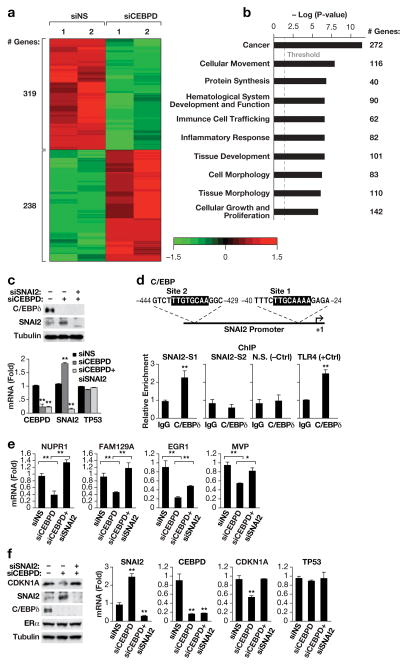
To identify the signaling pathways regulated by C/EBPδ we determined the effect of CEBPD-silencing on the transcriptome in MCF-7 cells. We prioritized MCF-7 cells because these cells have a significant basal level of C/EBPδ protein (Figure 2A) and express wild-type p53, a property of most ER+ breast cancers19. An exploratory mRNA-Seq approach revealed that C/EBPδ supports the expression of 319 genes and attenuates the expression of 238 genes (1.5– to 12–fold differential expression) (Figure 3a). About 90% of tested genes could be validated as C/EBPδ-regulated by QPCR with independent mRNA samples and by silencing CEBPD with either one of two siRNA sequences (Supplementary Figures S6a-b). Analysis of the association of these differentially expressed genes (DEGs) with biological pathways using the Ingenuity Pathway Analysis suite (IPA) showed that the top Canonical Pathway was “acute phase response signaling” (P value 5.26E-03), and the top Upstream Regulators were lipopolysaccharide (P =5.93E-08), IL1 (P =1.93E-07), and TNF (P =3.40E-07). These results are consistent with the well-known functions of C/EBPδ in inflammatory signaling pathways and immune responses5, 39. To assess whether C/EBPδ–regulated genes clustered with specific breast cancer subtypes we used the ONCOMINE™ platform, which identified datasets in which C/EBPδ–inhibited genes were enriched for genes that are upregulated in cancers that are ER-negative, metastatic or invasive compared to ER-positive, non-metastatic, and ductal carcinoma in situ, respectively (Supplementary Figure S6c). In contrast, C/EBPδ–activated genes were enriched for genes that are preferentially expressed in ER+ cancers compared to ER-negative cancers (Supplementary Figure S6d). These results are in agreement with the observation that C/EBPδ protein was predominantly expressed in ER+ and lower grade tumors (Figure 2 and Table 1) and indicate that C/EBPδ promotes the expression of genes that contribute to the ER+ tumor phenotype and attenuates the expression of genes that correlate with invasiveness, metastasis, and an ER− tumor phenotype.
Figure 3. C/EBPδ inhibits SNAI2 expression.
A) Heat map generated by Partek Genomics Suite showing the global changes in gene expression (fold change 1.5, P<0.05) upon C/EBPδ-silencing as determined by mRNA-Seq analysis. MCF-7 cells were transfected with siRNA against C/EBPδ or non-specific control and RNA was harvested 48 h later (n=2). B) Schematic generated by Ingenuity Pathway Analysis showing the top ten most significantly represented “biological functions” by the DEGs (P values: 3.08E-07 to 4.18E-12) from Supplementary Figure S7. Enrichment score is reported as the minus log transformation on the geometric mean of P-values from the enriched annotation terms associating with one or more of the gene group members. The genes are clustered into significantly enriched groups for specific biological functions. The threshold line indicates at P-value of 0.05. The number of genes in each group is indicated on the right. C) Western analysis (left) of C/EBPδ and SNAI2 protein levels and QPCR analysis (right) of C/EBPδ (CEBPD), SNAI2 and p53 (TP53) mRNA levels in MCF-7 cells 48 h after nucleofection with siRNAs against non-specific control (-,siNS), C/EBPδ (siCEBPD) or SNAI2 (siSNAI2) (n=3, * P<0.05 and ** P<0.01 when compared to siNS). D) Schematic (upper panel) showing the potential binding sites for C/EBP proteins in the SNAI2 promoter, and quantification of QPCR analyses (lower panels) of DNA fragments in chromatin immunoprecipitation (ChIP) assays with MCF-7 cells and C/EBPδ-specific antibodies or IgG as control. The primers were specific for regions encompassing sites 1 and 2 (SNAI2-S1,-S2), a genomic region without C/EBP binding motif (N.S.) as negative control, and the C/EBPδ binding site in the TLR4 promoter4 as positive control (n=3,** P<0.01). E) QPCR analysis of the mRNA expression levels of the indicated genes in MCF-7 cells as in panel A (n=3, * P<0.05, ** P<0.01). F) Western blot analysis (left) of C/EBPδ, SNAI2, CDKN1A and ERα protein levels and QPCR analysis (right) of C/EBPδ, SNAI2, CDKN1A and p53 mRNA levels in MCF-7 cells 48 h after nucleofection with siRNAs against non-specific control (-,siNS), C/EBPδ (siCEBPD) or SNAI2 (siSNAI2) (n=3,** P<0.01 when compared to siNS).
C/EBPδ regulates a subset of genes through direct inhibition of SNAI2 expression
Further analysis of the DEGs by IPA showed that the most significantly affected specific BioFunction was “cellular movement” (Figure 3b), consistent with the observation that C/EBPδ can attenuate cell migration and invasiveness47, 56, which we also confirmed in MCF-7 cells (Supplementary Figure S7a–b). Many molecular pathways that promote cell proliferation, migration and invasiveness in cultured cells, have been shown to correlate with or contribute to tumor aggressiveness in vivo45, 51. We therefore focused our attention on genes that are upstream regulators of cell motility, which led us to the transcriptional repressor SNAI2 (also known as SLUG) a well-known promoter of cell motility and invasiveness in different normal and cancer cells15. CEBPD-silencing induced the expression of SNAI2 mRNA and protein levels in MCF-7 (Figure 3c) and T47D cells (Supplementary Figure S8a). Expression of p53 is shown here and in subsequent Figures as a negative control because its mRNA levels were not affected by C/EBPδ or SNAI2 depletion. Chromatin binding assays showed that C/EBPδ bound directly to a site in the proximal SNAI2 promoter (Figure 3d) identifying SNAI2 as a direct target gene of C/EBPδ.
To address whether inhibition of the SNAI2 repressor by C/EBPδ was reflected in the C/EBPδ-dependent transcriptome, we assessed the overlap between genes that are repressed by ectopic SNAI2 in MCF-7 cells18 and genes that were downregulated in CEBPD-silenced cells. Several of these genes (NUPR1, FAM129A, EGR1 and MVP) were rescued to various degrees when SNAI2 was co-silenced along with C/EBPδ in MCF-7 cells (Figure 3e). Similar data were obtained for NUPR1, FAM129A, and EGR1 in T47D cells (Supplementary Figure S8b). These results demonstrate that C/EBPδ promotes expression of a subset of genes at least in part through inhibition of the SNAI2 repressor.
In addition to attenuating cell migration and invasion, C/EBPδ also decreased cell population growth (Supplementary Figure S7c). Analysis of the list of DEGs for potential regulators of the cell cycle led us to the cyclin-dependent kinase inhibitor 1A (CDKN1A, p21CIP1/WAF1), which was significantly reduced in CEBPD-silenced MCF-7 cells (Supplementary Figure S8c). CDKN1A attenuates MCF-7 cell growth29, and is a target gene of SNAI2 in mouse embryonic fibroblasts8. The co-silencing approach showed that C/EBPδ supports expression of CDKN1A in MCF-7 cells by inhibiting SNAI2 expression (Figure 3f), suggesting that this pathway may contribute to attenuation of MCF-7 cell growth by C/EBPδ.
C/EBPδ attenuates cell migration and proliferation through inhibition of SNAI2 expression
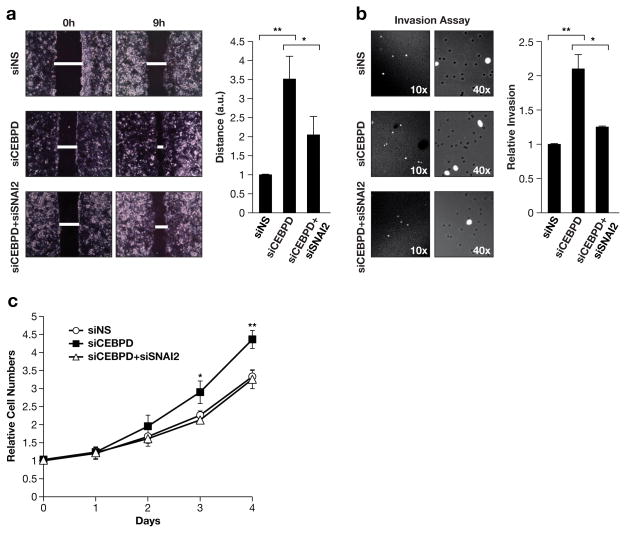
To determine if activation of SNAI2 was responsible for the phenotypes of CEBPD-depleted cells (Supplementary Figure 7), we silenced SNAI2 and assessed cell migration, invasiveness, and growth. Co-silencing of both SNAI2 and CEBPD indeed attenuated the increased migration by CEBPD-silenced cells (Figure 4a). To assess the role of SNAI2 in invasiveness we used the Boyden chamber assay because shorter incubation times make this assay amenable to the co-silencing approach. Although MCF-7 cells are not very capable of crossing the matrigel barrier, silencing of CEBPD doubled the number of invasive cells in a SNAI2-dependent manner (Figure 4b). Lastly, analysis of cell growth revealed that co-silencing of SNAI2 completely abolished the accelerated cell population growth of CEBPD-silenced MCF-7 cells (Figure 4c and Supplementary Figures S8d) along with rescue of CDKN1A expression (Figure 3f and Supplementary Figures S8e). Consistent with its very low basal levels, silencing of SNAI2 had no effect on control cells that express C/EBPδ. Taken together, these results identify inhibition of SNAI2 expression as one molecular mechanism by which C/EBPδ attenuates cell growth, migration, and invasion in MCF-7 cells. These results show that endogenous C/EBPδ attenuates malignant behaviors of MCF-7 cells in culture and are consistent with the observation that C/EBPδ expression was significantly associated with low tumor grade in patients.
Figure 4. Silencing of SNAI2 expression reverts the phenotype of C/EBPδ-depleted MCF-7 cells.
A) Cell migration assay. MCF-7 cells were transfected with the indicated siRNAs and grown with culture inserts until confluent. Representative images (left) are shown at the indicated time after removal of the insert along with quantification (right) of wound closure from three independent experiments each done in triplicates (*P<0.05, ** P<0.01). B) Transwell invasion assay. MCF-7 cells treated as in panel A were plated without serum in Matrigel invasion chambers. Cells that migrated to the side with serum-containing medium after 36 h were stained with DAPI and counted (n=3,* P<0.05, ** P<0.01). C) Analysis of cell population growth/viability by vital dye staining (Alamar blue) of MCF-7 cells with transient knockdown of C/EBPδ (siCEBPD), SNAI2 (siSNAI2), or control (siNS) (n=3, ** P<0.05). ** P<0.01).
C/EBPδ expression is associated with better outcome for ER+ breast cancer patients
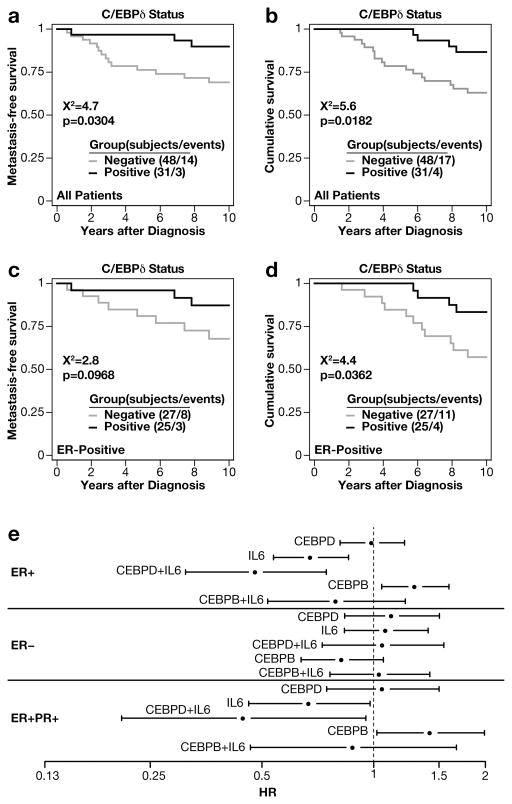
Lastly, we asked if C/EBPδ protein expression in breast tumors correlated with outcome for patients. For the TMA-1 cohort, across all patients, C/EBPδ immunostaining was associated with a longer time to distant metastasis (P = 0.03, Log-Rank test) and disease-specific patient survival (P = 0.018 for all patients; Log-rank test) (Figures 5a–b). Loss of C/EBPδ was a poor prognostic factor even within the 52 ER+ tumor cohort (hazard ratio for C/EBPδ positivity, 0.31; 95% confidence interval, 0.1 – 0.99; P = 0.048; P = 0.036 by Log-rank test) (Figure 5c–d). Multivariate analysis of survival data associated with the 299 patient cohort of TMA-2 with 192 ER+ cases also showed that a high nuclear C/EBPδ staining score was a significant (P=0.041) good prognostic marker independent of ER status (Table 2). Taken together, the results of both cohorts overall agree in that C/EBPδ expression correlates not only with lower tumor grade and steroid hormone receptor expression, but also with lower risk of progression
Figure 5. C/EBPδ expression correlates with longer survival of breast cancer patients.
Kaplan Meier survival plots of breast cancer patients represented on TMA-1. A) Metastasis-free survival of all patients. The number of patients and events (n/events) are indicated for the groups with and without C/EBPδ staining. B) Disease-specific survival for all patients as in panel A. C) Metastasis-free survival of patients with ER+ cancer. D) Disease-specific survival of patients with ER+ cancer. All P values are from log-rank tests. E) CEBPD, but not CEBPB, mRNA expression is associated with lower risk of progression for ER+ breast cancer patients when combined with IL6 mRNA. Analysis of CEBPD, CEBPB and IL6 gene expression levels alone or in combination and in association with disease free survival in the indicated patient populations. Meta-analysis of 26 data sets conducted with the BreastMark on-line tool (http://glados.ucd.ie/BreastMark/mRNA_custom.html) using high cut-off values and the indicated parameters. Shown are hazard ratios (HR) with confidence intervals (on the log scale with x-axis labels on the linear scale) according to the expression of the indicated gene(s) in the patient populations by ER and PGR (PR) status. The numbers of cases, events and P-values are shown in Table S1.
Table 2.
Multivariate Cox regression analysis indicating high C/EBPδ score as a significant good prognostic marker
| HR | 95% C.I. | P-value | ||
|---|---|---|---|---|
| Lower | Upper | |||
| Age | 0.981 | 0.947 | 1.017 | 0.299 |
| Treatment | 0.773 | 0.533 | 1.122 | 0.175 |
| Tumor Size | 1.009 | 0.991 | 1.028 | 0.316 |
| Tumor Grade | 1.467 | 0.952 | 2.26 | 0.082 |
| Node Status | 2.046 | 1.258 | 3.327 | 0.004** |
| ER Status | 0.577 | 0.384 | 0.865 | 0.008** |
| cyto.C/EBPδ | 0.890 | 0.596 | 1.327 | 0.567 |
| nucl.C/EBPδ | 0.621 | 0.393 | 0.982 | 0.041* |
Hazard ration (HR) with 95% Confidence Interval (C.I.) and P value.
Cyto.: cytoplasmic, nucl.: nuclear.
These findings are in stark contrast to the observation that C/EBPδ is associated with poor prognosis of glioblastoma, pancreatic and urothelial carcinoma, and its association with inflammatory signaling (see Introduction). In breast cancer, an inflammatory tumor signature and specifically systemic IL-6 are associated with poor outcome and metastasis17. To begin to address this conundrum, we interrogated the correlation of CEBPD and IL6 mRNA levels with patient outcome using the on-line tool BreastMark32. CEBPD alone did not show a specific correlation with disease progression (Figure 5e). This was not surprising given the known dissociation of CEBPD mRNA and protein expression levels (Figures 1 and 2). However, IL6 mRNA alone was associated with a lower hazard ratio (HR) specifically in ER+ breast cancer but not in ER-negative cancer. The combination of IL6 with CEBPD further improved outcome at the level of HR and statistical significance (Figure 5e and Supplementary Table S1). This pattern was also seen for ER+/PR+ cancers. In contrast, expression of the C/EBP family member CEBPB (C/EBPβ) alone was associated with worse outcome for ER+ cancer patients and negated any benefit of IL6 expression. These results further support a role for C/EBPδ as a marker of good prognosis in ER+ breast cancer and raise the possibility that C/EBPδ specifically contributes to a beneficial role of the IL-6 pathway in ER+ breast cancer.
DISCUSSION
In this study we describe expression of the C/EBPδ transcription factor in normal breast epithelial cells and in steroid hormone receptor (HR+) positive breast cancer with relatively indolent characteristics. This result was surprising for two reasons: 1) Large scale mRNA analyses had not predicted preferential expression of C/EBPδ in HR+ breast cancers; 2) C/EBPδ is best characterized as a pro-inflammatory factor and is associated with aggressiveness of other cancer types such as glioblastoma, pancreatic cancer, and urothelial carcinoma (see Introduction). Our results highlight the importance of tumor characterizations at the level of the protein and the significant role of cell type and context for the function of specific proteins.
ERα signaling promotes C/EBPδ protein stability at least in part through inhibition of the GSK-3β-SCFFBXW7 pathway, which is in fact a tumor suppressor pathway as it downregulates many oncoproteins 16, 53. Interestingly, GSK-3β has been shown to in turn contribute to ERα protein stability 20. On the other hand, C/EBPδ also downregulates expression of FBXW73. And Hence, these proteins engage in multiple circuits of cross talk, and the mechanisms regulating this delicate balance of tumor promoting and tumor suppressing proteins and pathways may be critical for the ultimate outcome of cancer cell fate.
Interestingly, among breast cancer subtypes, FBXW7 mRNA levels are lowest in luminal cancers54, which may also contribute to C/EBPδ’s expression in HR+ cancers. The co-expression of ERα and C/EBPδ raises the question if they coordinately regulate target genes. We could not positively identify physical interaction between C/EBPδ and ERα. However, C/EBP binding motives have been identified in ER target genes10. Although the presence of a C/EBP motif does not predict which of the C/EBP family proteins – if any - may bind, and analysis of our mRNA-Seq data did not indicate a significant effect of C/EBPδ on ERα pathways, we do not rule out a role of C/EBPδ in the regulation of specific ERα targets that may be relevant for breast cancer biology.
In tumor tissues, we also observed a significant correlation of C/EBPδ with PGR expression. Given that PGR is a direct target of ERα and that PGR+ tumors are typically also ER+23, we speculate that the considerable correlation of C/EBPδ with PGR in tissues reflects the ability of ERα to support both PGR gene expression as well as C/EBPδ protein stability. Loss of these arms of ER signaling may then lead to the development of PGR-/ C/EBPδ-cancers with worse prognosis.
The C/EBPδ protein harbors a classical activation domain but can also repress genes in association with co-repressors5. In this study we identified SNAI2 as C/EBPδ-repressed gene, whose activation mediates enhanced proliferation, migration and invasiveness of CEBPD-silenced MCF-7 cell, consistent with the reported roles of SNAI2 in motility and proliferation of breast cancer cell lines in culture (2 and references therein). In normal mammary epithelial cells, SNAI2 supports a basal phenotype and stemness while suppressing luminal differentiation; and loss of Snai2 in the mammary gland results in hyperproliferation of luminal cells37 and references therein). In breast cancers, SNAI2 expression is enriched in the triple-negative subtype, and tumors with lymph-node meatastsis and high grade1. Experimental model systems also support a role for SNAI2 in breast cancer cell stemness, basal phenotype, and metastasis18, 21. Thus, inhibition of SNAI2 expression by C/EBPδ may contribute to the development of luminal cancers while also attenuating their progression. For example, we found that C/EBPδ promotes expression of the CDK inhibitor CDKN1A through inhibition of SNAI2. CDKN1A has been correlated with hormone receptor positive and node-negative status of breast cancers41 and CDKN1A expression can predict recurrence-free survival of ER+ cancer patients31. Taken together, these pathways, which may also cooperate with other C/EBPδ regulated genes, provide a plausible mechanism for the correlation of C/EBPδ with better prognosis in ER+ breast cancer.
Our findings are in apparent conflict with C/EBPδ’s role as a pro-inflammatory molecule, in particular in the context of the cytokine IL-6. C/EBPδ and C/EBPβ activate the IL-6 gene and are in turn activated by IL-6 signaling39. Both systemic inflammation and tumor inflammation are associated with worse prognosis for breast cancer patients. Mechanistically, IL-6 promotes breast cancer stem cells, tumor escape from immune-surveillance, and treatment resistance28, 49. In contrast, IL-6 can also act as an intra-tumoral anti-inflammatory agent and promote the anti-tumor immune response17, and a few studies found a correlation of IL-6 in breast cancer with better prognosis27.
We presented data indicating that in ER+ breast cancer specifically, IL6 mRNA levels correlated with lower risk of progression, which was further reduced with the additional expression of CEBPD. This data agrees with reports that C/EBPδ mediates growth inhibition of a prostate cancer cell line by IL-646 and of a mouse mammary epithelial cell line by the IL-6 related cytokine oncostatin M24. In contrast, we observed that CEBPB was associated with greater risk of progression and abrogated any “benefit” of IL6 gene expression. This result agrees with many studies that have shown a role for C/EBPβ – and in particular of its truncated isoform – in the progression of breast cancer7, 57. The complexity of cell types within tumors and tumor cell heterogeneity are important aspects of tumor development and progression. We do not know which cells express IL6 and/or CEBPD in these tumors, nor whether the proteins are present. Therefore, these results do not permit conclusions on gene function at present, but indicate that pathways that allow more CEBPD than CEBPB gene activation in combination with IL-6 are beneficial to the patient. Given the current attempts to target IL-6 in cancer26, our results indicate that further investigations into the precise role of these molecules and their downstream effectors in the context of breast cancer subtypes are warranted.
Materials And Methods
Reagents and Cell lines
All reagents and antibodies were commercially available as described in Supplementary Materials and Methods.
Transient transfections and RNA interference
Cells were nucleofected with siRNAs according to the manufacturer’s instructions (Amaxa Biosystems/Lonza); see Supplementary Materials and Methods for details and sequences. The effect of siRNA on protein/RNA expression was assessed two days after nucleofection unless indicated otherwise
Cell behavior assays
Cell migration was quantified 8 h after removal of culture inserts (Ibidi). Invasive cells were quantified through migration across matrigel after 36 h of culture in Boyden chambers. Colony formation was assessed after two weeks of culture in 0.3% agarose. Cell proliferation/viability was assessed with a microplate reader after staining with AlamarBlue (Invitrogen). Data are presented as mean ± S.D.. For details see Supplementary Materials and Methods.
Protein and RNA Analysis, Chromatin Immunoprecipitation Assay, and Immunohistochemistry
Standard protocols were applied (see Supplementary Materials and Methods). Quantitative data are shown as the mean±S.D. unless indicated otherwise, and were analyzed by the two-tailed unequal variance t-test.
mRNA-Seq Analysis
Total RNAs from MCF-7 cells were purified after 48 h silencing with siRNAs against CEBPD or control. Library preparation was by standard protocol. Samples were run on a HiSeq2000 and analyzed as described in Supplementary Materials and Methods. The data are available at the NCBI Gene Expression Omnibus under accession number GSE69604: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=yvsjaqkgfdsrfmf&acc=GSE69604
Patient Cohorts and Tissue microarray analysis
TMA-1: A series of 140 invasive breast carcinomas, as described previously13, represented in a tissue microarray by a 0.6 mm core yielded data for C/EBPδ in 79 cases. The degree of nuclear C/EBPδ staining in carcinoma cells of each tissue core was assessed using an Allred scoring system encompassing staining intensity (0 = none, 1 = weak, 2 = moderate, 3 = strong) and the proportion of expressing cells (0 = 0%, 1 = <1%, 2 = 1–10%, 3 = 11–33%, 4 = 34–66%, 5 = >66%); the sum of the scores produced the final Allred score ranging from 0 to 8. Data relating to immunostaining of other markers was available as H-score, also a composite of staining intensity and the percentage of stained cells, ranging from 0 – 300. ER status was defined by the diagnostic assay at the source hospital.
Associations with dichotomous variables were assessed using Fisher’s exact test. C/EBPδ expression was dichotomized for some analyses; cases with an Allred score of >0 were deemed positive. Correlations between ordinal variables and ordinal versus continuous variables were assessed using Spearman’s correlation coefficient. Association with 10-year breast cancer-specific survival was assessed using a Cox-proportional hazards model providing a hazard ratio and 95% confidence interval. The Log-Rank test was used to compare survival probability between groups in Kaplan-Meier survival plots. All statistical analyses were conducted in Intercooled Stata version 11.1 (Stata Corp., Texas, USA).
TMA-2: The cohort investigated with TMA-2 was as described44: 299 tumors (mean size 25 mm, 84% ductal carcinoma in situ) were analyzed from pre-menopausal breast cancer patients (mean age 44 years, 72% lymph node positive) with no adjuvant treatment or 2-year tamoxifen treatment, and long follow-up time in the form of recurrence free survival. The frequency of nuclear and cytoplasmic C/EBPδ staining in carcinoma cells was evaluated separately as 0% (0), 50% (1), and >50% (2) and were distributed as follows for nuclear stain: score 0, 134 samples (45%); score 1, 83 samples, (28%); score 2, 82 samples (27%). After initial analyses, samples scoring 0 or 1 for C/EBPδ were grouped together for the survival analysis due to similar survival plots in Kaplan Meier analyses. All statistical tests were two-sided, and the calculations were done in Statistical Package for the Social Sciences version 17.0 (SPSS, Chicago, IL). ER+ status was defined as >10% nuclei positive for ER.
All human subject studies were approved by respective institutional review boards.
Supplementary Material
Acknowledgments
We are thankful to the Laboratory Animal Sciences Program (Leidos Biomedical Research, Inc., Frederick National Laboratory for Cancer Research) for excellent support, especially Donna Butcher and Glenn Summers for superb services, and Bao Tran and Jyoti Shetty (Leidos Biomedical Research, Inc.) for mRNA-Seq data. The authors thank Elise Nilsson for excellent technical assistance. We thank student interns Katherine L. Zhou and Yasmin Y. Lachir for their valuable contributions, Linda Miller for preparation of plasmids, and Allen Kane and Joseph Meyer (Leidos Biomedical Research, Inc.) for preparing the figures for publication.
This research was supported by the Intramural Research Program of the NIH, Frederick National Lab, National Cancer Institute and in part with Federal Funds from the Frederick National Laboratory (NIH) under contract no. HHSN261200800001E. D.Y.M.V. was supported in part by a scholarship from The National Council on Science and Technology (CONACYT), Mexico.
The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
SUPPLEMENTARY INFORMATION accompanies the paper on the Oncogene website: http//www.nature.com/onc.
References
- 1.Alves CC, Carneiro F, Hoefler H, Becker KF. Role of the epithelial-mesenchymal transition regulator Slug in primary human cancers. Front Biosci (Landmark Ed) 2009;14:3035–3050. doi: 10.2741/3433. [DOI] [PubMed] [Google Scholar]
- 2.Bailey CK, Mittal MK, Misra S, Chaudhuri G. High motility of triple-negative breast cancer cells is due to repression of plakoglobin gene by metastasis modulator protein SLUG. J Biol Chem. 2012;287:19472–19486. doi: 10.1074/jbc.M112.345728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balamurugan K, Wang JM, Tsai HH, Sharan S, Anver M, Leighty R, et al. The tumour suppressor C/EBPdelta inhibits FBXW7 expression and promotes mammary tumour metastasis. Embo J. 2010;29:4106–4117. doi: 10.1038/emboj.2010.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balamurugan K, Sharan S, Klarmann KD, Zhang Y, Coppola V, Summers GH, et al. FBXW7alpha attenuates inflammatory signalling by downregulating C/EBPdelta and its target gene Tlr4. Nature communications. 2013;4:1662. doi: 10.1038/ncomms2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balamurugan K, Sterneck E. The Many Faces of C/EBPdelta and their Implications in Inflammation and Cancer. Int J Biol Sci. 2013;9:917–933. doi: 10.7150/ijbs.7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baranda-Avila N, Mendoza-Rodriguez CA, Morimoto S, Camacho-Arroyo I, Guerra-Araiza C, Langley E, et al. Agonistic activity of ICI 182 780 on activation of GSK 3beta/AKT pathway in the rat uterus during the estrous cycle. Steroids. 2013;78:717–725. doi: 10.1016/j.steroids.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Begay V, Smink JJ, Loddenkemper C, Zimmermann K, Rudolph C, Scheller M, et al. Deregulation of the endogenous C/EBPbeta LIP isoform predisposes to tumorigenesis. J Mol Med (Berl) 2015;93:39–49. doi: 10.1007/s00109-014-1215-5. [DOI] [PubMed] [Google Scholar]
- 8.Bermejo-Rodriguez C, Perez-Caro M, Perez-Mancera PA, Sanchez-Beato M, Piris MA, Sanchez-Garcia I. Mouse cDNA microarray analysis uncovers Slug targets in mouse embryonic fibroblasts. Genomics. 2006;87:113–118. doi: 10.1016/j.ygeno.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Cardona-Gomez P, Perez M, Avila J, Garcia-Segura LM, Wandosell F. Estradiol inhibits GSK3 and regulates interaction of estrogen receptors, GSK3, and beta-catenin in the hippocampus. Mol Cell Neurosci. 2004;25:363–373. doi: 10.1016/j.mcn.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 11.Chaudary N, Hill RP. Hypoxia and metastasis in breast cancer. Breast Dis. 2006;26:55–64. doi: 10.3233/bd-2007-26105. [DOI] [PubMed] [Google Scholar]
- 12.Chen JC, Alvarez MJ, Talos F, Dhruv H, Rieckhof GE, Iyer A, et al. Identification of causal genetic drivers of human disease through systems-level analysis of regulatory networks. Cell. 2014;159:402–414. doi: 10.1016/j.cell.2014.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin SF, Teschendorff AE, Marioni JC, Wang Y, Barbosa-Morais NL, Thorne NP, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. doi: 10.1186/gb-2007-8-10-r215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cicenas J, Valius M. The CDK inhibitors in cancer research and therapy. J Cancer Res Clin Oncol. 2011;137:1409–1418. doi: 10.1007/s00432-011-1039-4. [DOI] [PubMed] [Google Scholar]
- 15.Cobaleda C, Perez-Caro M, Vicente-Duenas C, Sanchez-Garcia I. Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annu Rev Genet. 2007;41:41–61. doi: 10.1146/annurev.genet.41.110306.130146. [DOI] [PubMed] [Google Scholar]
- 16.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dethlefsen C, Hojfeldt G, Hojman P. The role of intratumoral and systemic IL-6 in breast cancer. Breast Cancer Res Treat. 2013;138:657–664. doi: 10.1007/s10549-013-2488-z. [DOI] [PubMed] [Google Scholar]
- 18.Dhasarathy A, Phadke D, Mav D, Shah RR, Wade PA. The transcription factors Snail and Slug activate the transforming growth factor-beta signaling pathway in breast cancer. PLoS One. 2011;6:e26514. doi: 10.1371/journal.pone.0026514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eroles P, Bosch A, Perez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38:698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Grisouard J, Medunjanin S, Hermani A, Shukla A, Mayer D. Glycogen synthase kinase-3 protects estrogen receptor alpha from proteasomal degradation and is required for full transcriptional activity of the receptor. Mol Endocrinol. 2007;21:2427–2439. doi: 10.1210/me.2007-0129. [DOI] [PubMed] [Google Scholar]
- 21.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagedorn M, Delugin M, Abraldes I, Allain N, Belaud-Rotureau MA, Turmo M, et al. FBXW7/hCDC4 controls glioma cell proliferation in vitro and is a prognostic marker for survival in glioblastoma patients. Cell Div. 2007;2:9. doi: 10.1186/1747-1028-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hefti MM, Hu R, Knoblauch NW, Collins LC, Haibe-Kains B, Tamimi RM, et al. Estrogen receptor negative/progesterone receptor positive breast cancer is not a reproducible subtype. Breast Cancer Res. 2013;15:R68. doi: 10.1186/bcr3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutt JA, DeWille JW. Oncostatin M induces growth arrest of mammary epithelium via a CCAAT/enhancer-binding protein delta-dependent pathway. Mol Cancer Ther. 2002;1:601–610. [PubMed] [Google Scholar]
- 25.Ikezoe T, Gery S, Yin D, O’Kelly J, Binderup L, Lemp N, et al. CCAAT/enhancer-binding protein delta: a molecular target of 1,25-dihydroxyvitamin D3 in androgen-responsive prostate cancer LNCaP cells. Cancer Res. 2005;65:4762–4768. doi: 10.1158/0008-5472.CAN-03-3619. [DOI] [PubMed] [Google Scholar]
- 26.Jones SA, Scheller J, Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J Clin Invest. 2011;121:3375–3383. doi: 10.1172/JCI57158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knupfer H, Preiss R. Significance of interleukin-6 (IL-6) in breast cancer (review) Breast Cancer Res Treat. 2007;102:129–135. doi: 10.1007/s10549-006-9328-3. [DOI] [PubMed] [Google Scholar]
- 28.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121:3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li YQ, et al. Estrogen receptor alpha mediates proliferation of breast cancer MCF-7 cells via a p21/PCNA/E2F1-dependent pathway. FEBS J. 2014;281:927–942. doi: 10.1111/febs.12658. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, Li J, Li H, Li A, Liu B, Han L. A comprehensive analysis of candidate genes and pathways in pancreatic cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:1849–1857. doi: 10.1007/s13277-014-2787-y. [DOI] [PubMed] [Google Scholar]
- 31.Lyng MB, Laenkholm AV, Tan Q, Vach W, Gravgaard KH, Knoop A, et al. Gene expression signatures that predict outcome of tamoxifen-treated estrogen receptor-positive, high-risk, primary breast cancer patients: a DBCG study. PLoS One. 2013;8:e54078. doi: 10.1371/journal.pone.0054078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madden SF, Clarke C, Gaule P, Aherne ST, O’Donovan N, Clynes M, et al. BreastMark: an integrated approach to mining publicly available transcriptomic datasets relating to breast cancer outcome. Breast Cancer Res. 2013;15:R52. doi: 10.1186/bcr3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinson HA, Jindal S, Durand-Rougely C, Borges VF, Schedin P. Wound healing-like immune program facilitates postpartum mammary gland involution and tumor progression. Int J Cancer. 2015;136:1803–1813. doi: 10.1002/ijc.29181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milde-Langosch K, Loning T, Bamberger AM. Expression of the CCAAT/enhancer-binding proteins C/EBPalpha, C/EBPbeta and C/EBPdelta in breast cancer: correlations with clinicopathologic parameters and cell-cycle regulatory proteins. Breast Cancer Res Treat. 2003;79:175–185. doi: 10.1023/a:1023929504884. [DOI] [PubMed] [Google Scholar]
- 35.Naderi A, Teschendorff AE, Barbosa-Morais NL, Pinder SE, Green AR, Powe DG, et al. A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene. 2007;26:1507–1516. doi: 10.1038/sj.onc.1209920. [DOI] [PubMed] [Google Scholar]
- 36.Pawar SA, Sarkar TR, Balamurugan K, Sharan S, Wang J, Zhang Y, et al. C/EBP{delta} targets cyclin D1 for proteasome-mediated degradation via induction of CDC27/APC3 expression. Proc Natl Acad Sci U S A. 2010;107:9210–9215. doi: 10.1073/pnas.0913813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips S, Prat A, Sedic M, Proia T, Wronski A, Mazumdar S, et al. Cell-State Transitions Regulated by SLUG Are Critical for Tissue Regeneration and Tumor Initiation. Stem Cell Reports. 2014;2:633–647. doi: 10.1016/j.stemcr.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter DA, Krop IE, Nasser S, Sgroi D, Kaelin CM, Marks JR, et al. A sage (serial analysis of gene expression) view of breast tumor progression. Cancer Res. 2001;61:5697–5702. [PubMed] [Google Scholar]
- 39.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranger JJ, Levy DE, Shahalizadeh S, Hallett M, Muller WJ. Identification of a Stat3-dependent transcription regulatory network involved in metastatic progression. Cancer Res. 2009;69:6823–6830. doi: 10.1158/0008-5472.CAN-09-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed W, Florems VA, Holm R, Hannisdal E, Nesland JM. Elevated levels of p27, p21 and cyclin D1 correlate with positive oestrogen and progesterone receptor status in node-negative breast carcinoma patients. Virchows Arch. 1999;435:116–124. doi: 10.1007/s004280050408. [DOI] [PubMed] [Google Scholar]
- 42.Resemann HK, Watson CJ, Lloyd-Lewis B. The Stat3 paradox: a killer and an oncogene. Mol Cell Endocrinol. 2014;382:603–611. doi: 10.1016/j.mce.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 43.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6:e17911. doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryden L, Jonsson PE, Chebil G, Dufmats M, Ferno M, Jirstrom K, et al. Two years of adjuvant tamoxifen in premenopausal patients with breast cancer: a randomised, controlled trial with long-term follow-up. Eur J Cancer. 2005;41:256–264. doi: 10.1016/j.ejca.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez-Garcia F, Villagrasa P, Matsui J, Kotliar D, Castro V, Akavia UD, et al. Integration of genomic data enables selective discovery of breast cancer drivers. Cell. 2014;159:1461–1475. doi: 10.1016/j.cell.2014.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanford DC, DeWille JW. C/EBPdelta is a downstream mediator of IL-6 induced growth inhibition of prostate cancer cells. Prostate. 2005;63:143–154. doi: 10.1002/pros.20159. [DOI] [PubMed] [Google Scholar]
- 47.Sarkar TR, Sharan S, Wang J, Pawar SA, Cantwell CA, Johnson PF, et al. Identification of a Src tyrosine kinase/SIAH2 E3 ubiquitin ligase pathway that regulates C/EBPdelta expression and contributes to transformation of breast tumor cells. Mol Cell Biol. 2012;32:320–332. doi: 10.1128/MCB.05790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamos JL, Weis WI. The beta-catenin destruction complex. Cold Spring Harb Perspect Biol. 2013;5:a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi K, Karin M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin Immunol. 2014;26:54–74. doi: 10.1016/j.smim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Thangaraju M, Rudelius M, Bierie B, Raffeld M, Sharan S, Hennighausen L, et al. C/EBPdelta is a crucial regulator of pro-apoptotic gene expression during mammary gland involution. Development. 2005;132:4675–4685. doi: 10.1242/dev.02050. [DOI] [PubMed] [Google Scholar]
- 51.van Roosmalen W, Le Devedec SE, Golani O, Smid M, Pulyakhina I, Timmermans AM, et al. Tumor cell migration screen identifies SRPK1 as breast cancer metastasis determinant. J Clin Invest. 2015;125:1648–1664. doi: 10.1172/JCI74440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang YH, Wu WJ, Wang WJ, Huang HY, Li WM, Yeh BW, et al. CEBPD amplification and overexpression in urothelial carcinoma: a driver of tumor metastasis indicating adverse prognosis. Oncotarget. 2015 doi: 10.18632/oncotarget.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, Inuzuka H, Zhong J, Wan L, Fukushima H, Sarkar FH, et al. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett. 2012;586:1409–1418. doi: 10.1016/j.febslet.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei G, Wang Y, Zhang P, Lu J, Mao JH. Evaluating the prognostic significance of FBXW7 expression level in human breast cancer by a meta-analysis of transcriptional profiles. J Cancer Sci Ther. 2012;4:299–305. doi: 10.4172/1948-5956.1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye Y, Xiao Y, Wang W, Yearsley K, Gao JX, Shetuni B, et al. ERalpha signaling through slug regulates E-cadherin and EMT. Oncogene. 2010;29:1451–1462. doi: 10.1038/onc.2009.433. [DOI] [PubMed] [Google Scholar]
- 56.Yu X, Si J, Zhang Y, Dewille JW. CCAAT/Enhancer Binding Protein-delta (C/EBP-delta) regulates cell growth, migration and differentiation. Cancer Cell Int. 2010;10:48. doi: 10.1186/1475-2867-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zahnow CA. CCAAT/enhancer-binding protein beta: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med. 2009;11:e12. doi: 10.1017/S1462399409001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang F, Phiel CJ, Spece L, Gurvich N, Klein PS. Inhibitory phosphorylation of glycogen synthase kinase-3 (GSK-3) in response to lithium. Evidence for autoregulation of GSK-3. J Biol Chem. 2003;278:33067–33077. doi: 10.1074/jbc.M212635200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.