Abstract
To delineate the genetic determinants associated with retinitis pigmentosa (RP), a hereditary retinal disorder, we recruited four large families manifesting cardinal symptoms of RP. We localized these families to regions on the human genome harboring the α and β subunits of phosphodiesterase 6 and identified mutations that were absent in control chromosomes. Our data suggest that mutations in PDE6A and PDE6B are responsible for the retinal phenotype in these families.
The diagnosis of retinitis pigmentosa (RP) encompasses a group of hereditary retinal disorders characterized by progressive degeneration of rod photoreceptors followed by the degeneration of cone photoreceptors. The initial loss of rod cells causes night blindness and constricts the visual field, while the subsequent loss of cone cells results in a total loss of vision.1 Though it affects one in 4,000, RP is clinically variable, so it is not surprising that it has been found to be genetically heterogeneous as well.2,3 Clinical manifestation of the disease includes pigmentary deposits in the retina, waxy disc pallor and attenuation of retinal blood vessels.1 In addition, affected individuals often have severely abnormal or undetectable electroretinographic responses, even in the early stage of the disease.1
RP is a genetically heterogeneous disorder that manifests as an autosomal dominant, autosomal recessive and X-linked trait. Phosphodiesterase 6 (PDE6) is one of the critical enzymes of the visual phototransduction cascade that facilitates the hydrolysis of cyclic guanine monophosphate, a reaction that maintains physiological levels of cyclic guanine monophosphate in the vertebrate retina. The PDE6 enzyme is composed of two catalytic subunits, PDE6A (88 kDa) and PDE6B (84 kDa), and a regulatory/inhibitory subunit, PDE6G (11 kDa).4 Reports on the pathogenesis of RP have previously implicated mutations in genes encoding PDE6A (OMIM: 180071) and PDE6B (OMIM: 180072) (Supplementary Tables S5 and S6).
In the pursuit of determining the genetic basis of autosomal recessive RP, we have collected a large cohort of volunteer participants from highly inbred families, all affected by retinal dystrophies. We previously reported causal mutations in PDE6A and PDE6B in multiple families that accounted for 2–3% of our large cohort, suggesting that mutations in these subunits of PDE6 are a significant contributor to the genetic load of autosomal recessive RP in the Pakistani population.5–7 Over the past 5 years, we have recruited additional familial cases of autosomal recessive RP, bringing the total number of our cohort to >350 families. Here we report novel mutations identified by our ongoing research.
The four families reported in this study were recruited from the Punjab province of Pakistan (Figure 1). Approval was granted from the Institutional Review Boards of the National Centre of Excellence in Molecular Biology in Lahore, Pakistan, the National Eye Institute in Bethesda, MD, USA, and Johns Hopkins University in Baltimore, MD, USA. Each individual participating in this study was informed and provided written consent in accordance with the Declaration of Helsinki. A detailed medical history was obtained by interviewing family members. Color vision was assessed with Ishihara charts. Fundoscopy was performed at the Layton Rehmatulla Benevolent Trust Hospital (Lahore, Pakistan). Electroretinographic measurements were recorded with equipment manufactured by LKC (Gaithersburg, MD, USA). Dark-adapted rod responses were measured with incident flash attenuated by −25 dB, whereas rod-cone responses were measured at 0 dB. The 30 Hz flicker responses were recorded at 0 dB with a background illumination of 17–34 cd/m2.
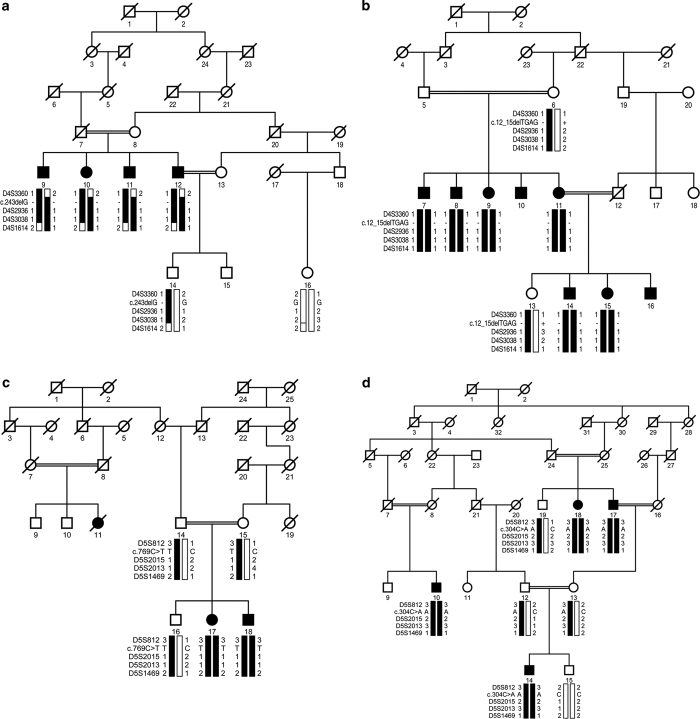
Figure 1.
Pedigree drawings with haplotypes formed from alleles of chromosome 4p and 5q microsatellite markers. (a) PKRP264, (b) PKRP336, (c) PKRP345 and (d) PKRP360. Alleles constituting the risk haplotype (segregating with the retinitis pigmentosa (RP) phenotype) are shaded black, whereas alleles not segregating with RP are shown in white. Square: male; circle: female; filled symbol: affected individual; double line between symbols: consanguineous mating; diagonal line through symbol: deceased individual.
Each affected member of the participating families was thoroughly evaluated to confirm that their ophthalmological symptoms were caused by RP (Supplementary Table S1). In early childhood, affected patients exhibited night blindness, a common indicator of RP. Fundus examinations documented arteriolar constriction, waxy pallor of the optic disc and pigmentation on the peripheral and central retina—all cardinal symptoms of RP that substantiated diagnoses of the affected subjects (Supplementary Figure S1). Furthermore, both rod and cone responses were absent when measured by electroretinography; scotopic and photopic electroretinographic responses were highly reduced or absent (Figure 2).
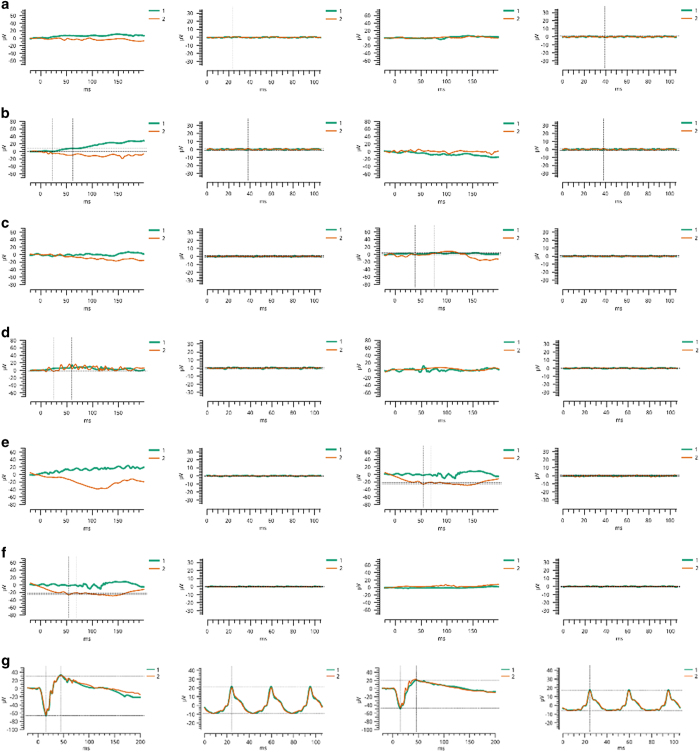
Figure 2.
Electroretinographic recordings of individuals diagnosed with retinitis pigmentosa. Panels (from left to right) are combined rod and cone response and cone flicker response of oculus dexter (OD) and oculus sinister (OS). (a) Affected individual 17 and (b) affected individual 18 of PKRP345, (c) affected individual 9, (d) affected individual 10, (e) affected individual 11, (f) affected individual 12 and (g) unaffected control individual 14 of PKRP264. Affected individuals exhibit no detectable electroretinographic responses, whereas the unaffected individual exhibits normal a- and b-waves, indicating normal rod and cone function.
The genomic DNA used in all experiments was extracted from the white blood cells of whole-blood samples with a modified procedure as described.8 We measured the concentration of the DNA with a SmartSpec Plus Spectrophotometer (Bio-Rad, Hercules, CA, USA). Short tandem repeat markers spanning either PDE6A or PDE6B were selected to perform exclusion analysis. We used Gene Mapper software (Applied Biosystems, Foster City, CA, USA) to assign genotypes and performed linkage analysis with alleles identified by exclusion analysis.
These analyses localized PKRP264 and PKRP336 to the PDE6B locus on chromosome 4p and PKRP345 and PKRP360 to the PDE6A locus on chromosome 5q. Maximum two-point logarithm of odds (LOD) scores of 2.04 and 3.10 were obtained with the alleles D4S2936 and D4S3038, respectively, at θ=0 for PKRP264, and scores of 3.04, 2.88 and 1.86 were obtained with D4S2936, D4S3038 and D4S1614, respectively, at θ=0 for PKRP336 (Supplementary Table S2). Likewise, LOD scores of 1.79, 2.02 and 1.21 were obtained with the alleles D5S2015, D5S2013 and D5S1469, respectively, at θ=0 for PKRP345, and scores of 2.33, 2.79, 2.21 and 1.42 were obtained with D5S812, D5S2015, D5S2013 and D5S1469, respectively, at θ=0 for PKRP360 (Supplementary Table S2).
Next we sequenced all coding exons, exon–intron boundaries and the 5′ and 3′ untranslated regions of PDE6B for PKRP264 and PKRP336 and PDE6A for PKRP345 and PKRP360. Primer pairs for individual exons of PDE6A and PDE6B were designed using Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/). The primer sequences and annealing temperatures are provided in Supplementary Tables S3 and S4. Sequencing reactions were completed as described,6,7 and sequencing results were assembled with the ABI PRISM sequencing analysis software, version 3.7, and analyzed with SeqScape software (Applied Biosystems).
Sanger sequencing identified novel deletion mutations in PDE6B: c.243delG (p.R82Afs68*) and c.12_15delTGAG (p.S4Rfs23*) in PKRP264 and PKRP336, respectively (Supplementary Figures S2 and S3). Likewise, previously reported nonsense and missense mutations were identified in PDE6A: c.769C>T (p.R257*) and c.304C>A (p.R102S) in PKRP345 and PKRP360, respectively (Supplementary Figures S4 and S5). All mutations segregated with the disease phenotype in their respective families and were absent from ethnically matched control chromosomes. Additionally, these variants were not present in the NHLBI exome variant server, 1000 Genomes or the dbSNP database.
Next we evaluated the evolutionary conservation of R102 in other PDE6A orthologs, which illustrated that the amino-acid residue R102 is highly conserved (data not shown). We next examined the possible impact on the PDE6A protein with the SIFT and PolyPhen-2 algorithms. SIFT prediction suggested that the R102S substitution would not be tolerated by the native three-dimensional structure of PDE6A. Likewise, position-specific score differences obtained from PolyPhen-2 suggested that the R102S substitution would be probably damaging, with a score of 0.993 (sensitivity: 0.70; specificity: 0.97).
In conclusion, we report four causal mutations present in consanguineous RP families recruited from the Punjab province of Pakistan. PDE6 is necessary for regulating the concentrations of cyclic guanine monophosphate in the retina and mutations in its subunits have been associated with RP, including a pathogenic connection between PDE6B and congenital stationary night blindness. We have previously reported mutations in both PDE6A and PDE6B that are responsible for RP in our cohort of Pakistani families.5–7 We have recruited >350 families, and of these, we have now localized 11 familial cases to PDE6 genes (PDE6A and PDE6B), suggesting that they contribute approximately 3% of the total genetic load of autosomal recessive RP in our cohort. Determining the genetic cause of RP contributes to our understanding of the disease itself and of the normal physiological function of the retina. A better understanding of the physiology and its underlying pathology will hopefully lead to improved diagnosis and treatment of RP.
Acknowledgments
We are thankful to all family members for their participation in this study. This study was supported in part by the Higher Education Commission, Islamabad, Pakistan and by the National Eye Institute Grant R01EY021237-01 (RA and SAR).
Footnotes
The authors declare no conflict of interest.
References
- Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet 2006; 368: 1795–1809. [DOI] [PubMed] [Google Scholar]
- Berson EL. Retinitis pigmentosa. The Friedenwald Lecture. Invest Ophthalmol Vis Sci 1993; 34: 1659–1676. [PubMed] [Google Scholar]
- Bird AC. Retinal photoreceptor dystrophies LI. Edward Jackson Memorial Lecture. Am J Ophthalmol 1995; 119: 543–562. [DOI] [PubMed] [Google Scholar]
- Fung BK, Young JH, Yamane HK, Griswold-Prenner I. Subunit stoichiometry of retinal rod cGMP phosphodiesterase. Biochemistry 1990; 29: 2657–2664. [DOI] [PubMed] [Google Scholar]
- Riazuddin SA, Zulfiqar F, Zhang Q, Yao W, Li S, Jiao X et al. Mutations in the gene encoding the alpha-subunit of rod phosphodiesterase in consanguineous Pakistani families. Mol Vis 2006; 12: 1283–1291. [PubMed] [Google Scholar]
- Ali S, Riazuddin SA, Shahzadi A, Nasir IA, Khan SN, Husnain T et al. Mutations in the beta-subunit of rod phosphodiesterase identified in consanguineous Pakistani families with autosomal recessive retinitis pigmentosa. Mol Vis 2011; 17: 1373–1380. [PMC free article] [PubMed] [Google Scholar]
- Khan SY, Ali S, Naeem MA, Khan SN, Husnain T, Butt NH et al. Splice-site mutations identified in PDE6A responsible for retinitis pigmentosa in consanguineous Pakistani families. Mol Vis 2015; 21: 871–882. [PMC free article] [PubMed] [Google Scholar]
- Khan SY, Vasanth S, Kabir F, Gottsch JD, Khan AO, Chaerkady R et al. FOXE3 contributes to Peters anomaly through transcriptional regulation of an autophagy-associated protein termed DNAJB1. Nat Commun 2016; 7: 10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Data Citations
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.