Abstract
Humans have more copies of amylase genes than other primates. It is still poorly understood, however, when the copy number expansion occurred and whether its spread was enhanced by selection. Here we assess amylase copy numbers in a global sample of 480 high coverage genomes and find that regions flanking the amylase locus show notable depression of genetic diversity both in African and non-African populations. Analysis of genetic variation in these regions supports the model of an early selective sweep in the human lineage after the split of humans from Neanderthals which led to the fixation of multiple copies of AMY1 in place of a single copy. We find evidence of multiple secondary losses of copy number with the highest frequency (52%) of a deletion of AMY2A and associated low copy number of AMY1 in Northeast Siberian populations whose diet has been low in starch content.
Alpha-amylases, hereafter referred to as amylases, are expressed in the salivary glands and pancreas by genes of the AMY gene family which in humans has a variable number of gene copies that co-localize as a tight cluster in a ~200 kb region on the reference assembly on chromosome 1. Copies of the AMY1 gene are expressed in the salivary glands and allow pre-ingestion breakdown of carbohydrates into simple sugars. AMY2 copies are expressed in the pancreas and serve the same purpose in the duodenum. Compared to other great apes the AMY locus is significantly expanded in humans, with high levels of copy number variation (CNV) found within and among populations1,2,3,4,5,6. Higher copy number of the AMY1 gene has been shown by multiple studies to cause increased expression of amylase in the saliva which is likely to enable more efficient digestion of starch-rich foods1,7. However, the broader phenotypic effect of amylase CNV is still poorly understood as the initial reports for AMY1 copy number association with body mass index and obesity risk7,8 have failed replication4.
It has been proposed that the AMY1 CNV expansion may be recent and associated with positive selection during the shifts from the low starch diet of hunter-gatherers to the starch-rich diets of Neolithic farmers1. The inference of positive selection was an observation of significantly higher differentiation of AMY1 copy number between high-starch diet Japanese and low-starch diet Yakutians, as compared to the genome-wide range of variation in other copy number and microsatellite loci. However, the qPCR method used in this earlier work to ascertain AMY1 copy numbers has been shown to lack sufficient accuracy3,4. Concerns also remained about the assignment of populations into groups of historically high versus low starch consumers9. Furthermore, ancient DNA evidence shows that an 8,000 year old Mesolithic hunter-gatherer from Loschbour, Luxembourg, already carried 13 copies of the AMY1 gene, which is at the high end of the variation observed in present-day Europeans2. This suggests that selection operating since the agricultural transition cannot fully explain the high number of amylase copies in humans. Despite the evidence for high differentiation between Japanese and Yakutian AMY1 copy numbers, scans of long-range haplotype homozygosity have failed so far to highlight the amylase locus as a significant target of recent positive selection in representative groups of human populations10,11. These varied lines of evidence suggest a complex evolutionary narrative of the AMY locus in humans that is not entirely captured by the model of recent selection during the Holocene period associated with agricultural subsistence.
Amylase copy number amplification may have started early in human evolutionary history, in the Pleistocene period. Because the effectiveness of salivary amylase is significantly enhanced when digesting cooked rather than raw starches12 it has been suggested that multiplication of amylase genes would have become selectively advantageous, as an energy source to sustain the growing brain size, only after cooking had become widespread. Even though the earliest use of fire by hominins has been predicted at 1.9 MYA13, the earliest evidence for consistent and repeated use of fire hearths is more recent and starts to emerge in Middle Pleistocene, only ca 300 KYA14. The relationship between cooked food and hominin brain size has been the focus of extensive anthropological research13,15,16,17. Considering archaeological, anthropological and genetic evidence available so far Hardy et al.9 have hypothesized that the spread of cooking of starch-rich tubers in combination with the gain of higher AMY1 copy numbers by our ancestors may have facilitated the rapid growth of brain size in the early Middle Pleistocene ca 800 KYA, before the split of modern humans and Neanderthals.
As both humans and Neanderthals are characterised by increased brain volumes at the species level, and given the potential overlap between the time depth of cooking and the divergence of these species, ancient DNA of extinct hominins provides a vital additional window into AMY evolution. While most humans today carry more copies of salivary AMY1 genes, Neanderthals, Denisovans, and a small number of present-day Europeans have been found to carry the ancestral number of two AMY1 copies per diploid genome18. With respect to the critical, unanswered questions as to when, where, and why amylase gene numbers increased, two possible scenarios can explain this: a) incomplete lineage sorting at the AMY locus, to explain the co-existence of low and high copy number lineages in two closely related species, and, early Middle Pleistocene amplification of AMY1 copy numbers, as proposed by Hardy et al., in the species that was ancestral to humans and Neanderthals to sustain the energetic needs of the growing brain and, potentially, secondary reduction of AMY1 copy numbers in Neanderthals enabled by their switch to high protein meat diet (Hardy et al.9); or b) late Middle Pleistocene selective sweep at the AMY1 locus after the human-Neanderthal split in conjunction with a divergence in associated food technologies and followed by secondary losses of AMY1 variation in some human populations. Neither of these scenarios precludes later selection associated with agricultural subsistence.
Patterns of genetic variation accumulated at the AMY locus and its flanks ascertained in a range of human populations can be informative for resolving these different possibilities. While previous studies1,3,4 have provided us with the first insights into the global distribution of CNV in the AMY locus, large regions such as Siberia, South East Asia and the Americas are still relatively poorly covered by data. In this study we use high coverage sequence data from 480 individuals sampled across the world to search for evidence of either recent or old signals of positive selection at the amylase locus. We confirm the robustness of estimating AMY1, AMY2A and AMY2B gene copy number from read depth data by genotyping a subset of 68 samples with paralogue ratio tests and microsatellite assay methods. By assessing the diversity, age and affinity to archaic humans of haplotypes associated with high and low AMY1 copy numbers we test whether the haplotypes associated with ancestral copy number in presently living humans can be explained by the retention of ancestral haplotypes, introgression from archaic hominins or convergence due to secondary deletions in the locus.
Results
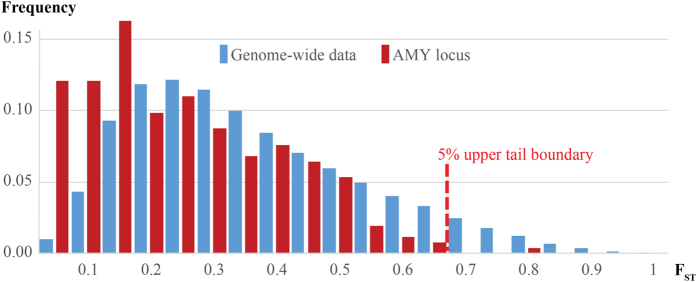
To cast further light on the question of whether the AMY locus has been subject to recent positive selection in humans we first examined the extent of genetic differentiation of human populations at the AMY locus in the context of other genetic loci in a global data set of 480 high coverage genomes19 (Table S1). We compared the maximum Fst values of SNPs in 200 kb regions immediately surrounding the AMY locus against the distribution of similar values obtained from genome wide data and found that genomic regions flanking the amylase genes are characterized globally by a significant (χ2–test, p < 0.001) deficit rather than enrichment of high Fst values: only one pairwise comparison, involving Africans vs Northeast Siberians, yielded an Fst score within the top 5% genome-wide ranks (Fig. 1). Furthermore, the distribution of the maximum Fst values by 200 kb windows from AMY flanks showed a relatively higher (50%) proportion of estimates of low differentiation (Fst < 0.2) compared to the genome-wide average of 26%. These results are unexpected under the scenario of recent selective sweeps and, in particular, do not support the model of Perry et al.1 which would predict high Fst values between our Southeast Asian and Northeast Siberian groups. Furthermore, none of the populations we examined showed signal of recent positive selection at p < 0.01 by the two commonly used haplotype homozygosity tests nSL and iHS (Table 1).
Figure 1. Distribution of estimates of maximum pairwise Fst per 200 kb non-overlapping segment among 12 human population groups.
‘AMY locus’ refers to data from three neighbouring 200 kb segments from the Chr1:103,800,000-104,400,000 region. The distribution of the maximum FST scores of the 200 kb regions is shown by bins of 0.05. The only 5% significant (red dotted line) FST estimate for the AMY locus comes from the African and Northeast Siberian comparison at the Chr1:104,000,000-104,200,000 segment.
Table 1. Genome-wide significance of the window containing AMY genes by three selection tests in 12 regional population groups.
| AMY WINDOW (Chr1 104-104.2 Mb) | iHS | nSL | TD |
|---|---|---|---|
| Africa | 0.99 | 0.97 | 0.0024** |
| Middle East | 0.02>* | 0.12 | 0.03>* |
| South & West Europe | 0.08 | 0.25 | 0.07 |
| East & North Europe | 0.93 | 0.06 | 0.08 |
| Volga Uralic | 0.19 | 0.28 | 0.14 |
| South Asia | 0.16 | 0.23 | 0.04>* |
| West Siberia | 0.88 | 0.41 | 0.24 |
| South Siberia & Mongolia | 0.21 | 0.10 | 0.11 |
| Central Siberia | 0.03>* | 0.12 | 0.19 |
| Northeast Siberia | 0.38 | 0.36 | 0.06 |
| Southeast Asia Mainland | 0.91 | 0.19 | 0.08 |
| Island Southeast Asia | 0.06 | 0.05>* | 0.18 |
*Indicates cases where the AMY genes containing window had the relevant selection test score ranking in the top 5% of the windows (p < 0.05), bold
**indicates cases where the AMY windows was within the top 1% of windows for the given positive selection test (p < 0.01).
To explore the possibility of an older selective sweep we used Tajima’s D20 test which highlighted the AMY region in Africans as a significant candidate of positive selection (p = 0.0024; Table 1). To further assess the magnitude of the signal revealed by the Tajima’s D test we scanned the genomes of the global set of 480 individuals by 50 kb non-overlapping windows for cumulative frequency of derived alleles at sites polymorphic in humans. We found that the 50 kb regions both upstream and downstream of the AMY locus are characterized by unusually low polymorphism in humans when considering the extent of divergence between humans and chimpanzee in the same regions (Fig. 2). The AMY-surrounding 50 kb regions belonged to the bottom 1% of the distribution in terms of the lowest cumulative frequency of derived alleles at polymorphic sites and also had a high concentration of derived alleles that are fixed in all human populations while being found in ancestral state both in the high coverage Altai Neanderthal and Denisovan data. The regions surrounding the AMY cluster of genes also belong to the small fraction of just 0.28% of the genome-wide 50 kb regions that in the 1000 Genomes sequence data21 have a coalescent date which is younger than the human-Neanderthal population split, estimated at 650 KYA18,22, and are characterized by the presence of multiple fixed derived alleles in all humans, which are homozygous and in ancestral state both in the high coverage Altai Neanderthal and Denisovan genomes. Altogether, these findings suggest that human AMY genes cluster in a genomic neighbourhood which has undergone a selective sweep in human ancestry following the split from the Neanderthals.
Figure 2. Distribution of genetic diversity on chromosome 1 in human populations.
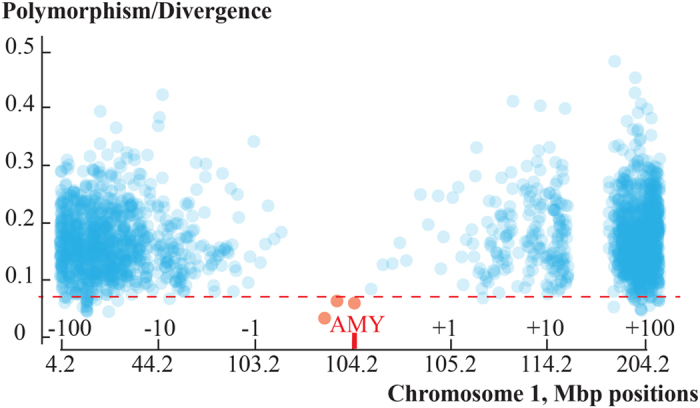
Presented on the y axis is the sum of derived allele frequency over all polymorphic loci in 1000 Genomes African data, estimated by 50,000 bp non-overlapping segments of chromosome 1, relative to the divergence of the human reference sequence from the ancestral sequence (determined by the 6 primate sequence consensus). Red dotted line indicates the 1% cut-off considering the empirical genome-wide distribution. Only those 50,000 bp segments which had >90% sites covered in human, Altai Neanderthal and Denisovan data were considered.
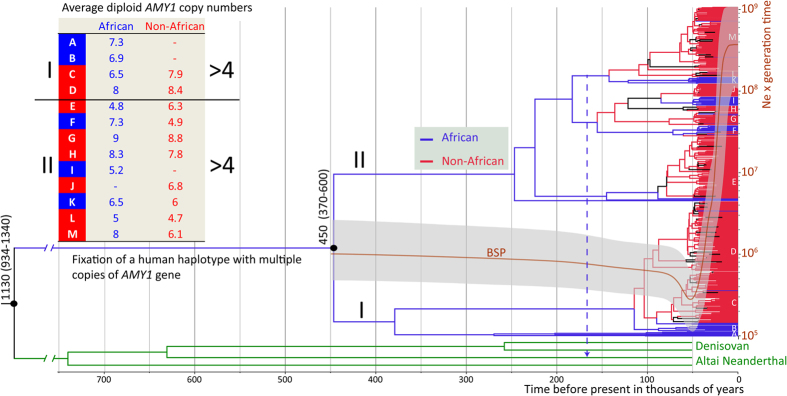
To further study the distribution of genetic variation at the AMY locus we subjected a 66 kb sequence from a high LD region, directly downstream of the AMY1 genes, to phylogenetic analyses using BEAST (Fig. 3). These analyses confirmed the existence of 21 sites where the derived allele is fixed in all humans while the ancestral allele is fixed in archaic humans. Both Altai Neanderthal and Denisovan sequences form a branch that shares a common ancestor with the human lineage at 1130 (95% CI 934–1340) KYA. We estimate the coalescent time of all human lineages at 450 (95% CI 370–600) KYA which postdates the population split of archaic and modern humans at 550–765 KYA18,22. Consistent with a previous report of ancient human introgression into Neanderthals23 we find a short chunk of sequence (Chr1:104,350,432–104,366,871) in the Altai Neanderthal genome which contains, in heterozygous state, derived alleles at four sites out of 11 defining a branch in the phylogeny that is ancestral to human haplotypes K, L and M. These haplotypes have a combined frequency of 24% in our global sample. Because none of the SNPs within this introgressed region that are private to K, L and M haplotypes were found to be shared with the Neanderthal it is likely that the donor haplotype was a distant relative of the K, L and M haplotypes rather than any one of them. The fact that human E-M haplotypes (Fig. 3) are associated each on average with >4 AMY1 copies per diploid genome makes it likely that at least one of the distant ancestors of the Altai Neanderthal also carried multiple copies of the AMY1 genes.
Figure 3. Phylogenetic tree and Bayesian Skyline Plot based on the analyses of a ~66 kb region downstream of the AMY locus with BEAST.
The analyses were restricted to bi-allelic SNP variants from a ~66 kb long high LD region (Chr1:104,303,310-104,369,301) which is downstream of (centromeric to) AMY1C gene (Figure S1). Coalescent time estimates are shown near branching points and assume mutation rate of 5.5 × 10−10 per bp per year34. BSP–Bayesian Skyline Plot; thick brown line shows the median estimates of human effective population size (Ne) x generation time (y-axis) over time (x-axis) as estimated from the ~66 kb region data in the global sample of 480 individuals. The grey shaded area around the brown line shows the 95% higher posterior density intervals of the BSP estimates. A dotted blue arrow highlights the branch of the human phylogeny that has been the likely source of an introgression of a chunk of ~16 kb (Chr1:104,350,432-104,366,871) into the genome of the Altai Neanderthal.
In contrast to the finding of human introgression to the Altai Neanderthal genome, none of the 480 individuals from the global set we have sampled showed evidence of either Neanderthal or Denisovan haplotypes, with evidence of a shared haplotype defined by the co-presence of derived alleles at two or more SNPs. The 16 individuals in our global sample, including 2 African and 14 non-African individuals, who carry the ancestral copy number of two AMY1 genes are therefore unlikely to be explained by the Neanderthal introgression. Furthermore, the sequences of these 16 individuals are not phylogenetically clustered as would be expected if they had low copy number due to the retention of an ancestral haplotype. Instead, they are associated with different haplotypes (D, E, F, I, and M) in the human phylogeny and cluster closely together with lineages characterized by higher AMY1 copy number in extant populations. The Bayesian Skyline Plot analysis (Fig. 3) reveals a notable drop of effective population size (Ne) at the AMY locus between 80–50 KYA followed by a major increase in global Ne consistent with the Out-of-Africa expansion. Given the finding of multiple copies of AMY1, typically higher than 4 per genome, being characteristic of all main branches of the phylogenetic tree, among African and non-African populations (Fig. 3) it is likely that human populations expanding out of Africa already carried multiple copies of the AMY1 genes per chromosome and the finding of occasional cases of two copies per diploid genome are due to secondary losses of AMY1 copy numbers rather than retention of the ancestral state at this locus. Apart from a small number of low frequency haplotypes that are by and large restricted to African (A, B, I, and K) or non-African (J, L) populations all common haplotypes (C-H, M), together accounting for 90% of the global variation, are widely spread in all major continental regions (Table 2, Tables S1 and S2).
Table 2. Geographic distribution of haplotypes in the Chr1:104,303 K-104,369 K region.
| N individuals | Average | Average diploid copy number (σ2) | Haplotype distribution (Heterozygote counting as 1, homozygote as 2 copies) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |latitude| | AMY1 | AMY2A | AMY2B | A | B | C | D | E | F | G | H | I | J | K | L | M | ||
| Africa | 33 | 3.6 | 6.4 (3.4) | 2.3 (0.2) | 2.1 (0.1) | 5 | 10 | 4 | 3 | 4 | 8 | 4 | 3 | 12 | 11 | 2 | ||
| West Asia/Caucasus | 59 | 39.2 | 7.2 (6.2) | 2.2 (0.5) | 2.3 (0.3) | 15 | 25 | 13 | 3 | 10 | 6 | 25 | 21 | |||||
| Southwest Europe | 23 | 47.3 | 6.5 (3.9) | 1.8 (0.2) | 2 (0) | 6 | 5 | 13 | 4 | 6 | 1 | 11 | ||||||
| Northeast Europe | 80 | 57.3 | 6.5 (6.5) | 1.8 (0.3) | 2.1 (0) | 25 | 40 | 36 | 8 | 9 | 5 | 11 | 4 | 22 | ||||
| Volga-Ural region | 25 | 54.9 | 7.4 (5.8) | 1.7 (0.6) | 2 (0) | 10 | 10 | 11 | 3 | 3 | 3 | 10 | ||||||
| South Asia | 29 | 23 | 7.2 (3.3) | 2.2 (0.3) | 2.1 (0.2) | 8 | 16 | 20 | 1 | 2 | 11 | |||||||
| Central Asia | 24 | 40.9 | 7.6 (6.5) | 2.2 (0.5) | 2.1 (0.2) | 9 | 7 | 12 | 2 | 5 | 1 | 12 | ||||||
| West Siberia | 18 | 63.7 | 5.9 (5.9) | 1.4 (0.6) | 2 (0) | 8 | 6 | 9 | 2 | 1 | 2 | 8 | ||||||
| South Siberia | 34 | 52.8 | 7.6 (4.2) | 1.9 (0.3) | 2 (0) | 16 | 20 | 15 | 1 | 2 | 14 | |||||||
| Central Siberia | 27 | 64.1 | 6.5 (13) | 2.1 (0.3) | 2 (0) | 11 | 9 | 16 | 2 | 16 | ||||||||
| Northeast Siberia | 23 | 62.6 | 6.2 (14.8) | 1 (0.6) | 2 (0) | 1 | 7 | 9 | 29 | |||||||||
| South America | 28 | 24.2 | 9.3 (9.3) | 1.9 (0.1) | 2 (0) | 10 | 31 | 15 | ||||||||||
| East Asia | 26 | 22.9 | 7.7 (4.3) | 2 (0) | 2 (0) | 12 | 20 | 11 | 9 | |||||||||
| Island Southeast Asia | 45 | 9.4 | 7.1 (2.8) | 2.1 (0.1) | 2 (0) | 19 | 26 | 16 | 1 | 28 | ||||||||
| Papua New Guinea | 6 | 7 | 6.3 (0.3) | 2.5 (0.3) | 2 (0) | 12 | ||||||||||||
| Total | 480 | 39.5 | 7.1 (6.5) | 1.9 (0.4) | 2.1 (0.1) | 5 | 10 | 154 | 225 | 197 | 21 | 33 | 16 | 12 | 55 | 12 | 12 | 208 |
| 0.01 | 0.01 | 0.16 | 0.23 | 0.21 | 0.02 | 0.03 | 0.02 | 0.01 | 0.06 | 0.01 | 0.01 | 0.22 | ||||||
The AMY copy numbers referred to above were based on the inferences made from the read depths in the high coverage whole genome sequences. The full set of AMY copy number predictions for the global set of 480 human high coverage sequences are presented in Table S1. We estimated the accuracy of the read depth based inference of AMY copy number using high-resolution DNA typing methods3 in a subset of 68 samples. We observed Pearson’s correlation coefficient r = 0.92 between the copy number predictions from read depth data and the paralogue ratio assay results for AMY1, and 0.85 for AMY2A (Table S1). At the same time, we observed errors up to 70% for making exactly the right inference of AMY1 copy when copy numbers were higher than 10, e.g. distinguishing 11 copies from 12. For AMY1 results correlation was at 92% for copies between 2 and 7 but dropped to 75% at copy numbers higher than this. These results show that we can, with reasonably high confidence, estimate the copy number of amylase genes from read depth data whereas the accuracy of distinguishing exact copy numbers in individual cases can be quite low. Nevertheless, consistent with previous studies based on accurate methods of AMY CNV inference3,4 we observe significant bias in our read depth based data towards even copy numbers of AMY1 across the globe (355 even versus 125 odd calls) in all regional populations (Fig. 4B). The excess of even copy number diploid genotype calls is a consequence of haplotypes having predominantly odd number of AMY1 copies. The parity between simultaneously even AMY1 and AMY2A copy numbers is also reproduced here, along with the global average AMY1 copy number of 7.1 in line with previously published mean read depth of 7.4 copies in the HapMap4 and 7.3 in the 1000 Genome Project data3.
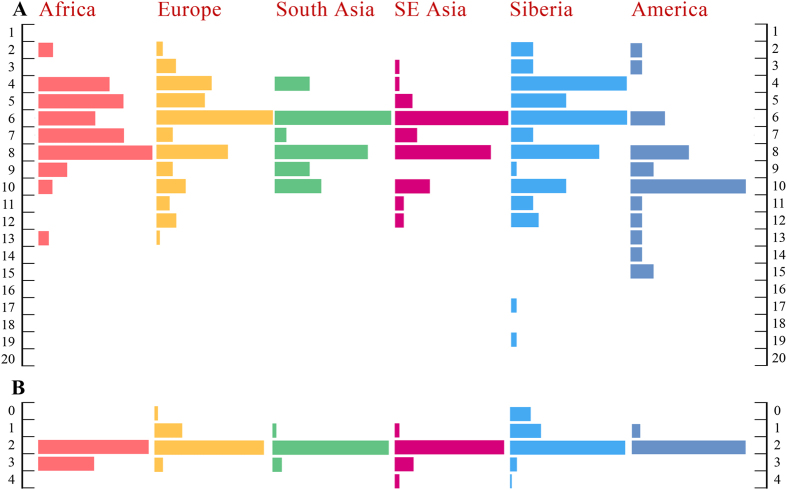
Figure 4. Distributions of AMY1 and AMY2A copy number across the major continental groups.
Pancreatic and salivary amylase genes show different ranges of copy number variation. While the minimum number of salivary AMY1 copies (Fig. 4B) we observed in our global sample was 2, in the case of pancreatic amylases we find a number of individuals (Fig. 4A, Table S1) with just a single or no copies of the AMY2A gene. However, all individuals sampled (Tables S1 and S2) carry at least two pancreatic copies of the AMY2B gene. We observe the highest concentration of the AMY2A deletion in Siberia where among the Northeast Siberians it reaches allele frequencies as high as 52% (Table 3). A large proportion (75/86) of the chromosomes carrying the AMY2A deletion world-wide is clustered within a restricted number of haplotypes, with haplotypes L and M accounting for the highest share (Table 3). These haplotypes are associated with derived alleles at SNP positions rs1930212 and rs72694406 and CNV-haplotype AH2 that has an even number (2 copies) of AMY1, a single copy of AMY2B and a deletion of the AMY2A copy4. Although the direct inference of the phase of AMY1 CNV-haplotype from our short-read data is not possible the presence and wide geographic spread of the same AH2 copy number haplotype in our data is supported by the observation that 34 out of the 40 (85%) carriers of the AMY2A deletion who are heterozygous for the rs72694406 SNP (Table S1) carry an odd number of AMY1 copies per diploid genome while in contrast the frequency of carriers of odd AMY1 copy number among individuals who are homozygous for the ancestral rs72694406-G allele is only 19.4%. These results suggest that majority of the AMY2A deletions in Europe, Central Asia, and Siberia has a single origin. High frequency of the AMY2A deletion in Siberians is also coupled with a notable deficit of individuals with pancreatic amylase duplications relative to populations from lower latitudes (e.g. South and Southeast Asia). Consistent with previous studies3,4 we find that the majority (24/25) of the duplication of both AMY2A and AMY2B genes occur on the background of rs12075086-T and rs79043596-C alleles that define haplotypes I and J (Table S1) which are common in African and West Eurasian populations, respectively (Table 2). We observe weak but significantly negative correlations with latitude both for AMY2A and AMY1 copy numbers (Table 4). These correlations remain highly significant after the exclusion of the Northeast Siberian group with the highest AMY2A deletion frequency (data not shown). Similarly to the trends observed previously in dog AMY2B copy number data24 we find that human populations from higher latitudes also show higher variance of amylase copy numbers (Table 2).
Table 3. Geographic distribution of the AMY2A deletion.
| N | AMY2A deletion | Haplotype distribution of AMY2A deletion by haplotypes | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | frequency | A | B | C | D | E | F | G | H | I | J | K | L | M | |||
| 1 | Africa | 36 | 0 | 0.00 | |||||||||||||
| 2 | West Asia/Caucasus | 110 | 5 | 0.04 | 1 | 4 | |||||||||||
| 3 | South Asia | 58 | 1 | 0.02 | 1 | ||||||||||||
| 4 | Southwest Europe | 46 | 5 | 0.11 | 1 | 1 | 1 | 2 | |||||||||
| 5 | Northeast Europe | 158 | 16 | 0.10 | 2 | 3 | 11 | ||||||||||
| 6 | Central Asia | 48 | 4 | 0.08 | 1 | 3 | |||||||||||
| 7 | Volga Ural region | 44 | 8 | 0.18 | 1 | 3 | 4 | ||||||||||
| 8 | West Siberia | 36 | 10 | 0.28 | 1 | 2 | 7 | ||||||||||
| 9 | South Siberia | 68 | 6 | 0.09 | 1 | 2 | 3 | ||||||||||
| 10 | Central Siberia | 54 | 3 | 0.06 | 3 | ||||||||||||
| 11 | Northeast Siberia | 46 | 24 | 0.52 | 1 | 23 | |||||||||||
| 12 | South America | 56 | 2 | 0.04 | 2 | ||||||||||||
| 13 | East Asia | 52 | 1 | 0.02 | 1 | ||||||||||||
| 14 | Island Southeast Asia | 90 | 1 | 0.01 | 1 | ||||||||||||
| 15 | Papua New Guinea | 6 | 0 | 0.00 | |||||||||||||
| Deletions | 0 | 0 | 8 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 11 | 64 | ||||
| Total count | 2 | 4 | 151 | 218 | 195 | 20 | 30 | 15 | 4 | 54 | 11 | 11 | 199 | ||||
| Deletion frequency | 0.00 | 0.00 | 0.05 | 0.00 | 0.01 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.32 | ||||
Note: N – number of phased chromosomes with information for AMY2A and AMY2B copy numbers.
Table 4. Correlations between amylase copy numbers and geographic latitude and longitude.
| Spearman rank-order | Copy number of | ||
|---|---|---|---|
| correlation coefficients rs | AMY1 | AMY2A | AMY2B |
| Absolute latitude | −0.19 (p = 2 × 10−5) | −0.33 (p < 10−6) | −0.05 (p = 0.29) |
| Longitude | 0.001 (p = 0.98) | 0.001 (p = 0.98) | −0.098 (p = 0.035) |
| AMY2A copy number | 0.21 (p = 9 × 10−6) | ||
| AMY2B copy number | −0.15 (p = 0.0015) | 0.48 (p < 10−6) | |
Note significance for the two-tailed Spearman rank-order correlation test is shown in parenthesis.
Discussion
In this study we have analysed genetic regions surrounding the human AMY cluster for evidence of natural selection and we have found: that human populations within and outside Africa are characterized by unusually low genetic diversity in the flanks of amylase locus relative to other genetic loci genome-wide; a young coalescent date postdating the human-Neanderthal population split; a significant Tajima’s D signal in Africans; and the lack of strong signal of recent positive selection in human population groups we studied. These results are generally in line with Middle Pleistocene9 rather than Holocene1 selection at the AMY locus although the significantly negative correlations between amylase copy numbers and latitude (Table 4), and the lower variance at low latitudes (Table 2) may point to the effect of long term and potentially recent selection that has maintained the copy numbers in populations relying on starch-based diets. Our finding that all human lineages at this locus coalesce to one ancestral lineage after the human-Neanderthal split, together with the evidence of both Neanderthals and Denisovans having the ancestral set of just two AMY1 copies18 gives more credence to the scenario that ties the multiplication of amylase copy numbers in association of a selective sweep specifically to the human ancestors25, rather than within the species ancestral to both humans and Neanderthals9. It also suggests longer food processing sequences, for example involving grinding, leaching and cooking of starch rich tubers in the middle and later Middle Pleistocene.
Our phylogenetic inferences suggest that people moving out of Africa already carried multiple copies of the AMY1 gene. The genome of the 45 KYA Ust’Ishim man from Siberia26 is homozygous for all the four SNPs that define haplotype M (Fig. 3). The core sequence of haplotype M shared by the Ust’Ishim man is identical over the 66 kb sequence range with haploid sequences from 60 modern individuals who carry on average 6.15 copies of AMY1 per diploid genome. The genome of the Mesolithic European from Loschbour has been previously2 estimated to carry 13 copies of AMY1. The analysis of SNP variation in the 66 kb region reveals that the Loschbour individual was heterozygous for SNPs defining haplotypes C and D, carriers of which have a combined haplotype frequency of 31.5% in present-day Western Europe with an average AMY1 copy number of 7 per diploid genome (Tables S1 and S2). The Neolithic farmer from Stuttgart, estimated to carry 16 AMY1 copies, is homozygous for three SNPs defining the G haplotype which has 8% frequency in present-day Europe. Individuals in the 1000 Genome Project data who are homozygous for the haplotype G defining rs74344448-C allele (Table S2) carry on average 10 copies of AMY1. So, for both the Mesolithic and Neolithic aDNA samples where the AMY1 copy number has been estimated the copy numbers in their related lineages show a trend in present-day populations to have decreased rather than increased on average over time.
A high range of variation in amylase copy numbers in present-day human populations could result from both copy number increases as well as deletions. The high frequency of AMY2A deletion we observed in our Siberian sample could be an outcome of neutral and/or selective processes. The possibility of neutrality is raised by the small long term effective population size of Northeast Siberians27, which implies high levels of genetic drift. Haplotypes containing deletions of the pancreatic AMY2A could have risen in frequency through random fluctuations in the absence of starch-digestion related selection. Such a relaxation in selective maintenance is supported by our observation that populations of higher latitudes are characterized by higher variance of amylase copy numbers (Table 2) and the dietary ecology argument: the diet of Siberian populations that have historically followed migrating herds of reindeer and woolly mammoth would have included relatively limited plant carbohydrates. A diet focussed on reindeer and woolly mammoth would have provided significant quantities of fat and hence the lipids needed, in the absence of bulky plant carbohydrates, to offset the nitrogen toxicity associated with a high protein diet28. One of the strongest selective sweeps in the human genome, the mutation of the CPT1A, involved in the oxidation of long-chain fatty acids, attests to this ecological adaptation in circum-Arctic populations27. Archaeological evidence for plant diet at these time depths is limited and the lower AMY2A numbers found in these regions suggest a less important role of starchy plants than in many other populations. Archaeological research in the Yana Valley (27 KYA) excavations concluded those who camped at the site hunted reindeer, horses and birds29. Archaeobotanical investigations at Dolní Vĕstonice II, Czech Republic, date to 30 KYA, at a latitude below the boreal zone, revealed extensive exploitation of dietary roots and tubers30 in addition to the hunting of reindeer and mammoth.
In conclusion, we find evidence for unusually low sequence diversity in regions flanking the amylase copy number locus in a global set of human populations. Significantly negative Tajima’s D scores in Africans, the presence of fixed differences between modern and archaic humans, and young coalescent date in the modern human lineage together suggest that the amylase locus has undergone a selective sweep after the separation of humans and Neanderthals. It is likely that this selective sweep that fixed multiple copies of AMY1 gene in modern human lineages was associated with a dietary shift and an elaboration of the processing sequences for starch-rich tubers, incorporating, for example, grinding, leaching and cooking.
Methods
We assessed the distribution of genetic diversity near the AMY locus from the Phase 1 of the 1000 Genomes panel21. Genetic diversity at the AMY locus was placed in the context of the patterns of genetic diversity observed in the rest of the genome by 50 kb non-overlapping windows. We applied the 1000 Genomes Project’s accessible genome strict mask (release 20141020) to retain information only from regions that can be uniquely mapped by Illumina short reads. As a consequence of applying these filters, 50 kb windows displaying less than 40 kb of accessible genome with available ancestral information were removed from downstream analyses. We used the Human-Chimpanzee ancestral sequence chimpanzee genome (inferred from the Sequences in Ensembl v64 EPO Compara 6 primate block18) as an outgroup and focused on two parameters: the cumulative frequency of derived alleles over polymorphic sites in Africans in a 50 kb window and the number of fixed derived sites in Africans where high coverage Altai Neanderthal18 and Denisovan31 sequences both carry the ancestral allele. The first of these parameters was estimated to serve as a proxy of the coalescent age of a given 50 kb window in humans while the second parameter was designed to distinguish cases of complete and incomplete lineage sorting among modern and archaic humans. We chose to focus on African genomes only, rather than the full 1000 Genomes panel, to minimize the effect on these statistics of archaic admixture that has been documented in non-Africans18,32.
To estimate the age of the haplotypes associated with ancestral copy number of the AMY1, AMY2A and AMY2B genes we used phased high coverage data for 480 whole genome sequences determined with the Complete Genomics platform19. We examined firstly the patterns of linkage disequilibrium, using the D’ statistic (Figure S1), in a region of ~100 kb both up and downstream of the AMY locus to define the boundaries of a region of extremely high linkage disequilibrium in our global data set for further haplotype based analyses. The D’ statistic was calculated for bi-allelic SNPs that had MAF >0.1 in our global sample. These analyses identified a ~66 kb region (Chr1:104,303,310–104,369,301) downstream of the AMY locus with high average (D’ = 0.992) LD among SNPs. Notably, the SNPs in the ~66 kb region show also relatively high average (D’ = 0.839) LD with the SNPs from the 150 kb region (Chr1:104,000,001–104,150,000) upstream of the AMY copy number variable locus, suggestive of a strong long-range association. We determined the phylogenetic relationships and ages of ancestral haplotypes in the ~66 kb region using BEAST version 1.833. In all age calculations we used the mutation rate of 5.5 × 10−10/bp/year34 with the relaxed lognormal clock and piecewise-linear Bayesian Skyline model with 10 groups. Maximum Likelihood trees inferred with RAxML v. 7.8.635 were provided as starting trees. Eight independent BEAST analyses were run for 100 million iterations, sampling every 5,000 steps. After inspection in Tracer v1.633, the results from the independent runs were merged using LogCombiner v1.8.0 with a burn-in of 20%. Then the Bayesian Skyline Plots were reconstructed in Tracer, confirming that the ESS values were above 200.
To infer the copy numbers of AMY1, AMY2A and AMY2B genes, we used high coverage sequence data for 480 individuals from 125 populations sampled worldwide19. A subset of 68 DNA samples from Siberia, Southeast Asia and the Andes were further subjected to paralogue ratio test and microsatellite analyses for determining copy number variation in the AMY locus using methods described elsewhere3. Informed consent had been obtained from all human subjects tested here, and the genome-scale work on their DNA had been approved by the ethics committees of the Institute of Biological Problems of the North of the Russian Academy of Sciences in Magadan (statement no. 001/011 from January 21, 2011) and by the AMIS-UPS Research Ethics Committee, University of Toulouse, Paul Sabatier (Ethical approval no. 005/011). All genetic analyses and data manipulations were performed in accordance with the relevant guidelines approved by the Cambridge Human Biology Research Ethics Committee (HBREC.2011.01).
To determine the AMY1, AMY2A and AMY2B copy numbers from the read depth data we used the ‘relative coverage’ (R) metric reported by CG. Considering the fact that the reference genome contains three haploid copies of AMY1 genes (AMY1A, AMY1B, AMY1C) the combined AMY1 copy number per diploid genome was determined by (RAMY1A/RAMY2B) × 6. This method yielded a Pearson correlation coefficient r = 0.92 with the results based on AMY1 paralogue ratio test. All 68 samples tested had two copies of AMY2B by the read depth and paralogue ratio tests. AMY2A copy numbers inferred from the CG output showed Pearson correlation coefficient r = 0.85 with the assay results.
In order to reveal the haplotypes on the background of which AMY2A deletion occurs (Table 3) we determined the phase of the AMY2A deletions in the context of the phased data for 891 binary SNPs in the Chr1:104,303,310–104,369,301 (hg19) region. Firstly, we estimated the genotype of AMY2A deletion for each sequenced individual by subtracting the read-depth inferred number of AMY2B copies from the AMY2A copy number. We assigned samples with equal or higher number of AMY2A copies as homozygous non-carriers of the deletion, individuals with single difference as heterozygotes and individuals with two more copies of AMY2B as homozygotes for the deletion. Beagle 4.136 was used to phase the heterozygous samples and to impute samples with missing AMY2A information.
Additional Information
How to cite this article: Inchley, C. E. et al. Selective sweep on human amylase genes postdates the split with Neanderthals. Sci. Rep. 6, 37198; doi: 10.1038/srep37198 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Richard W. Wrangham for his critical reading and helpful comments on the manuscript. This research was supported by European Research Council Starting Investigator grant FP7-261213 (T.K., C.E.I., L.P., T.E.), Estonian Research Infrastructure Roadmap grant no. 373 3.2.0304.11-0312, the European Union European Regional Development Fund Centre of Excellence in Genomics to the Estonian Biocentre and Estonian Institutional Research grant IUT24-1 (T.K., L.S., G.H., M.M. and R.V.); French Ministry of Foreign and European Affairs and French ANR grant number ANR-14-CE31-0013-01 (F.-X.R.); Program of the RAS Presidium “Basic research for the development of the Russian Arctic” (B.M.).
Footnotes
Author Contributions T.K. and C.E.I. designed the study and prepared the first draft; E.M., C.A.E., B.M., M.D., J.W., S.A., F.R., M.M., R.V., and M.Me. contributed samples or whole genome sequence data; C.E.I., C.D.A.L., N.A.A.S., L.P., L.S., T.A., G.J., G.H., M.Mi., R.M., M.K.J., J.A.L.A. and T.K. contributed to the analyses of the data and interpretation of the results; all authors contributed to the writing of the final manuscript.
References
- Perry G. H. et al. Diet and the evolution of human amylase gene copy number variation. Nature genetics 39, 1256–1260, doi: 10.1038/ng2123 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413, doi: 10.1038/nature13673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter D. et al. Obesity, starch digestion and amylase: association between copy number variants at human salivary (AMY1) and pancreatic (AMY2) amylase genes. Human molecular genetics 24, 3472–3480, doi: 10.1093/hmg/ddv098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher C. L. et al. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nature genetics 47, 921–925, doi: 10.1038/ng.3340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A. J. et al. Detection of large-scale variation in the human genome. Nature genetics 36, 949–951, doi: 10.1038/ng1416 (2004). [DOI] [PubMed] [Google Scholar]
- Redon R. et al. Global variation in copy number in the human genome. Nature 444, 444–454, doi: 10.1038/nature05329 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchi M. et al. Low copy number of the salivary amylase gene predisposes to obesity. Nature genetics 46, 492–497, doi: 10.1038/ng.2939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia-Benitez M. A. et al. Beneficial effect of a high number of copies of salivary amylase AMY1 gene on obesity risk in Mexican children. Diabetologia 58, 290–294, doi: 10.1007/s00125-014-3441-3 (2015). [DOI] [PubMed] [Google Scholar]
- Hardy K., Brand-Miller J., Brown K. D., Thomas M. G. & Copeland L. The Importance of Dietary Carbohydrate in Human Evolution. Q Rev Biol 90, 251–268 (2015). [DOI] [PubMed] [Google Scholar]
- Sabeti P. C. et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918, doi: 10.1038/nature06250 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K. et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res 19, 826–837, doi: 10.1101/gr.087577.108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth P. J., Warren F. J. & Ellis P. R. Human amylase and starch digestion: an interesting marriage. Starch 63, 395–405 (2011). [Google Scholar]
- Wrangham R. W., Jones J. H., Laden G., Pilbeam D. & Conklin-Brittain N. The Raw and the Stolen. Cooking and the Ecology of Human Origins. Current anthropology 40, 567–594 (1999). [PubMed] [Google Scholar]
- Shahack-Gross R. et al. Evidence for the repeated use of a central hearth at Middle Pleistocene (300 ky ago) Qesem Cave, Israel. Journal of Archaeological Science 44, 12–21 (2014). [Google Scholar]
- Aiello L. C. & Wheeler P. The expensive-tissue hypothesis: the brain and the digestive system in human and primate evolution. Current anthropology 36, 199–221 (1995). [Google Scholar]
- Fonseca-Azevedo K. & Herculano-Houzel S. Metabolic constraint imposes tradeoff between body size and number of brain neurons in human evolution. Proceedings of the National Academy of Sciences of the United States of America 109, 18571–18576, doi: 10.1073/pnas.1206390109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrangham R. W. & Carmody R. Human adaptation to the control of fire. Evolutionary Anthropology 19, 187–199 (2010). [Google Scholar]
- Prüfer K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49, doi: 10.1038/nature12886 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L. et al. Genomic analyses inform on migration events during the peopling of Eurasia. Nature 538, 238–242, doi: 10.1038/nature19792 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65, doi: 10.1038/nature11632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. et al. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature 531, 504–507, doi: 10.1038/nature17405 (2016). [DOI] [PubMed] [Google Scholar]
- Kuhlwilm M. et al. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530, 429–433, doi: 10.1038/nature16544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter T., Jagoda E. & Capellini T. D. Dietary Variation and Evolution of Gene Copy Number among Dog Breeds. PloS one 11, e0148899, doi: 10.1371/journal.pone.0148899 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G. H., Kistler L., Kelaita M. A. & Sams A. J. Insights into hominin phenotypic and dietary evolution from ancient DNA sequence data. J Hum Evol 79, 55–63 (2015). [DOI] [PubMed] [Google Scholar]
- Fu Q. et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449, doi: 10.1038/nature13810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente F. J. et al. A Selective Sweep on a Deleterious Mutation in CPT1A in Arctic Populations. American journal of human genetics 95, 584–589, doi: 10.1016/j.ajhg.2014.09.016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. K. In The Evolution of Hominin Diets: Integrating Approaches to the Study of Palaeolithic Subsistence Vol. 19 (eds Hublin J. J. & Richards M. P.) 171–180 (Springer Science & Business Media B. V., 2009). [Google Scholar]
- Pitulko V. V. et al. The Yana RHS site: humans in the Arctic before the last glacial maximum. Science 303, 52–56, doi: 10.1126/science.1085219 (2004). [DOI] [PubMed] [Google Scholar]
- Pryor A. J. E., Steele M., Jones M. K., Svoboda J. & Beresford-Jones D. G. Plant Foods in the Upper Palaeolithic at Dolní Vĕstonice – Parenchyma Redux. Antiquity 87, 971–984 (2013). [Google Scholar]
- Meyer M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226, doi: 10.1126/science.1224344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722, doi: 10.1126/science.1188021 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Suchard M. A., Xie D. & Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular biology and evolution 29, 1969–1973, doi: 10.1093/molbev/mss075 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M. et al. Calibrating the Human Mutation Rate via Ancestral Recombination Density in Diploid Genomes. PLoS Genet 11, e1005550, doi: 10.1371/journal.pgen.1005550 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690, doi: 10.1093/bioinformatics/btl446 (2006). [DOI] [PubMed] [Google Scholar]
- Browning B. L. & Browning S. R. Genotype Imputation with Millions of Reference Samples. American journal of human genetics 98, 116–126, doi: 10.1016/j.ajhg.2015.11.020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.