Abstract
A critical step in the HIV-1 lifecycle involves reverse transcription of the viral genomic RNA (gRNA). Human tRNALys3 serves as a primer for transcription initiation and is selectively enriched in virus particles. Human lysyl-tRNA synthetase (hLysRS) is also packaged into virions. Recently, a tRNA-like element (TLE) within the HIV-1 gRNA was shown to mimic the global tRNA fold and bind competitively to hLysRS, suggesting a mechanism of tRNA targeting to the primer binding site (PBS) and release from the synthetase. Here, we use NMR to investigate hLysRS anticodon-binding domain (ACB) binding to six RNA oligonucleotides, including a hairpin derived from the HIV-1 gRNA TLE. We show that ACB interacts with submicromolar affinity to U-rich RNA oligonucleotides—the tRNALys3 anticodon stem–loop (ACSL), the WT TLE, and a nonanucleotide, U9. In contrast, the ACB bound only weakly to two TLE loop mutants and a C9 nonanucleotide. NMR chemical shift perturbations induced by each RNA indicate that the ACSL and the WT TLE both interact with the ACB in a strikingly similar manner. Taken together, these findings support the conclusion that tRNA mimicry by the HIV-1 genome leads to a highly specific protein–RNA interaction that facilitates efficient primer release from hLysRS prior to reverse transcription.
Keywords: anticodon-binding domain, human lysyl-tRNA synthetase, tRNA-like element, HIV-1 genome, NMR, chemical shift perturbation
INTRODUCTION
All retroviruses reverse transcribe their viral RNA genomes into DNA prior to integration into the host genome. This process initiates from the 3′ end of a host cell tRNA, which is complementary to an 18-nucleotide (nt) region on the retroviral genome known as the primer binding site (PBS) (Telesnitsky and Goff 1997). Some retroviruses, including human immunodeficiency virus type 1 (HIV-1), selectively package specific host tRNAs into virions, which likely increases the efficiency of the initiation process. In the case of HIV-1, human tRNALys3 is the primer for reverse transcription and is selectively packaged into the virus along with the other major tRNALys isoacceptors in the cell, tRNALys1 and tRNALys2 (Jiang et al. 1993; Pavon-Eternod et al. 2010).
The selective incorporation of tRNALys isoacceptors is mediated by a packaging complex that involves interactions between tRNALys, human lysyl-tRNA synthetase (hLysRS), HIV-1 proteins Gag and GagPol, and genomic RNA (gRNA) (Kleiman et al. 2010). The U-rich anticodon of tRNALys specifically interacts with the anticodon-binding domain (ACB) of human LysRS (Stello et al. 1999). In contrast, interactions with a eukaryote-specific N-terminal extension domain of hLysRS are not believed to contribute to specific tRNA recognition, but increase overall tRNA binding affinity (Francin and Mirande 2006; Francin et al. 2002; Liu et al. 2013; S Liu, A Decker, M Refaei, P Tsang, C Jones, R Comandur, K Musier-Forsyth, J Hinerman, and A Herr, in prep.).
Mammalian LysRS binds its tRNA substrates with high affinity (Francin et al. 2002), and the mechanism of tRNALys3 release from hLysRS and targeting to the PBS during the formation of the annealing complex has recently been investigated. In particular, we reported that a U-rich tRNALys anticodon-like sequence in the gRNA proximal to the PBS interacts with hLysRS with relatively high affinity (Jones et al. 2013). Importantly, mutation of the U-rich sequence had a significant impact on tRNA primer placement and HIV-1 infectivity (Jones et al. 2013). Interestingly, the tRNA mimicry of the tRNA-like element (TLE) extends to the global fold of the PBS/TLE region of the HIV-1 genome, which resembles the L-shaped tRNA fold (Jones et al. 2014). Taken together, our previous data are consistent with a model wherein tRNALys3 is at least partially released from hLysRS through competition for hLysRS binding with the viral TLE (Jones et al. 2013).
The binding interaction between hLysRS and the gRNA TLE has not been investigated at the molecular level. We hypothesize that the ACB interacts with the U-rich TLE in the gRNA in a manner that mimics its interaction with the tRNALys anticodon domain. The high specificity of this interaction along with additional interactions between hLysRS and gRNA outside of the ACB interaction (Jones et al. 2013) contribute to primer release from LysRS and targeting to the PBS. As a first step toward testing this hypothesis, we used NMR to map the interaction between the hLysRS ACB and a 23-nt RNA stem–loop derived from the wild-type (WT) HIV-1 TLE (Fig. 1). We compared these results to mapping studies carried out with the 17-nt anticodon stem–loop of human tRNALys3 (ACSL), as well as two TLE variants wherein the four U's in the loop were mutated to A's (TLE 4A) or C's (TLE 4C) (Fig. 1). Finally, comparisons were also made to two unstructured single-stranded nonanucleotides containing all C's or all U's. The data support tRNA mimicry of the gRNA TLE that extends to its specific protein–RNA interactions with the hLysRS ACB.
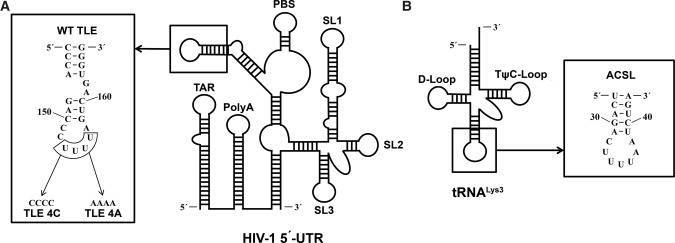
FIGURE 1.
The sequences and secondary structures of RNAs used in this study. (A) The location and sequence of the WT TLE in the 5′-UTR of the NL4-3 isolate of HIV-1. Two additional closing C-G base pairs were added to stabilize the stem–loop. The mutations of WT TLE to obtain TLE 4C and TLE 4A are indicated. (B) The location and sequence of the anticodon stem–loop in tRNALys3.
RESULTS AND DISCUSSION
Fluorescence anisotropy binding studies
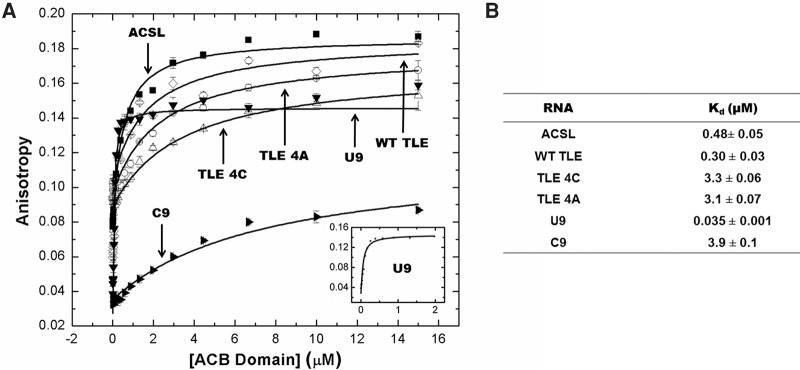
The equilibrium binding constants of ACB binding to all of the RNAs used in this study were determined via fluorescence anisotropy (FA). Figure 1 shows the sequences of the hairpin RNAs that were investigated (WT TLE, TLE 4C, TLE 4A, and ACSL). Although the native ACSL of tRNALys3 is modified at positions U34 (5-methoxycarbonylmethyl-2-thiouridine), A37 [2-methylthio-N(6)-threonylcarbamoyladenosine], and U39 (pseudouridine), these modifications are not critical for hLysRS recognition (Stello et al. 1999). The heavily modified ACSL is believed to ensure translation fidelity by stabilizing the relatively weak UUU anticodon pairing (Murphy et al. 2004; Vendeix et al. 2012). The global conformation of the TLE RNAs was first probed by circular dichroism spectroscopy and the spectra were consistent with A-form duplex regions (Supplemental Fig. S1). In addition to the hairpin RNAs, we also investigated binding to two nonanucleotides, C9 and U9. The relative affinity of ACB binding to all RNAs is summarized in Figure 2. The observed ACB binding trend was determined to be U9 >> TLE > ACSL > TLE 4C, TLE 4A > C9. The greatest binding difference involved the two nonanucleotides; ∼100-fold higher ACB binding affinity was observed to U9 versus C9. Interestingly, the flexible U9 oligonucleotide binds with higher affinity (∼35 nM) than either the ACSL (∼480 nM) or WT TLE (∼300 nM).
FIGURE 2.
FA binding assay results. RNAs were folded prior to FA measurements and binding was performed in 20 mM Tris–HCl, pH 8, 15 mM NaCl, 35 mM KCl, and 1 mM MgCl2, as described in Materials and Methods. (A) Binding of the ACB domain to various RNA oligonucleotides. The inset contains the binding curve of U9 with the concentration of the ACB domain up to only 2 µM to show the tight binding. (B) Table summarizing the apparent dissociation constants of the RNA oligonucleotides binding to the ACB domain measured by FA. The results are the average of two trials with the standard deviation indicated.
NMR studies of ACB titrated with different RNAs
Sharp, well-defined resonances in the 1H–15N HSQC NMR spectrum demonstrate that the ACB domain of hLysRS adopts a well-folded and stable structure at 25°C (Liu et al. 2012). To investigate how the various RNAs interact with the ACB on a residue-specific basis, a series of NMR titrations of ACB with the RNAs listed in Figure 2B was conducted. For these studies, 15N-labeled protein was used and each resonance in the HSQC spectrum corresponds to a specific backbone amide bond. Interactions between the ACB and the RNAs that result in perturbations of the ACB HSQC resonances are indicated by chemical shift changes and/or peak broadening. Formation of the ACB–RNA complex during titration results in a chemical exchange process whereby ACB undergoes exchange between its unbound and RNA-bound forms at equilibrium: free ACB ⇔ ACB-RNA. The main types of exchange include: fast (only one resonance is observed at an averaged chemical shift), intermediate (resulting in a shift as well as broadening of the resonance), or slow (a new resonance appears at a shifted frequency corresponding to the bound species) (Cavanagh et al. 2006). Increased interaction of the RNA with ACB protein will result in decreased or slower chemical exchange rates in general.
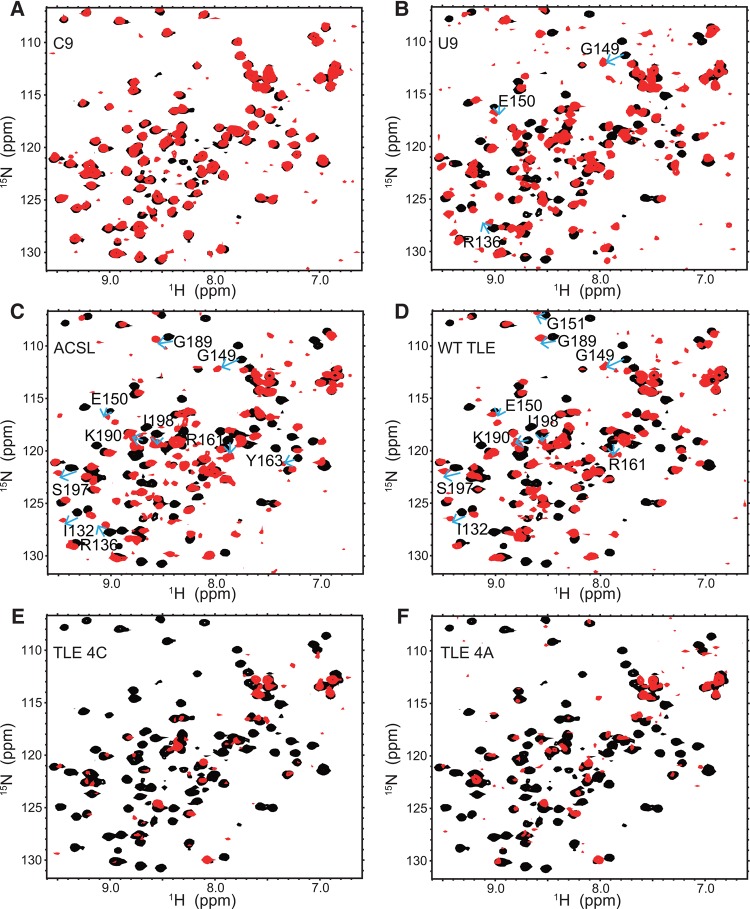
The HSQC spectra recorded for ACB titrated with each of the six RNAs are shown in Figure 3. To better compare and evaluate differences among these RNAs in terms of their effects on the protein, panels A–F of this figure consist of a superposition of the HSQC spectra obtained from the ACB alone or in the presence of RNA at an ACB:RNA ratio of 1:1.2 at 25°C, conditions under which all of the ACB protein is bound. In this way, both the pattern of chemical shift perturbation due to a given RNA, as well as differences in the patterns observed among the various RNAs, can be more clearly followed. Based upon the extent and type of NMR changes observed for the six RNAs studied, the types of ACB–RNA interactions were categorized as being minimal, specific, or nonspecific. The first category corresponds to RNAs whose influence upon the ACB spectrum is minimal since it consists of relatively small frequency shifts that arise exclusively from fast exchange. RNAs in the “specific” category introduce the most significant perturbations to the ACB spectrum in terms of both the number of resonances affected as well as the types of chemical exchange that result from formation of the ACB–RNA complex. Strong association between ACB and a given RNA would most likely result in a set of specific protein–RNA interactions resulting in slow exchange peaks. In the last category, the RNA and ACB interactions are nonspecific and this results in the extensive occurrence of fast and intermediate exchange resonances.
FIGURE 3.
(A–F) Overlays of HSQC spectra obtained for ACB alone (black) or ACB in the presence of C9, U9, ACSL, WT TLE, TLE 4C, and TLE 4A RNAs, respectively (red). These NMR spectra were collected using a Bruker DMX 500 spectrometer at 25°C. Protein concentration is 50 µM and all HSQC spectra with RNAs are at 1.2:1 RNA to protein ratio. In panels B, C, and D, blue arrows for the indicated ACB residues are shown to indicate the direction of shift and location of the slow exchange resonances observed in the titrations involving RNAs U9, ACSL, and WT TLE, respectively.
Minimal RNA–protein interaction: C9
Based on the extensive resonance overlap that exists between the free ACB versus the ACB-C9 HSQC resonances shown in Figure 3A, we conclude that C9 addition to ACB resulted in minor changes in the HSQC ACB spectrum. A relatively small number of resonances (∼15%) were shifted due to fast exchange during ACB titration with C9. Due to the small number of resonances affected and the relatively weak manner in which they are affected, i.e., fast exchange, ACB appears to be minimally influenced by C9.
Specific RNA–protein interactions: U9, ACSL, WT TLE
In contrast to C9, ACB titration with the second linear RNA, U9, resulted in more extensive effects on the protein. Many more protein resonances were perturbed (∼87%) and most of the observed changes involved intermediate and fast exchange effects (Fig. 3B). In addition, the most significant difference relative to C9 was the observation of slow exchange behavior for ACB residues R136, G149, and E150 as indicated by the arrows shown in Figure 3B.
Relative to U9, ACSL addition resulted in a more extensive pattern of chemical shift perturbations. Approximately 92% of the protein resonances were affected by titration with the ACSL RNA (Fig. 3C). While only three ACB residues exhibited slow exchange behavior with U9, 10 ACB resonances exhibited slow exchange effects upon titration with ACSL, as evidenced by the gradual appearance of new peaks at frequencies corresponding to the RNA-bound form of ACB for the following residues (see arrows in Fig. 3C): I132, R136, G149, E150, R161, Y163, G189, K190, S197, and I198.
The patterns of ACSL and WT TLE chemical shift changes observed upon ACB titration were very similar (Fig. 3D). The percentage of ACB NMR resonances perturbed by ACSL is quite comparable to those affected by WT TLE (92% versus 93%, respectively). The following nine ACB residues displayed slow exchange behavior (arrows in Fig. 3D) upon WT TLE titration: I132, G149, E150, G151, R161, G189, K190, S197, and I198. Eight out of 10 of the ACB-ACSL slow exchange residues were the same as those observed for the ACB–WT TLE complex.
Supplemental Figure S2 provides examples of slow and fast exchange resonances for three ACB residues (I132, G189, and S197) resulting from ACB titration with the U-rich RNAs (U9, ACSL, and WT TLE). All three residues give rise to slow exchange with the stem–loop RNAs (ACSL and WT TLE), but only fast exchange occurs when U9 is the titrant. The observation of slow exchange of ACB resonances when bound to these three U-rich RNAs suggests that they share a specific interaction surface on the ACB.
Nonspecific RNA–protein interactions: TLE 4A and TLE 4C
The chemical shift perturbation patterns observed upon titration of ACB with the two stem–loop mutant RNAs, TLE 4C and TLE 4A, differ substantially from those of all the previous RNAs. In contrast to WT TLE, most of the ACB resonances disappeared upon addition of these two RNAs. In the case of TLE 4C (Fig. 3E) and TLE 4A (Fig. 3F), ∼79% and ∼73% of the ACB resonances were broadened and lost when ACB was titrated with these RNAs, respectively. Since the rate of chemical exchange is temperature-dependent, HSQC spectra were recorded for both of these complexes at several additional temperatures to establish whether the titration and subsequent disappearance of ACB resonances at 25°C was a consequence of intermediate chemical exchange and resonance broadening. The spectra obtained at 15°C and 5°C did not significantly increase the total number of ACB resonances observed (S Liu and P Tsang, unpubl.). However, raising the sample temperature to 35°C for the TLE 4C and TLE 4A complexes resulted in resonance sharpening and recovery of nearly 80% of the observable ACB resonances (Supplemental Fig. S3). Therefore, the disappearance of ACB resonances during titration with TLE 4C (Fig. 3E) or TLE 4A (Fig. 3F) is consistent with intermediate exchange broadening of most protein resonances at 25°C. Resonance broadening upon RNA addition could result from nonspecific binding to the protein at multiple sites simultaneously (Auweter et al. 2007). It should be noted that the 35°C ACB-TLE 4C and ACB-TLE 4A HSQC spectra are very similar (Supplemental Fig. S3). Among the resonances that reappear at 35°C, ∼20% of these were unperturbed relative to the free ACB peaks while the remainder appear to undergo fast to moderately fast exchange corresponding to relatively minor chemical shift perturbations of <0.1 ppm on the average. Overall, these small changes reflect the relatively weak influence of the TLE4A and TLE4C RNAs on the ACB protein. Finally, no slow exchange behavior was observed during titration of ACB with either of these two RNAs at any of the temperatures studied (Fig. 3E,F; Supplemental Fig. S3).
Summary of NMR titration results
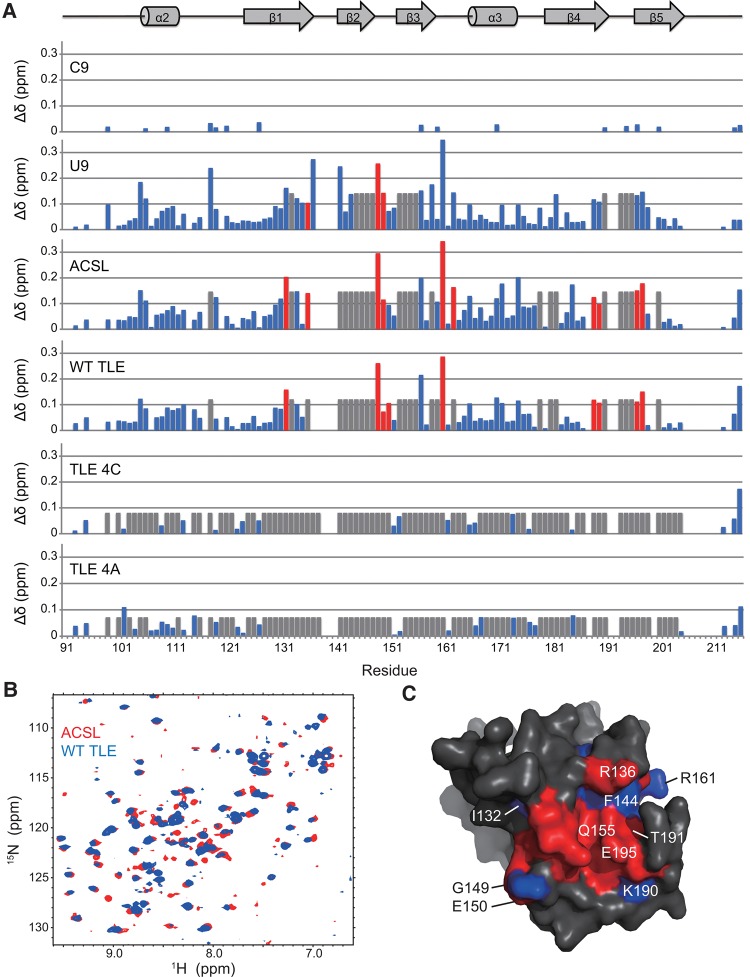
A summary of the resonance perturbations and chemical exchange behavior observed for ACB resonances due to titration with all six RNAs at 25°C is provided in Figure 4A. NMR titration of ACB with the U-rich RNAs (WT TLE, ACSL, and U9) resulted in the most significant changes based on the occurrence of slow exchange behavior for essentially the same set of ACB resonances and therefore similar regions of the ACB protein. Residues that exhibit such slow exchange behavior represent regions of the ACB that are the most strongly influenced by binding to these U-rich RNAs. The particular ACB residues involved were I132, R136, G149, E150, and R161 in β strands 1 to 3; G189, K190 at the end of β strand 4; and S197, I198 at the beginning of β strand 5 (red bars shown in Fig. 4A).
FIGURE 4.
Summary of ACB chemical shift perturbations according to magnitude and type of exchange at 25°C. (A) The colored bars correspond to fast exchange (blue), intermediate exchange (gray), and slow exchange (red). The heights of the bars reflect magnitude perturbation and were calculated as follows: Δδ = (ΔδH2 + ΔδN2/25)1/2, where ΔδH,N2 corresponds to ΔδH, N = δH,Nbound−δH,Nfree. In the case of intermediate exchange resonances, the heights of these gray bars are set to the mean chemical shift deviation of all the shifted resonances plus one standard deviation in each RNA titration experiment. The ACB residue number is indicated along the x-axis while the corresponding secondary structure is shown above the chart. (B) Overlay of HSQC spectra of ACB titrated with ACSL (red) and WT TLE (blue) at 1.2:1 RNA to protein ratio at 25°C. (C) Comparison of the ACB surface that was the most strongly affected (i.e., slow and intermediate exchange) by the U-rich RNAs as determined by NMR: ACSL, WT TLE, and U9. The important residues are indicated on the surface. Residues in red are those that are affected by all three RNAs. Blue residues are those that are affected by ACSL and WT TLE but not U9. The protein structure was generated using the hLysRS ACB domain coordinates (Guo et al. 2008) and rendered using a protein surface.
Based on the NMR data, the typical binding affinity ranges associated with the different regimes of chemical exchange are ∼0.5 to 250 nM (slow exchange), 400 to 2000 nM (intermediate exchange), and >15,000 nM (fast exchange) (Auweter et al. 2007). Slow exchange behavior was observed upon titration of ACB with only three of the RNAs studied—U9, ACSL, and WT TLE. For the remaining RNAs, only intermediate and fast exchange effects (TLE 4A, TLE 4C) or just a limited number of fast exchange (C9) examples was observed. Based upon this, there is a very good correlation between the FA binding and NMR titration trends; i.e., the RNAs that resulted in the greatest perturbation of the ACB resonances also correspond to the RNAs with the highest binding affinities for ACB according to FA.
Although ACB binds ∼10-fold more tightly to U9 than it does to either of the two structured U-rich hairpins (Fig. 2), the small size and greater flexibility of the linear U9 oligonucleotide may allow it to adopt a more optimal binding conformation in the U-binding pocket of ACB. Alternatively, the higher affinity binding observed for U9 could be explained by mainly nonspecific interaction. However, a competition binding assay wherein ACB was prebound to labeled ACSL revealed that both the WT TLE and U9 were able to effectively compete for ACB binding at similar concentrations of RNA (data not shown). Thus, as suggested by the slow and intermediate exchange behavior of a subset of the critical residues (Fig. 4), U9 is able to interact with the ACB via specific interactions. These data do not rule out the possibility that additional nonspecific interactions may be responsible for the higher-affinity binding.
ACB binds to WT TLE with submicromolar affinity and its affinity for ACSL is only slightly weaker. For the three remaining RNAs—C9, TLE 4A, and TLE 4C—FA measurements indicate ACB binds to these RNAs in the micromolar range so binding is significantly weaker relative to the U-rich RNAs. These affinities are consistent with the observation of either the lack of significant resonance perturbations (C9) or extensive resonance broadening (TLE 4A and TLE 4C) as a result of titration with these particular RNAs.
According to the extent and types of ACB chemical shift perturbation observed for the RNAs, the relative trend from greatest to lowest perturbation is ACSL/WT TLE > U9 > TLE 4C,TLE 4A > C9. From the NMR data, the set of ACB resonances that were most strongly affected by titration with the different RNAs provides essential criteria that can be used to map the ACB–RNA interface. Upon comparison of the different patterns of RNA-induced NMR chemical shift changes, the ACSL and WT TLE (Fig. 4B) were found to be the most similar to each other. This is based on the following: (i) there are 10 and nine ACB residues that exhibit slow exchange upon titration with the ACSL and WT TLE RNAs, respectively; eight out of these slow exchange residues are identical for both RNAs; (ii) all 32 ACB resonances that undergo slow and intermediate exchange upon titration with ACSL also exhibit the same patterns of exchange behavior in the case of WT TLE. Overall, these results indicate that there is extensive overlap of the ACB residues affected by both RNAs.
Since the ACSL corresponds to the anticodon stem–loop of tRNALys3, the pattern of ACB resonances that were most strongly affected by this RNA is likely to reflect the native binding surface of hLysRS with tRNALys. Based on the known LysRS-tRNALys structure of Thermus thermophilus LysRS and Escherichia coli tRNALys3, only interactions between the anticodon stem–loop and the ACB are observed (Cusack et al. 1996). Other interactions between LysRS and tRNALys3 (e.g., between the catalytic domain of LysRS and the tRNA CCA end) are unclear because only the ACSL is well ordered in the crystal structure (Cusack et al. 1996). The ACB residues most essential for specific anticodon “UUU” binding include residues in β strands 1 to 3 (R64, F71, and Q82 in T. thermophilus LysRS corresponding to R136, F144, and Q155 in human LysRS), as well as several near the end of β strand 4 and the beginning of β strand 5 (F112, T114, K115, T116, and E118 in T. thermophilus LysRS corresponding to G189, T191, K192, K193, and E195 in hLysRS). These residues correlate well with those showing slow exchange behavior upon ACSL and WT TLE binding to hLysRS ACB (Fig. 4A). Using these NMR data, the hLysRS ACB residues most strongly affected by NMR titration with the ACSL were mapped onto the surface of the human ACB domain using the X-ray coordinates for this domain (Guo et al. 2008). According to this map, the set of hLysRS ACB residues that undergo slow and intermediate chemical exchange effects upon titration with ACSL is consistent with the RNA binding interface of T. thermophilus LysRS. An overlay of the residues affected by all three of the most strongly binding RNAs (U9, ACSL, and WT TLE) is provided in Figure 4C. The stem–loop RNAs affect a slightly larger surface of ACB compared to U9. There is considerable overlap in terms of the regions of ACB affected by these RNAs, most strikingly in the case of ACSL and WT TLE.
Conclusions
In this study, we have shown that stem–loop RNAs derived from the tRNA-like element of the HIV-1 genome (i.e., WT TLE) and tRNALys3 (i.e., ACSL) interact specifically with the same region of the ACB of hLysRS, as observed by their similar affinities for ACB and their nearly identical patterns of slow and intermediate exchange effects on ACB resonances. These data support the conclusion that the protein binds to both of these RNAs in a very similar manner. Specific binding of hLysRS to its cognate tRNA relies primarily on ACB domain binding to the anticodon. Thus, these results are consistent with the model that the HIV-1 gRNA mimics the structure of tRNA to ensure specific and efficient primer placement. Additional interactions between hLysRS and gRNA elements proximal to the TLE are required for the highest affinity binding (Jones et al. 2013) and may be required to trigger the release of tRNA from the synthetase. These interactions have important implications for initiation of HIV-1 reverse transcription and represent novel targets for HIV-1 therapeutics.
MATERIALS AND METHODS
Preparation of proteins and nucleic acids
Plasmid GED_rrACBcs was used to overexpress the hLysRS ACB domain (residues 91–216) in E. coli BL21(DE3), and purification of the unlabeled protein was carried out as described previously (Liu et al. 2012). Uniformly 15N-labeled samples were produced by overexpression of this protein in E. coli BL21(DE3) cells grown in M9 minimal media containing 15N-labeled NH4Cl and unlabeled glucose as the sole sources of nitrogen and carbon, respectively. Purification was carried out as described previously (Liu et al. 2012).
The RNA molecules studied here include ACSL, a 17-nt RNA derived from the anticodon stem–loop of tRNALys3 (5′-UCAGACUUUUAAUCUGA-3′) (Stello et al. 1999), WT TLE, a 23-nt RNA derived from the HIV-1 genomic RNA TLE (5′-CCCAGACCCUUUUAGUCAGUGGG-3′), TLE 4C, a 23-nt RNA mutant of WT TLE with the four U's of the loop mutated to four C's (5′-CCCAGACCCCCCCAGUCAGUGGG-3′), TLE 4A, a 23-nt RNA mutant of WT TLE with the four U's of the loop mutated to four A's (5′-CCCAGACCCAAAAAGUCAGUGGG-3′) (Jones et al. 2013), and the nonanucleotides U9 and C9. All RNAs were purchased from Dharmacon and deprotected according to the supplier's protocol.
For the FA binding assays, RNAs were fluorescently labeled at the 3′-end with fluorescein-5-thiosemicarbazide (Invitrogen) using previously described methods (Pagano et al. 2007; Rye-McCurdy et al. 2015).
NMR titration
For the NMR titration experiments, a 50-µM stock solution of 15N-labeled ACB in a buffer consisting of 20 mM HEPES, pH 6.8, 15 mM NaCl, 35 mM KCl, 1 mM EDTA, 10% D2O (v/v) and 0.02% NaN3 was used. The RNAs were dissolved in diethylpyrocarbonate-treated double-distilled water. The concentrations of the stock RNA samples were determined by UV absorbance at 260 nm using the following extinction coefficients: ACSL, 173,400 M−1 cm−1; WT TLE, 220,700 M−1 cm−1; TLE 4C, 209,700 M−1 cm−1; TLE 4A, 229,700 M−1 cm−1; C9, 63,200 M−1 cm−1 and U9, 87,500 M−1 cm−1. RNA solutions were then made up to a final concentration of 1 mM RNA in 20 mM HEPES, pH 6.8, 15 mM NaCl, and 35 mM KCl. This RNA solution was refolded by incubating at 80°C for 2 min followed by heating at 60°C for 2 min and then adding Mg2+ to a final concentration of 10 mM before cooling on ice for at least 30 min.
Titrations of the ACB with the different RNAs were monitored via NMR 1H–15N HSQC experiments consisting of 1024 (1H) × 128 (15N) valid points with spectral widths of 10,000 × 1280 Hz. These NMR spectra were collected using a Bruker DMX 500 spectrometer (University of Cincinnati, Department of Chemistry) at 25°C and 35°C. The ratios of RNA to ACB protein used were 0:1, 0.1:1, 0.3:1, 0.6:1, 1.0:1, and 1.2:1 for all RNAs except for C9. The ratio of C9 to ACB was 0:1, 0.6:1, 1.2:1, 2.0:1, and 2.4:1. In general, the total NMR sample volume was increased by ≤6% by the conclusion of the titration and these dilution effects were taken into account during the NMR data processing steps performed using NMRPipe (Delaglio et al. 1995) and SPARKY 3 (Goddard and Kneller 2007, University of California, San Francisco, https://www.cgl.ucsf.edu/home/sparky/).
Classification of fast, intermediate, and slow exchange resonances was based on the relative shift (fast, moderately fast), broadening (intermediate), or appearance of a new resonance (slow) in the HSQC spectrum of ACB due to RNA addition. A resonance shift was deemed significant if the chemical shift difference observed between the free versus RNA-bound form of a resonance was >0.02 ppm (1H) and 0.05 ppm (15N) based on the experimental error of these measurements.
Using the previously assigned ACB resonance assignments (Liu et al. 2012), ACB residues that are affected upon titration with each of the six RNAs were determined. HSQC mapping of the ACB surface affected by titration with the U-rich RNAs (U9, WT TLE, and ACSL) involved use of the previously assigned ACB resonance assignments (Liu et al. 2012) and the published three-dimensional structure of residues 70–216 of the human ACB domain (Guo et al. 2008). PyMOL was employed to render the structure of this protein (ribbon or surface) and to highlight the residues that were the most strongly affected (i.e., strong and intermediate exchange) by titration with these particular RNAs.
FA binding assay
The FA binding assays were performed as previously described with some modifications (Jones et al. 2013). Fluorescently labeled folded RNAs (20 nM) were mixed with varying concentrations of ACB in 20 mM Tris–HCl, pH 8, 15 mM NaCl, 35 mM KCl, and 1 mM MgCl2. Following a 30 min incubation at 25°C, the samples were excited at 485 nm and the emission was measured at 525 nm. The data points were fit to a curve assuming 1:1 binding (Stewart-Maynard et al. 2008). The binding data were duplicated for each of the RNAs and the average with the standard deviation has been reported.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. William Cantara for critical reading of the manuscript. This work was funded by National Institutes of Health RO1 GM113887 (to K.M.-F.) and University of Cincinnati LEAF funding (to P.T.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.058081.116.
REFERENCES
- Auweter SD, Oberstrass FC, Allain FH. 2007. Solving the structure of PTB in complex with pyrimidine tracts: an NMR study of protein-RNA complexes of weak affinities. J Mol Biol 367: 174–186. [DOI] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother W, Palmer A III, Rance M, Skelton N. 2006. Protein NMR spectroscopy: principle and practice, 2nd ed. Academic Press, Burlington, MA. [Google Scholar]
- Cusack S, Yaremchuk A, Tukalo M. 1996. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNALys and a T. thermophilus tRNALys transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J 15: 6321–6334. [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Francin M, Mirande M. 2006. Identity elements for specific aminoacylation of a tRNA by mammalian lysyl-tRNA synthetase bearing a nonspecific tRNA-interacting factor. Biochemistry 45: 10153–10160. [DOI] [PubMed] [Google Scholar]
- Francin M, Kaminska M, Kerjan P, Mirande M. 2002. The N-terminal domain of mammalian lysyl-tRNA synthetase is a functional tRNA-binding domain. J Biol Chem 277: 1762–1769. [DOI] [PubMed] [Google Scholar]
- Guo M, Ignatov M, Musier-Forsyth K, Schimmel P, Yang XL. 2008. Crystal structure of tetrameric form of human lysyl-tRNA synthetase: implications for multisynthetase complex formation. Proc Natl Acad Sci 105: 2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Mak J, Ladha A, Cohen E, Klein M, Rovinski B, Kleiman L. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol 67: 3246–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CP, Saadatmand J, Kleiman L, Musier-Forsyth K. 2013. Molecular mimicry of human tRNALys anti-codon domain by HIV-1 RNA genome facilitates tRNA primer annealing. RNA 19: 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CP, Cantara WA, Olson ED, Musier-Forsyth K. 2014. Small-angle X-ray scattering-derived structure of the HIV-1 5′ UTR reveals 3D tRNA mimicry. Proc Natl Acad Sci 111: 3395–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman L, Jones CP, Musier-Forsyth K. 2010. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett 584: 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Howell M, Melby J, Tsang P. 2012. 1H, 13C and 15N resonance assignment of the anticodon binding domain of human lysyl aminoacyl tRNA synthetase. Biomol NMR Assign 6: 173–176. [DOI] [PubMed] [Google Scholar]
- Liu S, Decker A, Howell M, Caperelli C, Tsang P. 2013. 1H, 13C and 15N resonance assignment of the N-terminal domain of human lysyl aminoacyl tRNA synthetase. Biomol NMR Assign 7: 289–292. [DOI] [PubMed] [Google Scholar]
- Murphy FV IV, Ramakrishnan V, Malkiewicz A, Agris PF. 2004. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol 11: 1186–1191. [DOI] [PubMed] [Google Scholar]
- Pagano JM, Farley BM, McCoig LM, Ryder SP. 2007. Molecular basis of RNA recognition by the embryonic polarity determinant MEX-5. J Biol Chem 282: 8883–8894. [DOI] [PubMed] [Google Scholar]
- Pavon-Eternod M, Wei M, Pan T, Kleiman L. 2010. Profiling non-lysyl tRNAs in HIV-1. RNA 16: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rye-McCurdy T, Rouzina I, Musier-Forsyth K. 2015. Fluorescence anisotropy-based salt-titration approach to characterize protein-nucleic acid interactions. Methods Mol Biol 1259: 385–402. [DOI] [PubMed] [Google Scholar]
- Stello T, Hong M, Musier-Forsyth K. 1999. Efficient aminoacylation of tRNALys,3 by human lysyltRNA synthetase is dependent on covalent continuity between the acceptor stem and the anticodon domain. Nucleic Acids Res 27: 4823–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Maynard KM, Cruceanu M, Wang F, Vo MN, Gorelick RJ, Williams MC, Rouzina I, Musier-Forsyth K. 2008. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J Virol 82: 10129–10142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A, Goff SP. 1997. Reverse transcriptase and the generation of retroviral DNA. In Retroviruses (ed. Coffin JM, Hughes SH, Varmus HE), pp. 121–160. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Vendeix FA, Murphy FV IV, Cantara WA, Leszczynska G, Gustilo EM, Sproat B, Malkiewicz A, Agris PF. 2012. Human tRNALys3UUU is pre-structured by natural modifications for cognate and wobble codon binding through keto–enol tautomerism. J Mol Biol 416: 467–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.