Abstract
The myxobacteria are a family of soil bacteria that form biofilms of complex architecture, aligned multilayered swarms or fruiting body structures that are simple or branched aggregates containing myxospores. Here, we examined the structural role of matrix exopolysaccharide (EPS) in the organization of these surface-dwelling bacterial cells. Using time-lapse light and fluorescence microscopy, as well as transmission electron microscopy and focused ion beam/scanning electron microscopy (FIB/SEM) electron microscopy, we found that Myxococcus xanthus cell organization in biofilms is dependent on the formation of EPS microchannels. Cells are highly organized within the three-dimensional structure of EPS microchannels that are required for cell alignment and advancement on surfaces. Mutants lacking EPS showed a lack of cell orientation and poor colony migration. Purified, cell-free EPS retains a channel-like structure, and can complement EPS− mutant motility defects. In addition, EPS provides the cooperative structure for fruiting body formation in both the simple mounds of M. xanthus and the complex, tree-like structures of Chondromyces crocatus. We furthermore investigated the possibility that EPS impacts community structure as a shared resource facilitating cooperative migration among closely related isolates of M. xanthus.
Introduction
Multicellular life has evolved independently several times, giving rise to a variety of macroscopic organisms: land plants, animals, fungi and so on. This advance allows these organisms to coordinate multiple cellular responses cohesively (Blackstone, 2013). Examining the origins of multicellularity continues to provide new perspectives on life: for example, the transition of choanoflagellates from solitary to multicellular aggregates in response to predator prey relationships (Alegado et al., 2012). Likewise, quorum sensing and biofilm formation enable groups of bacterial cells to act as multicellular organisms that can extend their survival and competitive abilities beyond those of isolated individuals (Davey and O'toole, 2000; Waters and Bassler, 2005; Beauregard et al., 2013). In natural settings, biofilms are often heterogeneous, consisting of mixtures of different species that lack predictable organization. Even within single species biofilms, the structures that arise can be highly variable (Baum et al., 2009; Habimana et al., 2010). By contrast, the myxobacteria typically produce single species biofilms with species-specific architecture (Dawid, 2000). The myxobacteria are common soil bacteria that obtain nutrition through the degradation of prey bacteria, plant debris or macromolecules (Berleman and Kirby, 2009). As population levels rise and nutrition is reduced, individual cells aggregate to form 0.1–1-mm tall fruiting bodies. The fruiting bodies contain 105–107 cells of a single species; some species form simple mound shaped fruiting bodies while others form elaborate three-dimensional (3D) branched structures (Zhang et al., 2003; Hyun et al., 2008). Within fruiting bodies, starved vegetative cells convert to spores; the more complex branched structures of some species show spores within sporangia at branch tips.
Myxobacteria colonize surfaces as organized aggregates of cells. They move with a combination of single cell gliding (A-motility) and social (S)-motility (Pelling et al., 2006; Kraemer and Velicer, 2011; Sun et al., 2011; Nan et al., 2013). S-motility is powered by the extension and retraction of Type IV pili (T4P) that are extruded from one of the cell poles (Nudleman et al., 2006). Previous studies have shown that pili can bind to matrix exopolysaccharide (EPS) found on surfaces and presumed to coat individual cells (Li et al., 2003). Cells are pulled forward by pilus retraction, which exerts a force >100 pN (Clausen et al., 2009). According to current models, EPS forms a cell envelope that serves as an anchor for pilus binding from a neighboring cell; pilus retraction would then pull cells closer to each other (Wall and Kaiser, 1999; Mauriello et al., 2010). However, although a polysaccharide coat is common in bacteria such as Klebsiella and Azotobacter sp. (Sabra et al., 2001; Lin et al., 2013), there is no direct evidence for an EPS cell coat covering M. xanthus cells (Palsdottir et al., 2009; Remis et al., 2013), nor is there a clear explanation of how EPS-coated cells could promote movement away from other cells into new territory.
In this study, we used Ruthenium Red staining of M. xanthus biofilms to visualize the carbohydrate-rich EPS (Sutherland and Thomson, 1975). Our findings show that most of the EPS produced by M. xanthus is deposited on surfaces and sculpted into microchannel structures that guide cell movements. Our analysis indicates that EPS microchannels are important for the multicellular life of the myxobacteria by mediating the organization of cells during surface branch migration, fruiting body formation and intra-species interaction.
Materials and methods
Strains and growth conditions
M. xanthus strains were cultured according to previously established protocols, primarily using CYE liquid media for routine culturing and 0.5% agar CYE for motility assays. M. xanthus strains utilized that were all reported previously: cglB- (Rodriguez and Spormann, 1999), epsZ- (Berleman et al., 2011), pilT- (Wu et al., 1997) pilA::gfp (Bustamante et al., 2004). Environmental isolates of M. xanthus were enriched using previously described methods (Vos and Velicer, 2008) harvesting small quantities of local soils as a source for new strains. Strains were purified for isolation through routine restreaking on CYE and confirmation of species identification of each isolate was determined through 16S rDNA sequencing. Examination of each isolate for S-motility patterns was performed using a Nikon SMZ1500 stereo microscope (Nikon Instruments Inc., Melville, NY, USA)
Fluorescence microscopy
Separate M. xanthus cultures were harvested, washed and concentrated to a density of 300 Klett units. Non-fluorescent cultures were mixed with green fluorescent protein (GFP)-labeled cells at a ratio of 1:50 and 1 μl of the resulting suspension was spotted onto slides coated with 350 μl of 0.5% agar CYE. Several drops of water were spotted surrounding the coated medium to keep the humidity. Slides were incubated for 6–8 h at 32 °C to allow for motility branch formation. Image acquisition was performed on a DeltaVision Elite microscope set-up (Applied Precision, Issaquah, WA, USA) equipped with a CCD camera (CoolSnap HQ, Photometrics, Tucson, AZ, USA) using solid-state illumination at 461/489 nm (GFP). Time-lapses were performed for 20–30 min at 30–60-s intervals. Movies were compiled and analyzed with Image J software (NIH, http://rsbweb.nih.gov/ij/). For each assay condition, at least three time series were captured.
Cell tracking analysis
To quantify differences in migration efficiency among strains, quantitative analysis was performed to assess the ability of cells to travel in efficient, straight-line paths. For each strain, the step-to-step motion of at least six fluorescently labeled cells in the time-lapse series was graphed as trajectories (Microsoft Excel). For each cell, the most efficient path was calculated based on the shortest distance connecting the initial and terminal positions. Comparison of each cell's actual trajectory relative to the most efficient pathway was determined by integration using the Trapezoid Rule to calculate the total area of deviation, with larger areas indicative of a less efficient route of travel. Total areas for each cell were divided by the number of movements that each cell made to yield an average deviation. A Student's t-test (heteroscedastic, two-samples with unequal variance; one tailed) was then used to determine significant differences among the average areas of deviation for each species.
EPS purification and complementation
EPS was purified from M. xanthus cultures as described before (Berleman et al., 2011). Briefly, cell cultures were harvested and resuspended in 25 ml of TNE buffer (100 mM Tris pH 7.5, 100 mM NaCl, 5 mM EDTA). The cell suspensions were disrupted by sonication; then, sodium dodecyl sulfate was added to a final concentration of 0.1% to extract EPS. The extracts were centrifuged for 10 min at 8000 r.p.m. and the pellets were washed twice with 25 ml TNE+sodium dodecyl sulfate. The pellets were resuspended in 1 ml of 10 mM MOPS buffer and stored at −20 °C until use. For complementation, fresh cultures were prepared as before to a final cell density of 500 Klett units. Six-microliter aliquots of the 500 Klett units cell suspension were spotted onto 0.5% agar CYE plates. A 6-mm long line of EPS extract or buffer control was added 4 mm away from the cells. Plates were incubated at 32 °C for over several days. They were examined under a Nikon SMZ1500 stereo microscope (Nikon Instruments Inc., Melville, NY, USA; Table 1).
Table 1. Strains used in this study.
| Strain | Genotype or relevant properties | Source/reference |
|---|---|---|
| DZ2 | Wild type M. xanthus | Campos and Zusman (1975) |
| DZ10457 | pilA::gfp/pilA+ | Bustamante et al. (2004) |
| DZ4477 | DZ1622 cglB::mariner | Youderian et al. (2003) |
| DZ4831 | DZ2 epsZ::pGEM | Berleman et al. (2011) |
| DK10409 | DK1622 ΔpilT | Wu et al. (1997) |
| HP11 | M. xanthus isolate | This study |
| HP12 | M. xanthus isolate | This study |
| HP13 | M. xanthus isolate | This study |
| HP10 | M. xanthus isolate | This study |
| HP16 | M. xanthus isolate | This study |
| KYC221 | Wild type C. crocatus | Hyun et al. (2008) |
| TM12 | DZ2 ΔmglA | Mauriello et al. (2010) |
For EPS purification from agar-grown cultures, 5 ml of TNE buffer with 1 mg ml−1 Pronase E (Sigma–Aldrich, St Louis, MO, USA) was gently added directly to the agar surface and allowed to incubate at 37 °C for 2 h to remove protein. Colonies were then washed four times with TNE buffer + 0.1% sodium dodecyl sulfate to remove cells. For imaging, the remaining material was resuspended in 0.1 M sodium cacodylate buffer (pH 7.2) for 1 h, then fixed in glutaraldehyde and 1% osmium tetroxide was added and incubated for 1 h. Samples were then stained with 5 mM Ruthenium Red for 1 h and analyzed by transmission electron microscopy (TEM) as described below.
Electron microscopy
Transmission electron microscopy (TEM)
M. xanthus colonies and fruiting bodies were grown for 4 days on CYE agar plates in 40 mm diameter aluminum weighing pans to avoid damaging the samples by minimizing handling. Using a scalpel, areas around the colonies and fruiting bodies were removed leaving the samples in the tins. After each colony was cut into four smaller pieces for processing, samples were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) for 1 h, post-fixed on ice with 1% osmium tetroxide for 1 h and stained with 5 mM Ruthenium Red for 1 h.
Following fixation, post-fixation and staining, the samples were moved from the aluminum tins and placed into 20 ml scintillation vials and rinsed three times with buffer to wash off excess fixatives and stains. Dehydration was performed using a graded ethanol series (20%, 40%, 60%, 80%, 90%, and three times at 100%) for 15 min at each step. This was followed by a step-wise infiltration with epon resin at 1:2 resin:acetone for 4 h, 1:1 resin:acetone for 4 h, 2:1 resin:acetone for 4 h, 100% overnight and 100% resin with Benzyldimethylamine for 2 h then polymerized in an 80 °C oven. Sample blocks were sectioned at 90 nm using a Leica EM UC6 microtome (Leica Microsystems Inc., Buffalo Grove, IL, USA). Sections were collected on slot grids and stained with 2% uranyl acetate and Reynolds Lead Citrate. Imaging was carried out using a FEI Tecnai 12 transmission electron microscope (FEI, Inc., Hillsboro, OR, USA).
Focused ion beam/scanning electron microscopy (FIB/SEM)
For FIB/SEM analysis, biofilm samples were prepared similar to HPF-resin-section TEM samples, but were fixed at room temperature in a solution of 4% paraformaldehyde, 0.1% Ruthenium Red and 1% osmium in phosphate-buffered saline for 30 min before being rinsed three times with ddH2O prior to high-pressure freezing. This approach allows for sufficient sample contrast needed for FIB/SEM analysis, while avoiding dehydration artifacts typically encountered in conventionally prepared samples (McDonald and Auer, 2006). It is our experience that fixation and subsequent heavy metal staining causes minimal sample preparation artifacts and that it is mainly the artifacts stemming from dehydration that need to be guarded against. Therefore, by using ultra-rapid freezing and low temperature freeze-substitution sample preparation, dehydration of samples is held to a minimum. For FIB/SEM imaging, serial imaging was performed using a FEI Helios NanoLab DualBeam equipped with an in-lens detection system. Each slice of material was milled using a Ga2+ FIB operating at 30 keV and 460 pA. Image acquisition occurred after each milled slice using a 2 keV and 340 pA SEM probe in backscatter mode with the bias of the in-lens detector set to −50 V. Auto Slice & View (FEI, Inc.) was used for automated acquisition and the voxel size (3D pixel) for the 3D volume was 10 nm by 13 nm by 20 nm in the x, y and z axes. Image reconstruction was performed using Chimera software (UCSF, San Francisco, CA, USA).
Results
Visualization of microchannels in branching M. xanthus biofilms
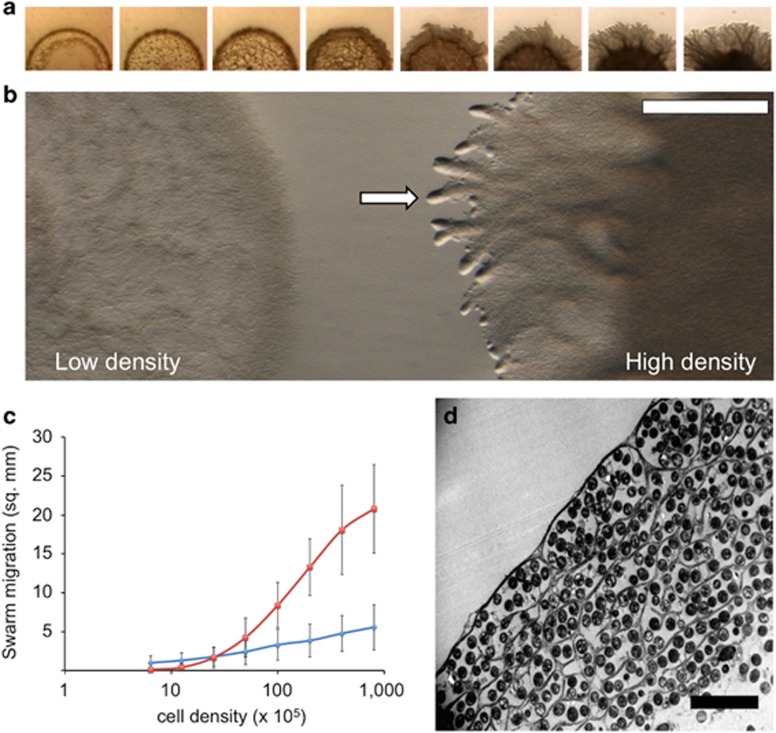
On 0.5% soft agar media, wild-type M. xanthus cells are non-motile at low cell density because A-motility does not function on soft surfaces and S-motility requires cell groups for movement. However, when cells are incubated at higher cell densities, S-motility powers the movement and spreading of groups of cells across surfaces (Rodriguez and Spormann, 1999; Berleman et al., 2011). To further examine the spreading of these vegetative cell swarms, cultures of M. xanthus strain DZ2 were plated on soft agar in a series of twofold dilutions ranging from low to high cell density and examined with time-lapse microscopy (Figure 1, Supplementary Movie S1). At low cell density, cells remain near the initial inoculum, but at higher cell densities, we observed outward migration. Close examination shows that migration correlates with the presence of organized branch-like structures that can be observed emanating radially from the initial inoculum (Figures 1a and b; arrow denotes branch). The timing of branch emanation depended on cell density (Supplementary Figure S1). Both branch formation and rapid migration were observed to follow a cooperative, sigmoidal function dependent on cell density, indicative of multicellular cooperation between cells in agreement with previous results (Figure 1c; Berleman et al., 2011). In contrast, when we examined cells on harder, 1.5% agar, surface migration followed a linear function with respect to cell density, indicative of cooperation-independent behavior during cell movement (Figure 1c). To examine the ultrastructure of these biofilm branches, swarming colonies were fixed with glutaraldehyde, treated with Osmium/Ruthenium Red to stain carbohydrate structures, ethanol-dehydrated, and then infiltrated and embedded in resin. Remarkably, electron micrographs of thin slices of branch sections cut tangentially showed near perfect cell alignment in the direction of migration, causing the rod shaped cells to look round in the cross-sectional view (Figure 1d). In addition, the sections revealed electron-dense material filling the spaces between small groups of cells, which only became apparent with the Ruthenium Red staining protocol, suggesting that it is indeed polysaccharide-rich, and presumably part of the so-called EPS matrix. The extracellular material observed appears continuous, forming a single ‘honeycomb'-like structure with multiple compartments. In cross-section, these compartments contained an average of ~5 cells/compartment (n=58 compartments examined, see Supplementary Figure S2 for detailed distribution).
Figure 1.
M. xanthus social swarms form organized branch structures. (a) M. xanthus cells were added to 0.5% agar CYE at cell densities ranging from 4 × 106 to 2.4 × 108 total cells in 3 μl aliquots. Surface migration correlates with higher cell densities and the formation of radial branch structures (Supplementary Movie S1). (b) A low density (left), non-migrating swarm, adjacent to a high density swarm with typical branch structures at the swarm edge (right). (c) Quantification of surface colonization by wild-type M. xanthus on soft and hard surfaces shows that migration on soft surfaces (red) is characterized by a cooperative sigmoidal function, whereas migration on hard surfaces (blue) is linear. (d) Resin-embedded and Ruthenium red-stained TEM cross-section of a branch reveals the organization of small groups of cells within microchannels. Scale bars are: white bar, 0.1 mm and black bar, 5 μm.
To visualize these structures in three dimensions, we analyzed our samples with FIB/SEM, a novel 3D imaging technique that has been used recently to study bacterial biofilms at ~10–20 nm-scale resolution (Remis et al., 2013). Using this technique on our fixed biofilm samples, we collected 250 serial images at 10-nm intervals along the z axis (Figure 2). These images confirmed that the extracellular honeycomb-like structures identified in two-dimensional TEM images constitute microchannels in a 3D space. Furthermore, the 3D images revealed the tubular character of the electron-dense material in microchannels that surround cells, as shown in Figure 2 and Supplementary Movie S3 (honeycomb-like structures are rendered in blue). Cells within the microchannels were clearly aligned in the direction of swarm migration, suggesting the possibility that these structures might serve to orient the movement of bacteria within the biofilms, thus promoting efficient colonization of new regions.
Figure 2.
Three-dimensional reconstruction of M. xanthus microchannel structure through FIB/SEM analysis. M. xanthus swarms were fixed in glutaraldehyde, resin-embedded and stained with Ruthenium Red for microchannel visualization. Blocks were analyzed by SEM that scans the block surface, then the focused ion beam (FIB) oblates 20 nm of material before scanning again. In total, 250 scans were obtained, spanning 5 μm in depth. (a) The entire 3D reconstruction, with cells shown in yellow and channels in blue pseudocolor (Supplementary Movie S3). (b) One of 250 slices captured during the FIB/SEM data collection shows an arrangement of cells within microchannels similar to observations with TEM. Panel (c) shows the 3D reconstruction of cells only and panel (d) shows the 3D reconstruction of microchannels only. Scale bar, 1 μm.
Genetic evidence that microchannels are composed of EPS
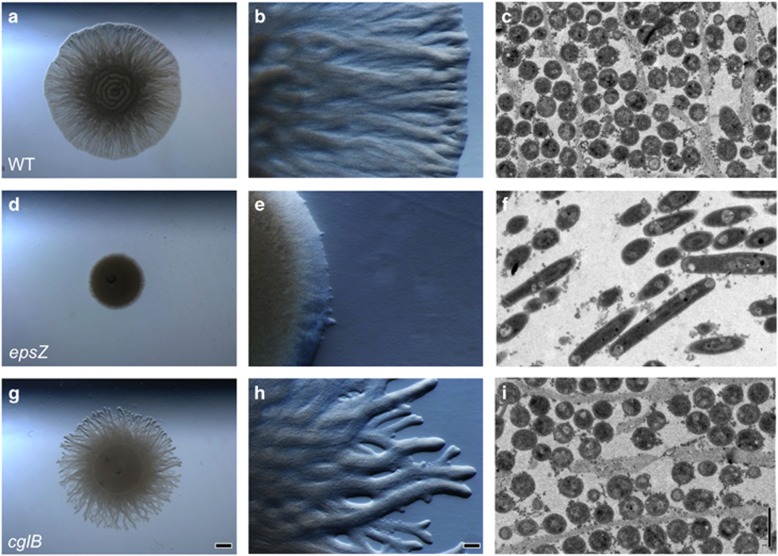
EPS has previously been proposed to form a cell coat that acts as the anchor facilitating T4P-driven social motility (Mauriello and Zusman, 2007). To further examine the possibility that the microchannels contained EPS, we examined the swarm migration pattern of an epsZ-mutant defective in EPS biosynthesis and S-motility, as well as an A-motility-defective cglB- mutant, normal with respect to EPS production and S-motility. The epsZ gene codes for a polyprenyl glycosylphosphotransferase homolog that belongs to the same family as E. coli WcaJ, Salmonella WbaP and Group B Streptococcus CpsE and provides a critical first step in polysaccharide export (http://www.ncbi.nlm.nih.gov). The cglB gene codes for a protein that contains a Von Willebrand factor type A domain, with a metal-dependent MIDAS site predicted to mediate ligand binding. Although the specific ligand bound CglB is unknown, it is an extracellular protein essential for A-motility (http://www.ncbi.nlm.nih.gov, Rodriguez and Spormann, 1999). As shown in Figure 3, vegetative swarms of wild-type cells showed typical S-motility movements and branch patterns (Figures 3a and b), whereas the epsZ- mutant was unable to effectively migrate across the surface or form the branch patterns (Figures 3d and e). epsZ- colonies showed some movement at the colony edge, leading to an overall broadening of the area colonized, but compared with the wild type, directional migration was not apparent. Interestingly, the cglB- mutant (Figures 3g and h), which migrates at a similar rate as the wild type, showed more distinct branch patterns (Figures 3g and h) compared with the wild type (Figures 3a and b), suggesting that while S-motility is responsible for branch formation, A-motility may fill in the spaces between the main branches.
Figure 3.
Genetic analysis of M. xanthus swarm architecture. Five-microliter aliquots of cells (~108 cells) were incubated 24 h on 0.5% agar CYE to promote social motility. (a–c) M. xanthus wild-type DZ2 swarms. (d–f) Social motility-defective epsZ-. (g–i) Adventurous motility-defective cglB-. Stereomicroscopy of swarms shows the migration of (a, b) wild type across the surface in striated branches, whereas (d, e) epsZ- migrates poorly and (g, h) cglB- migrates with distinct branches. For ultrastructural analysis (c, f, i), M. xanthus swarms were fixed, stained and resin-embedded to observe ultrastructure of cell organization by sectioning and TEM visualization. Panel (c) shows the highly organized pattern of wild-type cells and microchannels, whereas panel (f) shows the lack of channels, and a corresponding lack of cell orientation in the epsZ- background, panel (i) shows cglB- with a similar organization as the wild type. The epsZ- mutant also shows that cells do not pack tightly together, with large gaps between cells. Scale bars represent 1 mm, 100 μm and 1 μm.
Biofilms of each strain were fixed and Ruthenium Red-stained for TEM. TEM images of biofilms revealed that the epsZ- mutant lacked the microchannel structures observed in the wild-type branches (Figures 3c, f and i). epsZ- biofilms also showed large empty spaces between cells and no cell alignment. By contrast, microchannel structures and aligned cell organization were observed in both the cglB- mutant and the wild type as expected. Together, these results indicate that the microchannel structures are composed primarily of EPS and that they greatly influence cell organization within cooperative swarming branch structures.
Purification and analysis of EPS microchannels
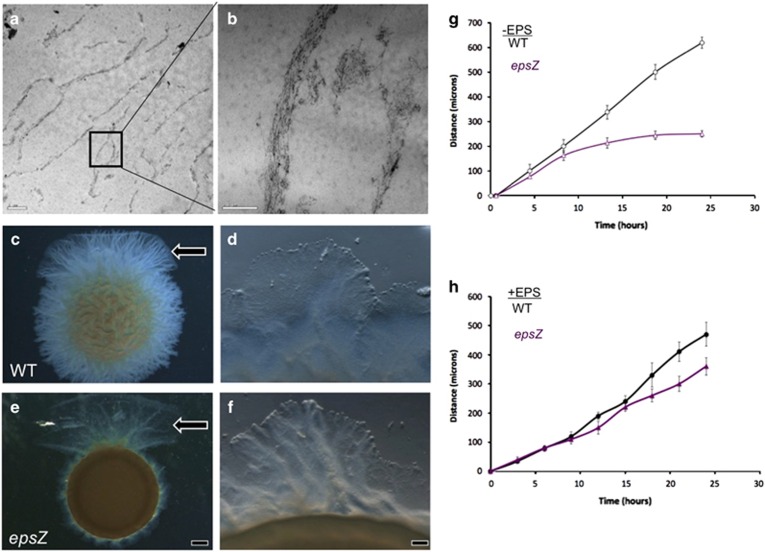
The presence of EPS-containing microchannels surrounding small groups of wild-type cells suggests that they may provide a scaffold for the anchoring of T4P, required for S-motility, thereby contributing to efficient cell movement. To test this possibility, wild-type M. xanthus biofilms were washed gently with sodium dodecyl sulfate to remove bacterial cells while preserving extracellular EPS and associated material (Chang and Dworkin, 1996). Thin section TEM imaging of the resulting cell-free material showed honeycomb-like compartmental structures with size (~300 nm thickness and ~5 μm inner diameter) and overall shape and dimensions similar to the structures observed by sectioning intact colonies (Figures 4a and b).
Figure 4.
Purification of EPS microchannels and rescue of swarm migration. (a, b) EM analysis of sodium dodecyl sulfate-purified EPS shows that the purified EPS has a filamentous structure similar to the native conformation of cells within microchannels. (c–f) M. xanthus cells were incubated on a 0.5% agar surface in 5 μl aliquots of cells (~108 cells) adjacent to EPS extracts (arrow, top). (c, d) Wild-type migration occurs in all directions, but is promoted in the direction of the added EPS. (e, f) epsZ- mutant migrates only in the direction of the added EPS. (d) Magnified view of wild-type migration into the EPS-coated surface (see Supplementary Movie S4), (f) epsZ- migration into the EPS-coated surface (see Supplementary Movie S5). (g) Quantification of swarm migration in the absence of EPS extracts shows a steady progression of wild-type swarms (black line), whereas epsZ- swarm migration (purple line) is non-processive. In the presence of EPS extracts (h), however, swarm migration of epsZ- is rescuable. Scale bars represent 1 μm, 0.5 μm, 1 mm and 100 μm.
We also assayed the ability of cell-free EPS to stimulate S-motility and branch formation by expanding on a previously described method (Berleman et al., 2011). We added purified EPS onto agar plates and allowed the transferred material to dry, followed by the addition of M. xanthus wild-type or epsZ- cells adjacent to the EPS and monitored biofilm migration with time-lapse microscopy. As expected, the wild-type strain migrated out in all directions, but in the area coated with EPS, we observed a marked increase in colonization speed and pronounced radial branch structures (Figures 4c and d, Supplementary Movie S4). The epsZ- mutant overall showed very limited migration across the surface, except in the area that had been coated with EPS, where a burst of organized movement was observed (see Figures 4e and f, Supplementary Movie S5). We noted that in addition to increased speed, swarms of both strains became very thin when cells moved over the EPS-coated surface, indicating that cells move across the purified EPS at a lower total cell density (see Supplementary Movies S4 and S5). Also of note is that migration was not stimulated outside of the EPS-coated area, nor did the purified EPS promote migration when mixed directly with cells. Quantification of swarm migration patterns showed that wild-type colonies, after a brief ~2-h lag, swarmed outward at a constant rate to a distance of ~600 microns from the initial edge in 24 h. The epsZ- mutant showed similar behavior for the first 8 h of the assay, then slowed dramatically so that after 24 h, colonies had only expanded ~200 μm outward (Figure 4g). EPS addition rescued epsZ- from migration stalling and restored migration overall to a level 75–90% of the wild type (Figure 4h).
Tracking cell movement within branch structures
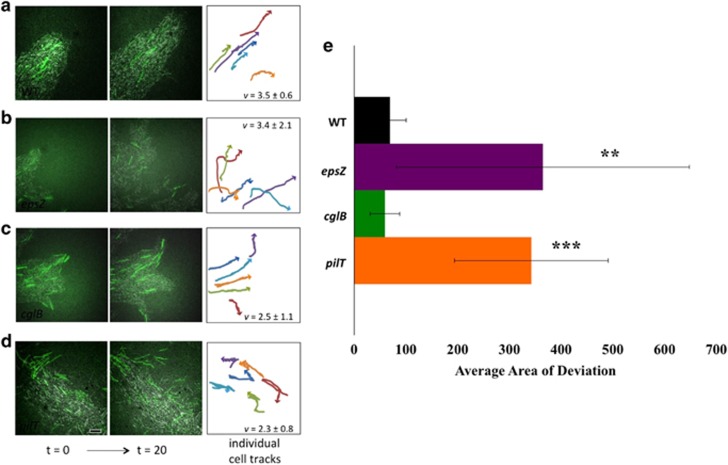
To examine the behavior of individual cells within large swarms, we mixed a small number of cells expressing GFP with a large majority of unlabeled cells (1:50) and plated them on 0.5% CYE agar-coated slides, conditions where only S-motility is utilized. We tracked the movement of the labeled cells by time-lapse fluorescence microscopy. We observed that wild-type cells moved at a velocity of 1.7±0.3 μm min−1 and remained aligned parallel to the direction of the branch as the branch structures elongated (Figure 5a, Supplementary Movie S6). While the overall direction of cell movement was maintained, cell migration was otherwise independent; for example, adjacent cells were observed to occasionally reverse or choose different directions at branch intersections. By contrast, when the epsZ- mutant was examined, branch-like structures were less common but occasionally observed; however, epsZ- structures were unstable and soon dissipated (Figure 5b, Supplementary Movie S7). Cell velocity of GFP tracker cells within EPS− swarms was 1.7±0.5 μm min−1, similar to wild-type velocities, but cell trajectories lacked a consistent direction: cell tracks curved and overlapped, cells traveled in different directions and cell groups dissociated readily. Examination of the cglB- strain showed extensive branching, with similar alignment and cohesiveness as the wild type, but overall branch structures were thinner, with more branch points (Figure 5c, Supplementary Movie S8). Cell movement was more constrained than that of the wild type, although similar cell velocities were observed. Together, these observations support a hypothesis that EPS helps to direct cell movement, with the interesting caveat that EPS is not required as an anchor for T4P movement as EPS− cells are fully motile under S-motility conditions, suggesting that EPS keeps cells aligned and moving together in the same direction and prevents them from dissociating from one another (Figure 5b).
Figure 5.
Tracking cell movement within branch structures. To observe the movement of cells within branch structures, M. xanthus cultures were added to agarose-coated microscope slides, incubated at 32 °C for 6 h to allow branch formation to occur and imaged with time-lapse fluorescence microscopy. Each series of images was captured at the swarm edge, specifically centered on branch tips when possible. Shown are merged differential interference contrast (DIC) microscopy and GFP channel images of 50:1 mixtures of each unlabeled strain with a constitutive GFP-labeled strain at times t=0 (left panels) and t=20 min (center panels). (a) M. xanthus wild-type cells, (b) epsZ-, (c) cglB- and (d) EPS overproducing pilT-. Branch structures are maintained over time in (a) wild type and (c) cglB-, whereas (b) epsZ- is unable to maintain branch organization despite cell movement and (d) pilT- shows aggregates of cells but no branch formation. Analysis of cell movement (right panels) is shown for six cells under each condition with average cell velocity indicated (in μm min−1). Parallel cell trajectories were observed only with wild type and cglB-. Scale bar represents 5 μm. (e) The average area of deviation between the observed path of a cell and its overall trajectory was determined to quantify the ability of labeled cells to maintain a consistent direction over time. P-value thresholds for statistical significance are shown as P⩽0.01:***, 0.01⩽P⩽0.05:**.
In M. xanthus, most T4P-defective mutants also lack EPS (Yang et al., 2011), with exceptions such as a pilT- mutant that overproduces EPS but is defective in T4P retraction and exhibits no S-motility (Wu et al., 1997; Berleman et al., 2011). To determine whether this additional EPS affects cell movements, we examined the behavior of a small population of GFP-labeled wild-type cells mixed with a pilT- mutant strain in a 1:50 ratio (Figure 5d, Supplementary Movie S9). Under these conditions, no coherent branch structures or colony migration were observed, cell alignment was not maintained, and cell movement of GFP tracker cells was characterized by lack of alignment in cell orientation and frequent reversal of gliding direction. Wild-type cells under the same conditions rarely reverse direction. To determine whether the cell tracks observed in Figures 5a–d are significantly different from each other, we further analyzed each cell track by comparing the actual path of movement to a shortest distance trajectory. Labeled cells in wild-type and cglB- conditions move with similarly efficient trajectories (for an explanation, see Supplementary Figure S11), with 68.9±31.6 for wild type and 59.7±28.4 for cglB- (Figure 5e). By comparison, epsZ- conditions resulted in 364.7±283.3 and pilT in 342.4±148.3. These results support the idea that EPS organization promotes efficient cell movement, and provides an important mechanistic clue to microchannel formation. It appears that EPS microchannels require a combination of both EPS synthesis and cell movement.
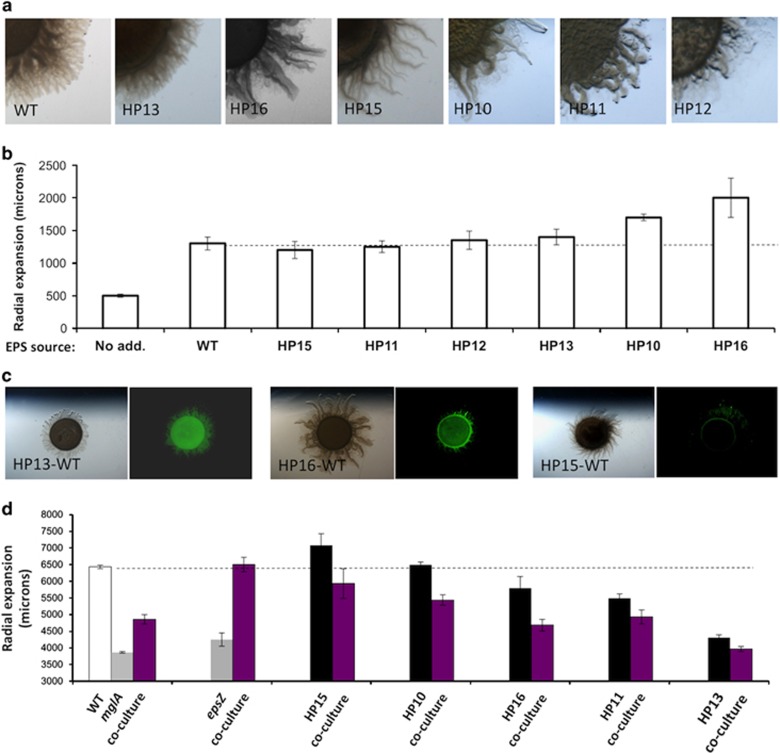
Examining the role of EPS in interactions between different M. xanthus isolates
Our analysis of the wild-type strain DZ2 and its derivative mutant strains strongly indicate that EPS has a critical role in the ability of cells to effectively colonize new surfaces. In soil environments, many M. xanthus strains co-exist within very small areas (Vos and Velicer, 2008). To test the possibility that extracellular EPS structures built by one strain or species could potentially be utilized by another strain, we isolated from local soils several wild M. xanthus strains that displayed social motility (Figure 6a). EPS extracts obtained from these strains were used to determine their effect on social swarming of our laboratory wild-type strain DZ2. As shown in Figure 6b, EPS extracts from all of the isolates tested stimulated S-motility, sometimes to an even greater degree than EPS extracts from DZ2 itself. This result suggests that EPS could be a shared resource, facilitating cooperation between closely related strains. Previous experiments have shown that DZ2 and its derivative mutants when co-cultured will either show additive motility or in some cases, complementation of social defects (Berleman et al., 2011). However, when GFP-labeled DZ2 was co-cultured in 1:1 ratio with wild isolates, we observed reduced motility relative to the migration of either strain in mono-culture, indicating that cooperation between closely related strains is not evident (Figures 6c and d). Fluorescence analysis indicated that in mixtures with isolate HP13, fluorescence signal of the lab wild type was observed to be confluent across the co-culture, but still produced less migration than either mono-culture (Figure 6d). In co-culture with HP16, the GFP signal was constrained to the initial inoculum, indicating that the lab wild type may not have access to the social swarming branches made by the HP16 isolate. In co-culture with HP15, fluorescence signal was greatly reduced, indicating that the GFP-labeled lab strain is either growth-inhibited or killed when cultured with the HP15 isolate. Quantitatively, co-culturing (purple bars) of wild-type DZ2 with derivative mutants such as the non-motile mglA- strain or S-motility-defective epsZ- (gray bars), results in additive or complementary swarming, respectively (Figure 6d). By contrast, every case of co-culturing with wild isolates (black bars) resulted in reduced migration of the co-culture relative to either of the two species cultured independently, suggesting that competition outweighs cooperation and that extracellular EPS microchannels are not automatically an open resource.
Figure 6.
The role of EPS in intra-species interactions. (a) Cultures of natural isolates of M. xanthus were added to 0.5% agar CYE and branch structures of various sizes and shapes were observed. (b) EPS was extracted from these strains and added adjacent to lab wild-type strain DZ2. Relative to a no addition control sample, DZ2 shows increased migration with all EPS fractions tested. (c) Light microscopy (left) and fluorescence microscopy (right) of GFP-labeled DZ2 with wild isolates resulted in several outcomes, from confluent signal (left) to confinement of GFP signal to the initial inoculum (center) to minimal detection of GFP signal (right). (d) Mono-cultures and 1:1 co-cultures with WT DZ2 (purple bars) were prepared and swarm migration quantified for M. xanthus DZ2 (white bar), two DZ2-based single mutants (gray bars) and five natural isolates (black bars). The rate of migration in DZ2 derivatives in co-culture with wild type results in an additive or complementary migration, whereas co-culturing with natural isolates resulted in an inhibitory impact on migration across all strains tested.
The role of EPS in building fruiting body structures
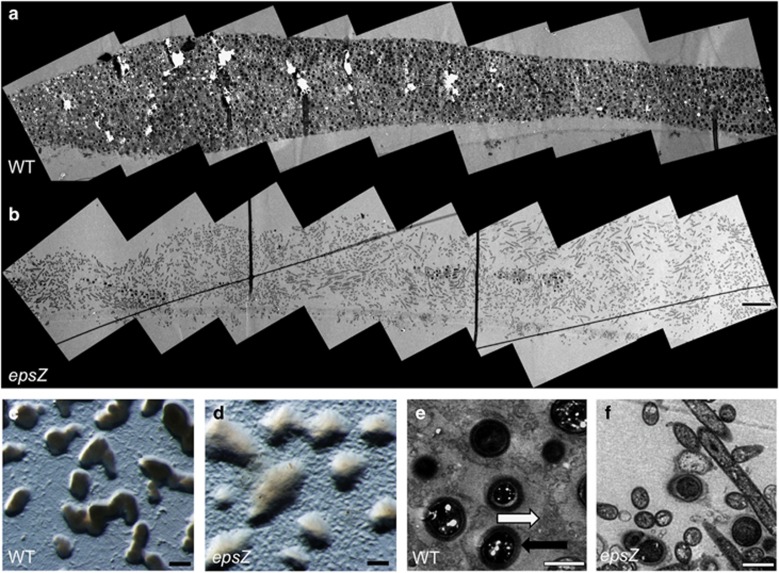
To determine whether EPS channels are involved in other multicellular processes in addition to swarming, we examined M. xanthus cells under conditions where they form fruiting bodies. Panoramic wide-field TEM montage analysis of wild-type and epsZ- strains allowed us to study a large field of view in detail, revealing dramatic differences between these two strains (Figure 7). While the wild-type formed dense, organized structures (Figures 7a, c and e), the epsZ- mutant showed a loose arrangement of cells, with only a few pockets of spores localized at the center of the fruiting aggregates (Figures 7b, d and f). At the multicellular level, the wild type forms dense mounds while epsZ- forms larger, translucent mounds (Figures 7c and d). As shown in Figure 7e, wild-type M. xanthus fruiting bodies displayed EPS (Ruthenium Red-stained) microcompartments (white arrow) surrounding the spores (black arrow). Examination of the epsZ mutant (Figures 7b and f) shows aggregates that are less compact and often less electron-dense. Interestingly, small, centrally located pockets of differentiating cells were observed in the epsZ aggregates (Figure 7b), but EM analysis showed greatly reduced sporulation, poor cellular alignment and a lack of any extracellular EPS structures.
Figure 7.
Panoramic TEM analysis of EPS structures in fruiting bodies of M. xanthus. Comparison of fruiting bodies in (a) M. xanthus wild type and (b) epsZ- mutant with TEM scanning to construct detailed panoramic images of multicellular structures, scale bar, 10 μm. Wild type shows rigid organization, dark staining of EPS and the presence of spores, whereas the epsZ- mutant has a disordered structure and only a few pockets of sporulation, observable in the center of the fruiting aggregate as dark patches. (c, d) Stereomicroscopy of fruiting bodies shows the typical dark, compact mounds of wild type (c) and the light, loose structures formed by epsZ, scale bar, 0.1 mm. (e, f) TEM analysis of cells shows microchannels are observed around spores in wild type, (e) whereas no microchannels are observed in epsZ- and cells show a wide variety of orientation and cell type. Scale bar, 1 μm.
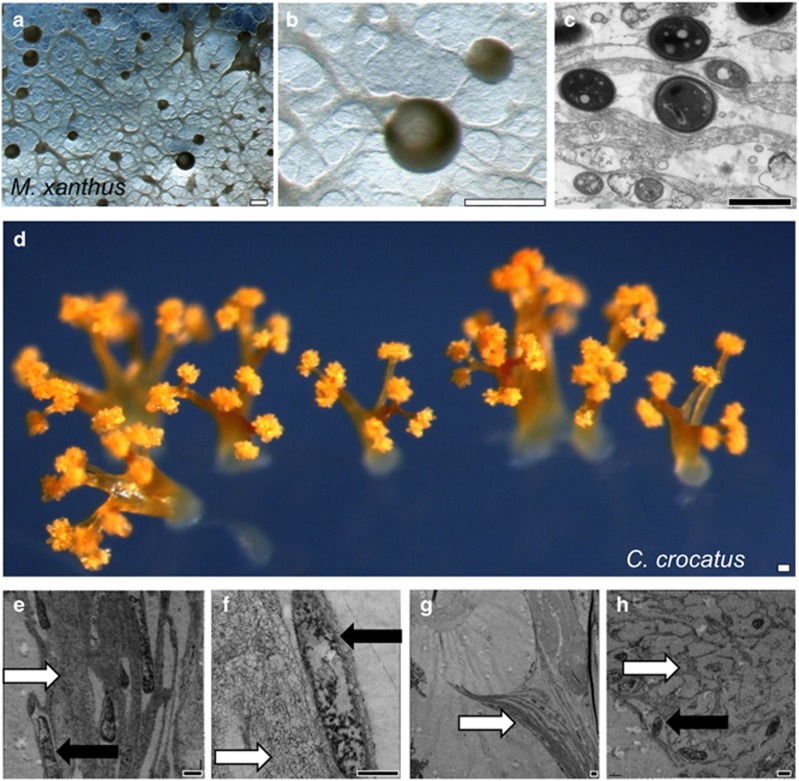
As typical fruiting conditions are distinctly different from swarming conditions, and to determine whether the two processes are at all connected, we devised a modified submerged culture method that would promote both swarming branches and fruiting bodies, by adding molten CYE agar to liquid CYE and producing a mixed environment of solid and liquid (Figures 8a–c). By growing cells in these submerged cultures supplemented with soft agar, both swarming branches and fruiting bodies form. Branches were observed between and connecting fruiting body structures (Figures 8a and b). TEM analysis of Ruthenium Red-stained fruiting bodies showed spores surrounded by thick EPS deposits (Figure 8c). These conditions stimulate M. xanthus to form more pronounced, raised fruiting bodies. Other myxobacterial species, such as Chondromyces crocatus, readily form raised structures of greater complexity that include a stalk, branches and spore-filled sporangioles (Figure 8d). Using ultra-rapid freezing approaches to minimize TEM sample preparation artifacts (McDonald and Auer, 2006), we examined C. crocatus fruiting bodies (Figures 8e–g) and found the stalk structures to be composed primarily of EPS channels (white arrows) with tightly embedded rod cells oriented within channels perpendicular to the agar surface (black arrows). Branches have long open channels all the way up to a concave curvature at their tips where they join to rounded sporangioles (Figure 8g). C. crocatus spores are localized in compartments similar to M. xanthus within the sporangioles (Figure 8h). The process of fruiting body formation in C. crocatus was observed with time-lapse microscopy (Supplementary Movie S10), where large globules were observed to diverge successively into smaller branches, then sporangioles. Swarm migration on the surface was also observed, with swarms entering nascent fruiting bodies and leaving completed structures. Our results show that the ability to build a 3D multicellular body plan can be mediated through well-crafted deposition of EPS material. Unfortunately, we do not have mutant strains of C. crocatus to better study the role of EPS in this organism, as genetic tools for working with this bacterium are unavailable.
Figure 8.
Presence of EPS channels in the 3D structures of Myxobacteria. Comparison of fruiting bodies in (a–c) M. xanthus and (d–h) C. crocatus. (a, b) Stereomicroscopy of M. xanthus grown under conditions that promote both branch migration and fruiting body formation, in which branches connect to spherical fruiting bodies. (c) TEM microscopy of M. xanthus cells within fruiting bodies, showing spores surrounded by layers of EPS. (d) Stereomicroscopy of a field of C. crocatus fruiting bodies, observable as intricately branched orange structures about ~1 mm in height and diameter. The thickness and number of branches varies widely, but the size of each spore sporangiole is similar. (e) TEM analysis of the base of the stalk structures reveals the presence of thick microchannels (white arrows) and several electron-dense cells (black arrows). (f) Further up the stalk, a single cell within a microchannel. (g) A branch tip, where the sporangiole has broken free of the supporting branch. (h) Within the sporangiole, a honeycomb-like organization of filaments surround spores. Scale bars: white, 0.1 mm and black, 1 μm.
Discussion
Biofilm formation has an important role in the life cycle and resistance to antibiotics of many bacterial species. Biofilms can form on practically any surface; however, the ability of bacteria to shape their EPS during biofilm formation is still underappreciated. In this paper, we used genetic and ultrastructural studies to reveal an unexpected level of complexity to EPS organization in M. xanthus. We found that for M. xanthus, EPS is a highly organized structural component of the bacterial biofilm, in microchannels that align cells and promote directed movement. These honeycomb-like channels surround small groups of cells, directing the passage of the bacteria towards new territory. Essentially, the EPS microchannels allow M. xanthus to act as a multicellular organism, accomplishing tasks that no single cell could manage alone. The microchannels guide cell movement and likely serve as convenient anchors for T4P binding and retraction, allowing even those cells that do not make direct contact with the substratum (for example, agar surface) a mechanism to travel forward. EPS does not appear to be required for cell movement, but rather for efficient multicellular migration as epsZ- cells were observed to move but not to maintain consistent orientation or efficient migration patterns (Figures 4 and 5). This helps explain the confounding result that epsZ- colonies can initially spread out on soft agar, but the migration gradually comes to a halt. The EPS microchannels then allow maintenance of cell alignment so that cellular movements are coordinated as cells move towards new territory during swarming or towards aggregation centers during fruiting body formation. Under the conditions tested, mutants that lack the ability to synthesize EPS do not exhibit the microchannels, lack cell alignment and show severe migration defects. However, swarming migration can be rescued by adding purified EPS to the surface. As it is unlikely for the 3D architecture of microchannels to be maintained throughout the purification process, purified EPS may provide a motility substrate rather than a defined radial orientation for cells. This is similar to other polysaccharides, like methylcellulose, which allow for T4P-based migration without showing social motility branch structures. This notion is supported by the fact that eps− strains migrate as a thin layer in regions where the EPS had been provided.
Our work here confirms previous studies that show that M. xanthus cells are typically aligned on surfaces, with the long axis of the cells in parallel to the orientation of the multicellular swarms (Wall and Kaiser, 1998). In those reports, end-to-end cell–cell signaling was proposed to provide the mechanism for maintaining cell orientation and organization. Other reports show that cell–cell packing can occur readily on hard surfaces, where individuals move freely through A-motility (Wei et al., 2011). Gliding movement on soft surfaces has different challenges, and the biophysical constraints of the EPS microchannels provide an alternative mechanism for multicellular organization. Cell–cell signaling may still have an important role, particularly in the area of regulating the construction process rather than in keeping cells aligned. We can hypothesize that microchannel construction occurs through a succession of events including: surface detection, EPS secretion and proper shaping of EPS by the movement of cells. The specific mechanism for correct localization of EPS construction will require further experimentation, but one prediction would be that extracellular enzymes are present on the outside surface of cells allowing dehydration reactions of polysaccharides into a cohesive microchannel structure. Cell migration through microchannels would allow efficient colonization of a surface as the crowded cells in the swarm interior push towards the low cell density/higher nutrient areas present at the swarm edge.
While swarm migration across surfaces allows for colonization of new territory, the synthesis of EPS microchannels may also provide a structural support for the raised structures that are prominent during fruiting body formation both in simple M. xanthus mounds as well as in the complex fruiting bodies of C. crocatus. Our data indicate EPS provides the primary structural component of the fruiting bodies as this material is not observed in fruiting bodies of an epsZ- strain or when carbohydrate-staining Ruthenium Red is not used in TEM analysis. Microchannels were observed within the stalk of C. crocatus and EPS microcompartments observed surrounding spores of both species within the fruiting bodies.
The transition from unicellular to multicellular life requires extracellular structural components that bind cells together. Plants use cellulose, hemicellulose, pectin and lignin to build cell walls (Yang et al., 2013); animals use collagen and other extracellular proteins such as laminin and glycoproteins (Chagnot et al., 2012). Here, we found that bacteria use EPS to help determine multicellular organization. Our analysis of intra-species interactions suggests that although purified EPS from various sources may serve as general substrate for migration of cells, there was no implicit sharing of EPS by co-culturing of closely related strains, but these interactions may also be greatly influenced by other factors such as cell–cell signals or antibiotics. This suggests that microbial community formation could depend on whichever organism has the keystone role in providing the predominating EPS structure. Given its importance in intra-species interactions, it will be of great importance in future experiments to study the role of EPS in inter-species interactions and microbial community structure.
Acknowledgments
We would like to thank Dr Kyungyun Cho for help with M. xanthus isolations and providing us with samples of C. crocatus. Thanks also to Ahmed Hassan for help with 3D visualization. This work was supported by the National Institutes of Health 5R01GM020509 and 3R01GM020509-36S1 to DRZ, and P01GM051487-15 to MA, by Lab directed research development funds from the Office of Biological and Environmental Research of the US Department of Energy under contract number DE-AC02-05CH11231 (to MA) and the US Department of Energy VFP program (to JEB).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alegado RA, Brown LW, Cao S, Dermenjian RK, Zuzow R, Fairclough SR et al. (2012). A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. eLife 1: e00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum MM, Kainović A, O'Keeffe T, Pandita R, McDonald K, Wu S et al. (2009). Characterization of structures in biofilms formed by a Pseudomonas fluorescens isolated from soil. BMC Microbiol 9: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. (2013). Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci USA 110: E1621–E1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Kirby JR. (2009). Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev 33: 942–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleman JE, Vicente JJ, Davis AE, Jiang SY, Seo YE, Zusman DR. (2011). FrzS regulates social motility in Myxococcus xanthus by controlling exopolysaccharide production. PloS One 6: e23920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone NW. (2013). Why did eukaryotes evolve only once? Genetic and energetic aspects of conflict and conflict mediation. Philos Trans R Soc Lond B Biol Sci 368: 20120266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante VH, Martínez-Flores I, Vlamakis HC, Zusman DR. (2004). Analysis of the Frz signal transduction system of Myxococcus xanthus shows the importance of the conserved C-terminal region of the cytoplasmic chemoreceptor FrzCD in sensing signals. Mol Microbiol 53: 1501–1513. [DOI] [PubMed] [Google Scholar]
- Campos JM, Zusman DR. (1975). Regulation of development in Myxococcus xanthus: effect of 3':5'-cyclic AMP, ADP, and nutrition. Proc Natl Acad Sci 72: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagnot C, Listrat A, Astruc T, Desvaux M. (2012). Bacterial adhesion to animal tissues: protein determinants for recognition of extracellular matrix components. Cell Microbiol 14: 1687–1696. [DOI] [PubMed] [Google Scholar]
- Chang BY, Dworkin M. (1996). Mutants of Myxococcus xanthus dsp defective in fibril binding. J Bacteriol 178: 697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen M, Jakovljevic V, Søgaard-Andersen L, Maier B. (2009). High-force generation is a conserved property of type IV pilus systems. J Bacteriol 191: 4633–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey ME, O'toole GA. (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64: 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid W. (2000). Biology and global distribution of myxobacteria in soils. FEMS Microbiol Rev 24: 403–427. [DOI] [PubMed] [Google Scholar]
- Habimana O, Moretro T, Langsrud S, Vestby LK, Nesse LL, Heir E. (2010). Micro ecosystems from feed industry surfaces: a survival and biofilm study of Salmonella versus host resident flora strains. BMC Vet Res 6: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun H, Chung J, Kim J, Lee JS, Kwon B-M, Son K-H et al. (2008). Isolation of Sorangium cellulosum carrying epothilone gene clusters. J Microbiol Biotechnol 18: 1416–1422. [PubMed] [Google Scholar]
- Kraemer SA, Velicer GJ. (2011). Endemic social diversity within natural kin groups of a cooperative bacterium. Proc Natl Acad Sci USA 108(Suppl 2): 10823–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun H, Ma X, Lu A, Lux R, Zusman D et al. (2003). Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc Natl Acad Sci USA 100: 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T-H, Huang S-H, Wu C-C, Liu H-H, Jinn T-R, Chen Y et al. (2013). Inhibition of Klebsiella pneumoniae growth and capsular polysaccharide biosynthesis by Fructus mume. Evid Based Complement Altern Med 2013: 621701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauriello EM, Zusman DR. (2007). Polarity of motility systems in Myxococcus xanthus. Curr Opin Microbiol 10: 624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauriello EM, Mignot T, Yang Z, Zusman DR. (2010). Gliding motility revisited: how do the myxobacteria move without flagella? Microbiol Mol Biol Rev 74: 229–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL, Auer M. (2006). High-pressure freezing, cellular tomography, and structural cell biology. Biotechniques 41: 137 139, 141 passim. [DOI] [PubMed] [Google Scholar]
- Nan B, Bandaria JN, Moghtaderi A, Sun I-H, Yildiz A, Zusman DR. (2013). Flagella stator homologs function as motors for myxobacterial gliding motility by moving in helical trajectories. Proc Natl Acad Sci USA 110: E1508–E1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudleman E, Wall D, Kaiser D. (2006). Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol Microbiol 60: 16–29. [DOI] [PubMed] [Google Scholar]
- Palsdottir H, Remis JP, Schaudinn C, O'Toole E, Lux R, Shi W et al. (2009). Three-dimensional macromolecular organization of cryofixed Myxococcus xanthus biofilms as revealed by electron microscopic tomography. J Bacteriol 191: 2077–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelling AE, Li Y, Cross SE, Castaneda S, Shi W, Gimzewski JK. (2006). Self-organized and highly ordered domain structures within swarms of Myxococcus xanthus. Cell Motil Cytoskeleton 63: 141–148. [DOI] [PubMed] [Google Scholar]
- Remis JP, Wei D, Gorur A, Zemla M, Haraga J, Allen S et al. (2013). Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ Microbiol 16: 598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AM, Spormann AM. (1999). Genetic and molecular analysis of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J Bacteriol 181: 4381–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabra W, Zeng AP, Deckwer WD. (2001). Bacterial alginate: physiology, product quality and process aspects. Appl Microbiol Biotechnol 56: 315–325. [DOI] [PubMed] [Google Scholar]
- Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. (2011). Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci USA 108: 7559–7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland IW, Thomson S. (1975). Comparison of polysaccharides produced by Myxococcus strains. J Gen Microbiol 89: 124–132. [DOI] [PubMed] [Google Scholar]
- Vos M, Velicer GJ. (2008). Natural variation of gliding motility in a centimetre-scale population of Myxococcus xanthus. FEMS Microbiol Ecol 64: 343–350. [DOI] [PubMed] [Google Scholar]
- Wall D, Kaiser D. (1998). Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc Natl Acad Sci USA 95: 3054–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, Kaiser D. (1999). Type IV pili and cell motility. Mol Microbiol 32: 1–10. [DOI] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21: 319–346. [DOI] [PubMed] [Google Scholar]
- Wei X, Pathak DT, Wall D. (2011). Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol 81: 315–326. [DOI] [PubMed] [Google Scholar]
- Wu SS, Wu J, Kaiser D. (1997). The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol Microbiol 23: 109–121. [DOI] [PubMed] [Google Scholar]
- Yang F, Mitra P, Zhang L, Prak L, Verhertbruggen Y, Kim JS et al. (2013). Engineering secondary cell wall deposition in plants. Plant Biotechnol J 11: 325–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hu W, Chen K, Wang J, Lux R, Zhou ZH et al. (2011). Alanine 32 in PilA is important for PilA stability and type IV pili function in Myxococcus xanthus. Microbiol Read Engl 157: 1920–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian P, Burke N, White DJ, Hartzell PL. (2003). Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol Microbiol 49: 555–570. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang H, Fang X, Stackebrandt E, Ding Y. (2003). Improved methods of isolation and purification of myxobacteria and development of fruiting body formation of two strains. J Microbiol Methods 54: 21–27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.