Abstract
In 1970, it was well accepted that the central role of lipids was in energy storage and metabolism, and it was assumed that amphipathic lipids simply served a passive structural role as the backbone of biological membranes. As a result, the scientific community was focused on nucleic acids, proteins, and carbohydrates as information-containing molecules. It took considerable effort until scientists accepted that lipids also “encode” specific and unique biological information and play a central role in cell signaling. Along with this realization came the recognition that the enzymes that act on lipid substrates residing in or on membranes and micelles must also have important signaling roles, spurring curiosity into their potentially unique modes of action differing from those acting on water-soluble substrates. This led to the creation of the concept of “surface dilution kinetics” for describing the mechanism of enzymes acting on lipid substrates, as well as the demonstration that lipid enzymes such as phospholipase A2 (PLA2) contain allosteric activator sites for specific phospholipids as well as for membranes. As our understanding of phospholipases advanced, so did the understanding that many of the lipids released by these enzymes are chiral information-containing signaling molecules; for example, PLA2 regulates the generation of precursors for the biosynthesis of eicosanoids and other bioactive lipid mediators of inflammation and resolution underlying disease progression. The creation of the LIPID MAPS initiative in 2003 and the ensuing development of the lipidomics field have revealed that lipid metabolites are central to human metabolism. Today lipids are recognized as key mediators of health and disease as we enter a new era of biomarkers and personalized medicine. This article is my personal “reflection” on these scientific advances.
Keywords: allosteric regulation, cell signaling, eicosanoid biosynthesis, enzyme mechanism, inflammation, lipid signaling, lipid-protein interaction, membrane enzyme, phospholipase, lipidomics
Introduction
Early on I did not know I was going to be a scientist. I was aware of science as a possible career as two of my maternal uncles were chemists. I also enjoyed my experiences in science; for example, my teachers in biology and chemistry at Senn High School in Chicago inspired me to successfully compete in the 1958 city science fair with a project on air pollution. However, I was equally inspired by my English teacher who encouraged me to consider organizational leadership directions. Thus, my intent when I matriculated at Yale was to obtain a liberal arts education. But in choosing a major I was influenced by William von Eggers Doering who first exposed me to the beauty of three-dimensional stereochemistry as a sophomore when I took organic chemistry for chemists—although I was not yet a chemistry major, I took the course and I was hooked. Doering's class stimulated me to major in chemistry and to carry out undergraduate research on the stereochemistry of the HCl elimination reaction with a great role model for me, Harry H. Wasserman.
My fascination with stereochemistry continued as I transitioned to graduate school in the Chemistry Department at Harvard University. During my second year, we were given a 1-week cumulative exam in which we were to make an “original” proposal. I thought about how carbon had four bonds and could form two isomers and wondered how many isomers would be possible for phosphorus, which could have five bonds. This led me to learn what inorganic chemists already knew well, namely that phosphorus bonds were arranged in a trigonal bipyramid or square pyramid with both equatorial and apical bonds. I then reasoned that in going from carbon to phosphorus, the number of isomers increases from 2 isomers to 30 isomers, so I proposed synthesizing a pentacoordinate phosphorus with different links so as to make “orbital isomers” of pentacoordinate phosphorus. I received a C (failing) grade with the professor's comment that orbital isomers could not be made. However, my graduate research with the father of enzyme mechanisms and their stereochemistry, Frank H. Westheimer (1) (Fig. 1), was on the mechanism of phosphate ester hydrolysis. As my thesis work proceeded and I realized that the intermediate or transition state in this process was pentacoordinate phosphorus, I was able to show that orbital isomers could exist and the concept of pseudorotation was key to understanding the mechanism (2). Later, I independently applied pseudorotation to understanding the mechanism of ribonuclease (3).
FIGURE 1.

Frank H. Westheimer (right) in his 90s pictured with R. Stephen Berry (left) who first described the concept of pseudorotation in the trigonal bipyramid structure of PF5.
My graduate work had kindled a strong interest in the enzymology of phosphate groups, so I stayed in Boston for a postdoctoral position in the Department of Biological Chemistry at Harvard Medical School on phospholipid biosynthesis (4) with Eugene P. Kennedy (5) (Fig. 2), who had discovered the metabolic pathways by which phospholipids are made. At that time, the prevailing view was that lipids played a central role in metabolism and especially in the storage and generation of energy; on the other hand, membranes were thought to be just a sea of unimportant lipids or “grease” in which membrane proteins carried out their supposedly important duties in “the viscous phospholipid bilayer solvent” (6). Thus, the scientific community was studying membrane structure but only by looking at the functional role of proteins in membranes. I had joined the lab with the intent of finding the enzyme that makes an unusual P–C bond in Tetrahymena phospholipids. As a precursor to this task, I started working with the more typical enzymes in the laboratory. As I began learning the lab's protocols, I was stunned to realize that scientists in the field were using detergents without having any idea at the chemical level why these lipids were needed! My recognition of this may have been related to the fact that I was an unusual addition to the group because I came from a chemistry background, and so I always wanted to know what was happening at the molecular level. The discovery that lipids were necessary for enzymatic function planted the seed of what was a radical view at the time that these detergents are lipids and they have essential functions.
FIGURE 2.
Symposium on Cellular Regulation and Membranes in honor of Eugene P. Kennedy on his 65th birthday organized by Edward Dennis on April 12–14, 1985 in Woods Hole, MA. A, Albert Lehninger; B, Fritz Lipmann; C, Edward Dennis; D, Marilyn Rumley; E, Adelaide Kennedy; F, Roberta Friedkin; G, Phyllis Elfman; H, Morris Friedkin; and I, Eugene Kennedy. Kennedy and Friedkin were Lehninger's first two graduate students, and Kennedy carried out postdoctoral studies with Lipmann. Rumley was a technician and Elfman was a secretary in the Kennedy laboratory. Both Lehninger and Lipmann passed away the following year.
I started my independent scientific career in 1970 as a new Assistant Professor at the University of California at San Diego (UCSD) intent on pursuing mechanistic questions about these enzymes. I believed that the optimal way to start was with an enzyme that acted in or on the membrane, and particularly one with activity toward membrane phospholipids. This decision started a theme that would continue throughout my research career—from developing the early methodology needed to study membrane-active enzymes, to tracking down the destinies and functions of lipids in the corresponding metabolic pathways, to creating conferences and consortia to harness and build on our increasing technological and conceptual breakthroughs in lipid enzymology, to seeing the specific implications of these scientific advances on understanding and improving human health. It has been very exciting to have contributed to these advances! Throughout these adventures, I have been very fortunate to work with fabulous colleagues, both internal and external to my lab, and I cannot begin to thank each of them appropriately within the space allotted me here; I have done my best to cite their contributions in the reference section, which only hints as to their importance in my scientific travels.
New Methods and Mechanistic Concepts for Membrane-active Enzymes
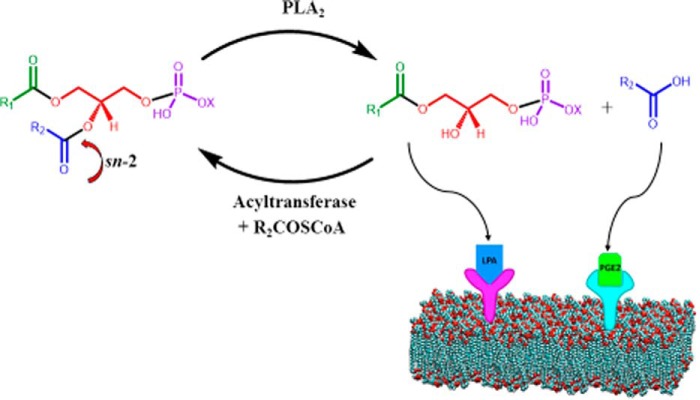
My first NIH RO1 grant was to study the reconstitution of phosphatidylserine decarboxylase—a holdover from my time in the Kennedy lab—in phosphatidylserine-containing surfactant mixed micelles. This work was designed to confirm that proteins were actually interacting with phospholipids in a productive manner. If the enzyme was active, CO2 would be released, which could be measured by a novel gas chromatography method (7). However, a serendipitous conversation with Dr. Beatriz M. Braganca, a scientist on sabbatical from India working in a neighboring lab, turned my attention to cobra venom as a commercially available and plentiful source of phospholipase A2 (PLA2) (8). This water-soluble enzyme hydrolyzes the fatty acid on the sn-2 position of membrane phospholipids to produce a lysophospholipid and a free fatty acid. It is more stable than phosphatidylserine decarboxylase, and its basic characteristics simplify several technical challenges. Finally, it also has many important biological functions including biosynthesis and generating lipid cell signals (9) (Fig. 3). We jumped at the opportunity to switch to this more advantageous model, and our studies on PLA2 led to my second RO1 grant, now in its 40th year.
FIGURE 3.
Phospholipases A2 function as degradative enzymes when they produce lysophospholipids and free fatty acids as products, as biosynthetic enzymes when the lysophospholipid product is reacylated with a PUFA to produce important phospholipids, and as signaling enzymes when the products are converted to agonists of GPCRs. Adapted from Ref. 9.
With PLA2 under study, our first challenge was to understand why detergents were necessary to obtain enzymatic activity. This led us to pursue NMR techniques to characterize the interaction of phospholipids with surfactants, principally the non-ionic detergent Triton X-100 (10–12). We proposed that one role of the detergent was to solubilize the phospholipid into mixed micelles, which provided a surface to present the substrate to the enzyme (8). Another possible function of the Triton X-100 would be to solubilize a membrane-localized enzyme so that it could be present in aqueous solution and be available to the substrate; however, the fact that PLA2 is already water-soluble simplified the kinetic problem enormously. This experimental system provided an ideal model for understanding protein-membrane interactions. But it was an uphill battle to convince those studying bilayer membranes that mixed micelles had any biological relevance! It was a battle worth fighting though, as surfactant micelles (and later bicelles) are now a mainstay in NMR studies of membrane proteins (13–16). Indeed, NMR work on mixed micelles of surfactants and phospholipids established the physical chemical principles that would allow the surfactants to be used to study a variety of phospholipids, including sphingolipids. Moreover, this established the generality of the mixed micelle system for studying protein-membrane interactions (17, 18).
A second critical step in our studies was formulating the concept of “surface dilution kinetics” (19, 20). This introduces two pre-catalytic steps into the reaction, first the enzyme binding to the membrane and then the enzyme separately finding and binding to its substrate in the two-dimensional interface (Fig. 4). This framework allowed us to translate existing thinking about enzyme mechanism to the study of enzymes acting in/on membranes. I presented this concept at the first California Membrane Conference, March 13–17, 1972, held in Squaw Valley (the first meeting in what became the Keystone Symposia) and also reported our first micelle studies (10). Eventually, the surface dilution concept was generalized (21) to apply to many other enzymes that act on phospholipids (22, 23).
FIGURE 4.
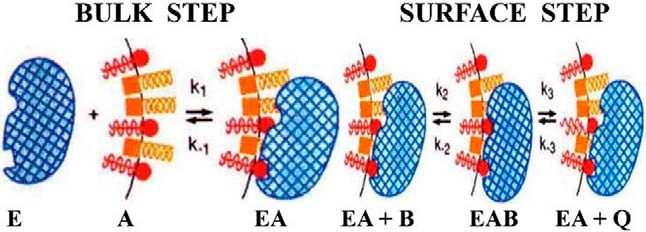
The concept of “surface dilution kinetics” requires two distinct steps for enzymes (E) acting on substrates (B) contained in lipid-water interfaces or surfaces (A) of membranes and micelles. Shown is an illustration of the application of “surface dilution kinetics” to the phospholipase A2- (blue) catalyzed hydrolysis of phospholipid substrate (red) contained in mixed micelles with nonionic surfactants such as Triton X-100 (yellow). The enzyme first associates with the mixed micelle surface (Bulk Step), and in a subsequent step (Surface Step), the enzyme associated with the micelle binds a phospholipid substrate molecule in its catalytic site and carries out hydrolysis, producing as products a lysophospholipid and a fatty acid, which may be released to solution or may be retained in the micelle surface. Reprinted from Ref. 21, adapted from Refs. 19 and 20.
Using the mixed micelle system and the surface dilution kinetic approach, it was now possible to pursue the mechanism of action of PLA2 (24). We first discovered that the enzyme was active only in the presence of certain phospholipids. This allowed application of the then underappreciated concept of allostery developed by Jean-Pierre Changeux (25), a concept I first learned about in the mid-1960s when Frank Westheimer returned from lecturing at the Pasteur Institute in Paris. Specifically, we hypothesized that PLA2 contained an “activator site,” although the enzyme functions as a monomer (a departure from Changeux's original concept), and that when the water-soluble enzyme associated with the mixed micelle (membrane), it underwent a conformational change (at that time, represented simply as a square changing to a circle) to enable binding of the substrate phospholipid at the catalytic site. Our data showed that the whole surface of the enzyme interacted with the micelle (membrane), perhaps sensing as many as 30 phospholipid head groups at once, and that one of those lipids had to be a phosphocholine-containing lipid that acted at a “specific site” that was distinct from the catalytic site (26, 27). This allosteric activator site was later confirmed by kinetics (28) and X-ray crystallography (29, 30) and verified by mutational studies (31) and gene synthesis (32).
Detailed binding (33) and kinetic studies on PLA2 (34–38) and the determination of the rate constants for acyl and phosphoryl migration in the lysophospholipid product (39) were critical for elucidating the kinetic analysis of PLA2, including its allosteric activation (40, 41) and varying activity on different membrane forms (42–44). The same approaches were also critical in my lab's study of the related mammalian lysophospholipases (45–49) and indeed many other phospholipases (22, 50–58). Along the way, we've also explored PLA2's evolution from snake venom to human forms (59) and its reported inhibition by lipocortin (annexin 1), which we demonstrated occurred via “surface dilution”; thus it was not true inhibition as claimed in the literature (60–62). We also realized early on that having tools to probe PLA2 function would be useful, and so we focused attention on the discovery, design, and characterization of a large variety of chemical inhibitors of PLA2 (63–76). Application of these inhibitors—in tandem with experiments probing enzyme aggregation (77)—not only provided insight into the catalytic mechanism, but also played a key role in advancing the understanding of PLA2 and its regulation in biological systems (78).
A Turning Point for Lipid Signaling
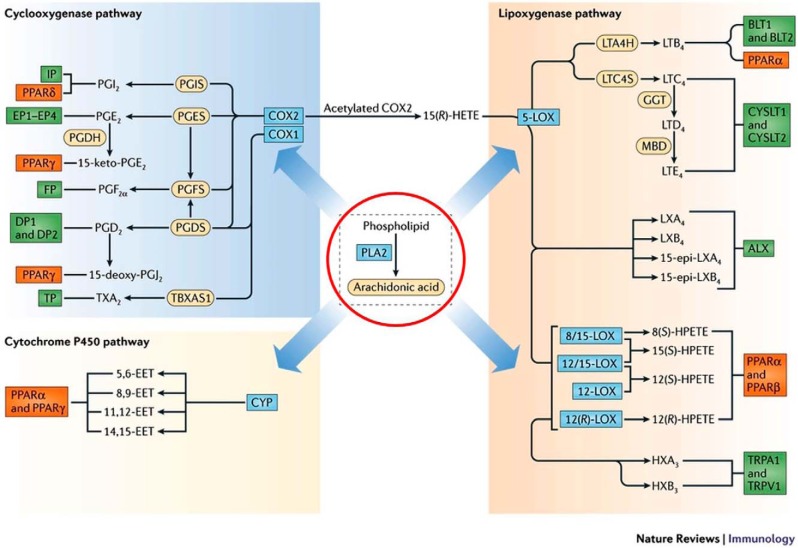
Despite the great progress in understanding PLA2 mechanisms we felt we were making, the biological implications for these studies remained obscure to the field at large. However, John Vane's discovery that aspirin and other non-steroidal anti-inflammatory drugs (NSAIDs) worked by acting on lipid biosynthesis (79) changed the landscape: suddenly, arachidonic acid metabolites—specifically prostaglandins—were important. Bengt Samuelsson's (Fig. 5, top) discovery of the first leukotrienes (80) further cemented the relevance of lipid mediators. This was recognized with the awarding of the 1982 Nobel Prize in Physiology or Medicine to Sune Bergström, John Vane, and Bengt Samuelsson. The 1990 Nobel Prize in Chemistry was received by E. J. Corey (Fig. 5, bottom), who had previously synthesized many of the key chiral eicosanoids. Of course, we were excited to see that the new questions raised by their discoveries pointed back to our favorite enzyme, PLA2. Specifically, free arachidonic acid is at very low levels in cells and is generated spontaneously as needed for prostaglandin/leukotriene biosynthesis; PLA2 is responsible for the initiation of this arachidonic acid cascade (9, 78, 81) (Fig. 6).
FIGURE 5.

Two key contributors to eicosanoid chemistry. Top, Edward Dennis and Bengt Samuelsson celebrating Samuelsson's 80th birthday at the first Lipid Mediators in Health and Disease Symposium held at the Nobel Forum, Stockholm, Sweden on August 27–29, 2014. Bottom, Edward Dennis introducing E. J. Corey as Keynote Speaker at the VI International Conference on Eicosanoids and Other Bioactive Lipids in Cancer, Inflammation, and Related Diseases held September 12–15, 1999 in Boston, MA.
FIGURE 6.
Central role of phospholipase A2 in eicosanoid biosynthesis and receptor signaling. Reprinted from Ref. 81.
These conceptual linkages led many investigators, myself included, to pursue the intracellular forms of PLA2 responsible for arachidonic acid release. The first two “FASEB Summer Conferences on Phospholipase A2,” which I organized with Moseley Waite, included watershed moments. In 1988, two groups announced the sequence and cloning of the human secreted sPLA2, and in 1992, two groups announced the sequencing and cloning of the human cytosolic cPLA2. In 1995, at the third FASEB Summer Conference, our lab reported on the identification, complete purification, and characterization of the Ca2+-independent iPLA2 (82). At that time, the explosion of PLA2s initiated a discussion among the conference attendees that led to the Group numbering system for PLA2s (83), which with several evolutions (84–86) is universally used today to distinguish the 16 groups and many subgroups of PLA2 (9). In the meantime, another unique form of mammalian sPLA2 secreted from macrophages was found, namely the Group V sPLA2 (87). And on the functional side, iPLA2 was sequenced and cloned (88, 89), and its remodeling function in macrophages (90–93) and unusual abundance in brain (94) were established.
In the 1980s, my laboratory turned its attention to the function of these enzymes in cellular metabolism and the generation of lipid cell signals—what I viewed as the most exciting new frontiers in cell biology and immunology. To kick start this new in vivo direction, I first needed to know more about these complex systems. Thus, in 1983, I received a Guggenheim Fellowship to spend a sabbatical with Manfred Karnovsky at Harvard Medical School, an expert on primary macrophage cells, and with Lawrence “Laurie” Levine at Brandeis University, the developer of radioimmune assays for eicosanoids. When I returned to UCSD, we decided to pursue macrophages as the archetypal mammalian cell—the “E. coli of mammalian cells”—in which to explore PLA2 function and eicosanoid mediator production. In macrophage cell lines, the numerous types of PLA2s that existed in a single cell type were characterized (95, 96) and the role of PLA2 in activating the arachidonic acid cascade by a variety of agonists was explored (97, 98). This led to characterizing the eicosanoids produced downstream of PLA2 and their autocrine and paracrine function in lipid signaling in the cellular milieu (99–101). This all became considerably more complex as we realized that the activation of eicosanoid synthesis could involve activation of multiple PLA2s (102–106) stimulated by the products of other PLA2s from distinct subcellular locations (107, 108).
Once the role of phospholipases in the field of signal transduction was established and the central role of the downstream eicosanoid mediators derived from arachidonic acid released by PLA2 was shown, our lab's interests naturally shifted to the emerging broader role of lipid mediators in signal transduction. Fred Fox invited me to organize the first Keystone Symposium on Cellular Activation and Signal Initiation in 1988, and we brought together Tony Hunter, Robin Irvine, and Michael Berridge for this conference, which launched great interest in this field. Chairing this conference also reminded me of my early interests in organizational leadership, as I greatly enjoyed organizing what I hoped would lead to a creative and engaging scientific meeting. It was no surprise that I had gravitated toward this kind of role in the past, and I made it a point to participate in similar opportunities in the future. Throughout the years, I have greatly valued these organizational leadership experiences, whether as president of the board of the Keystone Conferences, chair of the Gordon Research Conferences Board of Directors, chair of my department, or Editor-in-Chief of the Journal of Lipid Research. After the 1988 conference, I organized several more Keystone conferences on lipid signaling including an important one in 1994 on Lipid Second Messengers (109) and the 1989 Gordon Research Conference on Lipid Metabolism where this topical theme continued.
It was during this period that it became recognized that there are numerous lipid second messengers, many generated by phospholipases and lipid kinases/phosphatases, that play critical roles in cell signaling (51). It took a great deal of time and effort before the important roles of lipid cell signals were recognized by the broader biochemical community, which, at the time, was much more focused on signaling and regulation by phosphorylation and dephosphorylation of proteins and more traditional receptors. One challenge the field faced was in communicating the newly discovered roles of lipids in cell signaling. As a result, Ralph Bradshaw and I made it a priority to highlight the central role of lipid cell signaling and integration of lipid signaling with more familiar pathways (110) as we organized and edited the three-volume Handbook of Cell Signaling, which appeared in 2003 (111).
Back in my laboratory, lipid cell signaling had always been focused on the metabolites of arachidonic acid—the eicosanoids and related oxidized polyunsaturated fatty acids (PUFA)—that activate a large variety of G-coupled protein receptors (GPCRs) (81). We had already learned through our work on cobra venom sPLA2 that inhibitors could help us interrogate enzymes, but the inhibitors that worked on sPLA2 didn't necessarily translate well to the mammalian systems, especially the intracellular PLA2s that now occupied our attention. We thus expanded our initial inhibitor discovery work first to mammalian sPLA2s (112), including the use of antisense technology (113), and then to other PLA2 groups—especially the intracellular cPLA2 and iPLA2—by activated ketones (114, 115), fluorophosphonates (116), and other traditional inhibitors (98). In the last decade, this has expanded into a long collaboration with Professor George Kokotos from the University of Athens (9), in which we are designing and testing novel inhibitors including oxoamides (117–120), polyfluoroketones (121, 122), and thiazolyl ketones (123). Specifically, we've leaned on insights from molecular dynamics and simulations to inform rational drug design and medicinal chemistry efforts capped off with extensive biochemical characterization. Some of these inhibitors have shown specificity in important neurological disease models including experimental autoimmune encephalomyelitis, Wallerian degeneration, hyperalgesia, and spinal cord injury (124–127).
The Mechanism Comes Together
While these exciting changes and advances were occurring in the field at large and in my lab, we hadn't forgotten our goal to understand how these enzymes work at the molecular level. Research on enzymes that acted on soluble substrates had reached a high level of sophistication in their description of the catalytic mechanism, and I hoped that we might be able to do the same for our favorite enzyme! We therefore continued our detailed kinetic, mechanistic, crystallographic, NMR, and other structural studies on these enzymes. Many studies were carried out on iPLA2, especially on its aggregation using radiation inactivation approaches (82, 128) and its additional lysophospholipase and acyl transferase activities (129) and similar catalytic activities for cPLA2 (130, 131). We were particularly intrigued by the discovery of the dramatic and specific activation of cPLA2 by phosphatidylinositol 4,5-bisphosphate (PIP2) (132–134) at a uniquely defined allosteric activator site as it both generalized our previous discovery regarding allosteric sites but also established a novel mechanism to accomplish the task. In the last decade, the lab has also made use of a new approach utilizing deuterium exchange mass spectrometry (DXMS) (135). This approach has traditionally been used to study protein-protein and protein-ligand interactions, but we applied it to look at the interaction of water-soluble proteins with membranes and demonstrated this approach with PLA2 (136–138). We have now studied four of the major types of PLA2, including the secreted (139), cytosolic (140, 141), and Ca2+-independent forms (142), as well as lipoprotein-associated/platelet-activating factor (PAF) acetyl hydrolase (143) and the interactions of the latter with lipoproteins such as HDL (144). These DXMS methods also led to a better understanding of the catalytic action of an unusual form with a Cys in the catalytic site (145).
We also launched a collaboration with my UCSD colleague J. Andrew McCammon, leading to the incorporation of molecular dynamics and simulations guided by DXMS into our consideration of how the enzymes work. This technique was used to create structural complexes of each enzyme with a single phospholipid substrate molecule in its catalytic site to visualize substrate extraction. Simulations of the enzyme-substrate-membrane system revealed important information about the mechanisms by which enzymes associate with the membrane (146) and then extract and bind their phospholipid substrate (147) (Fig. 7). This allowed us to extend early observations of allosteric sites on PLA2s to include membranes as allosteric ligands. We also examined the effects of potent specific inhibitors of these PLA2s such as pyrophenone and oxoamides for cPLA2 (148) and various fluoroketones with iPLA2 (149–151) using these techniques.
FIGURE 7.
Cytosolic phospholipase A2 extracting, binding, and hydrolyzing phospholipid substrate releasing arachidonic acid for eicosanoid biosynthesis (147). Cover for July, 2015 Journal of Lipid Research issue introducing a Thematic Reviews series on Phospholipases (51).
These combined results led us to a complete model of the catalytic cycle of two different human PLA2s, cPLA2 and iPLA2 (147). After some forty years of studying PLA2, a most fulfilling moment for me was when our lab developed detailed simulations (and representative movies) depicting the association of PLA2 with a membrane during which the membrane acts as an allosteric ligand of the enzyme's interfacial surface by shifting its conformation from a closed (inactive) state in water to an open (active) state. This is followed by the extraction of a substrate phospholipid from the membrane into the active site of the enzyme, and then achievement of the energetically optimal conformation for catalysis and finally diffusion of the products into the membrane surface (147). My quest to match the quality of information available for “normal” water-soluble enzyme systems, the initial steps of which were recognized 15 years earlier by my receipt of an award from the ASBMB in lipid enzymology (Fig. 8), had been attained.
FIGURE 8.

Walter Shaw, President of Avanti Polar Lipids, presenting the ASBMB Avanti Award in Lipids to Edward Dennis on June 5, 2000 in Boston MA.
Even more exciting, this work led to a general hypothesis that “membranes serve as allosteric regulators of enzymes that act in or on membranes” (135, 147) (Fig. 9). Traditionally, allosteric regulators had been conceived of as small molecules binding to “topographically distinct sites” from the catalytic site, such as phosphatidylcholine for sPLA2 and PIP2 for cPLA2, and generally on oligomeric proteins exhibiting sigmoid kinetics. Here, we have an enormous ligand (when compared with the size of a typical enzyme) binding to the enzyme but effecting a significant conformational change that is essential for catalysis. We anticipate that this new concept will have broad applicability to many systems including membrane proteins and provide a new paradigm for understanding lipid-protein interactions.
FIGURE 9.
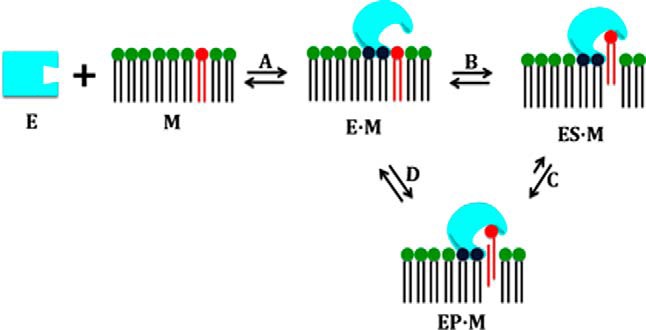
Membranes are allosteric activators of phospholipases. Reprinted from Ref. 147.
Charting a Course for Lipid Research
Even at the beginning of the 21st century—at the dawn of genomics and with interest in proteomics growing—little was known about lipids themselves, such as how many existed, what signaling roles they had, how they should be measured, and even how lipids should be classified and named in the expanding world of omics. Given my interests in organizational leadership, I recognized an opportunity. In 2002, I proposed to NIH an initiative titled LIPID MAPS, standing for “LIPID Metabolites And Pathways Strategies,” which would develop the field of “lipidomics.” This “omic” can be considered part of metabolomics, which aims at the identification and quantification of all of the metabolites in the cell including its nucleic acids, amino acids, sugars, and fats. The ultimate goal of biochemists is to follow the fluxes of molecules in biological pathways. But by far the largest number of distinct molecular species in cellular metabolism lies in the lipids, where tens of thousands of distinct molecular species exist. Thus, the initiative would undertake the identification, characterization, and quantification of all lipid metabolites. With a $25,000 “planning grant,” which was used to recruit the top academicians in each lipid type as well as bioinformatics experts, the eventual group of 12 principal investigators came from eight universities and a commercial company (Fig. 10). The resulting ten-year Large Scale Collaborative “Glue” Grant awarded by the National Institute of General Medical Sciences was charged with developing the field of lipidomics, and it produced over 400 publications (152). At last lipidomics could join the omics evolution (153)!
FIGURE 10.
LIPID MAPS Consortium leaders at annual meeting December, 2008. Left to right: Christian R. H. Raetz (Duke University), Joseph L. Witztum (UCSD), Nicholas Winograd (Penn State) who replaced Stephen White (University of California, Irvine), Alfred H. Merrill (Georgia Tech), Robert C. Murphy (University of Colorado), Michael S. VanNieuwenhze (Indiana University), Edward A. Dennis (UCSD), David W. Russell (UT Southwestern University), Walter A. Shaw (Avanti Polar Lipids), Christopher K. Glass (UCSD), Jean Chin (NIH representative), Shankar Subramaniam (UCSD), and H. Alex Brown (Vanderbilt University).
The success of LIPID MAPS over the years is one of the great highlights of my career. Together, we developed novel liquid chromatographic-mass spectrometric-based lipidomics techniques termed “CLASS” (138) using a quantitative approach based on over 500 LIPID MAPS synthetic standards and specific agonists such as Kdo2-Lipid A (154). This approach was applied to the overall omics analysis of some 500 lipid species in immunologically activated macrophages integrating transcriptomics, proteomics, and metabolomics of lipid metabolites (155). Novel methods for separating and assessing the purity of subcellular organelles (156) were developed and employed to determine the subcellular localization of many individual lipid molecular species and their changes during macrophage activation (157). A major finding from the integrated LIPID MAPS study of activated macrophages was the importance of desmosterol in linking lipid metabolism with inflammation (158). In another integrated study, a second sterol, 25-hydroxycholesterol, was shown to activate the stress response to reprogram transcription and translation (159).
We also profiled human plasma to quantify some 600 distinct lipid molecular species present across all mammalian lipid categories (160). We have high hopes for the potential implications of these data for clinical diagnostics and disease biomarkers as well as understanding the mechanisms of disease (161). However, these data also made a big impact on lipid research in another way. As we were sorting through all of the information, we realized that we didn't have sufficient classification, nomenclature, or structural drawing representations for lipids to be able to describe our results properly. So, we developed some (162, 163)! We've also gone further—the initiative includes a website (www.lipidmaps.org) and databases where over 40,000 individual molecular species of lipids can be searched; tutorials on lipid metabolism using the structural drawing tool can be found on this site as well. This standardization may be the most impactful and lasting achievement of the investigators of the LIPID MAPS Consortium and their international collaborators, the International Lipid Classification and Nomenclature Committee (ILCNC), who continue to work together so productively.
Lipidomics of Fatty Acids and Eicosanoids
Beyond these collaborative projects, my own laboratory's work under the LIPID MAPS initiative has continued its focus on the lipidomics of fatty acyls including fatty acids, eicosanoids and related oxidized fatty acids, and fatty amides. We like to think of this focus as looking to the future of the field (Fig. 11). So far, this focus has led to a robust and comprehensive approach to the lipidomics analysis of hundreds of fatty acids including their numerous metabolites arising from an array of cyclooxygenases, lipoxygenases, cytochrome P450s, and non-enzymatic oxidation reactions (164), such as the production of dihomoprostaglandins in stimulated macrophages resulting from the elongation of arachidonic acid to adrenic acid (165). We also used lipidomics to analyze very long chain fatty acids that are particularly abundant in the phospholipids of retinal membranes where they are critical for function (166) and demonstrated the essential role of ELOVL-4 in the synthesis of these fatty acids. Today over 200 fatty acyls can routinely be analyzed and quantified with internal standards (167) using the ultrahigh performance liquid chromatograph/mass spectrometric (UPLC/MS) techniques developed over the years for eicosanoids and related oxidized PUFAs (168) as well as free fatty acids (169) and complex lipids including phospholipids (170).
FIGURE 11.

Edward Dennis in his laboratory with mass spectrometer on the initiation of LIPID MAPS.
It is fascinating to see how this basic science has so many potential applications in nutrition and disease, and so we've continued to search for new biological areas relevant to lipids. One study demonstrated how a single immune cell can store a pro-resolving lipid precursor and then release it for bioactive maturation and secretion, conceptually similar to the production and inflammasome-dependent maturation of the pro-inflammatory IL-1 family of cytokines. Specifically, we examined the role of lipoxins as pro-resolution lipid mediators that inhibit phlogistic neutrophil recruitment and promote wound healing by macrophage recruitment via potent and specific signaling through the LXA4 receptor. Lipoxins are generated as a consequence of sequential activation of the Toll-like receptor 4 (TLR4), a receptor for endotoxin, and P2X7, a purinergic receptor for extracellular ATP (171). Initial activation of TLR4 results in accumulation of the COX-2-derived lipoxin precursor 15-hydroxyeicosatetraenoic acid (15-HETE) in esterified form within membrane phospholipids. 15-HETE production can be enhanced by aspirin. Subsequent activation of P2X7 results in efficient release of 15-HETE from membrane phospholipids by cPLA2 and the conversion of 15-HETE to bioactive lipoxins by 5-lipoxygenase (5-LOX). This study illustrated how aspirin in addition to inhibiting prostaglandin production also promotes pro-resolution eicosanoids. In its broader context, this result illustrates the importance of eicosanoids in the general response to infection, their relationship to inflammasome formation, and the consequent “eicosanoid storm” (81).
Once we had started examining the role of eicosanoids in inflammation, yet more questions surfaced. It was known that endotoxin-stimulated macrophages could serve as models for bacterial inflammation and infection, but the role of lipids in these processes was unclear. Using lipidomics analysis of eicosanoids, we were able to identify synergy in cellular lipid signaling of TLRs and purinergic receptors (172). This same approach was used to determine the course of infection-induced arthritic inflammation and its resolution in Lyme disease (173) and its broader implications for the cyclooxygenase pathway. We've also looked at lipids introduced from the environment. For example, we studied the effects of fish oil consumption on Lyme disease progression (174). In this research, novel eicosanoid mediators were found, including anti-inflammatory mediators that differentiate the pathogenicity of influenza strains in mouse and in human lavages (175). Lipidomic analysis of cells supplemented with small amounts of the fish oil-derived ω-3 fatty acids, eicosapentaenoic acid (EPA) or docosahexaenoic acid (DHA), clarified their effects on the inflammatory eicosadome in inhibiting COX-1 more than COX-2 and shunting arachidonate to LOX physiologically (176). Lipidomics analysis even allowed the elucidation of the role of liver X receptors (LXRs) in the biosynthesis of insulin-sensitizing ω-3 fatty acids (177).
As our understanding of biology has grown, our interests in applying those insights to questions with biomedical implications have grown in parallel. For example, inflammatory hyperalgesia and various forms of pain induce essential bioactive lipid production in the spinal cord and spinal fluid. Over the years with my UCSD colleague Tony Yaksh, we've characterized PLA2 enzymes in spinal cord and spinal fluid (178, 179) as well as examining the role of resulting eicosanoids (180). This included the discovery of the novel role of spinal 12-lipoxygenase-derived hepoxilins A3 and B3, which activate the TRPV1 and TRPA1 receptors, in inflammatory hyperalgesia (181). It was shown that intrathecal administration of these hepoxilins induces hyperalgesia in rats. The specific 12-lipoxygenase enzyme eLOX3 responsible for the generation of hepoxilins has also recently been identified (182) along with the particular glial cell source in spinal cord. In another application, persistent effects after the resolution of inflammation in serum-transferred arthritis were analyzed (183). Even the consequences for cancer resulting from the up-regulation of the cyclooxygenase in certain cell types are better understood by the incorporation of lipidomics analysis of the oxygenated PUFAs (184, 185). These results demonstrate the utility of a comprehensive lipidomics approach for identifying potential contributors to disease pathology, and we hope that they will facilitate the development of more precisely targeted treatment strategies.
During these years, we have also been reminded about the importance of thinking beyond enzymatic reactions. Specifically, we investigated the oxidation of LDL lipids to explain the effect on macrophage activation and its implications for atherosclerosis. The first critical finding was that when human LDL is oxidized, significant amounts of the PUFA in the sn-2 position of 1-stearoyl, 2-arachidonoyl phosphatidylcholine become oxidized and yield 1-stearoyl, 2-oxovaleroyl phosphatidylcholine as a major product. Furthermore, this product can easily undergo aldol condensations and also form Schiff bases with lysine side chains of apoproteins in the LDL (186). Such products are recognized by receptors and anti-phospholipid antibodies (187–189), and lipid complexes of phosphorylcholine were found to be pattern recognition ligands (190).
The LIPID MAPS lipidomics approaches enabled our discovery that oxidized PUFAs in LDL could also be found esterified to cholesterol (191), and lipidomics studies of zebrafish fed a high fat diet revealed oxidized phospholipids and cholesteryl esters (192). Subsequent lipidomics studies revealed that such oxidized phospholipids and cholesterol esters bind to apoproteins (193) and have multiple effects on macrophage signaling (194). Significantly, these oxidized species can be found in human coronary artery disease (195).
Finally, the power of lipidomics analysis has also been applied to fluxomics using unlabeled natural molecules to analyze the eicosanoid cascade. By taking advantage of the specificity of receptors to activate PLA2, the release of free arachidonic acid can be initiated (196). Fluxomics analysis allows the temporal course of metabolites in the eicosanoid cascade initiated by the activation of PLA2 to be followed quantitatively. In recent experiments, we've compared primary macrophages including tissue-resident, thioglycolate-elicited, and bone marrow-derived, with cell lines (197), leading to a general model describing the fluxes of eicosanoids initiated by priming with a TLR4 agonist followed by activation with a purinergic P2X7 agonist (198). Furthermore, this general approach has been integrated with transcriptional regulation and protein induction using a “directed proteomics” approach (199). The resultant competing eicosanoid fluxes of the multiple competing ω-3 and ω-6 fatty acid substrates under physiological conditions in the eicosanoid cascade can now be quantitatively compared (200).
Circling Back to Chirality
It is a delight to me that as we identify chiral eicosanoids, lipidomics studies have brought me back to my early fascination with stereochemistry. Enzymatically produced eicosanoids always turn out to be chiral compounds as one might expect, but interestingly, certain drugs can alter the chirality of enzymatically produced eicosanoids. A recent example is the effect of aspirin described above, which acetylates the cyclooxygenase enzyme and changes the chirality of a COX side product lipoxin A4 to epi-lipoxin A4, implicated in the resolution of inflammation (81, 171). Another example is the identification of a particular oxidized lipid that corrects a defect in peroxisome proliferator-activated receptor-γ (PPAR-γ) signaling in Cftr-deficient mice (201). Chirality determination is also important because oxidized lipids can be produced non-enzymatically in response to stress and are associated with many diseases including non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH), as characterized by LIPID MAPS (202). These products can be differentiated from the chirality that results from enzymatically produced products of cellular signaling pathways. The most recent work using quantitative UPLC/MS approaches resulted in the identity of a panel of eicosanoids in human plasma that can differentiate non-alcoholic fatty liver (NAFL) from NASH as a non-invasive biomarker important for NAFLD assessment and treatment (203).
When I was initially enticed by stereochemistry in college, I didn't expect it to occupy so much of my career. Indeed, my research trajectory has gone far beyond what I dreamed of then. I am humbled that the early connections that my colleagues and I described between PLA2 action and lipid mediators have helped build this exciting field, and I am grateful to my scientific colleagues—including many former students and scientific collaborators—who organized a recent conference in my honor (Fig. 12). Today my laboratory is focused on enzyme- and drug-induced stereochemical changes of bioactive lipid molecules involved in inflammation and its resolution as well as on membrane-induced allosteric effects on enzymes that act on lipids, and these studies are now enhanced by lipidomic approaches to specificity. And although I can't claim to have predicted the future, I can say that I was prepared from the beginning to question established scientific thought. By the time I defended my Ph.D. thesis, William Doering, the professor who had so stimulated my fascination with the enormous stereo-complexity of simple molecules in three-dimensional space, had moved from Yale to Harvard and served on my doctoral thesis committee. During my defense, he asked why I had included my failed Cumulative Exam proposal on phosphorus isomers in my thesis as an Appendix. I explained jokingly that I was still challenging my grade!
FIGURE 12.
Lipid Mediators in Health and Disease II: A Tribute to Edward Dennis and 7th International Conference on Phospholipase A2 and Lipid Mediators held May 19–20, 2016 at Scripps Seaside Forum in La Jolla, California. Honorary Chair Bengt Samuelsson and Organizing Committee Chair Nicolas G. Bazan (LSU Health New Orleans) and his international committee of Jerold Chun (Scripps Research Institute), Jesper Z. Haeggström (Karolinska Institutet), Timothy Hla (Weill Cornell Medical College), Charles N. Serhan (Harvard Medical School), and Takao Shimizu (University of Tokyo) are pictured along with invited speakers, chairs, and participants.
Acknowledgments
I thank my early scientific mentors Harry H. Wasserman (Yale), Frank H. Westheimer (Harvard), Eugene P. Kennedy (Harvard Medical School), and as a new faculty member especially Nathan O. Kaplan and Morris Friedkin (UCSD) as well as George Carman (Rutgers) and numerous other colleagues and collaborators at UCSD and around the world. I'm especially grateful to those who participated in my LIPID MAPS initiative with whom I have worked very closely in recent years. Special thanks go to the many undergraduates, over 30 graduate students, over 50 postdoctoral scholars, several project scientists, many staff research associates, and numerous visiting scientists and sabbatical visitors (many of whom are cited in the references) who graced my laboratory. I especially appreciate the continuity and wise counsel provided by the long-time presence of Ray Deems (over three decades) and now Oswald Quehenberger (almost a decade) in my laboratory. The NIH, NSF, and Lilly Research Laboratories and numerous other non-profit organizations and for-profit companies have provided the essential support for my laboratory. I want to explicitly express my appreciation to the National Institute of General Medical Sciences (NIGMS) for continuous individual predoctoral and postdoctoral fellowships, RO1 support for over 40 years (RO1 GM20501-40), and over 10 years of support for the LIPID MAPS “Glue” grant (U54 GM069338). I especially thank the Journal of Biological Chemistry, a journal in which over 70 publications on my work have appeared, for inviting me to write this Reflection.
References
- 1. Westheimer F. H. (2003) Musings. J. Biol. Chem. 278, 11729–11730 [DOI] [PubMed] [Google Scholar]
- 2. Dennis E. A., and Westheimer F. H. (1966) The geometry of the transition state in the hydrolysis of phosphate esters. J. Am. Chem. Soc. 88, 3432–3433 [DOI] [PubMed] [Google Scholar]
- 3. Roberts G. C., Dennis E. A., Meadows D. H., Cohen J. S., and Jardetzky O. (1969) The mechanism of action of ribonuclease. Proc. Natl. Acad. Sci. U.S.A. 62, 1151–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dennis E. A., and Kennedy E. P. (1972) Intracellular sites of lipid synthesis and the biogenesis of mitochondria. J. Lipid Res. 13, 263–267 [PubMed] [Google Scholar]
- 5. Kennedy E. P. (2001) Hitler's gift and the era of biosynthesis. J. Biol. Chem. 276, 42619–42631 [DOI] [PubMed] [Google Scholar]
- 6. Singer S. J., and Nicolson G. L. (1972) The fluid mosaic model of the structure of cell membranes. Science 175, 720–731 [DOI] [PubMed] [Google Scholar]
- 7. Warner T. G., and Dennis E. A. (1973) New sensitive assay for phosphatidylserine decarboxylase based on the detection of CO2 from nonradiolabeled phosphatidylserine. J. Lipid Res. 14, 595–598 [PubMed] [Google Scholar]
- 8. Dennis E. A. (1973) Kinetic dependence of phospholipase A2 activity on the detergent Triton X-100. J. Lipid Res. 14, 152–159 [PubMed] [Google Scholar]
- 9. Dennis E. A., Cao J., Hsu Y. H., Magrioti V., and Kokotos G. (2011) Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 111, 6130–6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dennis E. A., and Owens J. M. (1973) Studies on mixed micelles of Triton X-100 and phosphatidylcholine using nuclear magnetic resonance techniques. J. Supramol. Struct. 1, 165–176 [DOI] [PubMed] [Google Scholar]
- 11. Ribeiro A. A., and Dennis E. A. (1975) Proton magnetic resonance relaxation studies on the structure of mixed micelles of Triton X-100 and dimyristoylphosphatidylcholine. Biochemistry 14, 3746–3755 [DOI] [PubMed] [Google Scholar]
- 12. Robson R. J., and Dennis E. A. (1977) The size, shape, and hydration of nonionic surfactant micelles. Triton X-100. J. Phys. Chem. 81, 1075–1078 [Google Scholar]
- 13. Roberts M. F., and Dennis E. A. (1977) Proton nuclear magnetic resonance demonstration of conformationally nonequivalent phospholipid fatty acid chains in mixed micelles. J. Am. Chem. Soc. 99, 6142–6143 [DOI] [PubMed] [Google Scholar]
- 14. Roberts M. F., Bothner-By A. A., and Dennis E. A. (1978) Magnetic nonequivalence within fatty acyl chains of phospholipids in membrane models: 1H nuclear magnetic resonance studies of the α-methylene groups. Biochemistry 17, 935–942 [DOI] [PubMed] [Google Scholar]
- 15. De Bony J., and Dennis E. A. (1981) Magnetic nonequivalence of the two fatty acid chains in phospholipids of small unilamellar vesicles and mixed micelles. Biochemistry 20, 5256–5260 [DOI] [PubMed] [Google Scholar]
- 16. Whiles J. A., Deems R., Vold R. R., and Dennis E. A. (2002) Bicelles in structure-function studies of membrane-associated proteins. Bioorg. Chem. 30, 431–442 [DOI] [PubMed] [Google Scholar]
- 17. Robson R. J., and Dennis E. A. (1983) Micelles of nonionic detergents and mixed micelles with phospholipids. Acc. Chem. Res. 16, 251–258 [Google Scholar]
- 18. Lichtenberg D., Robson R. J., and Dennis E. A. (1983) Solubilization of phospholipids by detergents: structural and kinetic aspects. Biochim. Biophys. Acta 737, 285–304 [DOI] [PubMed] [Google Scholar]
- 19. Dennis E. A. (1973) Phospholipase A2 activity towards phosphatidylcholine in mixed micelles: surface dilution kinetics and the effect of thermotropic phase transitions. Arch Biochem. Biophys. 158, 485–493 [DOI] [PubMed] [Google Scholar]
- 20. Deems R. A., Eaton B. R., and Dennis E. A. (1975) Kinetic analysis of phospholipase A2 activity toward mixed micelles and its implications for the study of lipolytic enzymes. J. Biol. Chem. 250, 9013–9020 [PubMed] [Google Scholar]
- 21. Carman G. M., Deems R. A., and Dennis E. A. (1995) Lipid signaling enzymes and surface dilution kinetics. J. Biol. Chem. 270, 18711–18714 [DOI] [PubMed] [Google Scholar]
- 22. Eaton B. R., and Dennis E. A. (1976) Analysis of phospholipase C (Bacillus cereus) action toward mixed micelles of phospholipid and surfactant. Arch Biochem. Biophys. 176, 604–609 [DOI] [PubMed] [Google Scholar]
- 23. Warner T. G., and Dennis E. A. (1975) Action of the highly purified, membrane-bound enzyme phosphatidylserine decarboxylase Escherichia coli toward phosphatidylserine in mixed micelles and erythrocyte ghosts in the presence of surfactant. J. Biol. Chem. 250, 8004–8009 [PubMed] [Google Scholar]
- 24. Roberts M. F., Deems R. A., and Dennis E. A. (1977) Dual role of interfacial phospholipid in phospholipase A2 catalysis. Proc. Natl. Acad. Sci. U.S.A. 74, 1950–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Changeux J. P. (2013) The concept of allosteric interaction and its consequences for the chemistry of the brain. J. Biol. Chem. 288, 26969–26986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roberts M. F., Adamich M., Robson R. J., and Dennis E. A. (1979) Phospholipid activation of cobra venom phospholipase A2: 1. Lipid-lipid or lipid-enzyme interaction. Biochemistry 18, 3301–3308 [DOI] [PubMed] [Google Scholar]
- 27. Adamich M., Roberts M. F., and Dennis E. A. (1979) Phospholipid activation of cobra venom phospholipase A2: 2. Characterization of the phospholipid-enzyme interaction. Biochemistry 18, 3308–3314 [DOI] [PubMed] [Google Scholar]
- 28. Boegeman S. C., Deems R. A., and Dennis E. A. (2004) Phospholipid binding and the activation of Group IA secreted phospholipase A2. Biochemistry 43, 3907–3916 [DOI] [PubMed] [Google Scholar]
- 29. Fremont D. H., Anderson D. H., Wilson I. A., Dennis E. A., and Xuong N. H. (1993) Crystal structure of phospholipase A2 from Indian cobra reveals a trimeric association. Proc. Natl. Acad. Sci. U.S.A. 90, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Segelke B. W., Nguyen D., Chee R., Xuong N. H., and Dennis E. A. (1998) Structures of two novel crystal forms of Naja naja naja phospholipase A2 lacking Ca2+ reveal trimeric packing. J. Mol. Biol. 279, 223–232 [DOI] [PubMed] [Google Scholar]
- 31. Lefkowitz L. J., Deems R. A., and Dennis E. A. (1999) Expression of Group IA phospholipase A2 in Pichia pastoris: identification of a phosphatidylcholine activator site using site-directed mutagenesis. Biochemistry 38, 14174–14184 [DOI] [PubMed] [Google Scholar]
- 32. Kelley M. J., Crowl R. M., and Dennis E. A. (1992) Renaturation of cobra venom phospholipase A2 expressed from a synthetic gene in Escherichia coli. Biochim. Biophys. Acta 1118, 107–115 [DOI] [PubMed] [Google Scholar]
- 33. Roberts M. F., Deems R. A., and Dennis E. A. (1977) Spectral perturbations of the histidine and tryptophan in cobra venom phospholipase A2 upon metal ion and mixed micelle binding. J. Biol. Chem. 252, 6011–6017 [PubMed] [Google Scholar]
- 34. Plückthun A., and Dennis E. A. (1982) Role of monomeric activators in cobra venom phospholipase A2 action. Biochemistry 21, 1750–1756 [DOI] [PubMed] [Google Scholar]
- 35. Plückthun A., and Dennis E. A. (1985) Activation, aggregation, and product inhibition of cobra venom phospholipase A2 and comparison with other phospholipases. J. Biol. Chem. 260, 11099–11106 [PubMed] [Google Scholar]
- 36. Plückthun A., and Dennis E. A. (1981) 31P nuclear magnetic resonance study on the incorporation of monomeric phospholipids into nonionic detergent micelles. J. Phys. Chem. 85, 678–683 [Google Scholar]
- 37. Plückthun A., Rohlfs R., Davidson F. F., and Dennis E. A. (1985) Short-chain phosphatidylethanolamines: physical properties and susceptibility of the monomers to phospholipase A2 action. Biochemistry 24, 4201–4208 [DOI] [PubMed] [Google Scholar]
- 38. Lombardo D., Fanni T., Plückthun A., and Dennis E. A. (1986) Rate-determining step in phospholipase A2 mechanism: 18O isotope exchange determined by 13C NMR. J. Biol. Chem. 261, 11663–11666 [PubMed] [Google Scholar]
- 39. Plückthun A., and Dennis E. A. (1982) Acyl and phosphoryl migration in lysophospholipids: importance in phospholipid synthesis and phospholipase specificity. Biochemistry 21, 1743–1750 [DOI] [PubMed] [Google Scholar]
- 40. Hendrickson H. S., and Dennis E. A. (1984) Kinetic analysis of the dual phospholipid model for phospholipase A2 action. J. Biol. Chem. 259, 5734–5739 [PubMed] [Google Scholar]
- 41. Hendrickson H. S., and Dennis E. A. (1984) Analysis of the kinetics of phospholipid activation of cobra venom phospholipase A2. J. Biol. Chem. 259, 5740–5744 [PubMed] [Google Scholar]
- 42. Kensil C. R., and Dennis E. A. (1979) Action of cobra venom phospholipase A2 on the gel and liquid crystalline states of dimyristoyl and dipalmitoyl phosphatidylcholine vesicles. J. Biol. Chem. 254, 5843–5848 [PubMed] [Google Scholar]
- 43. Adamich M., and Dennis E. A. (1978) Exploring the action and specificity of cobra venom phospholipase A2 toward human erythrocytes, ghost membranes, and lipid mixtures. J. Biol. Chem. 253, 5121–5125 [PubMed] [Google Scholar]
- 44. Kensil C. R., and Dennis E. A. (1981) Alkaline hydrolysis of phospholipids in model membranes and the dependence on their state of aggregation. Biochemistry 20, 6079–6085 [DOI] [PubMed] [Google Scholar]
- 45. Jarvis A. A., Cain C., and Dennis E. A. (1984) Purification and characterization of a lysophospholipase from human amnionic membranes. J. Biol. Chem. 259, 15188–15195 [PubMed] [Google Scholar]
- 46. Zhang Y. Y., and Dennis E. A. (1988) Purification and characterization of a lysophospholipase from a macrophage-like cell line P388D1. J. Biol. Chem. 263, 9965–9972 [PubMed] [Google Scholar]
- 47. Stafford R. E., Fanni T., and Dennis E. A. (1989) Interfacial properties and critical micelle concentration of lysophospholipids. Biochemistry 28, 5113–5120 [DOI] [PubMed] [Google Scholar]
- 48. Wang A., Deems R. A., and Dennis E. A. (1997) Cloning, expression, and catalytic mechanism of murine lysophospholipase I. J. Biol. Chem. 272, 12723–12729 [DOI] [PubMed] [Google Scholar]
- 49. Wang A., Loo R., Chen Z., and Dennis E. A. (1997) Regiospecificity and catalytic triad of lysophospholipase I. J. Biol. Chem. 272, 22030–22036 [DOI] [PubMed] [Google Scholar]
- 50. Dennis E. A. (ed) (1991) Phospholipases, Methods in Enzymology, Vol. 197, 640 pages, Academic Press, Orlando [Google Scholar]
- 51. Dennis E. A. (2015) Introduction to Thematic Review Series: Phospholipases: central role in lipid signaling and disease. J. Lipid Res. 56, 1245–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roberts M. F., Otnaess A. B., Kensil C. A., and Dennis E. A. (1978) The specificity of phospholipase A2 and phospholipase C in a mixed micellar system. J. Biol. Chem. 253, 1252–1257 [PubMed] [Google Scholar]
- 53. Balboa M. A., Balsinde J., Dennis E. A., and Insel P. A. (1995) A phospholipase D-mediated pathway for generating diacylglycerol in nuclei from Madin-Darby canine kidney cells. J. Biol. Chem. 270, 11738–11740 [DOI] [PubMed] [Google Scholar]
- 54. Balboa M. A., Balsinde J., Dillon D. A., Carman G. M., and Dennis E. A. (1999) Proinflammatory macrophage-activating properties of the novel phospholipid diacylglycerol pyrophosphate. J. Biol. Chem. 274, 522–526 [DOI] [PubMed] [Google Scholar]
- 55. Johnson C. A., Balboa M. A., Balsinde J., and Dennis E. A. (1999) Regulation of cyclooxygenase-2 expression by phosphatidate phosphohydrolase in human amnionic WISH cells. J. Biol. Chem. 274, 27689–27693 [DOI] [PubMed] [Google Scholar]
- 56. Grkovich A., Johnson C. A., Buczynski M. W., and Dennis E. A. (2006) Lipopolysaccharide-induced cyclooxygenase-2 expression in human U937 macrophages is phosphatidic acid phosphohydrolase-1-dependent. J. Biol. Chem. 281, 32978–32987 [DOI] [PubMed] [Google Scholar]
- 57. Balsinde J., and Dennis E. A. (1996) Bromoenol lactone inhibits magnesium-dependent phosphatidate phosphohydrolase and blocks triacylglycerol biosynthesis in mouse P388D1 macrophages. J. Biol. Chem. 271, 31937–31941 [DOI] [PubMed] [Google Scholar]
- 58. Balboa M. A., Balsinde J., and Dennis E. A. (1998) Involvement of phosphatidate phosphohydrolase in arachidonic acid mobilization in human amnionic WISH cells. J. Biol. Chem. 273, 7684–7690 [DOI] [PubMed] [Google Scholar]
- 59. Davidson F. F., and Dennis E. A. (1990) Evolutionary relationships and implications for the regulation of phospholipase A2 from snake venom to human secreted forms. J. Mol. Evol. 31, 228–238 [DOI] [PubMed] [Google Scholar]
- 60. Davidson F. F., Dennis E. A., Powell M., and Glenney J. R. Jr. (1987) Inhibition of phospholipase A2 by “lipocortins” and calpactins: an effect of binding to substrate phospholipids. J. Biol. Chem. 262, 1698–1705 [PubMed] [Google Scholar]
- 61. Davidson F. F., and Dennis E. A. (1989) Biological relevance of lipocortins and related proteins as inhibitors of phospholipase A2. Biochem. Pharmacol. 38, 3645–3651 [DOI] [PubMed] [Google Scholar]
- 62. Davidson F. F., Lister M. D., and Dennis E. A. (1990) Binding and inhibition studies on lipocortins using phosphatidylcholine vesicles and phospholipase A2 from snake venom, pancreas, and a macrophage-like cell line. J. Biol. Chem. 265, 5602–5609 [PubMed] [Google Scholar]
- 63. Yu L., Deems R. A., Hajdu J., and Dennis E. A. (1990) The interaction of phospholipase A2 with phospholipid analogues and inhibitors. J. Biol. Chem. 265, 2657–2664 [PubMed] [Google Scholar]
- 64. Yu L., and Dennis E. A. (1991) Critical role of a hydrogen bond in the interaction of phospholipase A2 with transition-state and substrate analogues. Proc. Natl. Acad. Sci. U.S.A. 88, 9325–9329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yu L., and Dennis E. A. (1992) Defining the dimensions of the catalytic site of phospholipase A2 using amide substrate analogues. J. Am. Chem. Soc. 114, 8757–8763 [Google Scholar]
- 66. Plesniak L. A., Boegeman S. C., Segelke B. W., and Dennis E. A. (1993) Interaction of phospholipase A2 with thioether amide containing phospholipid analogues. Biochemistry 32, 5009–5016 [DOI] [PubMed] [Google Scholar]
- 67. Plesniak L. A., Yu L., and Dennis E. A. (1995) Conformation of micellar phospholipid bound to the active site of phospholipase A2. Biochemistry 34, 4943–4951 [DOI] [PubMed] [Google Scholar]
- 68. Lombardo D., and Dennis E. A. (1985) Cobra venom phospholipase A2 inhibition by manoalide: a novel type of phospholipase inhibitor. J. Biol. Chem. 260, 7234–7240 [PubMed] [Google Scholar]
- 69. Reynolds L. J., Morgan B. P., Hite G. A., Mihelich E. D., and Dennis E. A. (1988) Phospholipase A2 inhibition and modification by manoalogue. J. Am. Chem. Soc. 110, 5172–5177 [Google Scholar]
- 70. Reynolds L. J., Mihelich E. D., and Dennis E. A. (1991) Inhibition of venom phospholipases A2 by manoalide and manoalogue: stoichiometry of incorporation. J. Biol. Chem. 266, 16512–16517 [PubMed] [Google Scholar]
- 71. Washburn W. N., and Dennis E. A. (1990) A novel general approach for the assay and inhibition of hydrolytic enzymes: utilizing suicide inhibitory bifunctionally linked substrates (SIBLINKS) exemplified by a phospholipase A2 assay. J. Am. Chem. Soc. 112, 2040–2041 [Google Scholar]
- 72. Washburn W. N., and Dennis E. A. (1990) Suicide inhibitory bifunctionally linked substrates (SIBLINKS) as phospholipase A2 inhibitors. J. Am. Chem. Soc. 112, 2042–2043 [PubMed] [Google Scholar]
- 73. Washburn W. N., and Dennis E. A. (1991) Suicide-inhibitory bifunctionally linked substrates (SIBLINKS) as phospholipase A2 inhibitors: mechanistic implications. J. Biol. Chem. 266, 5042–5048 [PubMed] [Google Scholar]
- 74. Yu L., and Dennis E. A. (1993) Effect of polar head groups on the interactions of phospholipase A2 with phosphonate transition-state analogues. Biochemistry 32, 10185–10192 [DOI] [PubMed] [Google Scholar]
- 75. Barden R. E., Darke P. L., Deems R. A., and Dennis E. A. (1980) Interaction of phospholipase A2 from cobra venom with Cibacron Blue F3GA. Biochemistry 19, 1621–1625 [DOI] [PubMed] [Google Scholar]
- 76. Barlow P. N., Lister M. D., Sigler P. B., and Dennis E. A. (1988) Probing the role of substrate conformation in phospholipase A2 action on aggregated phospholipids using constrained phosphatidylcholine analogues. J. Biol. Chem. 263, 12954–12958 [PubMed] [Google Scholar]
- 77. Hazlett T. L., and Dennis E. A. (1985) Aggregation studies on fluorescein-coupled cobra venom phospholipase A2. Biochemistry 24, 6152–6158 [DOI] [PubMed] [Google Scholar]
- 78. Dennis E. A. (1987) Regulation of eicosanoid production: role of phospholipases and inhibitors. Nature Biotechnology 5, 1294–1300 [Google Scholar]
- 79. Vane J. R. (1971) Inhibition of prostaglandin synthesis as a mechanism of action for aspirin-like drugs. Nat. New Biol. 231, 232–235 [DOI] [PubMed] [Google Scholar]
- 80. Samuelsson B. (2012) Role of basic science in the development of new medicines: examples from the eicosanoid field. J. Biol. Chem. 287, 10070–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dennis E. A., and Norris P. C. (2015) Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 15, 511–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ackermann E. J., Kempner E. S., and Dennis E. A. (1994) Ca2+-independent cytosolic phospholipase A2 from macrophage-like P388D1 cells: isolation and characterization. J. Biol. Chem. 269, 9227–9233 [PubMed] [Google Scholar]
- 83. Dennis E. A. (1994) Diversity of group types, regulation, and function of phospholipase A2. J. Biol. Chem. 269, 13057–13060 [PubMed] [Google Scholar]
- 84. Dennis E. A. (1997) The growing phospholipase A2 superfamily of signal transduction enzymes. Trends Biochem. Sci. 22, 1–2 [DOI] [PubMed] [Google Scholar]
- 85. Six D. A., and Dennis E. A. (2000) The expanding superfamily of phospholipase A2 enzymes: classification and characterization. Biochim. Biophys. Acta 1488, 1–19 [DOI] [PubMed] [Google Scholar]
- 86. Schaloske R. H., and Dennis E. A. (2006) The phospholipase A2 superfamily and its group numbering system. Biochim. Biophys. Acta 1761, 1246–1259 [DOI] [PubMed] [Google Scholar]
- 87. Balboa M. A., Balsinde J., Winstead M. V., Tischfield J. A., and Dennis E. A. (1996) Novel Group V phospholipase A2 involved in arachidonic acid mobilization in murine P388D1 macrophages. J. Biol. Chem. 271, 32381–32384 [DOI] [PubMed] [Google Scholar]
- 88. Balboa M. A., Balsinde J., Jones S. S., and Dennis E. A. (1997) Identity between the Ca2+-independent phospholipase A2 enzymes from P388D1 macrophages and Chinese hamster ovary cells. J. Biol. Chem. 272, 8576–8580 [DOI] [PubMed] [Google Scholar]
- 89. Tang J., Kriz R. W., Wolfman N., Shaffer M., Seehra J., and Jones S. S. (1997) A novel cytosolic calcium-independent phospholipase A2 contains eight ankyrin motifs. J. Biol. Chem. 272, 8567–8575 [DOI] [PubMed] [Google Scholar]
- 90. Balsinde J., Barbour S. E., Bianco I. D., and Dennis E. A. (1994) Arachidonic acid mobilization in P388D1 macrophages is controlled by two distinct Ca2+-dependent phospholipase A2 enzymes. Proc. Natl. Acad. Sci. U.S.A. 91, 11060–11064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Balsinde J., Bianco I. D., Ackermann E. J., Conde-Frieboes K., and Dennis E. A. (1995) Inhibition of calcium-independent phospholipase A2 prevents arachidonic acid incorporation and phospholipid remodeling in P388D1 macrophages. Proc. Natl. Acad. Sci. U.S.A. 92, 8527–8531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Balsinde J., and Dennis E. A. (1997) Function and inhibition of intracellular calcium-independent phospholipase A2. J. Biol. Chem. 272, 16069–16072 [DOI] [PubMed] [Google Scholar]
- 93. Balsinde J., Balboa M. A., and Dennis E. A. (1997) Antisense inhibition of Group VI Ca2+-independent phospholipase A2 blocks phospholipid fatty acid remodeling in murine P388D1 macrophages. J. Biol. Chem. 272, 29317–29321 [DOI] [PubMed] [Google Scholar]
- 94. Yang H. C., Mosior M., Ni B., and Dennis E. A. (1999) Regional distribution, ontogeny, purification, and characterization of the Ca2+-independent phospholipase A2 from rat brain. J. Neurochem. 73, 1278–1287 [DOI] [PubMed] [Google Scholar]
- 95. Ross M. I., Deems R. A., Jesaitis A. J., Dennis E. A., and Ulevitch R. J. (1985) Phospholipase activities of the P388D1 macrophage-like cell line. Arch Biochem. Biophys. 238, 247–258 [DOI] [PubMed] [Google Scholar]
- 96. Balsinde J., and Dennis E. A. (1996) Distinct roles in signal transduction for each of the phospholipase A2 enzymes present in P388D1 macrophages. J. Biol. Chem. 271, 6758–6765 [DOI] [PubMed] [Google Scholar]
- 97. Lister M. D., Deems R. A., Watanabe Y., Ulevitch R. J., and Dennis E. A. (1988) Kinetic analysis of the Ca2+-dependent, membrane-bound, macrophage phospholipase A2 and the effects of arachidonic acid. J. Biol. Chem. 263, 7506–7513 [PubMed] [Google Scholar]
- 98. Lister M. D., Glaser K. B., Ulevitch R. J., and Dennis E. A. (1989) Inhibition studies on the membrane-associated phospholipase A2 in vitro and prostaglandin E2 production in vivo of the macrophage-like P388D1 cell: effects of manoalide, 7,7-dimethyl-5,8-eicosadienoic acid, and p-bromophenacyl bromide. J. Biol. Chem. 264, 8520–8528 [PubMed] [Google Scholar]
- 99. Glaser K. B., Asmis R., and Dennis E. A. (1990) Bacterial lipopolysaccharide priming of P388D1 macrophage-like cells for enhanced arachidonic acid metabolism: platelet-activating factor receptor activation and regulation of phospholipase A2. J. Biol. Chem. 265, 8658–8664 [PubMed] [Google Scholar]
- 100. Asmis R., Randriamampita C., Tsien R. Y., and Dennis E. A. (1994) Intracellular Ca2+, inositol 1,4,5-trisphosphate and additional signalling in the stimulation by platelet-activating factor of prostaglandin E2 formation in P388D1 macrophage-like cells. Biochem. J. 298, 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Balboa M. A., Balsinde J., Johnson C. A., and Dennis E. A. (1999) Regulation of arachidonic acid mobilization in lipopolysaccharide-activated P388D1 macrophages by adenosine triphosphate. J. Biol. Chem. 274, 36764–36768 [DOI] [PubMed] [Google Scholar]
- 102. Balsinde J., Balboa M. A., and Dennis E. A. (1998) Functional coupling between secretory phospholipase A2 and cyclooxygenase-2 and its regulation by cytosolic Group IV phospholipase A2. Proc. Natl. Acad. Sci. U.S.A. 95, 7951–7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Balsinde J., Balboa M. A., and Dennis E. A. (2000) Identification of a third pathway for arachidonic acid mobilization and prostaglandin production in activated P388D1 macrophage-like cells. J. Biol. Chem. 275, 22544–22549 [DOI] [PubMed] [Google Scholar]
- 104. Shinohara H., Balboa M. A., Johnson C. A., Balsinde J., and Dennis E. A. (1999) Regulation of delayed prostaglandin production in activated P388D1 macrophages by Group IV cytosolic and Group V secretory phospholipase A2s. J. Biol. Chem. 274, 12263–12268 [DOI] [PubMed] [Google Scholar]
- 105. Balsinde J., Shinohara H., Lefkowitz L. J., Johnson C. A., Balboa M. A., and Dennis E. A. (1999) Group V phospholipase A2-dependent induction of cyclooxygenase-2 in macrophages. J. Biol. Chem. 274, 25967–25970 [DOI] [PubMed] [Google Scholar]
- 106. Kessen U. A., Schaloske R. H., Stephens D. L., Killermann Lucas K., and Dennis E. A. (2005) PGE2 release is independent of upregulation of Group V phospholipase A2 during long-term stimulation of P388D1 cells with LPS. J. Lipid Res. 46, 2488–2496 [DOI] [PubMed] [Google Scholar]
- 107. Shirai Y., Balsinde J., and Dennis E. A. (2005) Localization and functional interrelationships among cytosolic Group IV, secreted Group V, and Ca2+-independent Group VI phospholipase A2s in P388D1 macrophages using GFP/RFP constructs. Biochim. Biophys. Acta 1735, 119–129 [DOI] [PubMed] [Google Scholar]
- 108. Balboa M. A., Shirai Y., Gaietta G., Ellisman M. H., Balsinde J., and Dennis E. A. (2003) Localization of Group V phospholipase A2 in caveolin-enriched granules in activated P388D1 macrophage-like cells. J. Biol. Chem. 278, 48059–48065 [DOI] [PubMed] [Google Scholar]
- 109. Liscovitch M., and Cantley L. C. (1994) Lipid second messengers. Cell 77, 329–334 [DOI] [PubMed] [Google Scholar]
- 110. Bradshaw R. A., and Dennis E. A. (2003) Cell signaling: yesterday, today and tomorrow. in HandBook of Cell Signaling (Dennis E. A., and Bradshaw R. A. eds), Vol. 1, pp. 1–3, Academic Press, an imprint of Elsevier Science, Orlando, FL [Google Scholar]
- 111. Bradshaw R. A., and Dennis E. A. (2003) Handbook of Cell Signaling, Vol 1, 747 pages; Vol 2, 899 pages; and Vol 3, 709 pages, Academic Press, an imprint of Elsevier Science, Orlando, FL [Google Scholar]
- 112. Balsinde J., Balboa M. A., Insel P. A., and Dennis E. A. (1999) Regulation and inhibition of phospholipase A2. Annu. Rev. Pharmacol. Toxicol. 39, 175–189 [DOI] [PubMed] [Google Scholar]
- 113. Barbour S. E., and Dennis E. A. (1993) Antisense inhibition of Group II phospholipase A2 expression blocks the production of prostaglandin E2 by P388D1 cells. J. Biol. Chem. 268, 21875–21882 [PubMed] [Google Scholar]
- 114. Ackermann E. J., Conde-Frieboes K., and Dennis E. A. (1995) Inhibition of macrophage Ca2+-independent phospholipase A2 by bromoenol lactone and trifluoromethyl ketones. J. Biol. Chem. 270, 445–450 [DOI] [PubMed] [Google Scholar]
- 115. Conde-Frieboes K., Reynolds L. J., Lio Y., Hale M., Wasserman H. H., and Dennis E. A. (1996) Activated ketones as inhibitors of intracellular Ca2+-dependent and Ca2+-independent phospholipase A2. J. Am. Chem. Soc. 118, 5519–5525 [Google Scholar]
- 116. Lio Y. C., Reynolds L. J., Balsinde J., and Dennis E. A. (1996) Irreversible inhibition of Ca2+-independent phospholipase A2 by methyl arachidonyl fluorophosphonate. Biochim. Biophys. Acta 1302, 55–60 [DOI] [PubMed] [Google Scholar]
- 117. Kokotos G., Kotsovolou S., Six D. A., Constantinou-Kokotou V., Beltzner C. C., and Dennis E. A. (2002) Novel 2-oxoamide inhibitors of human Group IVA phospholipase A2. J. Med. Chem. 45, 2891–2893 [DOI] [PubMed] [Google Scholar]
- 118. Kokotos G., Six D. A., Loukas V., Smith T., Constantinou-Kokotou V., Hadjipavlou-Litina D., Kotsovolou S., Chiou A., Beltzner C. C., and Dennis E. A. (2004) Inhibition of Group IVA cytosolic phospholipase A2 by novel 2-oxoamides in vitro, in cells, and in vivo. J. Med. Chem. 47, 3615–3628 [DOI] [PubMed] [Google Scholar]
- 119. Stephens D., Barbayianni E., Constantinou-Kokotou V., Peristeraki A., Six D. A., Cooper J., Harkewicz R., Deems R. A., Dennis E. A., and Kokotos G. (2006) Differential inhibition of Group IVA and Group VIA phospholipases A2 by 2-oxoamides. J. Med. Chem. 49, 2821–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Six D. A., Barbayianni E., Loukas V., Constantinou-Kokotou V., Hadjipavlou-Litina D., Stephens D., Wong A. C., Magrioti V., Moutevelis-Minakakis P., Baker S. F., Dennis E. A., and Kokotos G. (2007) Structure-activity relationship of 2-oxoamide inhibition of Group IVA cytosolic phospholipase A2 and Group V secreted phospholipase A2. J. Med. Chem. 50, 4222–4235 [DOI] [PubMed] [Google Scholar]
- 121. Baskakis C., Magrioti V., Cotton N., Stephens D., Constantinou-Kokotou V., Dennis E. A., and Kokotos G. (2008) Synthesis of polyfluoro ketones for selective inhibition of human phospholipase A2 enzymes. J. Med. Chem. 51, 8027–8037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Kokotos G., Hsu Y. H., Burke J. E., Baskakis C., Kokotos C. G., Magrioti V., and Dennis E. A. (2010) Potent and selective fluoroketone inhibitors of Group VIA calcium-independent phospholipase A2. J. Med. Chem. 53, 3602–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kokotos G., Feuerherm A. J., Barbayianni E., Shah I., Sæther M., Magrioti V., Nguyen T., Constantinou-Kokotou V., Dennis E. A., and Johansen B. (2014) Inhibition of Group IVA cytosolic phospholipase A2 by thiazolyl ketones in vitro, ex vivo, and in vivo. J. Med. Chem. 57, 7523–7535 [DOI] [PubMed] [Google Scholar]
- 124. Yaksh T. L., Kokotos G., Svensson C. I., Stephens D., Kokotos C. G., Fitzsimmons B., Hadjipavlou-Litina D., Hua X. Y., and Dennis E. A. (2006) Systemic and intrathecal effects of a novel series of phospholipase A2 inhibitors on hyperalgesia and spinal prostaglandin E2 release. J. Pharmacol. Exp. Ther. 316, 466–475 [DOI] [PubMed] [Google Scholar]
- 125. López-Vales R., Navarro X., Shimizu T., Baskakis C., Kokotos G., Constantinou-Kokotou V., Stephens D., Dennis E. A., and David S. (2008) Intracellular phospholipase A2 Group IVA and Group VIA play important roles in Wallerian degeneration and axon regeneration after peripheral nerve injury. Brain 131, 2620–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kalyvas A., Baskakis C., Magrioti V., Constantinou-Kokotou V., Stephens D., López-Vales R., Lu J. Q., Yong V. W., Dennis E. A., Kokotos G., and David S. (2009) Differing roles for members of the phospholipase A2 superfamily in experimental autoimmune encephalomyelitis. Brain 132, 1221–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. López-Vales R., Ghasemlou N., Redensek A., Kerr B. J., Barbayianni E., Antonopoulou G., Baskakis C., Rathore K. I., Constantinou-Kokotou V., Stephens D., Shimizu T., Dennis E. A., Kokotos G., and David S. (2011) Phospholipase A2 superfamily members play divergent roles after spinal cord injury. FASEB J. 25, 4240–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Reynolds L. J., Kempner E. S., Hughes L. L., and Dennis E. A. (1995) Inactivation of secretory phospholipase A2 by ionizing radiation. Biophys. J. 68, 2108–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lio Y. C., and Dennis E. A. (1998) Interfacial activation, lysophospholipase and transacylase activity of Group VI Ca2+-independent phospholipase A2. Biochim. Biophys. Acta 1392, 320–332 [DOI] [PubMed] [Google Scholar]
- 130. Pickard R. T., Chiou X. G., Strifler B. A., DeFelippis M. R., Hyslop P. A., Tebbe A. L., Yee Y. K., Reynolds L. J., Dennis E. A., Kramer R. M., and Sharp J. D. (1996) Identification of essential residues for the catalytic function of 85-kDa cytosolic phospholipase A2: probing the role of histidine, aspartic acid, cysteine, and arginine. J. Biol. Chem. 271, 19225–19231 [DOI] [PubMed] [Google Scholar]
- 131. Loo R. W., Conde-Frieboes K., Reynolds L. J., and Dennis E. A. (1997) Activation, inhibition, and regiospecificity of the lysophospholipase activity of the 85-kDa Group IV cytosolic phospholipase A2. J. Biol. Chem. 272, 19214–19219 [DOI] [PubMed] [Google Scholar]
- 132. Mosior M., Six D. A., and Dennis E. A. (1998) Group IV cytosolic phospholipase A2 binds with high affinity and specificity to phosphatidylinositol 4,5-bisphosphate resulting in dramatic increases in activity. J. Biol. Chem. 273, 2184–2191 [DOI] [PubMed] [Google Scholar]
- 133. Balsinde J., Balboa M. A., Li W. H., Llopis J., and Dennis E. A. (2000) Cellular regulation of cytosolic Group IV phospholipase A2 by phosphatidylinositol bisphosphate levels. J. Immunol. 164, 5398–5402 [DOI] [PubMed] [Google Scholar]
- 134. Six D. A., and Dennis E. A. (2003) Essential Ca2+-independent role of the Group IVA cytosolic phospholipase A2 C2 domain for interfacial activity. J. Biol. Chem. 278, 23842–23850 [DOI] [PubMed] [Google Scholar]
- 135. Cao J., Burke J. E., and Dennis E. A. (2013) Using hydrogen/deuterium exchange mass spectrometry to define the specific interactions of the phospholipase A2 superfamily with lipid substrates, inhibitors, and membranes. J. Biol. Chem. 288, 1806–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Burke J. E., and Dennis E. A. (2009) Phospholipase A2 structure/function, mechanism, and signaling. J. Lipid Res. 50, (suppl.) S237–S242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Burke J. E., and Dennis E. A. (2009) Phospholipase A2 biochemistry. Cardiovasc. Drugs Ther. 23, 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Harkewicz R., and Dennis E. A. (2011) Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 80, 301–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Burke J. E., Karbarz M. J., Deems R. A., Li S., Woods V. L. Jr., and Dennis E. A. (2008) Interaction of Group IA phospholipase A2 with metal ions and phospholipid vesicles probed with deuterium exchange mass spectrometry. Biochemistry 47, 6451–6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hsu Y. H., Burke J. E., Stephens D. L., Deems R. A., Li S., Asmus K. M., Woods V. L. Jr., and Dennis E. A. (2008) Calcium binding rigidifies the C2 domain and the intradomain interaction of GIVA phospholipase A2 as revealed by hydrogen/deuterium exchange mass spectrometry. J. Biol. Chem. 283, 9820–9827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Burke J. E., Hsu Y. H., Deems R. A., Li S., Woods V. L. Jr., and Dennis E. A. (2008) A phospholipid substrate molecule residing in the membrane surface mediates opening of the lid region in Group IVA cytosolic phospholipase A2. J. Biol. Chem. 283, 31227–31236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Hsu Y. H., Burke J. E., Li S., Woods V. L. Jr., and Dennis E. A. (2009) Localizing the membrane binding region of Group VIA Ca2+-independent phospholipase A2 using peptide amide hydrogen/deuterium exchange mass spectrometry. J. Biol. Chem. 284, 23652–23661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Cao J., Hsu Y. H., Li S., Woods V. L., and Dennis E. A. (2011) Lipoprotein-associated phospholipase A2 interacts with phospholipid vesicles via a surface-disposed hydrophobic α-helix. Biochemistry 50, 5314–5321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Cao J., Hsu Y. H., Li S., Woods V. L. Jr., and Dennis E. A. (2013) Structural basis of specific interactions of Lp-PLA2 with HDL revealed by hydrogen deuterium exchange mass spectrometry. J. Lipid Res. 54, 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Pang X. Y., Cao J., Addington L., Lovell S., Battaile K. P., Zhang N., Rao J. L., Dennis E. A., and Moise A. R. (2012) Structure/function relationships of adipose phospholipase A2 containing a Cys-His-His catalytic triad. J. Biol. Chem. 287, 35260–35274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bucher D., Hsu Y. H., Mouchlis V. D., Dennis E. A., and McCammon J. A. (2013) Insertion of the Ca2+-independent phospholipase A2 into a phospholipid bilayer via coarse-grained and atomistic molecular dynamics simulations. PLoS Comput. Biol. 9, e1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Mouchlis V. D., Bucher D., McCammon J. A., and Dennis E. A. (2015) Membranes serve as allosteric activators of phospholipase A2, enabling it to extract, bind, and hydrolyze phospholipid substrates. Proc. Natl. Acad. Sci. U.S.A. 112, E516–E525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Burke J. E., Babakhani A., Gorfe A. A., Kokotos G., Li S., Woods V. L. Jr., McCammon J. A., and Dennis E. A. (2009) Location of inhibitors bound to Group IVA phospholipase A2 determined by molecular dynamics and deuterium exchange mass spectrometry. J. Am. Chem. Soc. 131, 8083–8091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Hsu Y. H., Bucher D., Cao J., Li S., Yang S. W., Kokotos G., Woods V. L. Jr., McCammon J. A., and Dennis E. A. (2013) Fluoroketone inhibition of Ca2+-independent phospholipase A2 through binding pocket association defined by hydrogen/deuterium exchange and molecular dynamics. J. Am. Chem. Soc. 135, 1330–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Mouchlis V. D., Limnios D., Kokotou M. G., Barbayianni E., Kokotos G., McCammon J. A., and Dennis E. A. (2016) Development of potent and selective inhibitors for Group VIA calcium-independent phospholipase A2 guided by molecular dynamics and structure-activity relationships. J. Med. Chem. 59, 4403–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Mouchlis V. D., Morisseau C., Hammock B. D., Li S., McCammon J. A., and Dennis E. A. (2016) Computer-aided drug design guided by hydrogen/deuterium exchange mass spectrometry: a powerful combination for the development of potent and selective inhibitors of Group VIA calcium-independent phospholipase A2. Bioorg. Med. Chem. 24, 4801–4811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Dove A. (2015) Greasing the wheels of lipidomics. Science 347, 788–790 [Google Scholar]
- 153. Dennis E. A. (2009) Lipidomics joins the omics evolution. Proc. Natl. Acad. Sci. U.S.A. 106, 2089–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Raetz C. R., Garrett T. A., Reynolds C. M., Shaw W. A., Moore J. D., Smith D. C. Jr., Ribeiro A. A., Murphy R. C., Ulevitch R. J., Fearns C., Reichart D., Glass C. K., Benner C., Subramaniam S., Harkewicz R., Bowers-Gentry R. C., Buczynski M. W., Cooper J. A., Deems R. A., and Dennis E. A. (2006) Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J. Lipid Res. 47, 1097–1111 [DOI] [PubMed] [Google Scholar]
- 155. Dennis E. A., Deems R. A., Harkewicz R., Quehenberger O., Brown H. A., Milne S. B., Myers D. S., Glass C. K., Hardiman G., Reichart D., Merrill A. H. Jr., Sullards M. C., Wang E., Murphy R. C., Raetz C. R., Garrett T. A., Guan Z., Ryan A. C., Russell D. W., McDonald J. G., Thompson B. M., Shaw W. A., Sud M., Zhao Y., Gupta S., Maurya M. R., Fahy E., and Subramaniam S. (2010) A mouse macrophage lipidome. J. Biol. Chem. 285, 39976–39985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Andreyev A. Y., Shen Z., Guan Z., Ryan A., Fahy E., Subramaniam S., Raetz C. R., Briggs S., and Dennis E. A. (2010) Application of proteomic marker ensembles to subcellular organelle identification. Mol. Cell. Proteomics 9, 388–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Andreyev A. Y., Fahy E., Guan Z., Kelly S., Li X., McDonald J. G., Milne S., Myers D., Park H., Ryan A., Thompson B. M., Wang E., Zhao Y., Brown H. A., Merrill A. H., Raetz C. R., Russell D. W., Subramaniam S., and Dennis E. A. (2010) Subcellular organelle lipidomics in TLR-4-activated macrophages. J. Lipid Res. 51, 2785–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Spann N. J., Garmire L. X., McDonald J. G., Myers D. S., Milne S. B., Shibata N., Reichart D., Fox J. N., Shaked I., Heudobler D., Raetz C. R., Wang E. W., Kelly S. L., Sullards M. C., Murphy R. C., Merrill A. H. Jr., Brown H. A., Dennis E. A., Li A. C., Ley K., Tsimikas S., Fahy E., Subramaniam S., Quehenberger O., Russell D. W., and Glass C. K. (2012) Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 151, 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Shibata N., Carlin A. F., Spann N. J., Saijo K., Morello C. S., McDonald J. G., Romanoski C. E., Maurya M. R., Kaikkonen M. U., Lam M. T., Crotti A., Reichart D., Fox J. N., Quehenberger O., Raetz C. R., Sullards M. C., Murphy R. C., Merrill A. H. Jr., Brown H. A., Dennis E. A., Fahy E., Subramaniam S., Cavener D. R., Spector D. H., Russell D. W., and Glass C. K. (2013) 25-Hydroxycholesterol activates the integrated stress response to reprogram transcription and translation in macrophages. J. Biol. Chem. 288, 35812–35823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Quehenberger O., Armando A. M., Brown A. H., Milne S. B., Myers D. S., Merrill A. H., Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., Sullards C. M., Wang E., Murphy R. C., Barkley R. M., Leiker T. J., Raetz C. R., Guan Z., Laird G. M., Six D. A., Russell D. W., McDonald J. G., Subramaniam S., Fahy E., and Dennis E. A. (2010) Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51, 3299–3305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Quehenberger O., and Dennis E. A. (2011) The human plasma lipidome. N. Engl. J. Med. 365, 1812–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Fahy E., Subramaniam S., Brown H. A., Glass C. K., Merrill A. H. Jr., Murphy R. C., Raetz C. R., Russell D. W., Seyama Y., Shaw W., Shimizu T., Spener F., van Meer G., VanNieuwenhze M. S., White S. H., Witztum J. L., and Dennis E. A. (2005) A comprehensive classification system for lipids. J. Lipid Res. 46, 839–861 [DOI] [PubMed] [Google Scholar]
- 163. Fahy E., Subramaniam S., Murphy R. C., Nishijima M., Raetz C. R., Shimizu T., Spener F., van Meer G., Wakelam M. J., and Dennis E. A. (2009) Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 50, (suppl.) S9–S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Buczynski M. W., Dumlao D. S., and Dennis E. A. (2009) Thematic Review Series: Proteomics: an integrated omics analysis of eicosanoid biology. J. Lipid Res. 50, 1015–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Harkewicz R., Fahy E., Andreyev A., and Dennis E. A. (2007) Arachidonate-derived dihomoprostaglandin production observed in endotoxin-stimulated macrophage-like cells. J. Biol. Chem. 282, 2899–2910 [DOI] [PubMed] [Google Scholar]
- 166. Harkewicz R., Du H., Tong Z., Alkuraya H., Bedell M., Sun W., Wang X., Hsu Y. H., Esteve-Rudd J., Hughes G., Su Z., Zhang M., Lopes V. S., Molday R. S., Williams D. S., Dennis E. A., and Zhang K. (2012) Essential role of ELOVL4 protein in very long chain fatty acid synthesis and retinal function. J. Biol. Chem. 287, 11469–11480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Wang Y., Armando A. M., Quehenberger O., Yan C., and Dennis E. A. (2014) Comprehensive ultra-performance liquid chromatographic separation and mass spectrometric analysis of eicosanoid metabolites in human samples. J. Chromatogr. A 1359, 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Dumlao D. S., Buczynski M. W., Norris P. C., Harkewicz R., and Dennis E. A. (2011) High-throughput lipidomic analysis of fatty acid derived eicosanoids and N-acylethanolamines. Biochim. Biophys. Acta 1811, 724–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Quehenberger O., Armando A. M., and Dennis E. A. (2011) High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim. Biophys. Acta 1811, 648–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Baker P. R., Armando A. M., Campbell J. L., Quehenberger O., and Dennis E. A. (2014) Three-dimensional enhanced lipidomics analysis combining UPLC, differential ion mobility spectrometry, and mass spectrometric separation strategies. J. Lipid Res. 55, 2432–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Norris P. C., Gosselin D., Reichart D., Glass C. K., and Dennis E. A. (2014) Phospholipase A2 regulates eicosanoid class switching during inflammasome activation. Proc. Natl. Acad. Sci. U.S.A. 111, 12746–12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Buczynski M. W., Stephens D. L., Bowers-Gentry R. C., Grkovich A., Deems R. A., and Dennis E. A. (2007) TLR-4 and sustained calcium agonists synergistically produce eicosanoids independent of protein synthesis in RAW264.7 cells. J. Biol. Chem. 282, 22834–22847 [DOI] [PubMed] [Google Scholar]
- 173. Blaho V. A., Buczynski M. W., Brown C. R., and Dennis E. A. (2009) Lipidomic analysis of dynamic eicosanoid responses during the induction and resolution of Lyme arthritis. J. Biol. Chem. 284, 21599–21612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Dumlao D. S., Cunningham A. M., Wax L. E., Norris P. C., Hanks J. H., Halpin R., Lett K. M., Blaho V. A., Mitchell W. J., Fritsche K. L., Dennis E. A., and Brown C. R. (2012) Dietary fish oil substitution alters the eicosanoid profile in ankle joints of mice during Lyme infection. J. Nutr. 142, 1582–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Tam V. C., Quehenberger O., Oshansky C. M., Suen R., Armando A. M., Treuting P. M., Thomas P. G., Dennis E. A., and Aderem A. (2013) Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell 154, 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Norris P. C., and Dennis E. A. (2012) Omega-3 fatty acids cause dramatic changes in TLR4 and purinergic eicosanoid signaling. Proc. Natl. Acad. Sci. U.S.A. 109, 8517–8522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Li P., Spann N. J., Kaikkonen M. U., Lu M., Oh D. Y., Fox J. N., Bandyopadhyay G., Talukdar S., Xu J., Lagakos W. S., Patsouris D., Armando A., Quehenberger O., Dennis E. A., Watkins S. M., Auwerx J., Glass C. K., and Olefsky J. M. (2013) NCoR repression of LXRs restricts macrophage biosynthesis of insulin-sensitizing omega 3 fatty acids. Cell 155, 200–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Svensson C. I., Lucas K. K., Hua X. Y., Powell H. C., Dennis E. A., and Yaksh T. L. (2005) Spinal phospholipase A2 in inflammatory hyperalgesia: role of the small, secretory phospholipase A2. Neuroscience 133, 543–553 [DOI] [PubMed] [Google Scholar]
- 179. Lucas K. K., Svensson C. I., Hua X. Y., Yaksh T. L., and Dennis E. A. (2005) Spinal phospholipase A2 in inflammatory hyperalgesia: role of Group IVA cPLA2. Br J. Pharmacol. 144, 940–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Buczynski M. W., Svensson C. I., Dumlao D. S., Fitzsimmons B. L., Shim J. H., Scherbart T. J., Jacobsen F. E., Hua X. Y., Yaksh T. L., and Dennis E. A. (2010) Inflammatory hyperalgesia induces essential bioactive lipid production in the spinal cord. J. Neurochem. 114, 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Gregus A. M., Doolen S., Dumlao D. S., Buczynski M. W., Takasusuki T., Fitzsimmons B. L., Hua X. Y., Taylor B. K., Dennis E. A., and Yaksh T. L. (2012) Spinal 12-lipoxygenase-derived hepoxilin A3 contributes to inflammatory hyperalgesia via activation of TRPV1 and TRPA1 receptors. Proc. Natl. Acad. Sci. U.S.A. 109, 6721–6726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Gregus A. M., Dumlao D. S., Wei S. C., Norris P. C., Catella L. C., Meyerstein F. G., Buczynski M. W., Steinauer J. J., Fitzsimmons B. L., Yaksh T. L., and Dennis E. A. (2013) Systematic analysis of rat 12/15-lipoxygenase enzymes reveals critical role for spinal eLOX3 hepoxilin synthase activity in inflammatory hyperalgesia. FASEB J. 27, 1939–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Christianson C. A., Dumlao D. S., Stokes J. A., Dennis E. A., Svensson C. I., Corr M., and Yaksh T. L. (2011) Spinal TLR4 mediates the transition to a persistent mechanical hypersensitivity after the resolution of inflammation in serum-transferred arthritis. Pain 152, 2881–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Jiao J., Mikulec C., Ishikawa T. O., Magyar C., Dumlao D. S., Dennis E. A., Fischer S. M., and Herschman H. (2014) Cell-type-specific roles for COX-2 in UVB-induced skin cancer. Carcinogenesis 35, 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Jiao J., Ishikawa T. O., Dumlao D. S., Norris P. C., Magyar C. E., Mikulec C., Catapang A., Dennis E. A., Fischer S. M., and Herschman H. R. (2014) Targeted deletion and lipidomic analysis identify epithelial cell COX-2 as a major driver of chemically induced skin cancer. Mol. Cancer Res. 12, 1677–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Friedman P., Horkko S., Steinberg D., Witztum J. L., and Dennis E. A. (2002) Correlation of antiphospholipid antibody recognition with the structure of synthetic oxidized phospholipids. Importance of Schiff base formation and aldol condensation. J. Biol. Chem. 277, 7010–7020 [DOI] [PubMed] [Google Scholar]
- 187. Hörkkö S., Bird D. A., Miller E., Itabe H., Leitinger N., Subbanagounder G., Berliner J. A., Friedman P., Dennis E. A., Curtiss L. K., Palinski W., and Witztum J. L. (1999) Monoclonal autoantibodies specific for oxidized phospholipids or oxidized phospholipid-protein adducts inhibit macrophage uptake of oxidized low-density lipoproteins. J. Clin. Invest. 103, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Bird D. A., Gillotte K. L., Hörkkö S., Friedman P., Dennis E. A., Witztum J. L., and Steinberg D. (1999) Receptors for oxidized low-density lipoprotein on elicited mouse peritoneal macrophages can recognize both the modified lipid moieties and the modified protein moieties: implications with respect to macrophage recognition of apoptotic cells. Proc. Natl. Acad. Sci. U.S.A. 96, 6347–6352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Chang M. K., Bergmark C., Laurila A., Hörkkö S., Han K. H., Friedman P., Dennis E. A., and Witztum J. L. (1999) Monoclonal antibodies against oxidized low-density lipoprotein bind to apoptotic cells and inhibit their phagocytosis by elicited macrophages: evidence that oxidation-specific epitopes mediate macrophage recognition. Proc. Natl. Acad. Sci. U.S.A. 96, 6353–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Boullier A., Friedman P., Harkewicz R., Hartvigsen K., Green S. R., Almazan F., Dennis E. A., Steinberg D., Witztum J. L., and Quehenberger O. (2005) Phosphocholine as a pattern recognition ligand for CD36. J. Lipid Res. 46, 969–976 [DOI] [PubMed] [Google Scholar]
- 191. Harkewicz R., Hartvigsen K., Almazan F., Dennis E. A., Witztum J. L., and Miller Y. I. (2008) Cholesteryl ester hydroperoxides are biologically active components of minimally oxidized low density lipoprotein. J. Biol. Chem. 283, 10241–10251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Fang L., Harkewicz R., Hartvigsen K., Wiesner P., Choi S. H., Almazan F., Pattison J., Deer E., Sayaphupha T., Dennis E. A., Witztum J. L., Tsimikas S., and Miller Y. I. (2010) Oxidized cholesteryl esters and phospholipids in zebrafish larvae fed a high cholesterol diet: macrophage binding and activation. J. Biol. Chem. 285, 32343–32351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Leibundgut G., Scipione C., Yin H., Schneider M., Boffa M. B., Green S., Yang X., Dennis E., Witztum J. L., Koschinsky M. L., and Tsimikas S. (2013) Determinants of binding of oxidized phospholipids on apolipoprotein (a) and lipoprotein (a). J. Lipid Res. 54, 2815–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Choi S. H., Yin H., Ravandi A., Armando A., Dumlao D., Kim J., Almazan F., Taylor A. M., McNamara C. A., Tsimikas S., Dennis E. A., Witztum J. L., and Miller Y. I. (2013) Polyoxygenated cholesterol ester hydroperoxide activates TLR4 and SYK dependent signaling in macrophages. PLoS ONE 8, e83145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Ravandi A., Leibundgut G., Hung M. Y., Patel M., Hutchins P. M., Murphy R. C., Prasad A., Mahmud E., Miller Y. I., Dennis E. A., Witztum J. L., and Tsimikas S. (2014) Release and capture of bioactive oxidized phospholipids and oxidized cholesteryl esters during percutaneous coronary and peripheral arterial interventions in humans. J. Am. Coll. Cardiol. 63, 1961–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Gupta S., Maurya M. R., Stephens D. L., Dennis E. A., and Subramaniam S. (2009) An integrated model of eicosanoid metabolism and signaling based on lipidomics flux analysis. Biophys. J. 96, 4542–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Norris P. C., Reichart D., Dumlao D. S., Glass C. K., and Dennis E. A. (2011) Specificity of eicosanoid production depends on the TLR-4-stimulated macrophage phenotype. J. Leukoc. Biol. 90, 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Kihara Y., Gupta S., Maurya M. R., Armando A., Shah I., Quehenberger O., Glass C. K., Dennis E. A., and Subramaniam S. (2014) Modeling of eicosanoid fluxes reveals functional coupling between cyclooxygenases and terminal synthases. Biophys. J. 106, 966–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Sabidó E., Quehenberger O., Shen Q., Chang C. Y., Shah I., Armando A. M., Andreyev A., Vitek O., Dennis E. A., and Aebersold R. (2012) Targeted proteomics of the eicosanoid biosynthetic pathway completes an integrated genomics-proteomics-metabolomics picture of cellular metabolism. Mol. Cell. Proteomics 11, M111.014746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Gupta S., Kihara Y., Maurya M. R., Norris P. C., Dennis E. A., and Subramaniam S. (2016) Computational modeling of competitive metabolism between ω3- and ω6-polyunsaturated fatty acids in inflammatory macrophages. J. Phys. Chem. B 120, 8346–8353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Harmon G. S., Dumlao D. S., Ng D. T., Barrett K. E., Dennis E. A., Dong H., and Glass C. K. (2010) Pharmacological correction of a defect in PPAR-γ signaling ameliorates disease severity in Cftr-deficient mice. Nat. Med. 16, 313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Gorden D. L., Myers D. S., Ivanova P. T., Fahy E., Maurya M. R., Gupta S., Min J., Spann N. J., McDonald J. G., Kelly S. L., Duan J., Sullards M. C., Leiker T. J., Barkley R. M., Quehenberger O., Armando A. M., Milne S. B., Mathews T. P., Armstrong M. D., Li C., Melvin W. V., Clements R. H., Washington M. K., Mendonsa A. M., Witztum J. L., Guan Z., Glass C. K., Murphy R. C., Dennis E. A., Merrill A. H. Jr., Russell D. W., Subramaniam S., and Brown H. A. (2015) Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J. Lipid Res. 56, 722–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Loomba R., Quehenberger O., Armando A., and Dennis E. A. (2015) Polyunsaturated fatty acid metabolites as novel lipidomic biomarkers for noninvasive diagnosis of nonalcoholic steatohepatitis. J. Lipid Res. 56, 185–192 [DOI] [PMC free article] [PubMed] [Google Scholar]