Abstract
Background
This study characterized the genetic landscape of chemoradiation treated urothelial carcinoma of the bladder (UCB), with the goal of identifying potential correlates of response.
Methods
Primary tumors with (n=8) or without (n=40) matched recurrent tumor from 48 non-metastatic high grade UCB patients treated primarily with chemoradiation were analyzed using a next-generation sequencing assay enriched for cancer-related and canonical DNA damage response (DDR) genes. Protein expression of MRE11, a previously suggested biomarker, was assessed in 44 patients. We compared recurrent to primary tumors and evaluated clinical impact of likely deleterious DDR gene alterations.
Results
The profile of alterations approximated that of prior UCB cohorts. Within 5 pairs of matched primary-recurrent tumors, a median of 92% of somatic mutations was shared. A median 33% of mutations were shared between 3 matched bladder-metastasis pairs. Of 26 (54%) patients with DDR gene alterations, 12 (25%) harbored likely deleterious alterations. In multivariable analysis, these patients displayed a trend towards reduced bladder recurrence [hazard ratio (HR) 0.32; p=0.070] or any recurrence [HR 0.37; p=0.070]. The most common such alteration, ERCC2 mutations were associated with significantly lower 2-year metastatic recurrence (0% vs. 43%; log-rank p=0.044). We detected no impact of MRE11 protein expression on outcome.
Conclusions
A similar mutation profile between primary and recurrent tumors implies that pre-existing, resistant clonal populations represent the primary mechanism of chemoradiation treatment failure. Deleterious DDR genetic alterations, particularly recurrent alterations in ERCC2, may be associated with improved UCB chemoradiation outcomes. A small sample size necessitates independent validation of these observations.
Keywords: bladder chemoradiation, bladder preservation, ERCC2, DNA damage response, radiation resistance
Introduction
Transurethral resection of bladder tumor (TURBT) followed by chemoradiation (CRT) is an effective bladder-preservation alternative to chemotherapy and radical cystectomy (RC) for selected muscle-invasive bladder cancer (MBIC) patients 1-5. However, incomplete treatment response requiring immediate cystectomy and disease-related mortality occur in ∼30% of patients 6, 7. Further, theoretical concerns about “field cancerization” in UCB would imply inevitable local relapse following bladder preservation. Improved understanding of the biologic basis for chemoradiation response would help address its current poor acceptance.
Ionizing radiation induces cell death primarily through DNA double-strand breaks (DSB). Activation of the DNA-damage response (DDR) pathway by DSBs begins with recruitment of the MRE11-RAD50-NBS1 complex and ATM 8. Genes involved in this process include BRCA1, BRCA2, and PALB2 (homologous recombination, HR), FANCD2 and BRIP1 (Fanconi Anemia pathway), and mediators of non-homologous end joining (NHEJ). Concurrent CRT is superior to radiotherapy alone 3. DNA cross-links induced by cisplatin are repaired primarily via the nucleotide excision repair (NER) pathway, which includes ERCC2 9, a gene altered more frequently in urothelial carcinoma (12%) than in other malignancies analyzed by The Cancer Genome Atlas (TCGA) and associated with improved response to cisplatin-based chemotherapy 10.
Two studies assayed protein expression of mediators of DSB repair 11, 12 in UCB. Both studies correlated lowest quartile MRE11 expression and worse cancer-specific survival (CSS) in patients treated with RT or CRT but not surgery. The mechanistic basis for this observation remains unclear.
We hypothesized that molecular characterization of UCB chemoradiation patients with a focus on DDR alterations could identify correlates of treatment response and in-bladder tumor recurrence.
Methods
Sample collection and DNA preparation
Frozen and formalin-fixed paraffin-embedded (FFPE) tumor tissue was collected from patients with non-metastatic UCB treated with curative intent radiation or chemoradiation under IRB approved protocols. Pathologic review, germline control details and clinical annotation for each case are listed in Supplementary Tables 1-2. A representative H&E slide was reviewed by a board-certified genitourinary pathologist (H.A.A) to confirm histology. DNA was extracted from macro-dissected tissue as previously described 13.
Targeted capture and sequencing
Protein-coding exons of 341 cancer-associated genes were sequenced using an institutional capture-based sequencing assay (Memorial Sloan Kettering-Integrated Molecular Profiling of Actionable Cancer Targets, MSK-IMPACT) 14, with one modification: exons of 20 additional genes were added to enrich for canonical DDR pathways (Supplementary Table 3) 15. The two NimbleGen (Roche) panels were combined for capture proportionally by target territory and subjected to barcoding, capture, and sequencing on an Illumina HiSeq 2500 system as 2×100bp paired-end reads, as previously described13, 16. In one patient (DMP-RT1), the unmodified MSK-IMPACT assay was employed.
Sequence analysis
Sequence reads were aligned to the reference human genome (hg19) using the CASAVA (Illumina) pipeline, the Burrows-Wheeler Alignment tool v0.6.2, and GATK. Single-nucleotide variants were called using MuTect v1.0.27783 and indels using the SomaticIndelDetector tool 16. All candidate mutations and indels were reviewed manually (SNS). Minimum variant frequency to call an alteration was 2%. For copy number analysis in patients with matched germline DNA, the sequence coverage for each exon was compared in tumor and matched germline sample, following sample-wide LOESS (local regression) normalization for GC percentage and global differences in “on-target” sequence coverage. Changes in the tumor-to-germline coverage ratios were used to infer copy number alterations. Coverage ratios ≥2 or ≤0.5 were defined as amplifications or deletions, respectively.
We chose to include 6 patients without matched germline DNA (insufficient DNA/poor coverage) given the small cohort size. Somatic mutations in those patients were called after removing germline SNVs reported in dbSNP 17, the 1000 Genomes Project 18 and pooled normal from the other patients. Remaining SNVs in these patients with variant frequencies ∼50% were subjected to manual literature searches to rule out germline origin.
Statistical analysis
Given the unclear functional impact of many DDR gene alterations, correlation with clinical characteristics was focused on alterations likely to be deleterious. These were defined as nonsense, frameshift, or splice site mutations, or missense alterations at recurrent sites associated with chemotherapy response in prior work. Only ERCC2 was demonstrated to harbor such missense alterations, occurring at or near previously reported recurrent alteration sites within UCB 19, 20 or associated with chemotherapy response 10, as discussed further in the results. Bivariate comparisons of mutation frequency were performed using Fisher's exact test and comparisons of means with ANOVA testing.
Analyzed clinical endpoints were time to bladder recurrence (superficial or MIBC), metastatic recurrence, and any recurrence, as measured from the end of radiotherapy. Censoring was performed at last follow up or death. Bladder intact survival was not analyzed because not all patients were medically able to undergo salvage cystectomy. Kaplan-Meier method was used to estimate cumulative probability of each event, and the log-rank statistic tested differences between groups. Hazard ratios and confidence intervals were estimated using Cox regression. Analyses were conducted using SPSS v. 20 (IBM) and R software version 3.2.1 (R Core Development Team, Vienna, Austria). An alpha of 0.05 was used to define statistical significance, with two-sided testing.
Genomic and associated clinicopathologic data are publically available through the MSKCC cBioPortal (www.cbioportal.com) 21.
MRE11 assays
MRE11 immunohistochemistry (IHC) was performed on 4μM thick FFPE tissue sections using 2 different primary antibodies: mouse monoclonal Abcam [12D7] (Cat. No. ab214) and rabbit polyclonal #59-10 (Petrini laboratory, Sloan Kettering Institute, New York, NY). Staining was performed with the Discovery XT Automated System with DAB Map kit detection system with the appropriate secondary antibodies (Ventana Medical Systems, Tucson, AZ). Ab214 was used in 2 prior bladder cancer radiotherapy response studies 11, 12, while #59-10 was validated using positive and negative controls from an ERBB2 driven neoplasm in MRE11WT/WT and hypomorphic MRE11ATLD1/ATLD1 mouse models (Supplementary Figure 1) 22. Two pathologists (HAA, JH) used the semi-quantitative score (SQS) from prior biomarker studies of MRE11 (median % positive cells multiplied by modal intensity) to score samples.
Results
We identified 48 evaluable patients with radiotherapy or CRT treated UCB at our institution between 1994 and 2014. Baseline clinical and treatment data are summarized in Table 1. Triple modality therapy consisting of TURBT and CRT was prioritized; however, the typical co-morbidities and age of a UCB CRT population contributed to treatment heterogeneity. Thirty-one patients (71%) underwent visibly complete TURBT prior to treatment. Only three (6%) patients were deemed unsuitable for chemotherapy, while the remainder received chemoradiation (n=23, 48%) or neoadjuvant chemotherapy followed by chemoradiation (n=22, 46%). Twenty-four patients (50%) received platinum chemotherapy (22 neoadjuvant, 9 concurrent), while gemcitabine (n=20) or paclitaxel (n=1) based concurrent CRT was administered to 21 (44%) patients. Post-treatment cystoscopy evaluation was performed in 40/48 patients (83%). Details of chemotherapy regimen, radiotherapy, and outcome are provided in Supplementary Table 1.
Table 1. Clinical characteristics of cohort.
| Age, median (min-max) | 67 (37-87) |
| Gender | |
| Male | 36 (75%) |
| Female | 12 (25%) |
| Tobacco Exposure | 40 (83%) |
| Complete TURBT | 34 (71%) |
| Hydronephrosis | 12 (25%) |
| Chemotherapy | |
| Neoadjuvant/Concurrent | 22 (46%) |
| Concurrent Only | 23 (48%) |
| RT Alone | 3 (6%) |
| RT Dose, median (range) | 66 Gy (54-72) |
| Clinical T-stage | |
| T1 | 1 (2%) |
| T2 | 34 (71%) |
| T3 | 4 (8%) |
| T4 | 9 (19%) |
| Clinical N-stage | |
| cN0 | 40 (83%) |
| cN+ | 8 (17%) |
| Clinical Overall Stage | |
| I | 1 (2%) |
| II | 30 (62%) |
| III | 7 (15%) |
| IV | 10 (21%) |
Abbreviations: RT, Radiotherapy; TURBT, Transurethral resection of bladder tumor; Gy, Gray.
Targeted sequencing with MSK-IMPACT was performed on 56 tumors from 48 patients, including 5 primary-bladder recurrence pairs, 2 primary-metastasis pairs, and 1 bladder recurrence-metastasis pair (Supplementary Table 2). In 42/48 patients, germline DNA was available from peripheral blood or uninvolved tissue. Sufficient pre-treatment tumor tissue from diagnostic TURBT was available in 44/48 patients. In 4 patients, however, only locally recurrent tumor was analyzable. Average sequencing depth was 372×. The median number of somatic mutations per sample was 9 (range 1-34).
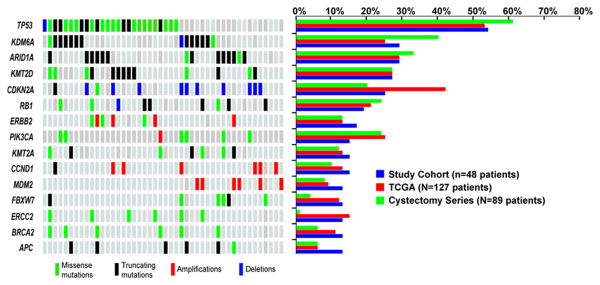
The most common alterations observed in this cohort are depicted in Figure 1. The alteration pattern was similar to those reported in treatment-naïve samples analyzed by TCGA and in an 89 patient UCB cohort treated with RC +/- chemotherapy from our institution 13. For instance, chromatin-modifying genes including ARID1A, KMT2D, and KDM6A were commonly mutated (Supplementary Figure 2A), and the prevalence of alterations within kinase signaling pathways was not substantially different (Supplementary Figure 2B).
Figure 1.
Representation of the most commonly altered genes in this series of 48 patients with urothelial carcinoma of the bladder (UCB) treated with radiation-based bladder-preservation (left) and comparison to observed frequencies in TCGA of UCB and an 89 patient cohort of patients treated with radical cystectomy (right). Mutations are categorized as missense mutations (green), or truncating nonsense, frameshift, or splice site mutations and indels (black). Copy number amplifications (red bars) or deletions (blue bars) are noted.
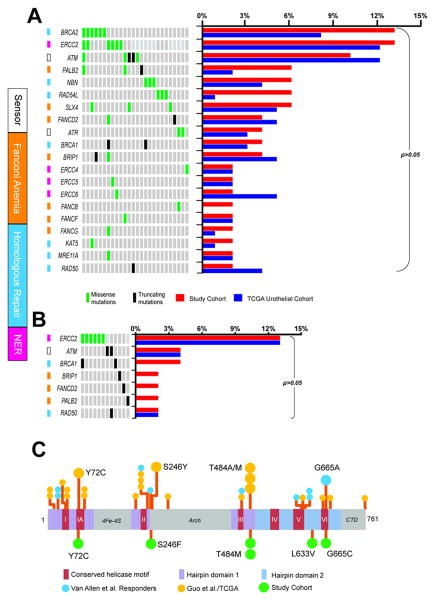
Twenty-six (54%) patients harbored DDR gene non-synonymous alterations (52 mutations in 20 genes), as illustrated in Figure 2A. Eight (15%) were truncating (nonsense, splice site, or frameshift alterations). Most DDR missense alterations (28/44, 64%) were not previously reported in a query of the Catalogue Of Somatic Mutations In Cancer (COSMIC), a literature search, or screening against a pan-cancer hotspot analysis of 11,119 tumors 23. These are of unclear significance. Thus, we focused on alterations likely to be deleterious to function, as previously defined. Deleterious truncating mutations (ATM n=2, BRCA1 n=2, BRIP1 n=1, FANCD2 n=1, PALB2 n=1, RAD50 n=1) or missense alterations (ERCC2, n=6) satisfying our criteria were noted in 12 patients (25%, Figure 2B and Supplementary Table 4). ERCC2 was the only DDR gene with missense alterations detected in multiple patients that have also been previously linked to chemotherapy response in UCB. Figure 2C illustrates the close relationship of the ERCC2 alterations noted in the current cohort as compared to recurrently altered sites.. Deleterious DDR alterations were associated with a higher tumor mutation burden (mean 18.8 vs. 8.7 mutations; ANOVA p<0.001).
Figure 2.
A, Oncoprint (left) of DNA damage response (DDR) gene alterations and comparison to frequencies in TCGA (right). Only altered cases are shown. Functional annotation of genes is displayed (colored squares) B, Oncoprint (left) showing only those alterations predicted to be deleterious and comparison to TCGA deleterious alteration frequencies (right). C, ERCC2 non-synonymous somatic missense mutations in the current study (bottom) are mapped on a stick plot and compared to 3 previously reported whole exome sequencing cohorts (top), including a cisplatin responder study (Van Allen et al.) and studies by Guo et al,(superficial and muscle-invasive UCB included) and TCGA for urothelial MIBC. Prior alterations in close proximity to currently observed alterations are detailed. CTD- Carboxy-terminus domain.
We evaluated the association of these deleterious DDR alterations with bladder recurrence (24 events), metastatic recurrence (22 events), and any recurrence (28 events). We did not analyze CSS, due to an insufficient number of events (n=13). Median follow up of surviving patients was 26 months (range 5-115). Nearly half of bladder recurrences were ≤T1 (10/24, 42%), with 50% remaining disease-free following TURBT. Salvage cystectomy was required for MIBC recurrence in 8 patients.
On univariable analysis, the presence of a DDR deleterious alteration corresponded to a trend towards reduced cumulative incidence of bladder recurrence (2-year cumulative incidence 28% vs. 63%, p=0.071) or any recurrence (2-year cumulative incidence 36% vs. 63%, p=0.067; Supplementary Figure 3). Given the known influence of clinical stage and complete TURBT prior to therapy, we further examined the prognostic effect of DDR deleterious alterations in a multivariable model (Table 2). This demonstrated again a trend towards reduced risk of bladder recurrence (hazard ratio, HR 0.32, 95% CI 0.10-1.10; p=0.070), metastatic recurrence (HR 0.35, 95% CI 0.10-1.20; p=0.094), and any recurrence (HR 0.37, 95% CI 0.13-1.08; p=0.070) in patients with deleterious DDR mutations.
Table 2. Cox regression multivariable analysis for outcomes.
| Variable | Bladder Recurrence | Metastatic Recurrence | Any Relapse | |||
|---|---|---|---|---|---|---|
|
|
||||||
| HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | |
| DDR Deleterious Alteration | 0.32 (0.10-1.10) | 0.070 | 0.35 (0.10-1.20) | 0.094 | 0.37 (0.13-1.08) | 0.070 |
| Stage (IV vs. I-III) | 2.01 (0.79-5.10) | 0.144 | 3.56 (1.39-9.14) | 0.008 | 2.12 (0.90-5.00) | 0.084 |
| Complete TURBT | 0.47 (0.19-1.15) | 0.098 | 0.40 (0.16-0.98) | 0.045 | 0.67 (0.29-1.55) | 0.348 |
Patient outcomes according to each deleteriously altered DDR gene are detailed in Supplementary Table 4. Recurrences (any site) were noted in those with ATM (2/2), BRIP1 (1/1), FANCD2 (1/1), and RAD50 (1/1) deleterious alterations. In contrast, only one of 8 patients with deleterious alterations in ERCC2, BRCA1, or PALB2 experienced a recurrence (bladder). ERCC2 alterations were associated with lower 2-year cumulative incidence of bladder recurrence (20% vs. 55%, p=0.154), metastatic recurrence (0% vs. 43%, p=0.044), and any recurrence (20% vs. 61%, p=0.085) in univariable analysis. Only one of the 6 patients with ERCC2 missense alterations recurred, with a T1 lesion successfully treated by TURBT. The cohort size and the genomic heterogeneity of bladder cancers prohibited multivariable modeling of the specific contribution of ERCC2 or other genes.
Prior investigations have identified a correlation between low MRE11 protein expression and worse outcomes in radiotherapy treated UCB patients 11, 12. We sought to validate this finding in 44/48 of our patients with sufficient tissue for MRE11 IHC. Using two different antibodies (Abcam ab214 and #59-10), we found no correlation between any outcome measure and quartile cut-offs of MRE11 SQS (all p>0.5). We also observed a poor correlation in SQS between the two different antibodies (Supplementary Figure 1C, R2 = 0.068).
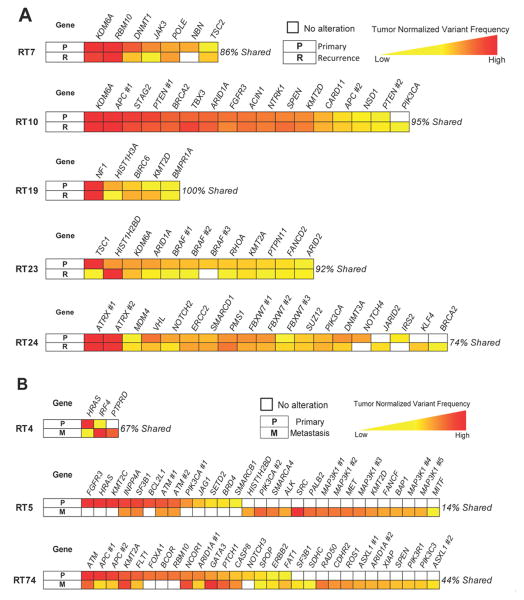
Lastly, in order to better understand treatment failure, we compared the mutation profiles of bladder recurrence tumors to their paired pre-treatment primary bladder tumors in 5 patients. We found that a median of 92% (range, 74-100%) of mutations was shared between the pre- and post-treatment bladder tumors. Alterations unique to the bladder recurrences were rare (mean 4%, range 0-16% of a patient's alterations). The relative allele frequencies of alterations remained generally the same (Figure 3A and Supplementary Table 5.) The same analysis was carried out in 3 patients with paired bladder tumors and distant metastases (Figure 3B and Supplementary Table 6; median 33% mutations shared, range 14-44%; mean 41% of alterations unique to the metastasis).
Figure 3.
Comparison of mutation profiles between paired tumor samples from individual patients. A, Recurrent bladder tumors share a majority of mutations with pre-radiation primary bladder tumors. B, Metastatic lesions also share alterations with bladder primaries, albeit with less overlap. Variant frequencies for mutations are normalized within each tumor to give relative values, as depicted on the color scales, which allow comparison across a patient's samples. Gene alterations are ordered for each patient based upon relative variant frequency in the reference bladder tumor in the first row of each diagram. For genes with multiple mutations, each shared alteration is represented separately; specific alterations corresponding to the genes listed are given in Supplementary Tables 5-6. Alterations were counted if present at ≥2% frequency.
Discussion
Chemoradiation is an effective bladder preservation strategy for selected UCB patients; however, uncertainties in predicting response and physician biases have limited its widespread use. In the present study, we characterized the genetic landscape of tumors from patients who underwent primarily definitive CRT, with the goal of identifying correlates of response and mechanisms of treatment failure.
While most patients in our cohort harbored DDR pathway alterations, only a small fraction were functionally deleterious based upon pre-defined criteria. Subsequently, we included only truncating mutations and recurrent missense alterations at or near sites previously linked to chemotherapy response (only seen in ERCC2 in this cohort) within our exploratory analysis of the prognostic impact of DDR pathway alterations. We found a trend towards reduced cumulative incidence of bladder recurrence and any recurrence in the 25% of patients who harbored deleterious alterations in DDR genes, namely ATM, BRCA1, BRIP1, ERCC2, FANCD2, PALB2, and RAD50.
Improved outcomes in MIBC patients with DDR alterations to surgery and/or chemotherapy have been previously described. Yap et al. demonstrated significantly improved recurrence-free survival following RC with or without chemotherapy in patients with non-synonymous alterations in a similar gene set (ATM, ERCC2, FANCD2, PALB2, BRCA1 and BRCA2) 24. Plimack et al. identified a significant association between alterations in any one of ATM, FANCC, or RB1 and complete response to neoadjuvant cisplatin-based chemotherapy 25. Of the DDR mutations identified in these studies, a minority was truncating (5/33 in Yap et al., 11/30 in Plimack et al.), and the functionality of missense alterations was assessed by computational modeling or recurrent nature. As noted, our analysis was even more restrictive in defining deleterious alterations to minimize the influence of passenger alterations and did not employ recursive algorithms such as clustering typically used in RNA-based biomarker discovery. For this reason and the fundamental differences in treatment approach, while noting the interesting overlap of our findings with the noted prior work, we did not presume to attempt validation analyses of the prognostic gene sets reported by these two groups.
With that said, it is probable that the deleterious DDR alterations in this cohort do not impact upon therapeutic response equally. The main contributor of DDR deleterious alterations in our cohort was ERCC2. ERCC2 alterations correlated with a significantly lower cumulative incidence of metastatic recurrence and with a trend towards lower cumulative incidence of any recurrence. ERCC2 alterations occur at a higher rate in UCB than in other neoplasms and have been correlated significantly with pathologic complete response at RC following cisplatin-based neoadjuvant chemotherapy 10. The alterations in our study were clustered near conserved helicase domains, which are predicted to impact response primarily to adduct-forming agents such as cisplatin. However, only 3/6 patients with ERCC2 alterations received cisplatin as part of their treatment. Indeed, the single patient with ERCC2 alteration and local recurrence retained a pre-treatment ERCC2 G665C alteration in the recurrent tumor, following cisplatin-based neoadjuvant chemotherapy and gemcitabine CRT. This recurrence may be due to suboptimal effect from gemcitabine-based CRT or other factors. While our study is too small to assess the specific influence of ERCC2 or its interaction with chemotherapy regimen, these observations nevertheless support further investigation into its potential role in regimen selection.
In contrast, we were unable to confirm an association between patient outcomes and MRE11 expression by IHC, previously suggested as a predictive biomarker of RT response in UCB 11, 12. The reasons for our differing findings may be related to our smaller cohort size, a focus on recurrence events in our study vs. CSS in prior work, or variability in the performance characteristics of different antibody lots.
A primary study goal was to assess changes during tumor progression. A commonly held tenet of the “field effect” theory of UCB is that carcinogenic exposures imbue the urothelium with a predisposition to de novo tumor formation, challenging the concept of bladder-preservation. However, our study's finding that paired pre-treatment and recurrent bladder tumors share near identical genetic profiles implies that insufficient log-kill, possibly related to intrinsic low CRT sensitivity, drives local treatment failure. This highlights the importance of identifying baseline sensitivity factors to CRT and the relevance of pre-treatment actionable alterations.
Limitations of this study include its retrospective nature, small size, heterogeneous treatments and tissue and the lack of a validation set for the exploratory analysis of the prognostic impact of DDR alterations. From a technical standpoint, the rarity of available tissue necessitated inclusion of samples without paired normal, risking overcalling of alterations, or paired with normal-appearing areas of bladder tissue that may in fact harbor genetic alterations, risking the exclusion of true somatic alterations identified within tumor tissue during the mutation calling process. The current work is the first effort to our knowledge to profile the genetic landscape of tumors from UCB patients undergoing bladder preservation intent definitive chemoradiation. In the future, validation and homogenization of treatment/tissue input will require more extensive, multi-institution cohorts, which are likely only accessible from collaborative group trials, such as those of the RTOG. In the interim, we believe this work provides insights into the mechanism of treatment failure and the potential relationship between DDR alterations, specifically in ERCC2, to clinical outcomes in this population. Independent validation in larger CRT cohorts is needed.
Conclusion
In this next-generation sequencing analysis of a primarily CRT bladder preservation UCB cohort, we found that deleterious DDR alterations, most commonly found in ERCC2, may be associated with lower recurrence risk. Further, recurrent bladder tumors largely resembled pre-treatment tumors, arguing for pre-existing treatment resistance as the primary mechanism of failure. While these data require validation, they provide insight into potential predictors of therapeutic response and the biology of recurrence failures in UC managed with chemoradiation.
Supplementary Material
Acknowledgments
Funding acknowledgments: This work was supported by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology, in part by the Integrated Genomics Operation Core, funded by National Cancer Institute Cancer Center Core Grant No. P30 CA008748 (core grant provides funding to institutional cores [e.g., biostatistics], which were used in this study), and a Cycle for Survival grant.
Footnotes
Authorship Contributions: Conceptualization: NBD, MAK, HAA, DBS, GI
Methodology: NBD, SNS, ECZ, EKC, MFB, HAA, DBS, GI; Software: IO
Formal Analysis: NBD, SNS, ECZ, EKC, JH, RR, JER, DFB, MFB, BHB, MJZ, MAK, IO, HAA, DBS, GI
Investigation: NBD, SNS, ECZ, EKC, JH, JPS, RR, AB, GI
Resources: JER, DFB, MFB, BHB, MJZ, MAK, HAA, DBS, GI
Data curation: SNS, ECZ, EKC, JH, RR, IO, HAA, MFB
Writing (initial): NBD, SNS, ECZ, EKC, JER, HAA, DBS, GI
Reviewing/Editing: all
Visualization: NBD, SNS, ECZ, EKC, JH, JPS, RR, AB, MFB, HAA, GI
Supervision: NBD, JER, DFB, MFB, BHB, MJZ, MAK, HAA, DBS, GI
Administration: NBD, MFB, DBS, GI
Guarantor/Author responsible for data and authorship verification: NBD
Funding: DBS, GI
Conflicts of Interest: The authors declare no applicable conflicts of interest.
References
- 1.Perdona S, Autorino R, Damiano R, et al. Bladder-sparing, combined-modality approach for muscle-invasive bladder cancer: a multi-institutional, long-term experience. Cancer. 2008;112:75–83. doi: 10.1002/cncr.23137. [DOI] [PubMed] [Google Scholar]
- 2.Rodel C. Combined-Modality Treatment and Selective Organ Preservation in Invasive Bladder Cancer: Long-Term Results. Journal of Clinical Oncology. 2002;20:3061–3071. doi: 10.1200/JCO.2002.11.027. [DOI] [PubMed] [Google Scholar]
- 3.James ND, Hussain SA, Hall E, et al. Radiotherapy with or without chemotherapy in muscle-invasive bladder cancer. N Engl J Med. 2012;366:1477–1488. doi: 10.1056/NEJMoa1106106. [DOI] [PubMed] [Google Scholar]
- 4.Shipley WU, Kaufman DS, Tester WJ, Pilepich MV, Sandler HM, Radiation Therapy Oncology G. Overview of bladder cancer trials in the Radiation Therapy Oncology Group. Cancer. 2003;97:2115–2119. doi: 10.1002/cncr.11282. [DOI] [PubMed] [Google Scholar]
- 5.Hagan MP, Winter KA, Kaufman DS, et al. RTOG 97-06: Initial report of a Phase I–II trial of selective bladder conservation using TURBT, twice-daily accelerated irradiation sensitized with cisplatin, and adjuvant MCV combination chemotherapy. International Journal of Radiation Oncology*Biology*Physics. 2003;57:665–672. doi: 10.1016/s0360-3016(03)00718-1. [DOI] [PubMed] [Google Scholar]
- 6.Ploussard G, Daneshmand S, Efstathiou JA, et al. Critical analysis of bladder sparing with trimodal therapy in muscle-invasive bladder cancer: a systematic review. Eur Urol. 2014;66:120–137. doi: 10.1016/j.eururo.2014.02.038. [DOI] [PubMed] [Google Scholar]
- 7.Mak RH, Hunt D, Shipley WU, et al. Long-term outcomes in patients with muscle-invasive bladder cancer after selective bladder-preserving combined-modality therapy: a pooled analysis of Radiation Therapy Oncology Group protocols 8802, 8903, 9506, 9706, 9906, and 0233. J Clin Oncol. 2014;32:3801–3809. doi: 10.1200/JCO.2014.57.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res. 2012;751:158–246. doi: 10.1016/j.mrrev.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 10.Van Allen EM, Mouw KW, Kim P, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4:1140–1153. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laurberg JR, Brems-Eskildsen AS, Nordentoft I, et al. Expression of TIP60 (tat-interactive protein) and MRE11 (meiotic recombination 11 homolog) predict treatment-specific outcome of localised invasive bladder cancer. BJU Int. 2012;110:E1228–1236. doi: 10.1111/j.1464-410X.2012.11564.x. [DOI] [PubMed] [Google Scholar]
- 12.Choudhury A, Nelson LD, Teo MT, et al. MRE11 expression is predictive of cause-specific survival following radical radiotherapy for muscle-invasive bladder cancer. Cancer Res. 2010;70:7017–7026. doi: 10.1158/0008-5472.CAN-10-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim PH, Cha EK, Sfakianos JP, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol. 2015;67:198–201. doi: 10.1016/j.eururo.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer. 2012;12:587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- 16.Won HH, Scott SN, Brannon AR, Shah RH, Berger MF. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Exp. 2013:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genomes Project C, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo G, Sun X, Chen C, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45:1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta GP, Vanness K, Barlas A, Manova-Todorova KO, Wen YH, Petrini JH. The Mre11 complex suppresses oncogene-driven breast tumorigenesis and metastasis. Mol Cell. 2013;52:353–365. doi: 10.1016/j.molcel.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2015 doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yap KL, Kiyotani K, Tamura K, et al. Whole-exome sequencing of muscle-invasive bladder cancer identifies recurrent mutations of UNC5C and prognostic importance of DNA repair gene mutations on survival. Clin Cancer Res. 2014;20:6605–6617. doi: 10.1158/1078-0432.CCR-14-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plimack ER, Dunbrack RL, Brennan TA, et al. Defects in DNA Repair Genes Predict Response to Neoadjuvant Cisplatin-based Chemotherapy in Muscle-invasive Bladder Cancer. Eur Urol. 2015;68:959–967. doi: 10.1016/j.eururo.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.