Abstract
Extrahepatic disease manifestations are common in chronic hepatitis C virus (HCV) infection. The mechanism of HCV-related lymphoproliferative disorders is not fully understood. Recent studies have found that HCV in peripheral blood mononuclear cells (PBMCs) from chronically infected patients is mainly associated with CD19+ B cells. To further elucidate this preferential association of HCV with B cells, we used in vitro cultured virus and uninfected PBMCs from healthy blood donors to investigate the necessary serum components that activate the binding of HCV to B cells. First, we found that the active serum components were present not only in HCV carriers, but also in HCV recovered patients and HCV negative healthy blood donors and that the serum components were heat labile. Second, the preferential binding activity of HCV to B cells could be blocked by anti-complement C3 antibodies. In experiments with complement-depleted serum and purified complement proteins, we demonstrated that complement proteins C1, C2, and C3 were required to activate such binding activity. Complement protein C4 was partially involved in this process. Third, using antibodies against cell surface markers, we showed that the binding complex mainly involved CD21 (complement receptor 2), CD19, CD20, and CD81; CD35 (complement receptor 1) was involved but had lower binding activity. Fourth, both anti-CD21 and anti-CD35 antibodies could block the binding of patient-derived HCV to B cells. Fifth, complement also mediated HCV binding to Raji cells, a cultured B cell line derived from Burkitt’s lymphoma.
Conclusion
In chronic HCV infection, the preferential association of HCV with B cells is mediated by the complement system, mainly through complement receptor 2 (CD21), in conjunction with the CD19 and CD81 complex.
Keywords: HCV, complement, B cell, CD21, Raji
Introduction
An estimated 180 million individuals are infected with hepatitis C virus (HCV) worldwide (1). More than 75% of HCV infections become persistent and may eventually lead to cirrhosis and hepatocellular carcinoma in 20% - 40%. A broad spectrum of extrahepatic manifestations is also associated with chronic HCV infection, including mixed cryoglobulinemia, cutaneous vasculitis, and B-cell non-Hodgkin’s lymphoma (B-NHL) (2–5). Although there are several reports suggesting extrahepatic replication of HCV, particularly in PBMCs, even in some patients who appear to have resolved their infection (6–9), the existence of extrahepatic reservoirs of HCV replication remains highly controversial (10, 11).
Although the major site of HCV replication is in the liver, HCV RNA has been detected in association with peripheral blood mononuclear cells (PBMCs) (7, 10), and is particularly enriched in CD19+ B cells (10, 12, 13). Several factors have been shown to play important roles in HCV entry into susceptible cells (14–18), including CD81, scavenger receptor SR-BI, and the tight-junction protein Claudin-1. All three entry factors are required for infection of Huh-7.5 cells by cell culture-produced infectious HCV particles (16). However, all PBMC subsets lack at least one of these known HCV entry factors (16). CD81 is a widely distributed cell-surface tetraspanin that participates in different molecular complexes on various cell types, including B, T, and natural killer cells (16) and has been proposed as the primary mediator of HCV binding to B cells (17, 18). In human B cells, CD81 is known to form a costimulatory complex with CD19 and complement receptor 2 (CD21) (19). Co-ligation of the B cell antigen receptor (BCR) with this costimulatory complex can lower the threshold required for BCR-mediated B cell proliferation (20). Because all major subsets of PBMCs have high level expression of CD81 (18), CD81 alone could not account for the preferential association of HCV with B cells.
In this study, we used cell culture-produced HCV particles, pre-incubated with serum components plus various factors, followed by mixing with PBMCs from healthy blood donors. After separating PBMC into B cell and non-B cell fractions by CD19 magnetic microbeads, quantification of HCV RNA was carried out by real-time reverse-transcription polymerase chain reaction (qPCR) to measure the quantities of HCV binding to B cells. We sought to investigate the mechanism of preferential association of HCV with B cells in PBMCs from chronic HCV carriers, and to identify specific cell surface receptors involved in the binding process.
Patients and Methods
Patients
Consenting blood donors identified as anti-HCV positive by enzyme immunoassay at the time of routine blood donation at the Department of Transfusion Medicine, National Institutes of Health (NIH) or the Greater Chesapeake and Potomac Region of the American Red Cross were enrolled in an NIH prospective study of the natural history of HCV infection (10). Ethics committees of the American Red Cross and the NIH approved the study protocol in accordance with the Declaration of Helsinki and the study has been reviewed annually by an NIH Institutional Review Board (NIH Protocol 91-CC-0017). All subjects gave written informed consent to participate in the study.
Methods
A detailed description of methods used for cell culture, in vitro RNA synthesis, HCV production in cell culture, conversion of plasma to serum, PBMC isolation, part of in vitro HCV binding assay, and HCV detection can be found in Supporting Information.
In vitro HCV binding assay
In a standard binding assay, 3 ml of virus (1×107 genomic copies for HCV genotype 1a virus) was mixed with 100 µl of serum sample (about 20–25 CH50 units) to initiate complement activation and incubated at 25°C for 1 h or at 37°C for 0.5 h, followed by adding 2 ml of PBMCs (4–5×107 cells) to the mixture and incubating for 1 h. The reaction was carried out in 50 ml sterile tubes with occasional mixing. After incubation, the cells were pelleted by centrifugation, washed once with 10 ml 1× PBS, pH7.4, centrifuged again, and re-suspended in 400 µl of MACS buffer (Miltenyi Biotec Inc., Auburn, CA) supplemented with 0.5% bovine serum albumin. The cells were mixed with 100 µl of mouse anti-human CD19 magnetic microbeads and incubated at 4°C for 20 min. The cells were then diluted with 10 ml MACS buffer, collected by centrifugation, and re-suspended in 1 ml MACS buffer. The cell suspension was applied to a MS column (Miltenyi Biotec Inc., Auburn, CA) on a magnetic field separator. The column was washed three times with 1 ml MACS buffer each time. The column was separated from the magnetic stand and placed on a 2 ml sterile tube. The CD19+ B cells were flushed from the column with 1 ml MACS buffer, collected by centrifugation, and re-suspended in 650 µl of RNeasy Plus lysis buffer (Qiagen, Chatsworth, CA) supplemented with 1% 2-mercaptoethanol. RNA isolation was carried out by using RNeasy Plus mini kit (Qiagen, Chatsworth, CA) according to the manufacturer’s instructions, and each sample was eluted from the column with 50 µl of nuclease-free water (Thermo Fisher Scientific, Waltham, MA) supplemented with 1.0 mM dithiothreitol and 200 unit/ml RNAsin (Promega, Madison, WI).
Statistical analysis
Quantitative analysis of HCV genome copy number assessed by qPCR is expressed as the mean ± standard deviation. An unpaired Student’s t-test was used to determine the statistical significance. Values of P < 0.05 were judged significant. Data analysis and graphs were performed with GraphPad Prism 5 (GraphPad Software, La Jolla, CA).
RESULTS
Serum components from both HCV recovered patients and healthy blood donors can promote HCV binding to B cells
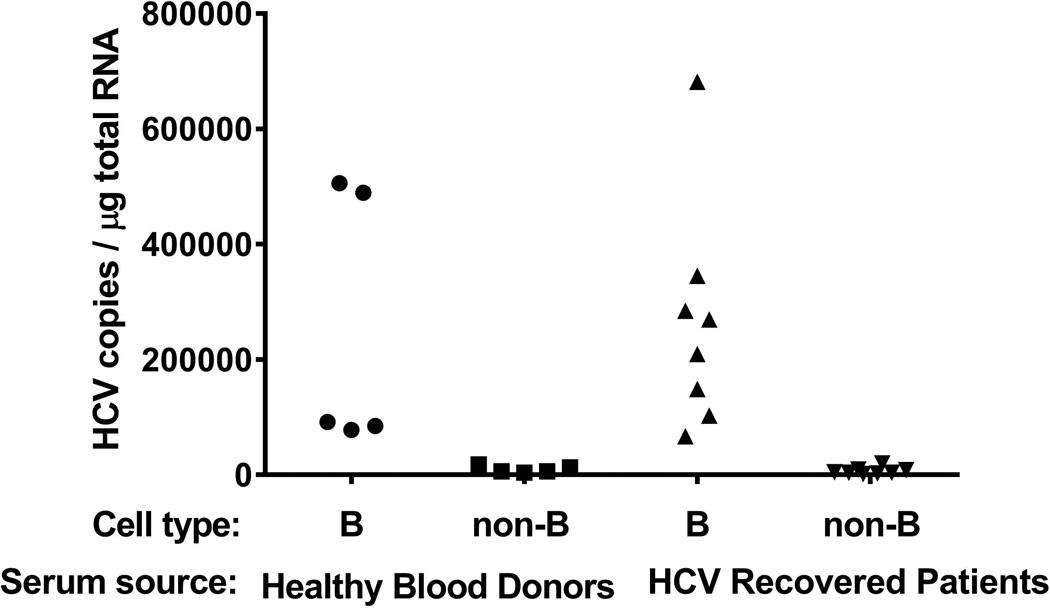
To investigate the mechanism of preferential association of HCV with B cells in PBMC from chronic patients, we used in vitro cultured HCV virus (H77s, HCV genotype 1a) and B-cell enriched fractions from healthy donors to determine which serum components are necessary for promoting HCV binding to B cells. In the absence of serum, binding of HCV particles derived from in vitro cell culture was minimal in our in vitro assay system (data shown in Fig. 1 legend and Fig. 2). When cell culture-produced HCV particles were pre-incubated with human serum samples, the viral particles attached to B cells with more than 100-fold efficiency as compared to that without serum treatment. As shown in Fig. 1, serum samples from both HCV recovered patients and heathy blood donors contained such enhancing activity. This result indicated that the enhancement of HCV binding to B cells by serum was independent of HCV infection and inherent in normal human serum. We also found significant variation among individuals of the enhancing activity present in their serum samples.
Fig. 1.
Serum samples from both healthy blood donors and HCV recovered subjects can promote HCV binding to B cells. Ten million genomic copies of HCV 1a (H77s) in 3 ml medium were incubated with 100 µl serum sample at room temperature for 1 h, followed by mixing with 2 ml PBMCs (2.5×107 cells/ml) in complete RPMI medium. The reaction was carried out at 37°C for 2 h. The cells then were processed for separation into B and non-B fractions by using CD19 magnetic microbead column purification as described in Methods section. A negative control that did not incubate virus with serum was included in this study but not plotted in this figure; this control had an HCV viral load on B cells of 411 copies per µg total RNA. Each value represents the mean of triplicate determinations.
Fig. 2.
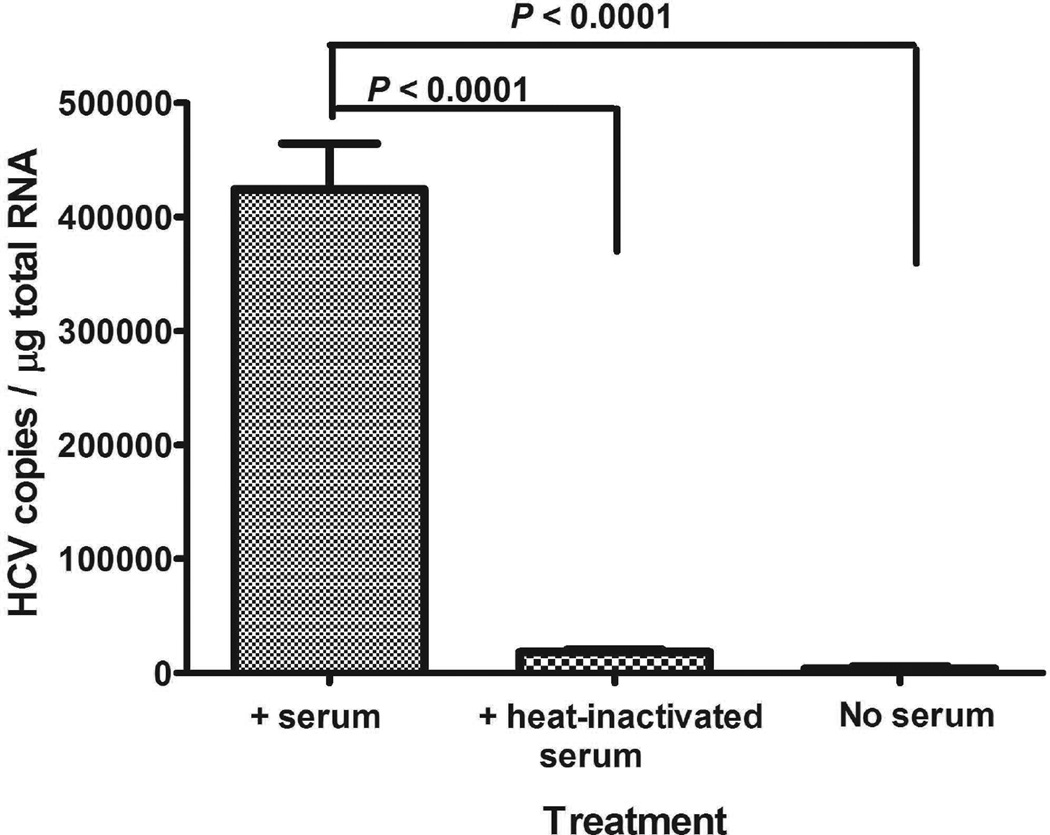
Heat-labile components in human serum promote the binding of HCV to B cells. Ten million genomic copies of HCV 1a (H77s) in 3 ml medium were incubated with 100 µl serum sample or heat-inactivated serum sample (56°C for 30 min) at room temperature for 1 h, followed by mixing with 2 ml PBMCs (2.5×107 cells/ml) in complete RPMI medium. The reaction was carried out at room temperature (25°C) for 1 h. The cells then were processed for HCV quantification as described in Methods section. Each value represents the mean ± SD of 9 determinations. The experiments were repeated twice with similar results using PBMCs from two different donors.
Heat-labile components in human serum promote the binding of HCV to B cells
During the investigation period, we observed that the activity promoting HCV binding to B cells present in the serum samples was quickly lost even when the serum samples were stored at 4°C. Therefore, we measured the sensitivity of HCV binding activity to B cells by incubating serum samples at 56°C for 30 min first before mixing with virus. About 90% of HCV binding activity to B cells was lost after heat treatment as compared to the untreated serum samples (Fig. 2).
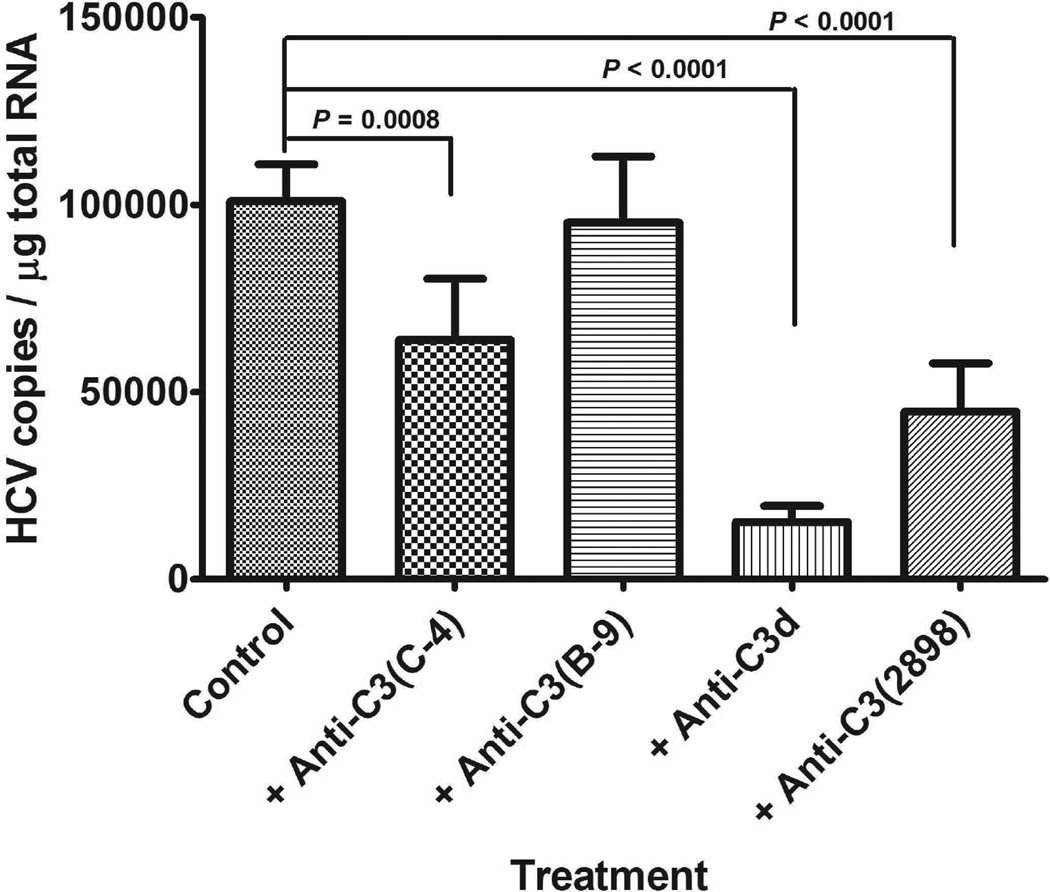
Antibodies against complement C3 protein can block HCV binding to B cells
Since complement activity is well known for heat sensitivity, we tested whether the available antibodies against complement C3 protein could block the binding of HCV to B cells in our assay system. As shown in Fig. 3, several anti-C3 antibodies, such as anti-C3 (C-4), anti-C3 (2898), and anti-C3d, had significant blocking activity against HCV binding to B cells, particularly, anti-C3d showed the strongest blocking activity. However, because of the abundance of C3 protein in serum, when lowering the amount of C3d antibody, the blocking activity was gradually decreased (Supporting Fig. S1). Anti-C3 (B-9) antibody did not show blocking activity against HCV binding to B cells in our assay system. This may be because the epitope recognized by this antibody was not accessible in this assay.
Fig. 3.
Antibodies against complement C3 protein can block HCV binding to B cells. Twenty-five µl serum sample was incubated with 20 µg of the indicated mouse monoclonal antibody (clones C4, B9, and 2898) against complement C3 protein and C3d (clone 003-05) at 25°C for 30 min, followed by mixing with virus and incubating at 25°C for 1 h. After mixing with PBMC, the mixture was incubated at 25°C for one more hour. The cells then were processed for HCV quantification as described in Methods section. The most potent inhibitory of binding was observed with anti-C3d. Each value represents the mean ± SD of 6 determinations. The experiments were repeated three times with similar results using PBMCs from three different donors.
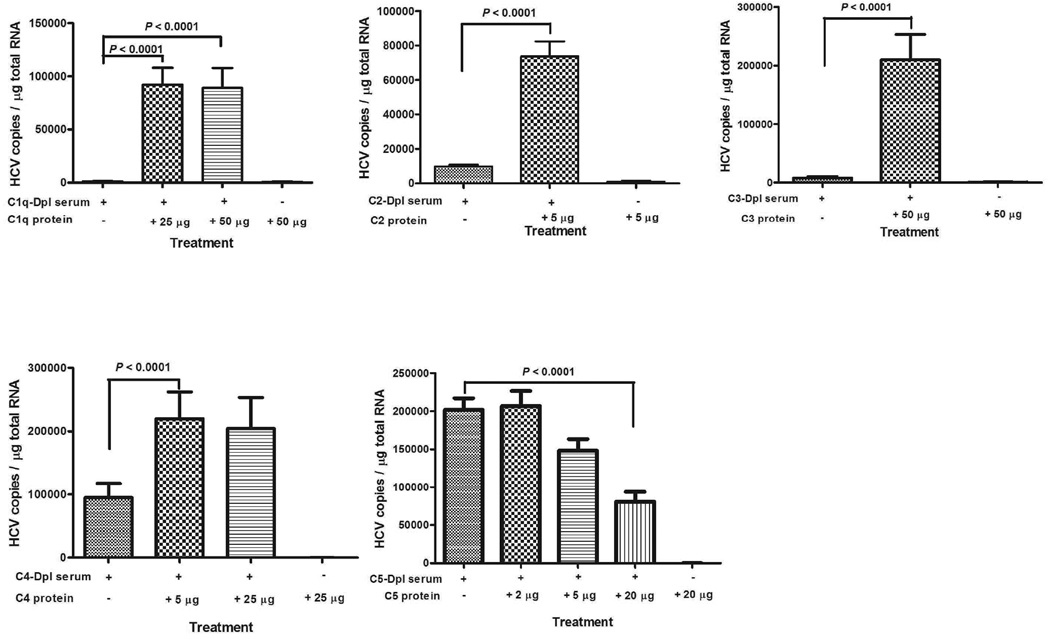
Purified complement proteins added to the complement-depleted serum can restore HCV binding to B cells
To further demonstrate the complement system is responsible for promoting the binding of HCV to B cells, we obtained several commercially available complement-depleted serum samples and purified complement proteins to reconstitute the binding experiment. As shown in Fig. 4, when binding of HCV was performed with C1, C2 or C3 depleted serum samples, the binding activity could hardly be detected. After adding the respective purified complement protein back to the reaction mixture, the binding activity of HCV to B cells was restored. In the absence of serum, none of the purified complement proteins alone had the ability to promote HCV binding to B cells. These results suggested that complement C1, C2, and C3 proteins are absolutely required for activating HCV to attach to B cells. When complement C4 depleted serum was tested, about 40% binding activity was detected as compared to the complete system with C4 depleted serum plus purified C4 protein (Fig. 4). Purified C4 protein alone did not promote HCV binding to B cells. Therefore, complement C4 plays a lessor role in the binding of HCV to B cells compared to complement C1, C2 and C3. These results indicated that the classical complement activation pathway is involved in promoting the binding of HCV to B cells. When we mixed purified proteins of C1, C2, C3, and C4 together in the absence of serum, we were able to show enhancement of HCV binding to B cells (Supporting Fig. S2). Using complement C5 depleted serum and purified C5 protein to assay the HCV binding activity to B cells, demonstrated that C5 is not required for HCV binding to B cells. Indeed, when excess of C5 protein was added to the C5-depleted serum, the binding of HCV to B cells was significantly decreased (Fig. 4).
Fig. 4.
Purified complement proteins C1q, C2, C3, and C4 added to the indicated complement-depleted (Dpl) serum can restore HCV binding to B cells. Ten million genomic copies of HCV 1a (H77s) in 3 ml medium were incubated with 100 µl of the indicated complement-depleted serum sample or complement-depleted serum sample plus the indicated purified complement protein, or purified complement protein only. After 1h incubation at 25°C, 2 ml PBMCs (2.5×107 cells per ml) in complete RPMI medium was added. The reaction was carried out at room temperature (25°C) for 1 h. The cells then were processed for HCV quantification as described in Methods section. All experiments were repeated twice with similar results using PBMCs from two different donors. Each value represents the mean ± SD of 6 determinations.
Receptors involved in complement-mediated HCV binding to B cells
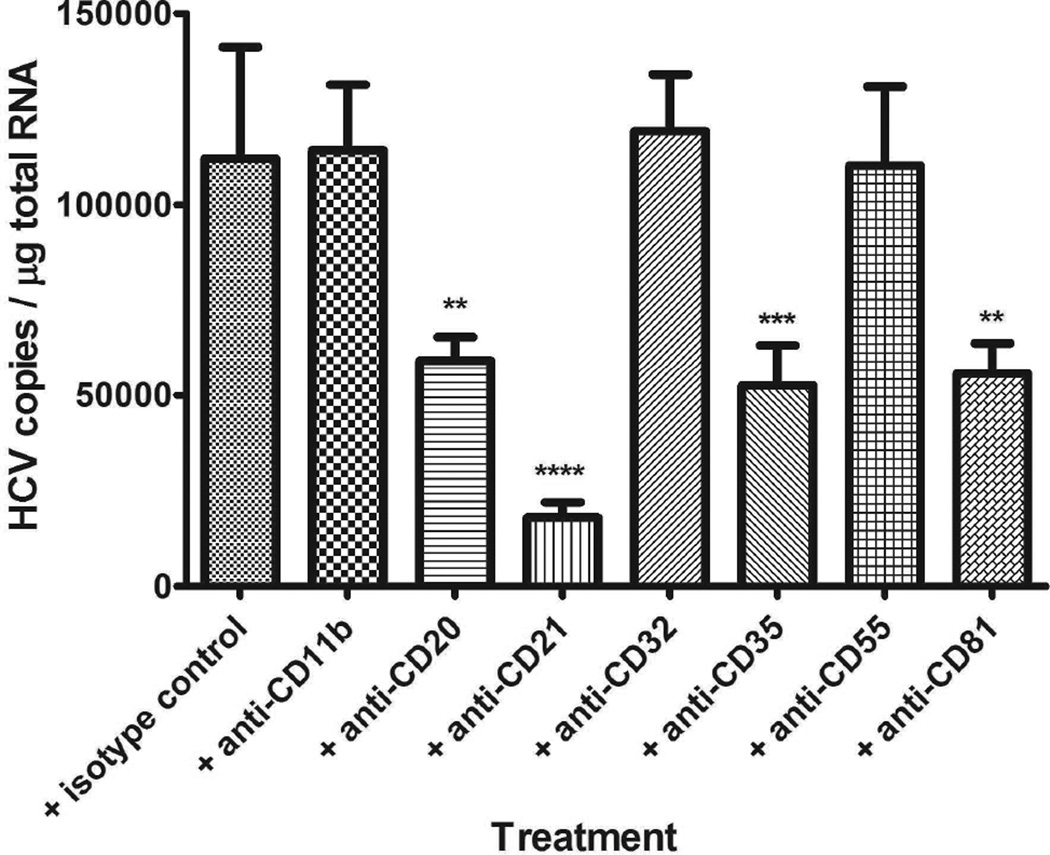
The above results indicated that the preferential association of HCV with B cells in PBMC from chronically infected patients is likely mediated by the complement system. In B cells, complement receptor 2 (CD21) is most abundant and usually forms a complex with CD19 and CD81, followed by complement receptor 1 (CD35). Using a battery of antibodies against complement-related receptors, we demonstrated that anti-CD21 had the strongest blocking activity for HCV binding to B cells (Fig. 5); this was consistent at varying concentrations of antibody ranging from 1 µg to 10 µg (Supporting Fig. S3 and S4). In addition, we showed that CD19 (Supporting Fig. S5) or CD20 (Fig. 5) are also involved in HCV binding to B cells. For CD81, about 60% (6 out of 10) of PBMCs isolated from healthy blood donors showed inhibitory activity upon treatment of PBMC with anti-CD81.
Fig. 5.
Receptors involved in complement-mediated HCV binding to B cells. Two ml of PBMCs (2.5×107 cells/ml) were incubated with 10 µg of the indicated antibody at 25°C for 0.5 h; followed by mixing with 3 ml of activated virus (pre-incubation with serum for 1 h at 25°C, 1×107 genomic copies total), and incubating at 25°C for 1 h. The cells then were processed for RNA isolation and HCV quantification as described in Methods section. **: P < 0.01, ***: P < 0.001, ****: P < 0.0001; when the reduction of HCV binding to B cells by the indicated antibody was compared to antibody isotype control. Each value represents the mean ± SD of 6 determinations. The experiments were repeated three times with similar results using PBMCs from three different donors. CD11b is part of complement receptor 3, CD20 is one of major B cell markers, CD21 is also known as complement receptor 2, CD32 is receptor for Fc fragment of immunoglobulin G, CD35 is complement receptor 1, and CD55 is membrane-bound decay-accelerating factor for convertases of complement system.
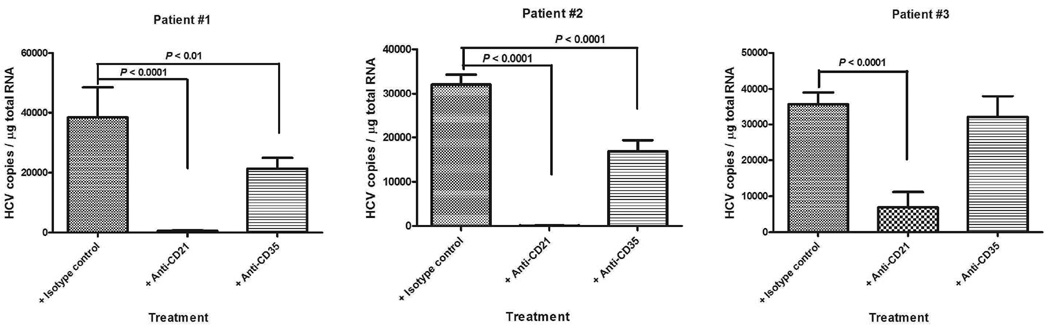
Anti-CD21 and anti-CD35 antibodies can block B cell binding of HCV viruses derived from chronically infected patients
To demonstrate that the preferential binding of HCV to B cells mediated by the complement system is not limited to cell culture derived virus, we repeated the B-cell binding experiments using the serum from three chronically infected patients as the source of virus and complement. In patients #1 and #2, both anti-CD21 and anti-CD35 antibodies inhibited HCV binding to B cells with anti-CD21 showing stronger activity. In patient #3, only anti-CD21 showed significant inhibitory activity (Fig. 6).
Fig. 6.
Anti-CD21 and anti-CD35 antibodies can block the binding of HCV viruses from chronically infected patients to B cells. Two ml of PBMCs (2.5×107 cells/ml) were incubated with 10 µg of the indicated antibody at 25°C for 0.5 h; followed by mixing with 3 ml of the indicated 15-fold diluted virus (pre-incubation for 0.5 h at 25°C, 3–5×106 genomic copies total), and incubating at 25°C for 1 h. The cells then were processed for RNA isolation and HCV quantification as described in Methods section. Each value represents the mean ± SD of 6 determinations. The experiments were repeated twice with similar results using PBMCs from two different donors.
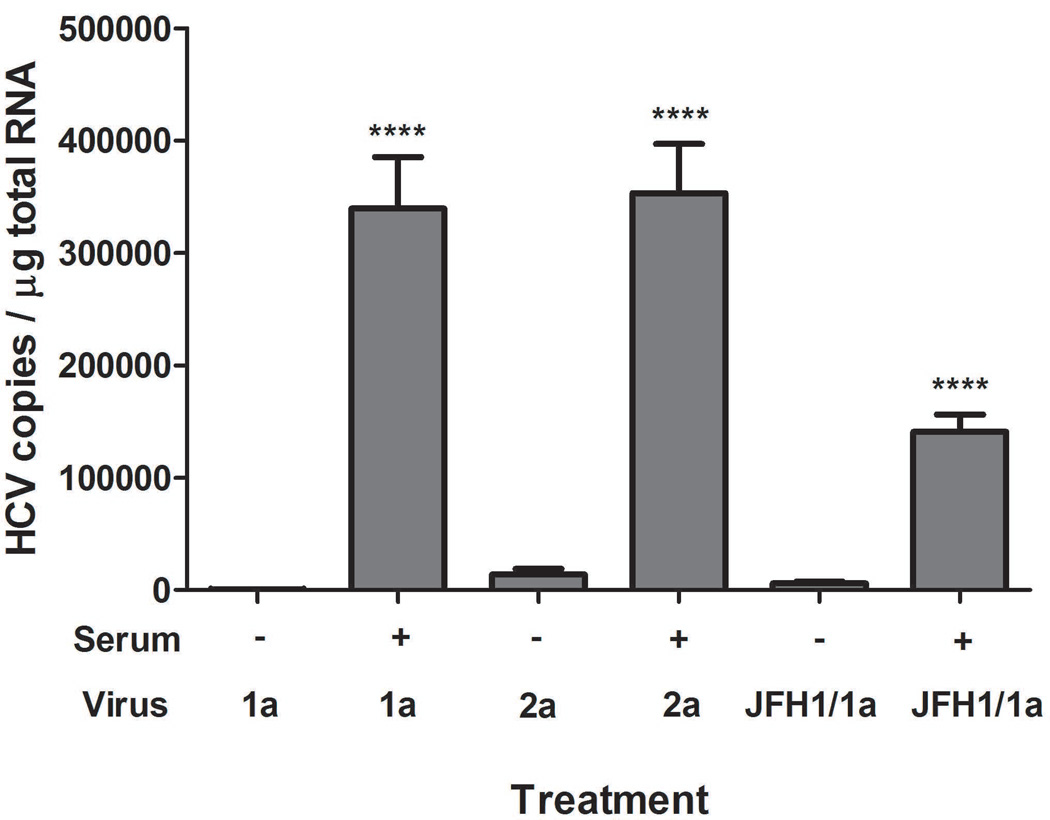
Complement-mediated HCV binding to B cells using in vitro cultured HCV 1a and HCV 2a viruses
We further investigated whether complement-mediated HCV binding to B cells could be demonstrated using other commonly used cell culture-produced viruses, specifically JFH1 virus (HCV genotype 2a) and chimeric JFH1/1a virus. As shown in Fig. 7, both these well-characterized viral strains became much more efficiently bound to B cells (P < 0.0001) after mixing with complement-active serum samples.
Fig. 7.
Complement-mediated HCV binding to B cells worked on in vitro cultured HCV 1a and HCV 2a viruses. Three ml of the indicated virus (HCV1a, 1×107 copies total; HCV2a, 1×108 copies total; JFH1/1a, 3×107 total) were treated either with 100 µl medium only as control or 100 µl serum at 25°C for 1 h, followed by mixing with 2 ml PBMCs (2.5×107 cells per ml) in complete RPMI medium. The reaction was carried out at room temperature (25°C) for 2 h. The cells then were processed for HCV quantification as described in Methods section. ****: P < 0.0001 when compared to without serum treatment for each virus. Each value represents the mean ± SD of 6 determinations.
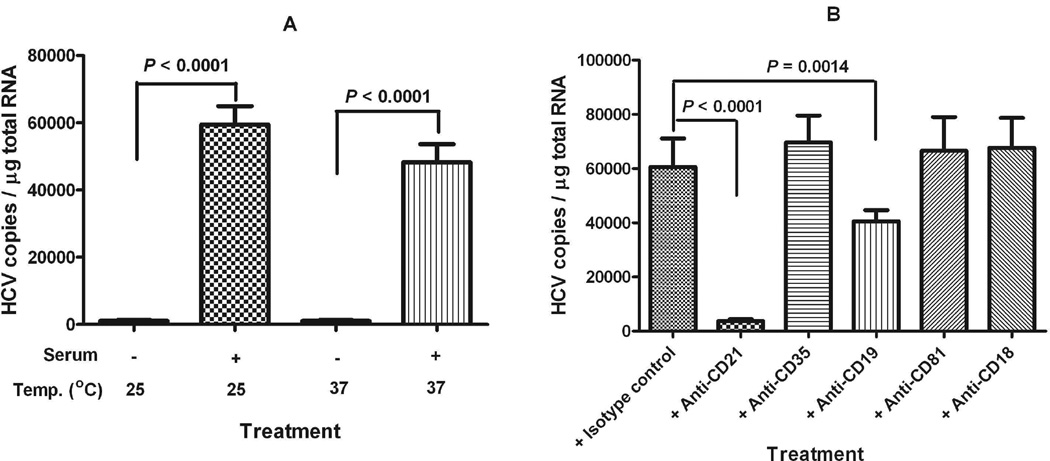
Enhanced HCV binding to Raji cells was also mediated by complement system
We also investigated whether complement-mediated HCV binding to B cells could be demonstrated by replacing human B cells with Raji cells, a cultured B cell line derived from Burkitt’s lymphoma. As shown in Fig. 8A, cell culture-produced HCV became much more efficiently bound to Raji cells after treatment of virus with complement-active serum samples either at 25°C or at 37°C as compared to the untreated samples. We also demonstrated that pre-incubation of Raji cells with anti-CD21 or anti-CD19 antibody could block the subsequent binding of complement-activated HCV to the cells (Fig. 8B). Pre-incubation of Raji cells with anti-CD81 antibody did not prevent complement-activated HCV binding to the cells (Fig. 8B), as we had also observed in PBMCs isolated from 40% of healthy blood donors.
Fig. 8.
Complement-mediated HCV binding to Raji cells. (A) HCV genotype 1a virus in 3 ml medium was incubated with 100 µl serum sample or medium only at 25°C for 1 h, followed by mixing with 2 ml of Raji cells (1×106 cells/ml) and incubating at the indicated temperature for 1 h. (B) 2 ml of Raji cells (1×106 cells/ml) were incubated with 10 µg each of the indicated antibody at room temperature for 30 min, followed by mixing with 3 ml of complement-activated HCV genotype 1a virus (pre-incubation with serum for 1 h at 25°C, 1×107 genomic copies per reaction), and incubating at 25°C for 1 h. After incubation, the cells were pelleted, washed two times with complete RPMI medium, and processed for RNA isolation and HCV quantification as described in Methods section. Each value represents the mean ± SD of 6 determinations.
Discussion
In our previous study of the association of HCV with PBMCs, we found no evidence that HCV replicates in PBMCs (10), but rather that HCV binds to PBMCs and preferentially to B cells. Further, we found that normal PBMCs could be made to simulate PBMCs from HCV infected patients simply by mixing them with HCV-positive plasma. Thus, it appeared that HCV could adhere to PBMC/B-cells without necessarily infecting them, suggesting that the binding was a more generalized phenomenon of B-cell interactions rather than a specific effect of HCV.
To elucidate the mechanism of preferential association of HCV with B cells, we began the investigation by using in vitro cell culture produced HCV particles mixed with PBMCs from healthy donors for in vitro binding assays, followed by fractionation of CD19+ B cells and HCV quantification by qPCR. In the absence of exogenous serum factors, there was negligible binding of culture-produced HCV particles to B cells. However, upon pre-incubation of culture-produced HCV particles with serum, HCV binding to B cells was increased by more than two orders of magnitude compared to virus in the absence of serum. The essential serum factors were found to be heat-labile and present in both healthy blood donors and HCV recovered patients, suggesting that complement might be integral to the binding process.
By using various antibodies against complement C3 proteins in the B-cell binding assays, we demonstrated that several anti-C3 mouse monoclonal antibodies, particularly anti-C3d antibody, effectively blocked the binding of HCV to B cells. C3d is the surface-bound end–product of the proteolytic cleavage of C3b deposited on invading microorganisms during the complement activation process that splits C3 to C3a and C3b (21–25). In reconstitution experiments with complement-depleted serum samples and the respective purified complement proteins, we demonstrated that the preferential binding of HCV particles to B cells in PBMCs is indeed mediated by the complement system as complement components involved in the classical pathway (C1q, C2, C3 and C4) were required for the binding of HCV particles to B cells. In addition, the alternative pathway and the mannose-binding lectin pathway of complement activation could also be involved in this process (22). The generation of C3d is essential for the opsonization of HCV particles leading to their attachment to B cells. We propose that C3d attached to viral particles will have high affinity for complement receptor 2 (CR2; CD21) on the surface of B cells. CR2 is highly expressed in B cells and follicular dendritic cells (22, 24). In our current assay system, there was no significant change of HCV genomic copy numbers in the culture supernatant before and after complement treatment as determined by qPCR (data not shown), which implies that HCV particles are not lysed, but rather opsonized by complement activation. Although previous reports have shown that HCV core protein can bind to gC1q receptor (gC1qR) specific for the globular heads of the C1q protein (26), this mechanism would not explain why HCV particles are preferentially bound to B cells since this receptor is widely distributed in all leukocyte subsets and platelets (27). Further, the preferential binding of HCV to B cells only happens when exogenous serum with complement activity is added to the virus supernatant and thus is not related to a virus-specific protein per se. In the C5-depleted serum reconstitution experiment, we found that C5 protein is not required for the preferential binding of HCV to B cells. Again this indicates that HCV particles are opsonized, and not lysed through a membrane attack complex. However, when excess of the purified C5 protein was present in reconstituted C5-depleted serum, the binding of HCV to B cells was significantly decreased suggesting that complement-mediated viral lysis may have occurred (Fig 4).
There are at least ten receptors for complement components, fragments, and complexes (22, 27). To further elucidate the mechanism of preferential binding of HCV to B cells, we used various antibodies against cell surface receptors in the assay system to assess the effectiveness of each antibody in blocking HCV binding to B cells. As shown in Fig. 5, anti-CD21 displayed the strongest activity in blocking HCV binding to B cells, followed by anti-CD20, anti-CD35, and anti-CD81. CD21 is complement receptor 2 (CR2), and the receptor for C3d (27). CD21 usually forms a complex with CD19 and CD81 on the surface of B cells (28). CD35 is complement receptor 1 (CR1), and it has dual roles as complement receptor and complement regulator (27). The ligands for CR1 are C3b and C4b. In this study, we found that anti-CD81 antibody only blocked HCV binding to B cells in 60% of PBMCs tested from 10 individuals. This could be due to polymorphic CD81 antigen expression on B cells from different individuals. Anti-CD81 antibody also did not block HCV binding to Raji cells (Fig. 8B). The preferential binding of HCV to B cells was shown not only for HCV particles produced in cell culture, but also for HCV particles derived directly from three chronically infected patients. Whether using culture-derived or patient-derived HCV, binding to B cells was blocked more efficiently by anti-CD21 (complement receptor 2) than by anti-CD35 (complement receptor 1). Similar finding of human immunodeficiency virus 1 (HIV-1) binding to B cells through CD21-complement interactions has been reported (29).
The complement system plays a central role in innate immune defense and consists of both soluble factors and cell surface receptors that interact to sense and respond to a wide range of invading microorganisms. Activation of complement system has been demonstrated during infection by several enveloped viruses including HIV-1 (24), herpesvirus (30), Ebola virus (31), and influenza virus (32). However, these viruses have also evolved escape mechanisms to evade the complement system by incorporating host regulators of complement activation into their viral envelopes and, as a result, escape antibody-dependent complement-mediated virus lysis (30, 33). Complement activation upon HCV infection has been implicated in several studies (34–36). In addition, CD55 and CD59, regulators of complement activation, were shown to be incorporated into HCV virions (37, 38), which led to resistance to complement-mediated antibody-dependent virolysis. In the present study, we demonstrated that HCV particles either produced by in vitro cell culture system or directly from chronic HCV patients were opsonized, but not lysed by complement activation, which eventually led to preferential association of HCV particles with B cells in PBMC. Although not investigated in this study, in chronic HCV patients, complement receptor 1/CD35 on erythrocytes may also play a key role in transporting complement-mediated, opsonized HCV immune-complexes. In this study, we mainly investigated antibody-independent activation of the complement system by HCV. In chronic HCV infection, antibody-dependent complement activation may play a more complex role and interplay with many other types of cells, as seen in HIV infection (24).
Epidemiological studies have demonstrated an increased risk of developing B-cell non-Hodgkin lymphoma (B-NHL) in patients with chronic HCV infection (39, 40). B-NHL subtypes frequently associated with HCV are marginal zone lymphoma, lymphoplasmacytic lymphoma, and diffuse large B-cell lymphoma. The most convincing evidence for a causal relationship between HCV infection and lymphoma development is the observation of B-NHL regression after HCV eradication with interferon-α monotherapy or in combination with ribavirin (39, 40). However, the strength of the association shows great geographic discrepancies, with higher relative risk in countries with high HCV prevalence. Three general hypotheses have been proposed to elucidate HCV-induced lymphomagenesis (39): 1) Continuous external stimulation of lymphocyte receptors by viral antigens throughout persistent infection; 2) HCV replication in B cells with oncogenic effect mediated by intracellular viral proteins; 3) permanent B-cell damage by mutation of tumor suppressor genes caused by a transiently present intracellular virus (41). The lymphoma subtypes associated with HCV infection originate from germinal center or post-germinal center lymphocytes, suggesting a possible antigen-driven proliferation (40). Indeed, a monoclonal/oligoclonal B-cell expansion has been shown in circulating B cells, as well as in the bone marrow or intrahepatic B cells of chronic HCV patients (42). However, the strongly biased immunoglobulin gene usage by abnormal B cells in HCV-associated mixed cryoglobulinemia (43) and in HCV-associated lymphomas (44, 45), as well as the inability of lymphoma BCRs from patients with B-NHL and chronic HCV infection to bind HCV antigens (46), may indicate that lymphoma formation in HCV infection is a complex process in which persistent antigenic stimulation through complement- mediated binding plays only an indirect, and as yet undefined role. Our results support the notion that chronic HCV infection can potentially promote lymphomagenesis through complement-mediated preferential binding of HCV to B cells, which generates a complex by combining C3d-tagged HCV with CD19 and CD81. This complex can then bring B-cell antigen receptors together into lipid rafts and lower the threshold for B-cell activation and proliferation (47, 48). Further investigation of signaling pathways involved in complement-mediated HCV binding to B cells is warranted.
Supplementary Material
Acknowledgments
We thank Dr. Francis Chisari for providing us with Huh-7.5.1 cells, Dr. Stanley Lemon for providing us with H77S plasmids, and Dr. Jake Liang for providing us with JFH-1 virus and JFH1/1a chimeric virus.
Financial support: This research was supported by the Intramural Research Program of Warren G. Magnuson Clinical Center, NIH.
List of Abbreviations
- HCV
hepatitis C virus
- PBMCs
peripheral blood mononuclear cells
- PBS
phosphate-buffered saline
- B-NHL
B-cell non-Hodgkin’s lymphoma
- Dpl
depleted
- CR1/CD35
complement receptor 1
- CR2/CD21
complement receptor 2
- qPCR
quantitative real-time reverse-transcription polymerase chain reaction
Footnotes
Potential conflict of interest: Nothing to report.
References
- 1.Mohamed AA, Elbedewy TA, El-Serafy M, El-Toukhy N, Ahmed W, Ali El Din Z. Hepatitis C virus: A global view. World J Hepatol. 2015;7:2676–2680. doi: 10.4254/wjh.v7.i26.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackard JT, Kemmer N, Sherman KE. Extrahepatic replication of HCV: insights into clinical manifestations and biological consequences. Hepatology. 2006;44:15–22. doi: 10.1002/hep.21283. [DOI] [PubMed] [Google Scholar]
- 3.Ferri C, Sebastiani M, Giuggioli D, Colaci M, Fallahi P, Piluso A, Antonelli A, et al. Hepatitis C virus syndrome: A constellation of organ- and non-organ specific autoimmune disorders, B-cell non-Hodgkin's lymphoma, and cancer. World J Hepatol. 2015;7:327–343. doi: 10.4254/wjh.v7.i3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charles ED, Dustin LB. Hepatitis C virus-induced cryoglobulinemia. Kidney Int. 2009;76:818–824. doi: 10.1038/ki.2009.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenthal E, Cacoub P. Extrahepatic manifestations in chronic hepatitis C virus carriers. Lupus. 2015;24:469–482. doi: 10.1177/0961203314556140. [DOI] [PubMed] [Google Scholar]
- 6.MacParland SA, Pham TN, Guy CS, Michalak TI. Hepatitis C virus persisting after clinically apparent sustained virological response to antiviral therapy retains infectivity in vitro. Hepatology. 2009;49:1431–1441. doi: 10.1002/hep.22802. [DOI] [PubMed] [Google Scholar]
- 7.Pham TN, King D, Macparland SA, McGrath JS, Reddy SB, Bursey FR, Michalak TI. Hepatitis C virus replicates in the same immune cell subsets in chronic hepatitis C and occult infection. Gastroenterology. 2008;134:812–822. doi: 10.1053/j.gastro.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 8.Pham TN, Michalak TI. Occult persistence and lymphotropism of hepatitis C virus infection. World J Gastroenterol. 2008;14:2789–2793. doi: 10.3748/wjg.14.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zignego AL, Giannini C, Monti M, Gragnani L. Hepatitis C virus lymphotropism: lessons from a decade of studies. Dig Liver Dis. 2007;39(Suppl 1):S38–S45. doi: 10.1016/s1590-8658(07)80009-0. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara K, Allison RD, Wang RY, Bare P, Matsuura K, Schechterly C, Murthy K, et al. Investigation of residual hepatitis C virus in presumed recovered subjects. Hepatology. 2013;57:483–491. doi: 10.1002/hep.25921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Welker MW, Zeuzem S. Occult hepatitis C: how convincing are the current data? Hepatology. 2009;49:665–675. doi: 10.1002/hep.22706. [DOI] [PubMed] [Google Scholar]
- 12.Boisvert J, He XS, Cheung R, Keeffe EB, Wright T, Greenberg HB. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J Infect Dis. 2001;184:827–835. doi: 10.1086/323391. [DOI] [PubMed] [Google Scholar]
- 13.Zehender G, Meroni L, De Maddalena C, Varchetta S, Monti G, Galli M. Detection of hepatitis C virus RNA in CD19 peripheral blood mononuclear cells of chronically infected patients. J Infect Dis. 1997;176:1209–1214. doi: 10.1086/514114. [DOI] [PubMed] [Google Scholar]
- 14.Meertens L, Bertaux C, Cukierman L, Cormier E, Lavillette D, Cosset FL, Dragic T. The tight junction proteins claudin-1û, and-9 are entry cofactors for hepatitis C virus. J Virol. 2008;82:3555–3560. doi: 10.1128/JVI.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset FL, Wakita T, et al. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46:1722–1731. doi: 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- 16.Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, et al. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology. 2008;48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosa D, Saletti G, De Gregorio E, Zorat F, Comar C, D'Oro U, Nuti S, et al. Activation of naive B lymphocytes via CD81, a pathogenetic mechanism for hepatitis C virus-associated B lymphocyte disorders. Proc Natl Acad Sci U S A. 2005;102:18544–18549. doi: 10.1073/pnas.0509402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stamataki Z, Shannon-Lowe C, Shaw J, Mutimer D, Rickinson AB, Gordon J, Adams DH, et al. Hepatitis C virus association with peripheral blood B lymphocytes potentiates viral infection of liver-derived hepatoma cells. Blood. 2009;113:585–593. doi: 10.1182/blood-2008-05-158824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fearon DT, Carter RH. The CD19/CR2/TAPA-1 complex of B lymphocytes: linking natural to acquired immunity. Annu Rev Immunol. 1995;13:127–149. doi: 10.1146/annurev.iy.13.040195.001015. [DOI] [PubMed] [Google Scholar]
- 20.Carter RH, Fearon DT. CD19: lowering the threshold for antigen receptor stimulation of B lymphocytes. Science. 1992;256:105–107. [PubMed] [Google Scholar]
- 21.Fujita T, Matsushita M, Endo Y. The lectin-complement pathway--its role in innate immunity and evolution. Immunol Rev. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- 22.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stoermer KA, Morrison TE. Complement and viral pathogenesis. Virology. 2011;411:362–373. doi: 10.1016/j.virol.2010.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoiber H, Banki Z, Wilflingseder D, Dierich MP. Complement-HIV interactions during all steps of viral pathogenesis. Vaccine. 2008;26:3046–3054. doi: 10.1016/j.vaccine.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Thurman JM, Kulik L, Orth H, Wong M, Renner B, Sargsyan SA, Mitchell LM, et al. Detection of complement activation using monoclonal antibodies against C3d. J Clin Invest. 2013;123:2218–2230. doi: 10.1172/JCI65861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao ZQ, Eisen-Vandervelde A, Waggoner SN, Cale EM, Hahn YS. Direct binding of hepatitis C virus core to gC1qR on CD4+ and CD8+ T cells leads to impaired activation of Lck and Akt. J Virol. 2004;78:6409–6419. doi: 10.1128/JVI.78.12.6409-6419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan BP. The complement system: an overview. Methods Mol Biol. 2000;150:1–13. doi: 10.1385/1-59259-056-X:1. [DOI] [PubMed] [Google Scholar]
- 28.Rickert RC. Regulation of B lymphocyte activation by complement C3 and the B cell coreceptor complex. Curr Opin Immunol. 2005;17:237–243. doi: 10.1016/j.coi.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Moir S, Malaspina A, Li Y, Chun TW, Lowe T, Adelsberger J, Baseler M, et al. B cells of HIV-1-infected patients bind virions through CD21-complement interactions and transmit infectious virus to activated T cells. J Exp Med. 2000;192:637–646. doi: 10.1084/jem.192.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee MS, Jones T, Song DY, Jang JH, Jung JU, Gao SJ. Exploitation of the complement system by oncogenic Kaposi's sarcoma-associated herpesvirus for cell survival and persistent infection. PLoS Pathog. 2014;10:e1004412. doi: 10.1371/journal.ppat.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason CP, Tarr AW. Human lectins and their roles in viral infections. Molecules. 2015;20:2229–2271. doi: 10.3390/molecules20022229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monsalvo AC, Batalle JP, Lopez MF, Krause JC, Klemenc J, Hernandez JZ, Maskin B, et al. Severe pandemic 2009 H1N1 influenza disease due to pathogenic immune complexes. Nat Med. 2011;17:195–199. doi: 10.1038/nm.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu W, Yu Q, Hu N, Byrd D, Amet T, Shikuma C, Shiramizu B, et al. A high-affinity inhibitor of human CD59 enhances complement-mediated virolysis of HIV-1: implications for treatment of HIV-1/AIDS. J Immunol. 2010;184:359–368. doi: 10.4049/jimmunol.0902278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown KS, Keogh MJ, Owsianka AM, Adair R, Patel AH, Arnold JN, Ball JK, et al. Specific interaction of hepatitis C virus glycoproteins with mannan binding lectin inhibits virus entry. Protein Cell. 2010;1:664–674. doi: 10.1007/s13238-010-0088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamed MR, Brown RJ, Zothner C, Urbanowicz RA, Mason CP, Krarup A, McClure CP, et al. Recombinant human L-ficolin directly neutralizes hepatitis C virus entry. J Innate Immun. 2014;6:676–684. doi: 10.1159/000362209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim H, Meyer K, Di Bisceglie AM, Ray R. Hepatitis C virus suppresses C9 complement synthesis and impairs membrane attack complex function. J Virol. 2013;87:5858–5867. doi: 10.1128/JVI.00174-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amet T, Ghabril M, Chalasani N, Byrd D, Hu N, Grantham A, Liu Z, et al. CD59 incorporation protects hepatitis C virus against complement-mediated destruction. Hepatology. 2012;55:354–363. doi: 10.1002/hep.24686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazumdar B, Kim H, Meyer K, Bose SK, Di Bisceglie AM, Ray RB, Diamond MS, et al. Hepatitis C virus infection upregulates CD55 expression on the hepatocyte surface and promotes association with virus particles. J Virol. 2013;87:7902–7910. doi: 10.1128/JVI.00917-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peveling-Oberhag J, Arcaini L, Hansmann ML, Zeuzem S. Hepatitis C-associated B-cell non-Hodgkin lymphomas. Epidemiology, molecular signature and clinical management. J Hepatol. 2013;59:169–177. doi: 10.1016/j.jhep.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 40.Vannata B, Zucca E. Hepatitis C virus-associated B-cell non-Hodgkin lymphomas. Hematology Am Soc Hematol Educ Program. 2014;2014:590–598. doi: 10.1182/asheducation-2014.1.590. [DOI] [PubMed] [Google Scholar]
- 41.Machida K, Cheng KT, Sung VM, Shimodaira S, Lindsay KL, Levine AM, Lai MY, et al. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc Natl Acad Sci U S A. 2004;101:4262–4267. doi: 10.1073/pnas.0303971101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weng WK, Levy S. Hepatitis C virus (HCV) and lymphomagenesis. Leuk Lymphoma. 2003;44:1113–1120. doi: 10.1080/1042819031000076972. [DOI] [PubMed] [Google Scholar]
- 43.Carbonari M, Caprini E, Tedesco T, Mazzetta F, Tocco V, Casato M, Russo G, et al. Hepatitis C virus drives the unconstrained monoclonal expansion of VH1-9-expressing memory B cells in type II cryoglobulinemia: a model of infection-driven lymphomagenesis. J Immunol. 2005;174:6532–6539. doi: 10.4049/jimmunol.174.10.6532. [DOI] [PubMed] [Google Scholar]
- 44.Chan CH, Hadlock KG, Foung SK, Levy S. V(H)1-9 gene is preferentially used by hepatitis C virus-associated B cell lymphomas and by normal B cells responding to the E2 viral antigen. Blood. 2001;97:1023–1026. doi: 10.1182/blood.v97.4.1023. [DOI] [PubMed] [Google Scholar]
- 45.Marasca R, Vaccari P, Luppi M, Zucchini P, Castelli I, Barozzi P, Cuoghi A, et al. Immunoglobulin gene mutations and frequent use of VH1-69 and VH4-34 segments in hepatitis C virus-positive and hepatitis C virus-negative nodal marginal zone B-cell lymphoma. Am J Pathol. 2001;159:253–261. doi: 10.1016/S0002-9440(10)61691-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng PP, Kuo CC, Wang S, Einav S, Arcaini L, Paulli M, Portlock CS, et al. B-cell receptors expressed by lymphomas of hepatitis C virus (HCV)-infected patients rarely react with the viral proteins. Blood. 2014;123:1512–1515. doi: 10.1182/blood-2013-10-532895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niemann CU, Wiestner A. B-cell receptor signaling as a driver of lymphoma development and evolution. Semin Cancer Biol. 2013;23:410–421. doi: 10.1016/j.semcancer.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierce SK. Lipid rafts and B-cell activation. Nat Rev Immunol. 2002;2:96–105. doi: 10.1038/nri726. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.