Abstract
BRD4 is an epigenetic regulator and transcription cofactor whose phosphorylation by CK2 and dephosphorylation by PP2A modulates its function in chromatin targeting, factor recruitment, and cancer progression. While the bromodomains of BET family proteins, including BRD4, BRD2, BRD3 and BRDT, have been the primary targets of small compounds such as JQ1, I-BET and MS417 that show promising anticancer effects against some hematopoietic cancer and solid tumors, drug resistance upon prolonged treatment necessitates a better understanding of alternative pathways underlying not only the resistance but also persistent BET protein dependence for identifying new targets and effective combination therapy strategies.
Introduction
Bromodomain-containing protein 4 (BRD4) is a member of bromodomain (B) and extraterminal (ET) family proteins (BET) that in humans also include BRD2, BRD3 and BRDT [1]. BET proteins are evolutionarily conserved epigenetic readers featuring the presence of two tandem bromodomains (BD1 and BD2), which bind to select acetyl-lysine residues in nucleosomal histones [2] or nonhistone proteins like NFκB p65/RelA [3,4], Cyclin T1 [5], Twist [6] and GATA1 [7], and a structurally defined ET domain [8] that interacts with specific chromatin regulators, including NSD3, JMJD6, CHD4, GLTSCR1, and ATAD5 [9,10]. The C-terminal motif (CTM) is uniquely found in the long but not short isoform of BRD4 and BRDT, nor in BRD2 and BRD3 [1,11,12]. Both positive transcription elongation factor b (P-TEFb) and human papillomavirus (HPV) E2 protein target primarily the CTM of BRD4 to regulate human immunodeficiency virus (HIV) and HPV transcription [13–17].
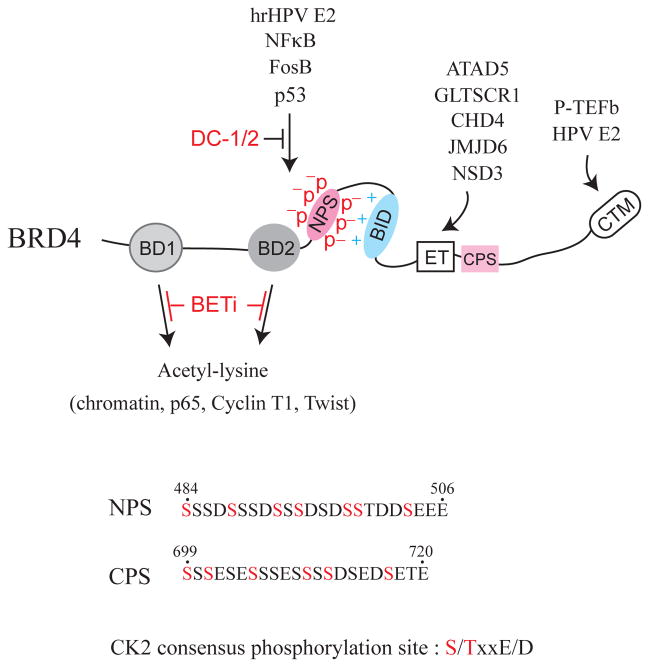
Besides BD1, BD2 and CTM, BET proteins also contain two clusters of casein kinase II (CK2) phosphorylation sites and a basic residue-enriched interaction domain (BID) [18]. In human BRD4, the N-terminal phosphorylation sites (NPS, amino acids 484–506) situated at the 3′ side of BD2 contain seven CK2 phosphorylation sequences (S/TxxD/E), and the C-terminal phosphorylation sites (CPS, amino acids 699–720) located downstream of the ET domain have six putative CK2 sites [Figure 1]. Phosphorylation of NPS by CK2 is crucial for BRD4 binding to acetylated chromatin and facilitating sequence-specific transcription factors, such as p53 tumor suppressor protein [18], high-risk HPV-encoded E2 protein, the FosB member of AP-1, and the p50 and p65 subunits of NFκB [19], in regulating specific sets of gene transcription. The extent of BRD4 phosphorylation has been shown to be important for learning and memory [20] and for triple-negative breast cancer progression [21].
Figure 1.
BRD4 domain features and select interacting factors. Conserved domains shown include bromodomain I (BD1), bromodomain II (BD2), N-terminal cluster of CK2 phosphorylation sites (NPS), basic residue-enriched interaction domain (BID), extraterminal (ET) domain, C-terminal cluster of CK2 phosphorylation sites (CPS), and C-terminal motif (CTM). P indicates phosphorylation, whereas + and − depict positive and negative charge distributions surrounding NPS and BID, respectively. Arrows show factors interacting with individual domains that can be selectively blocked by BET inhibitors (BETi) or DC-1 and DC-2 peptoids. The amino acid sequences of NPS, CPS, and the CK2 consensus phosphorylation site are also listed with potential CK2-phosphorylated serine residues indicated in red. Numbers correspond to the residue positions in the full-length BRD4 protein. hrHPV is an abbreviation for high-risk human papillomavirus.
Phospho-switch mechanism
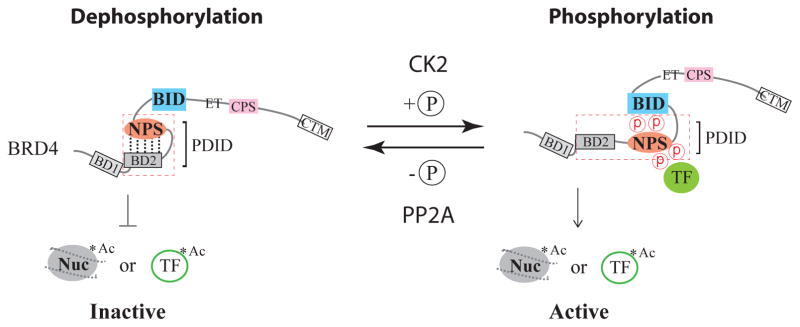
In addition to the putative CK2 phosphorylation sites in NPS and CPS [Figure 1], global phosphopeptide mapping in HeLa cells has identified phosphorylation of BRD4 at serine 470 (S470), BRD3 at S263 and S563, and BRD2 at S298 and S301 [22]. S470 in human BRD4 is situated between BD2 and NPS in a regulatory region defined as the phosphorylation-dependent interaction domain (PDID) spanning amino acids 287–530 [18]. The PDID has kinase-undefined S470, a putative CK2 phosphorylation site in BD2 at S405 (consensus: SKLE) and seven CK2 sites in NPS, and is highly phosphorylated by multiple kinases including CK2, PKA, and IKKε [18]. Hyper-phosphorylation of BRD4 NPS by decreased protein phosphatase 2A (PP2A) activity correlates with bromodomain-independent BRD4 association with the MED1 component of the human Mediator coactivator complex and drug resistance in triple-negative breast cancer [21]. When dephosphorylated, BD2 is masked by NPS that also functionally impairs the chromatin-binding activity of BD1 via allosteric regulation of NPS-modulated BD2-BD1 interaction [18], thereby preventing BRD4 from binding to acetylated nucleosomes and presumably acetylated transcription factors as well [Figure 2]. CK2-mediated phosphorylation of NPS switches its intramolecular contact from upstream BD2 to downstream BID and, concurrently, generates a new contact surface for recruitment of sequence-specific transcription factors [18,19], leading to two active configurations of BRD4 (i.e., unmasking of bromodomains and factor recruitment) allowing for site-specific regulation of target gene transcription [Figure 2]. Conceivably, modulation of the switch between dephosphorylated and phosphorylated states of BRD4 is an effective way to control the function of BRD4 in gene targeting and disease progression, recruitment of NFκB to its chromatin target site, and HPV origin replication [19].
Figure 2.
Phosphorylation-induced contact switch in BRD4 regulated by CK2 and PP2A. PDID represents a phosphorylation-dependent interaction domain that encompasses BD2 and NPS as bracketed in dashed lines. Nuc and TF are abbreviations for nucleosome and transcription factors, respectively, and asterisks indicate the acetyl-lysine (Ac) in nucleosome or transcription factors. Dashed lines connecting BD2 and NPS in the dephosphorylation state as well as circled P between NPS and BID in the phosphorylation state show intra-molecular contacts within BRD4 dictated by the extent of phosphorylation.
Pharmacological inhibitors of BRD4 and BET proteins
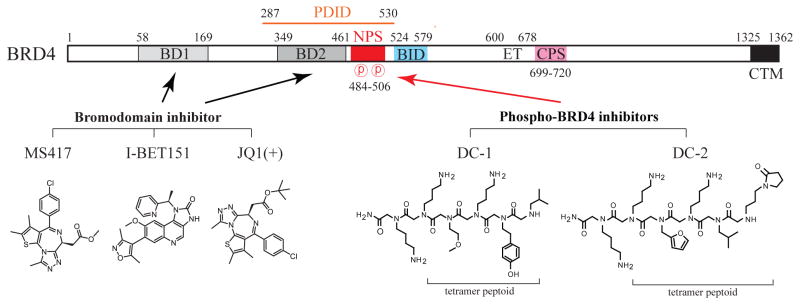
The identification of acetyl-lysine-mimicking compounds, such as JQ1 [23], I-BET [24] and MS417 [4], with high affinity (in nM range) and specificity towards BET bromodomains [Figure 3] that show promising effects as anticancer and anti-inflammatory agents has raised significant interest and enthusiasm for developing pharmacological inhibitors against BET family proteins. A number of BET bromodomain inhibitors with distinct chemical scaffolds and selectivity for individual or both bromodomains [12,25], of which some are in clinical trials, have been recently reviewed [26]. These inhibitors generally work by blocking BET proteins, particularly BRD4 that exhibits highest affinity, from binding acetylated chromatin in “super-enhancers”, consisting of clusters of enhancer elements [27]. Super-enhancers, unlike classical enhancers, are frequently activated in cancer-associated oncogenes [27] or cell fate-determining genes [28] that are driven by an ensemble of transcription factors working in a combinatorial and cell type-specific manner [29,30]. Since the action of DNA-binding transcription factors and chromatin regulators is typically enhanced by BRD4 [1,31], this mutual dependency among components in a BRD4-mediated enhanceosome complex assembled on a super-enhancer is far more sensitive to disruption by BET bromodomain inhibitors compared to that formed on a regular enhancer [27]. This differential sensitivity to compound disruption between super-enhancers and enhancers partly explains why treatment of cancer cells with low concentrations of JQ1 or I-BET could reverse cancer phenotypes without globally impairing normal gene activity. Although MYC is often the oncogenic driver in many cancer cells, overexpression of other transcription factors, such as the FOSL1/FRA-1 component of AP-1 [32,33], could also lead to cancer phenotypes via MYC-independent pathways. Accordingly, identifying the transcription factors and pathways specifically regulated by BRD4 and/or other BET family proteins is critical for developing appropriate regimens for effective cancer therapy.
Figure 3.
Chemical structures of BD1/BD2-targeting bromodomain inhibitors (JQ1, I-BET151, and MS417) and phospho-NPS/BRD4-targeting inhibitors (DC-1 and DC-2). Numbers indicate the boundaries of individual domains in BRD4.
Other than these well-known BET bromodomain inhibitors, a different class of small compounds targeting the phospho region of BRD4 has been described [34]. Two hits, DC-1 (Nlys-Nlys-Nmea-Nlys-Ntyr-Nleu) and DC-2 (Nlys-Nlys-Nffa-Nlys-Nleu-Napp), were isolated from a combinatorial library of ~38,500 peptoids (i.e., N-substituted oligoglycines) and showed specific binding to phosphorylated PDID/NPS of human BRD4 [Figure 3]. Peptoids are protein-binding ligands similar to peptides but are protease/peptidase-resistant and more cell-permeable compared to peptides alone [35]. The affinity of peptoids to the protein target, however, is usually low (in μM range), which may somewhat limit their applications. Nevertheless, the isolation of DC-1 and DC-2 indicates that targeting a phosphorylated protein is conceptually and technically feasible. Given the importance of phospho-NPS in contacting BID and recruiting transcriptional regulators such as p53 and high-risk HPV E2, targeting this regulatory region [Figure 1] offers a unique strategy to inhibit the gene-specific function of BRD4 and factor-regulated transcriptional programs and pathways without non-selectively shutting down the universal chromatin-binding activity of BRD4 as seen with the BET bromodomain inhibitors.
Therapeutic window for drug targeting on phospho-BRD4
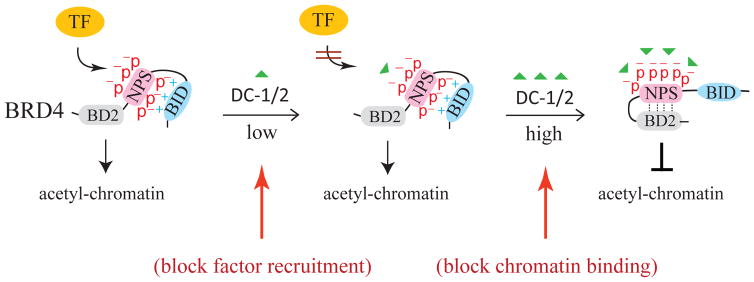
Effective blocking of a specific transcription pathway requires a careful titration of compound concentrations that elicit minimal off-target effects. Since phosphorylation of NPS is necessary for intra-molecular contact with BID and also for inter-molecular interaction of BRD4 with certain transcription factors, it is conceivable to find a therapeutic window in which low concentrations of DC-1 and DC-2 block phospho-NPS interaction with transcription factors without perturbing phosphorylation-mediated NPS-BID contact [Figure 4]. Within this therapeutic window, phospho-NPS-targeting compounds would only inhibit specific pathways regulated by the transcription factor. Since the BID-anchored NPS is not free to mask the bromodomain, binding of BRD4 to acetylated chromatin remains unperturbed. Hence, the untargeted BRD4 function in mediating other chromatin dynamics and transcriptional programs would not be affected. At high concentrations of DC-1 and DC-2, the intra-molecular contact between phospho-NPS and BID would also be disrupted, leading to NPS masking of BD2 that is incapable of binding to acetylated chromatin thus mimicking the action of BET bromodomain inhibitors [Figure 4]. Conceivably, properly titrated DC-1 and DC-2 are able to inhibit the action of specific transcription factors without stripping BRD4 off its chromatin target sites. Indeed, DC-1 proves to be effective in inhibiting high-risk HPV E2-enhanced MMP-9 gene transcription upon keratinocyte differentiation by preventing NFκB from binding to its cognate site in the MMP-9 promoter-proximal region without affecting non-phospho-BRD4-mediated recruitment of other transcription factors [19], highlighting a more selective inhibition of BRD4-regulated transcription programs via targeting phospho-NPS contacting surfaces with specific transcription factors. In the case of p53, phospho-NPS-mediated p53 recruitment might provide cytoprotection to normal cells/tissues surrounding radiotherapy-targeted tumor sites by transiently inducing cell cycle arrest. It is also likely that phospho-BRD4 could potentiate p53 mutant gain-of-function in certain cancer cells, which remains an interesting hypothesis to be experimentally tested. This new class of compounds targeting phospho-BET proteins will be of great value for future development of therapeutic agents.
Figure 4.
Concentration-dependent DC-1 and DC-2 inhibition of BRD4-mediated transcription factor recruitment (at low concentrations) and chromatin binding (at high concentrations). The concentration of DC-1 and DC-2 is indicated by green triangle.
Transcription plasticity and drug resistance
An important issue in cancer therapy is the development of drug resistance and cancer relapse in patients undergoing prolonged treatment. While genetic heterogeneity and gene mutation may partly or frequently contribute to the outcomes of drug treatment and disease progression, an emerging view and understanding of the plasticity (or flexibility) in transcription programming point to remarkable cellular adaptations to highly fluctuated environments. Inhibition of a primary oncogenic driver, e.g., MYC, by BET inhibitors (BETi) in a specific cell type or cancer is often rescued by activation of alternative transcriptional programs or signaling pathways. This compensatory mechanism has recently been described in acute myeloid leukemia where suppression of MYC oncogene expression by JQ1 or I-BET can be overcome by triggering the activation of alternative enhancers (or a super-enhancer) driving the same MYC gene expression or by the Wnt/β-catenin pathway, along with additional mechanisms such as inhibition of polycomb repressor complex 2 or stimulation of the TGF-β pathway [36,37]. In breast cancer, depending on genetic and phenotypic subtypes, BETi resistance could be attributed to factors (e.g., PP2A) modulating BRD4 phosphorylation [21] or PIK3CA mutation [38]. In four subgroups of pediatric medulloblastoma brain tumors, distinct classes of enhancers and super-enhancers that recruit different sets of transcription factors have been identified via genomic profiling of 28 primary medulloblastoma specimens [30].
Clearly, oncogenic drivers are potentially variable among diverse cancer types/subtypes and are often the familiar factors controlling our normal physiology, as seen with BRD4-potentiated estrogen receptor (ER) function in ER-positive breast cancer [39] and androgen receptor/BRD4-linked prostate cancer [40]. An important note to mention is that even though BET inhibitors may not work effectively upon prolonged treatment, BRD4 is persistently required for oncogenic phenotypes in many BETi-resistent cells, which may also be caused by switching BRD4 chromatin association from bromodomain-dependent acetyl-lysine recognition to bromodomain-independent factor recruitment, e.g., via interaction with Mediator [21] or sequence-specific DNA-binding factors [29]. Indeed, BRD4 association with transcription factors, Mediator, and P-TEFb represent three major steps of transcriptional regulation targeting, respectively, template/gene commitment, initiation, and elongation steps [31], which are crucial for RNA polymerase II-driven transcription [41].
Concluding remarks
The identification of BRD4 as the primary target for BET inhibitors in cancer therapy and the discovery of CK2/PP2A-regulated BRD4 phosphorylation as the critical entity linked to gene regulation, drug resistance, and cancer progression highlight the importance of developing new BET inhibitors targeting the phospho region and other domains of BRD4 mediating protein-protein interaction. Since phosphorylation at NPS regulates adjacent bromodomain binding to acetylated chromatin, it is possible that phosphorylation at CPS likewise controls ET domain-mediated interaction with NSD3 or other ET-associated factors, which will need further investigation. In addition, understanding alternative and compensatory transcriptional programs and signaling pathways activated or suppressed in BETi-resistant cells and patient populations should be of high priority in designing effective combination therapy for cancer treatment.
Acknowledgments
I thank Dr. Shwu-Yuan Wu for illustration and discussion. Research in my lab is currently supported by grants from the National Institutes of Health (CA103867), Cancer Prevention Research Institute of Texas (RP110471 and RP140367), and the Welch Foundation (I-1805).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wu S-Y, Chiang C-M. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 2.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-κB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang G, Liu R, Zhong Y, Plotnikov AN, Zhang W, Zeng L, Rusinova E, Gerona-Nevarro G, Moshkina N, Joshua J, Chuang PY, Ohlmeyer M, He JC, Zhou MM. Down-regulation of NF-κB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem. 2012;287:28840–28851. doi: 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schröder S, Cho S, Zeng L, Zhang Q, Kaehlcke K, Mak L, Lau J, Bisgrove D, Schnölzer M, Verdin E, Zhou MM, Ott M. Two-pronged binding with bromodomain-containing protein 4 liberates positive transcription elongation factor b from inactive ribonucleoprotein complexes. J Biol Chem. 2012;287:1090–1099. doi: 10.1074/jbc.M111.282855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, Lin Y, Li J, Kang T, Tao M, Rusinova E, Zhang G, Wang C, Zhu H, Yao J, Zeng YX, Evers BM, Zhou MM, Zhou BP. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, Cheng Y, Billin AN, Hardison RC, Mackay JP, Blobel GA. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci USA. 2011;108:E159–168. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin YJ, Umehara T, Inoue M, Saito K, Kigawa T, Jang MK, Ozato K, Yokoyama S, Padmanabhan B, Güntert P. Solution structure of the extraterminal domain of the bromodomain-containing protein BRD4. Protein Sci. 2008;17:2174–2179. doi: 10.1110/ps.037580.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR. NSD3-Short Is an Adaptor Protein that Couples BRD4 to the CHD8 Chromatin Remodeler. Mol Cell. 2015;60:847–859. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, Fydrych A, Ho R, Greenberger BA, Chen GC, Maffa A, Del Rosario AM, Root DE, Carpenter AE, Hahn WC, Sabatini DM, Chen CC, White FM, Bradner JE, Yaffe MB. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498:246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang C-M. Nonequivalent response to bromodomain-targeting BET inhibitors in oligodendrocyte cell fate decision. Chem Biol. 2014;21:804–806. doi: 10.1016/j.chembiol.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc Natl Acad Sci USA. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 16.Wu S-Y, Lee AY, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang C-M. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 2006;20:2383–2396. doi: 10.1101/gad.1448206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee AY, Chiang C-M. Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J Biol Chem. 2009;284:2778–2786. doi: 10.1074/jbc.M805835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu S-Y, Nin DS, Lee A-Y, Simanski S, Kodadek T, Chiang C-M. Phosphorylation of BRD4 regulates HPV E2-mediated viral transcription and origin replication and cellular MMP-9 gene transcription. Cell Reports. 2016 doi: 10.1016/j.celrep.2016.07.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korb E, Herre M, Zucker-Scharff I, Darnell RB, Allis CD. BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat Neurosci. 2015;18:1464–1473. doi: 10.1038/nn.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, Doherty E, Mohammed H, Guo H, Stover DG, Ekram MB, Peluffo G, Brown J, D’Santos C, Krop IE, Dillon D, McKeown M, Ott C, Qi J, Ni M, Rao PK, Duarte M, Wu SY, Chiang CM, Anders L, Young RA, Winer EP, Letai A, Barry WT, Carroll JS, Long H, Brown M, Liu XS, Meyer CA, Bradner JE, Polyak K. Response and resistance to BET bromodomain inhibitors in triple negative breast cancer. Nature. 2016;529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villén J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gacias M, Gerona-Navarro G, Plotnikov AN, Zhang G, Zeng L, Kaur J, Moy G, Rusinova E, Rodriguez Y, Matikainen B, Vincek A, Joshua J, Casaccia P, Zhou MM. Selective chemical modulation of gene transcription favors oligodendrocyte lineage progression. Chem Biol. 2014;21:841–854. doi: 10.1016/j.chembiol.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang G, Smith SG, Zhou MM. Discovery of Chemical Inhibitors of Human Bromodomains. Chem Rev. 2015;115:11625–11668. doi: 10.1021/acs.chemrev.5b00205. [DOI] [PubMed] [Google Scholar]
- 27.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell. 2015;58:1028–1039. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin CY, Erkek S, Tong Y, Yin L, Federation AJ, Zapatka M, Haldipur P, Kawauchi D, Risch T, Warnatz HJ, Worst BC, Ju B, Orr BA, Zeid R, Polaski DR, Segura-Wang M, Waszak SM, Jones DT, Kool M, Hovestadt V, Buchhalter I, Sieber L, Johann P, Chavez L, Gröschel S, Ryzhova M, Korshunov A, Chen W, Chizhikov VV, Millen KJ, Amstislavskiy V, Lehrach H, Yaspo ML, Eils R, Lichter P, Korbel JO, Pfister SM, Bradner JE, Northcott PA. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016;530:57–62. doi: 10.1038/nature16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang C-M. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 Biol Rep. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker EK, Taylor S, Gupte A, Sharp PP, Walia M, Walsh NC, Zannettino AC, Chalk AM, Burns CJ, Walkley CR. BET inhibitors induce apoptosis through a MYC independent mechanism and synergise with CDK inhibitors to kill osteosarcoma cells. Sci Rep. 2015;5:10120. doi: 10.1038/srep10120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai D, Lee AY, Chiang C-M, Kodadek T. Peptoid ligands that bind selectively to phosphoproteins. Bioorg Med Chem Lett. 2011;21:4960–4964. doi: 10.1016/j.bmcl.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan NC, Yu P, Kwon Y-U, Kodadek T. High-throughput evaluation of relative cell permeability between peptoids and peptides. Bioorg Med Chem. 2008;16:5853–5861. doi: 10.1016/j.bmc.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D, Stanley K, Sinha D, Yeh P, Morison J, Giotopoulos G, Lugo D, Jeffrey P, Lee SC, Carpenter C, Gregory R, Ramsay RG, Lane SW, Abdel-Wahab O, Kouzarides T, Johnstone RW, Dawson SJ, Huntly BJ, Prinjha RK, Papenfuss AT, Dawson MA. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M, Deswal S, Cerny-Reiterer S, Peter B, Jude J, Hoffmann T, Boryń ŁM, Axelsson E, Schweifer N, Tontsch-Grunt U, Dow LE, Gianni D, Pearson M, Valent P, Stark A, Kraut N, Vakoc CR, Zuber J. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, Virtanen C, Bradner JE, Bader GD, Mills GB, Pe’er D, Moffat J, Neel BG. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagarajan S, Hossan T, Alawi M, Najafova Z, Indenbirken D, Bedi U, Taipaleenmäki H, Ben-Batalla I, Scheller M, Loges S, Knapp S, Hesse E, Chiang C-M, Grundhoff A, Johnsen SA. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 2014;8:460–469. doi: 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, Iyer MK, Jing X, Wu YM, Cao X, Qin ZS, Wang S, Feng FY, Chinnaiyan AM. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas MC, Chiang C-M. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]