Abstract
Silver nanoparticles (AgNPs) demonstrating good antimicrobial activity are widely used in many fields. However, the impact of AgNPs on the community structures of marine biofilms that drive biogeochemical cycling processes and the recruitment of marine invertebrate larvae remains unknown. Here, we employed MiSeq sequencing technology to evaluate the bacterial communities of 28-day-old marine biofilms formed on glass, polydimethylsiloxane (PDMS), and PDMS filled with AgNPs and subsequently tested the influence of these marine biofilms on plantigrade settlement by the mussel Mytilus coruscus. AgNP-filled PDMS significantly reduced the dry weight and bacterial density of biofilms compared with the glass and PDMS controls. AgNP incorporation impacted bacterial communities by reducing the relative abundance of Flavobacteriaceae (phylum: Bacteroidetes) and increasing the relative abundance of Vibrionaceae (phylum: Proteobacteria) in 28-day-old biofilms compared to PDMS. The settlement rate of M. coruscus on 28-day-old biofilms developed on AgNPs was lower by >30% compared to settlement on control biofilms. Thus, the incorporation of AgNPs influences biofilm bacterial communities in the marine environment and subsequently inhibits mussel settlement.
Recently, the application of nanomaterials such as copper, titanium and silver has expanded considerably in various fields1. Due to their efficacious antimicrobial actions against bacteria, diatoms and viruses1,2,3,4, silver nanoparticles (AgNPs) have been applied in the food sector5, medical devices6, textile finishings7 and water treatment systems8. While the impact of nanoparticles (NPs) on aquatic microbes has been investigated9, few studies10,11,12 have been conducted in the marine environment to examine the impact of AgNPs on natural biofilms that play key roles in biogeochemical cycling processes and larval and spore recruitment13,14.
In the marine environment, biofilms are ubiquitous on submerged substrata15. Marine biofilms comprise many species of bacteria and unicellular eukaryotes (e.g., diatoms and flagellates)15 but are dominated by multiple bacterial and diatom species15,16. Marine biofilms are crucial for larval recruitment for some invertebrates14,15,16,17,18, and the bacteria in biofilms are viewed as sources of settlement-inducing cues for invertebrate larvae18,19.
Bacterial strains isolated from biofilms have been shown to induce the larval recruitment of invertebrates, such as the coral Pocillopora damicornis20, the tubeworm Hydroides elegans21, and the mussel Mytilus galloprovincialis22. However, the presence of such bacterial strains in natural biofilms does not always explain the recruitment activity of biofilms23,24. Previous studies have demonstrated the importance of complex microbial communities during invertebrate larval settlement, such as that of H. elegans23,25,26, Balanus amphitrite24,26,27, B. trigonus28, and Rhopaloeides odorabile28. In the case of mussels, variances in biofilm composition result in differential larval settlement29. Therefore, it is important to investigate changes in marine biofilm communities and subsequent effects on mussel settlement to understand the possible environmental impact of NPs.
The mussel Mytilus coruscus is an important aquaculture and fouling bivalve in China30,31,32. In this study, MiSeq sequencing technology was employed to ascertain the influence of AgNPs on the formation of marine biofilms on glass and polydimethylsiloxane (PDMS) substrata in a marine environment as well as the subsequent influence of these bacterial communities on M. coruscus settlement. Glass is widely used to investigate biofilm formation in marine environments20,21,29,33, and PDMS is a known fouling release material employed in antifouling studies with marine invertebrates34,35,36. The purpose of this study was to determine whether biofilm communities differ on substrata with and without AgNPs and if variations in bacterial communities alter settlement-inducing activity.
Results
Settlement bioassay
No differences were observed in the settlement of M. coruscus plantigrades on surfaces lacking biofilms (Fig. 1, p = 0.98). Similar tendencies were also observed for 7-day-old biofilms. For 14-day-old biofilms, the plantigrade settlement rate on AgNP-filled PDMS was similar to the settlement rates on glass and PDMS (controls), with the exception of PDMS filled with 10 weight% (wt%) AgNPs (p < 0.0001). After 21 days, the biofilms that developed on PDMS filled with 10 wt% AgNPs induced significantly lower settlement than those on glass (p < 0.0001). After 28 days, the settlement rates were significantly lower for biofilms on PDMS filled with 1 wt% and 10 wt% AgNPs compared with those on glass and PDMS (p < 0.0001).
Figure 1. Percentage of M. coruscus plantigrades on non-biofilm and biofilm substrata.
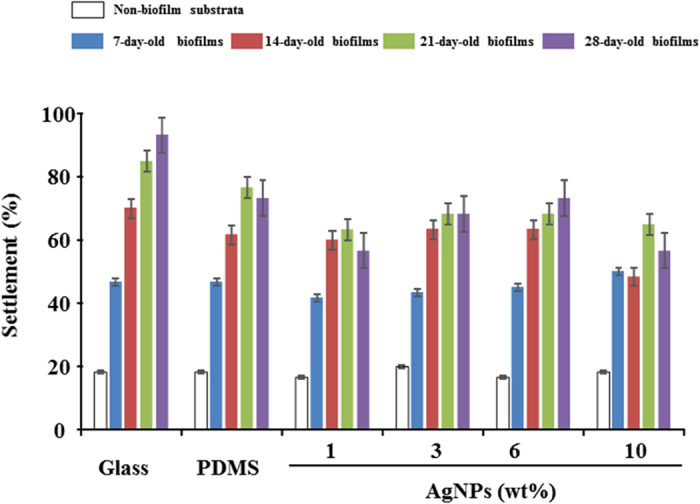
Non-biofilm substrata included glass, PDMS, and PDMS filled with AgNPs. Data are the mean ± standard error (SE) (n = 6).
Biomass of marine biofilms
There were no differences between the dry weights of 7-day-old biofilms on differentially treated surfaces (Fig. 2a). The dry weights of biofilms that developed after 14 d on PDMS filled with 3 wt% and 6 wt% AgNPs, after 21 d on PDMS filled with 3 wt% AgNPs and after 28 d on PDMS filled with 1 wt% and 10 wt% AgNPs were significantly lower (p < < 0.0001) than those on glass and PDMS (Fig. 2a).
Figure 2.
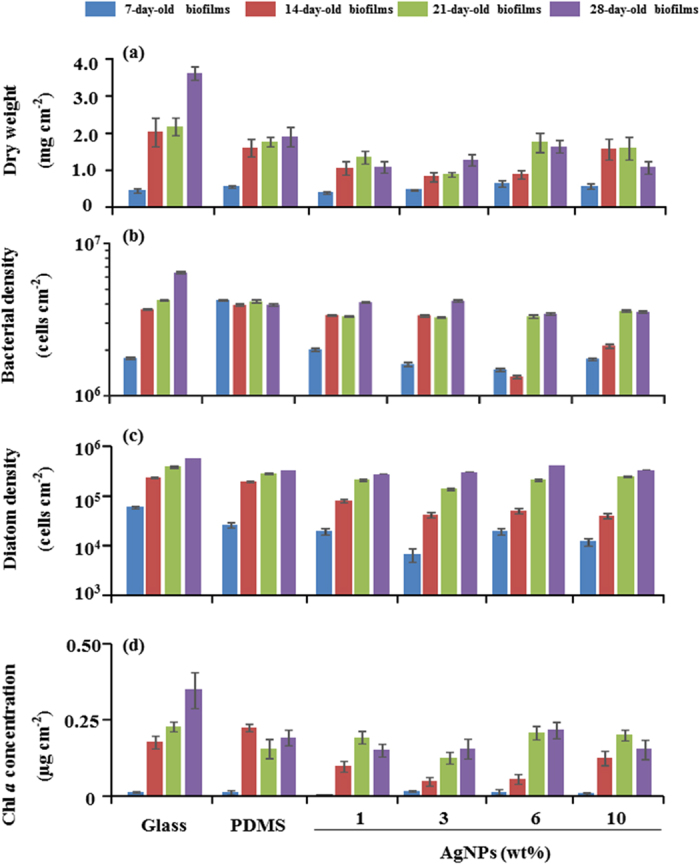
Dry weights (a), bacterial densities (b), diatom densities (c) and chl a concentrations (d) of natural biofilms on different substrata. Data are the mean ± SE (n = 6).
After 7 days, bacterial densities were significantly lower (p < 0.0001) for biofilms that developed on PDMS filled with 6 wt% AgNPs (Fig. 2b). After 14 and 21 days, bacterial densities were significantly lower (p < 0.0001) for biofilms on PDMS filled with 1–10 wt% AgNPs. After 28 days, bacterial densities for biofilms on PDMS filled with 1–10 wt% AgNPs were significantly lower (p < 0.0001) compared to the densities of biofilms on glass but were similar to those on PDMS (p > 0.05).
Significantly lower diatom density was observed for 7-day-old biofilms on PDMS filled with 3 wt% AgNPs compared to that of controls (Fig. 2c, p < 0.0001). After 14 days, diatom densities were significantly lower for biofilms on PDMS filled with 1–10 wt% AgNPs (p < 0.0001). Furthermore, diatom densities were significantly lower for 21-day-old biofilms on PDMS filled with 1–6 wt% AgNPs (p < 0.0001) and for 28-day-old biofilms on PDMS filled with 1 wt% AgNPs (p < 0.0001) compared to that of controls.
After 7, 21 and 28 days, no significant differences in chlorophyll a (chl a) concentrations were observed (Fig. 2d, p > 0.05). After 14 days, significantly lower concentrations of chl a were detected in biofilms that developed on PDMS filled with 1–6 wt% AgNPs (p < 0.0001).
Bacterial community analysis based on MiSeq sequencing
A total of 286,793 valid reads and 5,231 operational taxonomic units (OTUs) were obtained from 28-day-old biofilms on glass, PDMS and PDMS filled with 1 wt% AgNPs, for which the OTU numbers were 2,261, 1,612 and 1,358, respectively. At a 3% dissimilarity level, Good’s coverage estimations suggested 99.1% to 99.9% of bacterial species were present for all treatments.
At the phylum level, one-way analysis of similarity (ANOSIM) analysis revealed significant differences between the bacterial communities that developed on glass, PDMS and PDMS filled with 1 wt% AgNPs; the R value was 0.802 (p < 0.05). Proteobacteria was the most dominant phylum (>45% of all sequences) present in all samples (Fig. 3, Table S1). There was no significant difference in the relative abundance of Proteobacteria between samples (p > 0.05, Fig. 3, Table S1). The second most dominant phylum, Bacteroidetes, accounted for 6–40% of the sequences in all biofilm samples, and the relative abundance of Bacteroidetes was significantly higher in biofilms on PDMS than in biofilms on glass and PDMS filled with 1 wt% AgNPs (p < 0.05, Table S1). The relative abundance of Firmicutes accounted for 3–23% of the sequences in all samples, and the relative abundance of Firmicutes was significantly lower in biofilms on PDMS than in biofilms on glass and PDMS filled with 1 wt% AgNPs (p < 0.05, Table S1). The abundances of bacteria belonging to all other phyla were lower (< 17%).
Figure 3. Relative abundance of bacterial phyla in 28-day-old biofilms on glass, PDMS and PDMS filled with 1 wt% AgNPs.
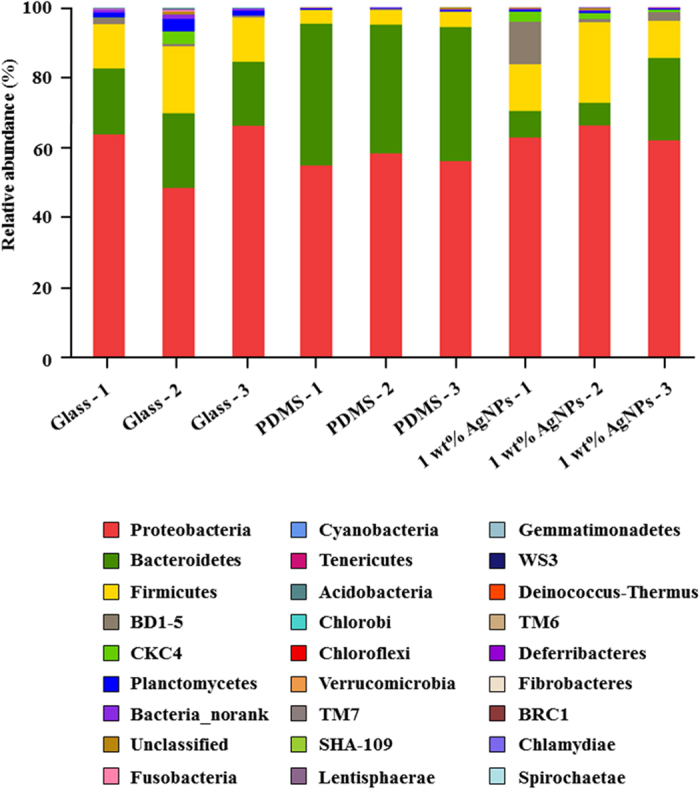
Three replicates for each treatment are labelled with the numbers 1, 2 and 3.
At the family level, significant differences were observed in bacterial communities on different substrata, with an R value of > 0.9 (p < 0.05). Pseudoalteromonadaceae, Vibrionaceae and Flavobacteriaceae were the most dominant families present for all three treatments (45–72% of all sequences, Fig. 4, Table S2). No significant difference in the relative abundance of Pseudoalteromonadaceae was observed in bacterial communities in all samples (p > 0.05, Fig. 4, Table S2); however, the relative abundance of Vibrionaceae was higher in bacterial communities on PDMS filled with 1 wt% AgNPs than in those on glass and PDMS (p < 0.05, Fig. 4, Table S2). Additionally, the relative abundance of Flavobacteriaceae in bacterial communities on PDMS filled with 1 wt% AgNPs was significantly lower than that on PDMS (p < 0.05). Based on similarity of percentage (SIMPER) analysis, the families Vibrionaceae, Flavobacteriaceae and BD1–5 explained >5% of the variance between bacterial communities on glass and PDMS filled with 1 wt% AgNPs (Table 1). Vibrionaceae and BD1–5 explained >5% of the variance between marine biofilm communities on PDMS and PDMS filled with 1 wt% AgNPs (Table 1).
Figure 4. Heatmap revealing the top 20 bacterial families (%) in each biofilm sample on glass, PDMS and PDMS filled with 1 wt% AgNPs.
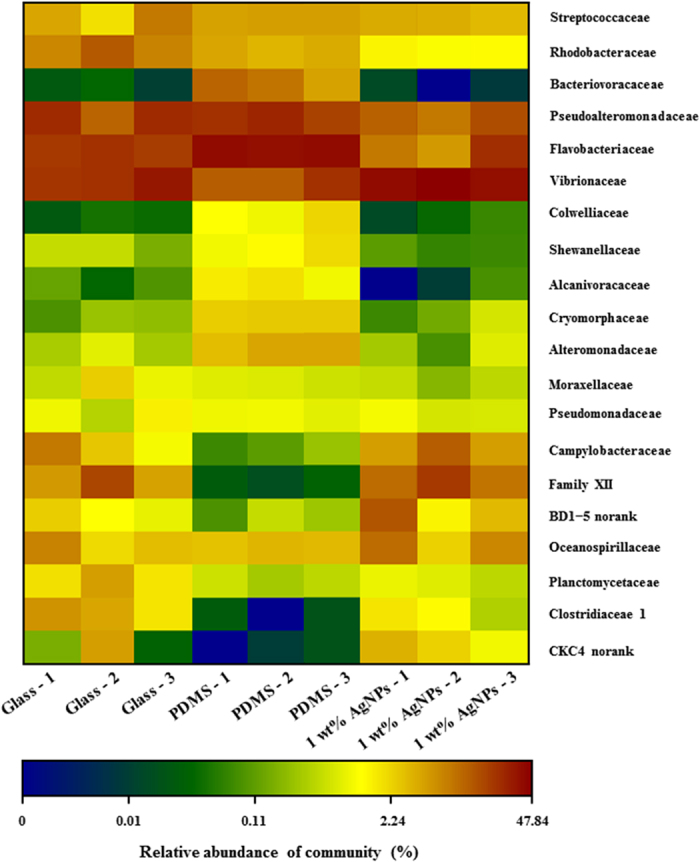
Three replicates for each treatment are labelled with the numbers 1, 2 and 3.
Table 1. SIMPER analysis showing the % contribution of specific bacterial families to the total dissimilarity among bacterial communities that developed on glass, PDMS, and PDMS filled with 1 wt% AgNPs.
| Family | Comparison with PDMS filled with 1 wt% AgNPs | |
|---|---|---|
| Glass | PDMS | |
| Vibrionaceae | 6.45 | 7.58 |
| Flavobacteriaceae | 6.72 | |
| BD1-5 | 5.28 | 5.43 |
| Others | 88.27 | 80.27 |
Note: The contributions are averaged over all significant pair-wise treatment comparisons. Bacteria that contributed less than 5% are not shown.
At the genus level, there was a significant difference in bacterial communities on different substrata, with an R value of > 0.9 (p < 0.05). Vibrio, Pseudoalteromonas, Fusibacter and Tenacibaculum were the four major genera present in all samples (>40% of all sequences, Fig. 5, Table S3). The average relative abundance of Vibrio species increased by more than 0.73-fold and 1.9-fold in bacterial communities that developed on PDMS filled with 1 wt% AgNPs compared to those on glass and PDMS, respectively (p < 0.05, Fig. 5, Table S3). Based on SIMPER analysis, the genera Vibrio explained >3% of the variance between communities that developed on glass and PDMS filled with 1 wt% AgNPs as well as between communities that developed on PDMS and PDMS filled with 1 wt% AgNPs (Table 2).
Figure 5. Heatmap revealing the top 10 bacterial genera (%) in each biofilm sample on glass, PDMS and PDMS filled with 1 wt% AgNPs.
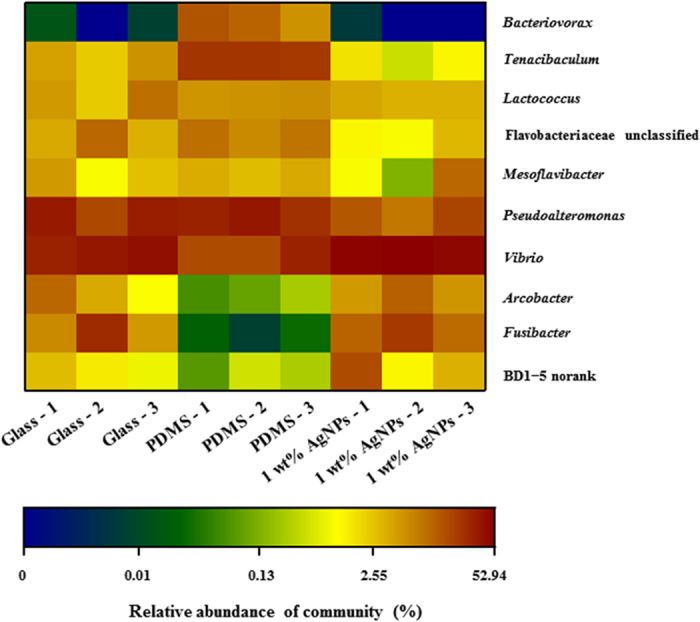
Three replicates for each treatment are labelled with the numbers 1, 2 and 3.
Table 2. SIMPER analysis showing the % contribution of specific bacterial genera to the total dissimilarity among bacterial communities that developed on glass, PDMS, and PDMS filled with 1 wt% AgNPs.
| Genus | Comparison with PDMS filled with 1 wt% AgNPs |
|
|---|---|---|
| Glass | PDMS | |
| Vibrio | 3.24 | 4.47 |
| Tenacibaculum | 1.22 | 4.59 |
| Shewanella | 1.24 | |
| Others | 95.54 | 89.70 |
Note: The contributions are averaged over all significant pair-wise treatment comparisons.
Principal component analysis (PCA) revealed large differences between biofilm communities that developed on glass and PDMS compared to those that developed on PDMS filled with 1 wt% AgNPs (PC1–70.24% of the variance) (Fig. 6).
Figure 6. PCA of three biofilm samples on glass, PDMS and PDMS filled with 1 wt% AgNPs.
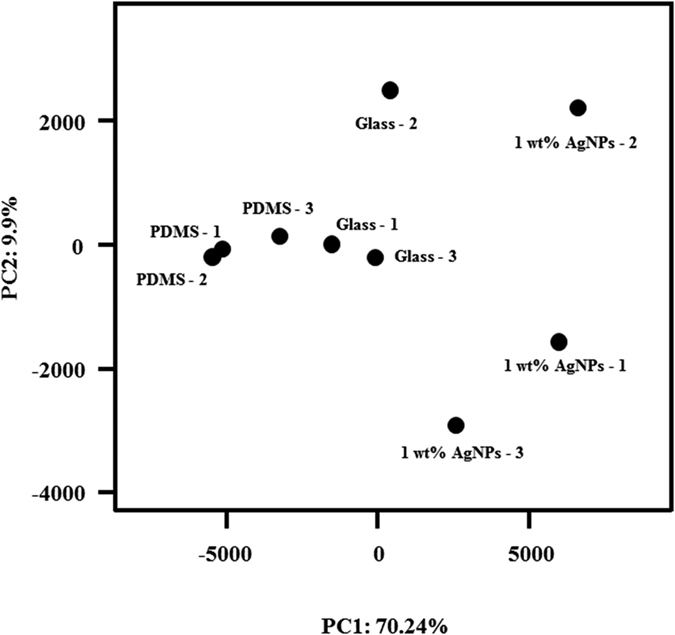
Three replicates for each treatment are labelled with the numbers 1, 2 and 3.
Discussion
In marine ecosystems, microbial community succession within a biofilm has been reported24,37,38,39,40,41. However, the mechanisms by which biofilm communities mediate the settlement of marine invertebrate larvae remain poorly constrained23,24. Additionally, although AgNPs possess excellent anti-biofilm activity1,40, how AgNPs impact biofilm communities in complex marine environments remains unknown. The present study investigated the impact of AgNPs on biofilm bacterial communities in a marine environment and explored the subsequent effects of biofilms on the settlement of M. coruscus plantigrades.
AgNPs are efficient antimicrobial agents that combat the growth of a broad spectrum of bacterial species1,3. Inbakandan et al.11 collected marine biofilms that formed at the air-seawater interface on the bottom of the hull of a fishing vessel, isolated 11 bacterial strains and examined the influence of AgNPs on these bacterial species under laboratory conditions. AgNPs exhibited good antibacterial properties, and inhibition depended on the concentration of AgNPs and the bacterial species11. Similarly, the experiments in this study revealed the ability of PDMS filled with AgNPs to decrease bacterial densities in marine biofilms after 14 and 21 days compared with controls (glass and PDMS). Thus, in the natural environment, AgNPs exert inhibitory effects on marine bacteria during biofilm formation. The antimicrobial activity of AgNPs may be attributable to the Ag+ ions released from the nanoparticles as a result of their exposure to reactive entities generated intracellularly1.
Moreover, the results of the present study reveal variations in bacterial communities among biofilms that formed in the presence and absence of AgNPs, indicating colonization by bacterial communities in the marine environment is impacted by AgNPs. This finding is consistent with our hypothesis that AgNPs may affect microbial community formation. According to previous studies, bacterial communities are impacted by many factors, such as surface characteristics, substratum materials and environmental conditions23,39,42,43. In addition, variations in biofilm communities on surfaces with and without antifouling properties were recently reviewed11. Prior to this study, no study has investigated marine biofilm communities that form on substrata containing AgNPs.
According to our results, Proteobacteria was the most dominant phylum in all marine biofilms, similar to previous investigations23,44,45,46. The incorporation of AgNPs reduced the relative abundance of Bacteroidetes (Flavobacteriaceae) and increased the relative abundance of Vibrionaceae (Gammaproteobacteria) bacteria in biofilms. Thus, AgNPs may specifically inhibit Bacteroidetes cell attachment. In contrast, in activated sludge microbial communities, AgNPs increase the abundance of Gammaproteobacteria and Bacteroidetes47. These differing results may be explained by the presence of different microbial communities in different environments. Based on further metagenomic analysis at the genus level, AgNPs increased the relative abundance of Vibrio and reduced the relative abundances of Tenacibaculum and Shewanella compared with that of control PDMS (Table 2, Table S3). In previous studies, Vibrio spp. inhibited the larval settlement of certain invertebrate larvae, including H. elegans48,49, Bugula neritina48,50, and B. amphitrite51. However, Tenacibaculum spp. and Shewanella spp. promote the larval settlement of various invertebrates, including M. coruscus31 and B. neritina50. Therefore, the high abundance of Vibrio spp. and the lower abundance of Tenacibaculum spp. and Shewanella spp. in biofilms on PDMS filled with AgNPs may explain the lower mussel settlement observed in the present study.
The antibacterial properties of AgNPs are relatively well documented52, while their activities against diatoms have not been well characterized. In the laboratory, AgNPs inhibit the growth of the marine diatom Thalassiosira pseudonana53. In this study, the densities of marine diatoms in biofilms formed on PDMS filled with AgNPs were lower than controls (glass and PDMS), even after 28 days, suggesting AgNPs effectively inhibit the attachment of diatoms during the formation of natural biofilms in the marine environment. Although the mechanism underlying the anti-diatom action of AgNPs remains unclear, previous studies have described toxic effects of NPs and released Ag+ ions on diatoms53. AgNPs penetrate the cell wall of the diatom Cylindrotheca fusiformis and cause local damage inside the cell without disintegrating the cell wall54. Additionally, AgNPs reach the cytoplasm of the diatom T. pseudonana and cause toxicity55.
In summary, the findings of this study revealed the impact of AgNPs on the formation of marine biofilms, which subsequently inhibited the plantigrade settlement of M. coruscus. Thus, AgNPs possess antifouling properties, and it is necessary to perform proper toxicological studies for NPs before their practical application in the marine environment.
Methods
Preparation of tested surfaces
AgNPs (CW-Ag-001; particle size: 20 nm; specific surface area: 42 m2/g; purity: >99%) were purchased from Shanghai Chaowei Nano Co., Ltd. (China). PDMS, a common fouling released material, was selected as a coating control for our assays. In previous studies, PDMS has been employed to investigate the interactions between substratum properties and the settlement of marine invertebrates34,35,36. As a PDMS support substratum, a glass slide (38 mm × 26 mm, AS ONE Shanghai Corporation, Shanghai, China) was used as an additional control and conveniently submerged during biofilm formation. PDMS (Sylgard 184; Dow Corning, Shanghai, China) was made according to the method of Carl et al.34. AgNPs were incorporated into the PDMS matrix (PDMS filled with AgNPs) at concentrations of 0 (PDMS only), 1, 3, 6 and 10 weight% (wt%). The bubbles created from mixing PDMS with AgNPs were removed with a vacuum desiccator. Then, the mixture was poured onto a glass slide and cured for up to 10 h in a hot oven at 90 °C.
Development of natural biofilms
Coated substrata were randomly placed on PVC holders and submerged vertically at a depth of approximately 1.0 m at Gouqi Island (122°77′E; 30°72′N), Zhoushan, China for 7, 14, 21 and 28 days in August 2013. Biofilm samples were collected and transported on ice to Shanghai Ocean University, and macrofouling organisms were removed.
Biofilm dry weight
The dry weight of biofilms was evaluated following the method of Bao et al.33. In brief, biofilms from six treatment replicates (n = 24) were scraped off substrata surfaces into 100-ml beakers containing autoclaved filter seawater (AFSW). An additional 50 ml of 0.22-μm AFSW was used to wash the biofilms attached to the beakers. Then, the suspensions were filtered using a pre-weighed Whatman GF/C filter (pore size: 1.2 μm, Whatman International Ltd, Maidstone, England). Each GF/C filter paper was dried at 80 °C for 48 h. The dry weight of each biofilm sample was calculated based on the weight difference between the weight of the filter before and after filtration.
Bacterial and diatom density
Bacterial densities were determined according to the method of Yang et al.56. Natural biofilms from each treatment (n = 24) were separately scraped off using sterile glass slides, suspended in AFSW, and fixed in formalin (final concentration = 5%). Suspensions containing bacteria and diatoms were vortexed for 60 s to ensure the homogeneous distribution of microorganisms before counting cell densities. Bacteria were stained with 0.1% acridine orange (Sigma Chemical Co., St Louis, Mo) solution. The suspended bacteria were filtered and collected on polycarbonate Nucleopore filters (pore size: 0.2 μm, Whatman 4.9 cm2, Whatman International Ltd, Maidstone, England). The bacterial densities of each stained sample (n = 3) were immediately determined with an Olympus BX51 epifluorescence microscope (Olympus, Tokyo, Japan) at 1000× magnification. The cell densities of suspended diatoms (n = 3) were evaluated using a hemocytometer under an Olympus BX51 epifluorescence microscope (Olympus, Tokyo, Japan) with 200× magnification. Ten random fields of view for each biofilm sample were counted, and the mean densities of bacteria and diatoms were calculated.
Chlorophyll a (chl a) concentration in biofilms
Biofilms from each treatment (n = 24) were scraped, filtered and preserved at −20 °C. A 90% acetone solution was used to extract chl a at 4 °C for 14 h in the dark. After centrifugation, chl a concentrations in the supernatants were evaluated spectrophotometrically (UNIC 2100 spectrophotometer, UNIC (Shanghai) Instruments, Shanghai, China) according to the method of Wang et al.29. Six independent glass slides were examined for each treatment.
MiSeq sequencing analysis of bacterial communities
Bacterial DNA was extracted from each 28-day-old biofilm sample (glass, PDMS, PDMS filled with 1 wt% AgNPs) with 3 biological replicates according to Li et al.45. The V4-V5 region of the 16 S rRNA gene was amplified using previously described primers and condition45. PCR products were extracted, purified and quantified following Li et al.45. The purified amplicons were pooled in equimolar concentrations and sequenced on the Illumina MiSeq platform at Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China. The generated metagenomic data were deposited in the NCBI sequence read archive (accession number: SRP 048906).
Mussel settlement assay
Mussel plantigrades were supplied by the Institute of Marine Science and Technology in Zhoushan, China. The culture conditions were as described by Yang et al.57. A plantigrade settlement assay was conducted in Petri dishes containing 20 ml of AFSW and one biofilm-covered (7, 14, 21 and 28 days) substratum (glass [control], PDMS [control], 1 wt% AgNPs, 3 wt% AgNPs, 6 wt% AgNPs and 10 wt% AgNPs). Ten plantigrades were added into each Petri dish (AS ONE Shanghai Corporation, Shanghai, China). The number of settled plantigrades was counted after 12 h and later calculated as a percentage. The effects of substrata without biofilms (glass, PDMS, PDMS filled with AgNPs) on plantigrade settlement were examined. Plantigrades attached to the coating via byssal threads were considered settled. The mussel settlement assay was performed according to the method of Yang et al.57. Six replicates were examined for each biofilm and non-biofilm treatment.
Statistical and bioinformatics analysis
To ensure data normality, the percentages of biofilm-inducing activity were arcsine-square root transformed. The data were analyzed using JMPTM software (SAS Institute (Shanghai) Co., LTD, Shanghai, China). A p value of 0.05 (p < 0.05) was considered significant. Settlement rate, dry weight, chl a and bacteria and diatom density analyses were conducted by performing Kruskal-Wallis analysis of variance followed by the Steel-Dwass All Pairs test.
ANOSIM (PRIMER 6)58 was performed with substratum as the factor. Specifically, ANOSIM was conducted to compare similarities in bacterial species composition between substrata with and without AgNPs. ANOSIM computes a test statistic (R), where R = 1 if all replicates within a treatment are more similar to each other than any replicates from different treatments. R is approximately zero if the null hypothesis that similarities between and within treatments are the same is true. ANOSIM analysis was performed for each biofilm sample (n = 3). SIMPER analysis (Primer 6 software; Primer-E Ltd) was performed to compare the major species contributing to the dissimilarity between bacterial communities that developed on substrata with and without AgNPs.
Raw fastq data were analyzed using QIIME (version 1.17)59. The processing steps were as follows. The raw mate-paired fastq files were demultiplexed and quality-filtered as described previously45. The UPARSE pipeline (version 7.1 http://drive5.com/uparse/) was applied to choose the OTUs with a similarity threshold of 97%. The phylogenetic affiliation of each 16 S rRNA gene sequence was evaluated with RDP classifier (http://rdp.cme.msu.edu/) against the Silva (SSU115) 16 S rRNA database using a confidence threshold of 70%60.
Additional Information
How to cite this article: Yang, J.-L. et al. Silver Nanoparticles Impact Biofilm Communities and Mussel Settlement. Sci. Rep. 6, 37406; doi: 10.1038/srep37406 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We acknowledge Mr. Zhongqi Gu for friendly help in providing the mussel plantigrades. This study was supported by the National Natural Science Foundation of China (No. 41476131), the Innovation Program of Shanghai Municipal Education Commission (14ZZ143), the Shanghai Universities Plateau Discipline Project of Marine Sciences and the Peak Discipline Program for Fisheries from the Shanghai Municipal Government to J.L.Y., as well as an award from the Shanghai Ocean University Doctoral Research Foundation (A2-0203-00-100320) to Y.F.L.
Footnotes
Author Contributions J.L.Y., X.L. and W.Y.B. conceived and designed the experiments. Y.F.L. and X.P.G. performed the experiments, Y.F.L., X.P.G. and S.X.Z. analyzed the data. J.L.Y., D.W.D., D.M.Z., W.Y.B., N.B. and S.D. wrote the manuscript. All authors reviewed the manuscript.
References
- Martinez-Gutierrez F. et al. Anti-biofilm activity of silver nanoparticles against different microorganisms. Biofouling 29, 651–660 (2013). [DOI] [PubMed] [Google Scholar]
- Gong P. et al. Preparation and antibacterial activity of Fe3O4@Ag nanoparticles. Nanotechnology 18, 604–611 (2007). [Google Scholar]
- Sharma V. K., Yngard R. A. & Lin Y. Silver nanoparticles: green synthesis and their antimicrobial activities. Adv. Colloid. Interfac. 145, 83–96 (2009). [DOI] [PubMed] [Google Scholar]
- Sanyasi S. et al. Polysaccharide-capped silver Nanoparticles inhibit biofilm formation and eliminate multi-drug-resistant bacteria by disrupting bacterial cytoskeleton with reduced cytotoxicity towards mammalian cells. Sci. Rep. 6, 24929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry Q. et al. Applications and implications of nanotechnologies for the food sector. Food. Addit. Contam. A. 25, 241–258 FOOD ADDIT CONTAM A (2008). [DOI] [PubMed] [Google Scholar]
- Maki D. G. In vitro studies of a novel antimicrobial luer-activated needleless connector for prevention of catheter-related bloodstream infection. Clin. Infect. Dis. 50, 1580–1587 (2010). [DOI] [PubMed] [Google Scholar]
- Kramer A. et al. Hygienic relevance and risk assessment of antimicrobial-impregnated textiles. Curr. Probl. Derm-US 33, 78–109 (2006). [DOI] [PubMed] [Google Scholar]
- Dror-Ehre A., Adin A., Markovich G. & Mamane H. Control of biofilm formation in water using molecularly capped silver nanoparticles. Water. Res. 44, 2601–2609 (2010). [DOI] [PubMed] [Google Scholar]
- Battin T. G., Kammer F. V. D., Weilhartner A., Ottofuelling S. & Thilo Hofmann T. Nanostructured TiO2: transport behavior and effects on aquatic microbial communities under environmental conditions. Environ. Sci. Technol. 43, 8098–8104 (2009). [DOI] [PubMed] [Google Scholar]
- Fabrega J., Zhang R., Renshaw J. C., Liu W. T. & Lead J. R. Impact of silver nanoparticles on natural marine biofilm bacteria. Chemosphere 85, 961–966 (2011). [DOI] [PubMed] [Google Scholar]
- Inbakandan D. et al. Silver nanoparticles with anti microfouling effect: A study against marine biofilm forming bacteria. Colloid. Surfaces. B. 111, 636–643 (2013). [DOI] [PubMed] [Google Scholar]
- Fabrega J., Luoma S. N., Tyler C. R., Galloway T. S. & Lead J. R. Silver nanoparticles: behaviour and effects in the aquatic environment. Environ. Int. 37, 517–531 (2011). [DOI] [PubMed] [Google Scholar]
- Qian P. Y., Lau S. C. K., Dahms H. U., Dobretsov S. & Harder T. Marine biofilms as mediators of colonization by marine macroorganisms: implications for antifouling and aquaculture. Mar. Biotech. 9, 399–410 (2007). [DOI] [PubMed] [Google Scholar]
- Dobretsov S. In Biofouling (eds Dürr S. & Thomason J. C.) 123–136 (Wiley-Blackwell 2010). [Google Scholar]
- Dobretsov S., Abed R. M. M. & Teplitski M. Mini-review: inhibition of biofouling by marine microorganisms. Biofouling 29, 423–441 (2013). [DOI] [PubMed] [Google Scholar]
- Wieczorek S. K. & Todd C. D. Inhibition and facilitation of settlement of epifaunal marine invertebrate larvae by microbial biofilm cues. Biofouling 12, 81–118 (1998). [Google Scholar]
- Dobretsov S. In Marine and industrial biofouling (eds Flemming H. C. et al.) 293–313 (Springer 2009). [Google Scholar]
- Hadfield M. G. Biofilms and marine invertebrate larvae: What bacteria produce that larvae use to choose settlement sites. Annu. Rev. Mar. Sci. 3, 453–470 (2011). [DOI] [PubMed] [Google Scholar]
- Shikuma N. J. et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like Structures. Science 343, 529–533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran C. & Hadfield M. G. Larvae of Pocillopora damicornis (Anthozoa) settle and metamorphose in response to surface-biofilm bacteria. Mar. Ecol. Prog. Ser. 433, 85–96 (2011). [Google Scholar]
- Unabia C. R. C. & Hadfield M. G. Role of bacteria in larval settlement and metamorphosis of the polychaete Hydroides elegans. Mar. Biol. 133, 55–64 (1999). [Google Scholar]
- Bao W. Y., Yang J. L., Satuito C. G. & Kitamura H. Larval metamorphosis of the mussel Mytilus galloprovincialis in response to Alteromonas sp. 1: evidence for two chemical cues? Mar. Biol. 152, 657–666 (2007). [Google Scholar]
- Chung H. C. et al. Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. ISME J. 4, 817–828 (2010). [DOI] [PubMed] [Google Scholar]
- Lee O. O. et al. In situ environment rather than substrate type dictates microbial community structure of biofilms in a cold seep system. Sci. Rep. 4, 3587 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. & Hadfield M. G. Composition and density of bacterial biofilms determine larval settlement of the polychaete Hydroides elegans. Mar. Ecol. Prog. Ser. 260, 161–172 (2003). [Google Scholar]
- Lau S. C. K., Thiyagarajan V., Cheung S. C. K. & Qian P. Y. Roles of bacterial community composition in biofilms as a mediator for larval settlement of three marine invertebrates. Aquat. Microb. Ecol. 38, 41–51 (2005). [Google Scholar]
- Qian P. Y., Thiyagarajan V., Lau S. C. K. & Cheung S. C. K. Relationship between bacterial community profile in biofilm and attachment of the acorn barnacle Balanus amphitrite. Aquat. Microb. Ecol. 33, 225–237 (2003). [Google Scholar]
- Whalan S. & Webster N. S. Sponge larval settlement cues: the role of microbial biofilms in a warming ocean. Sci. Rep. 4, 4072 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. et al. Larval settlement and metamorphosis of the mussel Mytilus coruscus in response to natural biofilms. Biofouling 28, 249–256 (2012). [DOI] [PubMed] [Google Scholar]
- Yang J. L. et al. Effects of neuroactive compounds, ions and organic solvents on larval metamorphosis of the mussel Mytilus coruscus. Aquaculture 396–399, 106–112 (2013). [Google Scholar]
- Yang J. L. et al. Larval settlement and metamorphosis of the mussel Mytilus coruscus in response to monospecific bacterial biofilms. Biofouling 29, 247–259 (2013). [DOI] [PubMed] [Google Scholar]
- Zeng Z. et al. Characterization of self-generated variants in Pseudoalteromonas lipolytica biofilm with increased antifouling activities. Appl. Microb. Biot. 99, 10127–10139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao W. Y., Satuito C. G., Yang J. L. & Kitamura H. Larval settlement and metamorphosis of the mussel Mytilus galloprovincialis in response to biofilms. Mar. Biol. 150, 565–574 (2007). [Google Scholar]
- Carl C. et al. Enhancing the efficacy of fouling-release coatings against fouling by Mytilus galloprovincialis using nanofillers. Biofouling 28, 1077–1091 (2012). [DOI] [PubMed] [Google Scholar]
- Martinelli E. et al. Poly(dimethylsiloxane) (PDMS) network blends of amphiphilic acrylic copolymers with poly(ethylene glycol)-fluoroalkyl side chains for fouling-release coatings. II. Laboratory assays and field immersion trials. Biofouling 28, 571–582 (2012). [DOI] [PubMed] [Google Scholar]
- Vucko M. J. et al. Using textured PDMS to prevent settlement and enhance release of marine fouling organisms. Biofouling 30, 1–16 (2014). [DOI] [PubMed] [Google Scholar]
- Tujula N. A. et al. Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. ISME J. 4, 301–311 (2010). [DOI] [PubMed] [Google Scholar]
- Qian P. Y. et al. Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J. 5, 507–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione J. F. et al. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc. Natl. Acad. Sci. USA 109, 7633–17638 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp K. H., Distel D. & Paul V. J. Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 6, 790–801 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe P., Richter J., Myint M. T. Z., Dobretsov S. & Dutta J. Self-decontaminating photocatalytic zinc oxide nanorod coatings for prevention of marine microfouling: a mesocosm study. Biofouling 32, 383–395 (2016). [DOI] [PubMed] [Google Scholar]
- Burke C., Thomas T., Lewis M., Steinberg P. & Kjelleberg S. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5, 590–600 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salta M., Wharton J. A., Blache Y., Stokes K. R. & Briand J. F. Marine biofilms on artificial surfaces: Structure and dynamics. Environ. Microbiol. 15, 2879–2893 (2013). [DOI] [PubMed] [Google Scholar]
- Jones P. R., Cottrell M. T., Kirchman D. L. & Dexter S. C. Bacterial community structure of biofilms on artificial surfaces in an estuary. Microb. Ecol. 53, 153–162 (2007). [DOI] [PubMed] [Google Scholar]
- Li Y. F. et al. Effects of bacterial biofilms on settlement of plantigrades of the mussel Mytilus coruscus. Aquaculture 433, 434–441 (2014). [Google Scholar]
- Muthukrishnan T., Abed R. M. M., Dobretsov S., Kidd B. & Finnie A. A. Long-term microfouling on commercial biocidal fouling control coatings. Biofouling 30, 1155–1164 (2014). [DOI] [PubMed] [Google Scholar]
- Yang Y. et al. Pyrosequencing reveals higher impact of silver nanoparticles than Ag+ on the microbial community structure of activated sludge. Water. Res. 48, 317–325 (2014). [DOI] [PubMed] [Google Scholar]
- Dobretsov S. & Qian P. Y. The role of epibotic bacteria from the surface of the soft coral Dendronephthya sp. in the inhibition of larval settlement. J. Exp. Mar. Biol. Ecol. 299, 35–50 (2004). [Google Scholar]
- Dobretsov S. V. & Qian P. Y. Effect of bacteria associated with the green alga Ulva reticulata on marine micro- and macrofouling. Biofouling 18, 217–228 (2002). [Google Scholar]
- Dobretsov S. & Qian P. Y. Facilitation and inhibition of larval attachment of the bryozoan Bugula neritina in association with mono-species and multi-species biofilms. J. Exp. Mar. Biol. Ecol. 333, 263–274 (2006). [Google Scholar]
- Lau S. C. L., Thiyagarajan V. & Qian P. Y. The bioactivity of bacterial isolates in Hong Kong waters for the inhibition of barnacle (Balanus amphitrite Darwin) settlement. J. Exp. Mar. Biol. Ecol. 282, 43–60 (2003). [Google Scholar]
- Rai M. et al. Broad-spectrum bioactivities of silver nanoparticles: the emerging trends and future prospects. Appl. Microbiol. Biot. 98, 1951–1961 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchardt A. D. et al. Effects of silver nanoparticles in diatom Thalassiosira pseudonana and cyanobacterium Synechococcus sp. Environ. Sci. Technol. 46, 11336–11344 (2012). [DOI] [PubMed] [Google Scholar]
- Pletikapić G., Vera Žutić V., Vinković Vrček V. & Svetličić V. Atomic force microscopy characterization of silver nanoparticles interactions with marine diatom cells and extracellular polymeric substance. J. Mol. Recognit. 25, 309–317 (2012). [DOI] [PubMed] [Google Scholar]
- Pascual García C. et al. Detection of silver nanoparticles inside marine diatom Thalassiosira pseudonana by electron microscopy and focused ion beam. PLoS One 9, e96078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. L., Satuito C. G., Bao W. Y. & Kitamura H. Larval settlement and metamorphosis of the mussel Mytilus galloprvincialis on different macroalgae. Mar. Biol. 152, 1121–1131 (2007). [Google Scholar]
- Yang J. L. et al. Effects of natural biofilms on settlement of plantigrades of the mussel Mytilus coruscus. Aquaculture 424–425, 228–233 (2014). [Google Scholar]
- Clarke K. R. & Warwick R. M. Change in marine communities: an approach to statistical analysis and interpretation. Plymouth: Plymouth Marine Laboratory (2001). [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of highthroughput community sequencing data. Nat. Meth. 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato K. R. et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 7, 1344–1353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


