Abstract
The A1 subunits of Shiga toxin 1 (Stx1A1) and Shiga toxin 2 (Stx2A1) interact with the conserved C termini of ribosomal-stalk P-proteins to remove a specific adenine from the sarcin/ricin loop. We previously showed that Stx2A1 has higher affinity for the ribosome and higher catalytic activity than Stx1A1. To determine if conserved arginines at the distal face of the active site contribute to the higher affinity of Stx2A1 for the ribosome, we mutated Arg172, Arg176, and Arg179 in both toxins. We show that Arg172 and Arg176 are more important than Arg179 for the depurination activity and toxicity of Stx1A1 and Stx2A1. Mutation of a single arginine reduced the depurination activity of Stx1A1 more than that of Stx2A1. In contrast, mutation of at least two arginines was necessary to reduce depurination by Stx2A1 to a level similar to that of Stx1A1. R176A and R172A/R176A mutations eliminated interaction of Stx1A1 and Stx2A1 with ribosomes and with the stalk, while mutation of Arg170 at the active site reduced the binding affinity of Stx1A1 and Stx2A1 for the ribosome, but not for the stalk. These results demonstrate that conserved arginines at the distal face of the active site are critical for interactions of Stx1A1 and Stx2A1 with the stalk, while a conserved arginine at the active site is critical for non-stalk-specific interactions with the ribosome. Arginine mutations at either site reduced ribosome interactions of Stx1A1 and Stx2A1 similarly, indicating that conserved arginines are critical for ribosome interactions but do not contribute to the higher affinity of Stx2A1 for the ribosome.
INTRODUCTION
Shiga toxin (Stx)-producing Escherichia coli (STEC) is an emerging foodborne and waterborne pathogen responsible for hemolytic uremic syndrome (HUS) and hemorrhagic colitis (HC), which are the leading causes of acute renal failure and mortality in children in the United States (1). STEC serotypes, such as E. coli O157:H7, are associated with severe disease (2). Antibiotics are known to exacerbate the disease symptoms, and at present, there are no FDA-approved vaccines or therapeutics against STEC infection (3–5). STEC produces a family of structurally and functionally related virulence factors called Shiga toxins, the most predominant ones being Shiga toxin 1 (Stx1) and Shiga toxin 2 (Stx2) (6). Stx2 and Stx1 have one prototype (Stx1a and Stx2a) and several subtypes. Stxs are type II ribosome-inactivating proteins (RIPs) with a catalytically active A subunit attached to a pentamer of B subunits. The B subunits facilitate the endocytosis of the toxins into the cell by binding to a common receptor globotriaosylceramide (GB3 or CD77). The toxin travels in a retrograde manner from the endosome to the endoplasmic reticulum (ER) via the Golgi network (7). In order to intoxicate the cell, the A subunit is cleaved into the A1 fragment and the A2 fragment, which remain together via a disulfide bond. After reduction of the disulfide bond, the A1 fragment is translocated into the cytosol from the ER, where it refolds into an active conformation, binds to the ribosome, and removes a specific adenine (A4324 in Saccharomyces cerevisiae) from the highly conserved sarcin/ricin loop (SRL) of the 28S rRNA (8). Structural studies using bacterial ribosomes showed that the SRL is the key ribosomal element involved in GTPase activation of elongation factor G (EF-G) (9). Depurination of A2660 in E. coli ribosomes prevents binding of GTPase factors and GTP hydrolysis by EF-G, resulting in arrest of protein synthesis at the translocation step (10, 11).
Stx1, as well as other structurally and functionally related RIPs, such as ricin and trichosanthin (TCS), interact with the ribosomal P-protein stalk to depurinate the SRL (12–15). The ribosomal stalk and the SRL are part of the GTPase-associated center (GAC) of the ribosome, which is involved in binding of elongation factors and stimulation of translation factor-dependent GTP hydrolysis (16, 17). In eukaryotes, the stalk is organized as a pentamer with ribosomal uL10 protein (former name, P0 [18]), which constitutes the base of the stalk and anchors two eukaryotic-unique P1/P2 heterodimers (19). Lower eukaryotes, such as S. cerevisiae, possess two P1/P2 protein forms, P1A, P1B, P2A, and P2B, which form two dimers, P1A/P2B and P1B/P2A, bound to two specific contiguous sites on the uL10 protein (20). The most prominent feature of the eukaryotic stalk proteins is the highly conserved motif present in the C-terminal part of the uL10 and P1/P2 proteins, consisting of a stretch of highly acidic and hydrophobic amino acids involved in interaction with translational GTPases (trGTPases) and RIPs (21). We established that the ribosomal stalk is the primary docking site for the ricin A chain (RTA) on the ribosome (13) and showed that multiplication of P-proteins is a critical factor accelerating the recruitment of RTA to the ribosome (22). Using the yeast model, we showed that the two P1A/P2B and P1B/P2A stalk dimers on the ribosome do not contribute equally to the interaction of ribosomes with RTA (23).
The crystal structure of the P11 peptide (SDDDMGFGLFD), corresponding to the conserved last 11 residues of P-proteins in a complex with TCS, a single-chain RIP, showed that the acidic amino acids at the amino end of P11 (DDD) form electrostatic interactions with the positively charged Lys173, Arg174, and Lys177 of TCS, while the LF motif in the hydrophobic carboxyl end of P11 (FGLF) is inserted into a hydrophobic pocket in the C-terminal domain of TCS (15). Stx1A1 has been shown to interact with the P11 peptide using Arg172, Arg176, and Arg179 located on the distal face of the active site (14, 24). Since P-proteins exist as a pentameric complex on the ribosome, these studies provided a valuable view but did not address the interactions with P11 in the context of the intact ribosome or the pentameric stalk complex. The interactions of Stx2A1 with P11 or with the ribosome were not investigated.
We previously showed that the ribosomal P-protein stalk is required for ribosome depurination and toxicity of the A subunits of Stx1 and Stx2 (25). The depurination activity of Stx1A on the ribosome was greatly reduced if P1/P2 binding sites on P0 were deleted, while the depurination activity of Stx2A was not affected as much, indicating that although stalk P-proteins were important for ribosome depurination by both toxins, Stx2A was less dependent on the stalk proteins for depurination of the SRL than Stx1A (25). We recently showed that Stx2A1 has a higher affinity for the ribosome and higher catalytic activity and toxicity than Stx1A1 in both yeast and mammalian cells (26). The interaction of RTA with the ribosome depended on electrostatic interactions and followed a two-step binding model, where the slower, non-stalk-dependent electrostatic interactions concentrated RTA molecules on the ribosome and allowed the faster electrostatic interactions with the stalk P-proteins (27, 28). Analysis of the electrostatic surface of the A1 subunits revealed differences in charge distribution around the active site and on the distal face of the active site in a region shown to be important for the interaction of Stx1A1 with the P11 peptide (26). Surface-exposed conserved arginines in both regions contribute to differences in surface charge. These arginines or surface charge differences in other regions of the A1 subunits could lead to the differences in their affinities for the ribosome. In the present study, we mutated the conserved arginines on the distal face of the active site and at the active site in both Stx1A1 and Stx2A1 to determine if they contribute to the higher affinity of Stx2A1 for the ribosome. Our results demonstrate for the first time that conserved arginines at the distal face of the active site are critical for the interaction of Shiga toxins with the ribosome and with the ribosomal stalk and their depurination activity and toxicity, while a conserved arginine at the active site affects non-stalk-specific interactions with the ribosome. We show that conserved arginines in neither site contribute to the higher affinity of Stx2A1 for the ribosome.
MATERIALS AND METHODS
Yeast strains and plasmids.
Point mutations of arginines to alanines were introduced into mature Stx1A1 (plasmid NT1643) and Stx2A1 (NT1644) in pBluescript by site-directed mutagenesis using a Q5 High-Fidelity DNA polymerase kit (New England BioLabs, Ipswich, MA). All mutations were confirmed by DNA sequencing. The wild-type (WT) Stx1A1 (K1 to R251) (NT1651) and Stx2A1 (R1 to R250) (NT1652) and the mutated genes for Stx1A1 E167K (NT1660), R170A (NT1661), R172A (NT1704), R176A (NT1706), and R172A/R176A (NT1730) and for Stx2A1, E167K (NT1664), R170A (NT1663), R172A (NT1696), R176A (NT1699), and R172A/R176A (NT1731) were subcloned into a yeast expression vector, NT1617, carrying a URA3 marker and a GAL1 promoter with V5 and a 10×His epitope at their C termini. S. cerevisiae strain W303 (MATa ade2-1 trp1-1 ura3-1 leu2-3,112 his-3-11,15 can1-100) was transformed with each of the constructs or the empty vector.
Yeast cell viability assay.
The W303 cells containing Stx constructs were grown at 30°C in synthetic dropout (SD) medium supplemented with 2% glucose overnight, and toxin expression was induced by transferring them to SD medium with 2% galactose. Cells were collected at 0 and 4 h postinduction (hpi), and serial dilutions to an optical density at 600 nm (OD600) of 0.1 were plated on SD-uracil plates containing 2% glucose. The plates were then grown at 30°C for 2 to 3 days.
Total-protein extraction and immunoblot analysis.
The W303 strain expressing WT and mutant forms of Stx was collected at 6 h postinduction, and total protein was extracted from yeast cells at an OD600 of 5, as described previously (29). Stx1A1 and Stx2A1 expression was detected using monoclonal antibodies against V5 (Invitrogen, Carlsbad, CA). Monoclonal antibodies against 3-phosphoglycerate kinase (Pgk1p) (Life Technologies, Grand Island, NY) were used as a loading control.
Purification of 10×His-tagged and untagged Stx1A1 and Stx2A1.
The 10×His-tagged WT (NT1570) and R172A (NT1761), R176A (NT1741), and R172A/R176A (NT1765) mutants in Stx1A1 and the WT (NT1567) and R172A (NT1762), R176A (NT1742), and R172A/R176A (NT1766) mutants in Stx2A1 were purified by Karen Chave at the Northeast Biodefense Center protein expression core facility using the Impact protein expression system (New England BioLabs, Ipswich, MA), as described previously (26). Briefly, forward and reverse primers with NdeI and SapI restriction sites were synthesized to enable in-frame cloning of the PCR fragments into the polylinker of the pTXB1 vector (New England BioLabs, Ipswich, MA), resulting in a C-terminal fusion of the Mycobacterium xenopi intein tag and a chitin binding domain. E. coli strain Rosetta2(DE3)(pLysS) transformed with the constructs was grown in 2× yeast extract-tryptone medium overnight at 16°C. The fusion proteins were purified from E. coli lysates using chitin beads, and the proteins were eluted by thiol-induced cleavage.
Analysis of depurination.
W303 cells expressing WT and mutant forms of Stx1A1 and Stx2A1 were collected at 1 h postinduction. An RNeasy minikit (Qiagen, Valencia, CA) with on-column DNase treatment was used to extract the total RNA. cDNA was obtained from the RNA using a High Capacity cDNA reverse transcription kit (Applied Biosystems, Grand Island, NY), and depurination was detected with a quantitative real-time PCR method using the StepOnePlus real-time PCR system (Applied Biosystems, Grand Island, NY). The 25S reference primers were 5′-AGA CCG TCG CTT GCT ACA AT-3′ and 5′-ATG ACG AGG CAT TTG GCT AC-3′, and the depurination primers were 5′-CTA TCG ATC CTT TAG TCC CTC-3′ and 5′-CCG AAT GAA CTG TTC CAC A-3′, respectively. The comparative threshold (ΔΔCT) method was used to calculate the depurination levels, and data were expressed as a fold change of depurination in Stx-treated RNA over depurination in control nontreated RNA, as described previously (30).
Monomeric ribosomes were isolated as previously described (26). The presence of the 10×His tags did not affect the activity of the A1 subunit of Stx1 or Stx2 (26). The 10×His-tagged WT Stx1A1 and Stx2A1 (1 nM) and 3-fold dilutions (9 nM, 3 nM, and 1 nM) of 10×His-tagged mutant proteins (R170A, R176A, and R172A/R176A) were added to 1× RIP buffer (60 mM KCl, 10 mM Tris-HCl [pH 7.4], 10 mM MgCl2) in a final volume of 100 μl. The reaction was started by adding 7 pmol of monomeric ribosomes and incubated at 30°C for 10 min. One hundred microliters of 2× extraction buffer (120 mM NaCl, 25 mM Tris-HCl [pH 8.8], 10 mM EDTA, 1% SDS) was added to stop the reaction. RNA was extracted with phenol and then phenol-chloroform and precipitated overnight with ethanol. Depurination was determined using quantitative reverse transcriptase PCR (qRT-PCR) (30).
Total RNA (1 μg) from yeast cells isolated using an RNeasy minikit (Qiagen, Valencia, CA) was incubated with 250 nM WT and mutant Stx1A1and Stx2A1 (R170A, R176A, and R172A/R176A) in a final volume of 20 μl in 20 mM citrate buffer (pH 5) at 37°C for 15 min. RNA was purified with phenol and then phenol-chloroform, and the depurination of rRNA was quantified using a qRT-PCR assay. The ΔΔCT method was used to calculate the depurination level, and data were expressed as the fold change of depurination in Stx-treated RNA over depurination in nontreated RNA, as described previously (30).
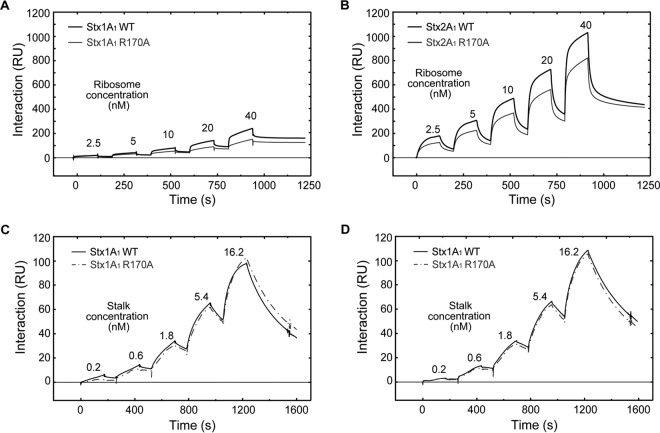
Interaction of Stx1A1 and Stx2A1 with yeast ribosomes and with the ribosomal-stalk pentamer.
Biacore T200 (GE Healthcare Bio-Sciences, Pittsburgh, PA) was used to study the interaction of 10×His-tagged A1 subunits with the ribosome using a nitrilotriacetic acid (NTA) chip. The amount of captured toxin monitored in real time by Biacore T200 was 800 ± 20 resonance units (RU). Ribosomes at different concentrations were passed over the surface at 30 μl/min for 2 min using the single-injection kinetic method. Dissociation was for 5 min. The running buffer contained 10 mM HEPES, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 50 μM EDTA, and 0.003% surfactant P20 (GE Healthcare Bio-Sciences, Pittsburgh, PA). The surface was freshly captured at each cycle. Interaction of the A1 subunits with the isolated ribosomal-stalk pentamer was analyzed by capturing 1,000 RU of 10×His-tagged toxins on an NTA chip and the same amount of 10×His-tagged enhanced green fluorescent protein (EGFP) on the reference channel. The running buffer was the same as the ribosome interaction buffer, except that it contained 10 mM MgCl2. The yeast stalk pentamer was passed over the surface at different concentrations at 30 μl/min for 3 min, and the final dissociation was for 5 min.
Transfection of Vero cells with the A1 subunits.
WT Stx1A1 (K1 to R251) (NT1776) and Stx2A1 (R1 to R250) (NT1777); Stx1A1 E167K (NT1796), R170A (NT1784), R172A (NT1804), R176A (NT1774), and R172A/R176A (NT1806) variants; and Stx2A1 E167K (NT1797), R170A (NT1785), R172A (NT1805), R176A (NT1775), and R172A/R176A (NT1807) variants with V5 and 10×His tags were cloned into the mammalian expression vector pCAGGs at the SacI-XhoI sites and cotransfected into Vero cells with an EGFP expression plasmid in pCAGGs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), as previously described (26). A Biotek plate reader was used to measure the EGFP fluorescence at 22 h after DNA exposure from the bottom of the plate with a 485/20-nm bandpass excitation filter and a 530/25-nm bandpass emission filter. Assays were performed in quadruplicate. The fluorescence measured in cells cotransfected with EGFP and empty vector was considered 100%, and the fluorescence in control cells lacking the EGFP plasmid was considered background.
Analysis of ribosome depurination in Vero cells.
Vero cells containing WT Stx1A1 (K1 to R251) (NT1776) and Stx2A1 (R1 to R250) (NT1777); Stx1A1 E167K (NT1796), R170A (NT1784), R172A (NT1804), R176A (NT1774), and R172A/R176A (NT1806) variants; or Stx2A1 E167K (NT1797), R170A (NT1785), R172A (NT1805), R176A (NT1775), and R172A/R176A (NT1807) variants were grown as described above. Cells were collected 23 h after DNA exposure. Total RNA was extracted using the RNeasy minikit (Qiagen, Valencia, CA) with on-column DNase treatment. A qRT-PCR assay was used to quantify depurination (30).
Isolation of rat liver ribosomes.
Monomeric ribosomes were isolated from rat livers as previously described (26). Briefly, livers were dissected from rats, rinsed in cold buffer A (20 mM HEPES-KOH, pH 7.6, 5 mM Mg acetate [MgOAc], 50 mM KCl, 10% glycerol) with 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), and protease inhibitor cocktail for mammalian tissue culture (Sigma-Aldrich, St. Louis, MO) and frozen in liquid N2. Thawed livers were homogenized in cold buffer A plus 1 mM PMSF, 1 mM DTT, and protease inhibitor cocktail for mammalian tissue culture (Sigma-Aldrich, St. Louis, MO) and centrifuged at 20,000 × g for 20 min. The supernatant was treated with 1% sodium deoxycholate for 10 min with constant stirring on ice and sedimented at 150,000 × g for 90 min. The pellet was rinsed twice in buffer B (20 mM HEPES-KOH, pH 7.6, 20 mM MgOAc, 0.5 M KCl, 10% glycerol) with 1 mM PMSF and 1 mM DTT and stored overnight at 4°C in a small amount of the same buffer. The ribosomes were resuspended with a Dounce homogenizer in buffer B with 1 mM DTT and 1 mM PMSF and incubated for 30 min at 30°C with 1 mM puromycin and 1 mM GTP. The supernatant was clarified by centrifugation at 20,000 × g for 15 min and layered over a cushion of buffer B with glycerol raised to 25%. Ribosomes were sedimented at 150,000 × g for 2 h. The pellets were rinsed in buffer C (50 mM HEPES-KOH, pH 7.6, 5 mM MgOAc, 50 mM NH4Cl, 0.1 mM DTT, and 25% glycerol), resuspended in 5 ml buffer C, and stored at −80°C. All animal experiments were conducted with the approval of the Rutgers University Animal Care and Use Committee.
Depurination of rat liver ribosomes.
Depurination analysis of rat liver ribosomes was performed as described for yeast. One nanomolar WT Stx1A1 and Stx2A1 and 3-fold dilutions (9 nM, 3 nM, and 1 nM) of mutant proteins (R170A, R176A, and R172A/R176A) were used in the treatment of 7 pmol of rat ribosomes. Depurination was analyzed by qRT-PCR (30).
Interaction of Stx1A1 and Stx2A1 with rat liver ribosomes.
The interaction of 10×His-tagged WT and Stx1A1 and Stx2A1 variants with rat liver ribosomes was examined using Biacore T200 (GE Healthcare Bio-Sciences, Pittsburgh, PA). The amount of toxin captured on an NTA chip monitored in real time was 1,800 ± 50 RU. Ribosomes at different concentrations were passed over the surface at 30 μl/min for 2 min using the single-injection kinetic method. Dissociation was for 5 min. The running buffer contained 10 mM HEPES, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 50 μM EDTA, and 0.01% surfactant P20. The surface was freshly captured at each cycle.
Statistical analysis.
Data (see Fig. 2 to 7) were analyzed by Student's two-sample t test using Origin software (OriginLab v.8.0; OriginLab, Northampton, MA). Statistical analyses of data (see Fig. S2A and C in the supplemental material) were conducted by using SAS 9.4 (SAS Institute, Inc., Cary, NC). Data were analyzed by generalized mixed linear models using PROC GLM (Procedure for Generalized Linear Models) to test for statistical differences between treatments. Least-square means were calculated, and specific preplanned contrasts (31) were computed to compare treatment means between Stx1A1 and Stx2A1 for each treatment.
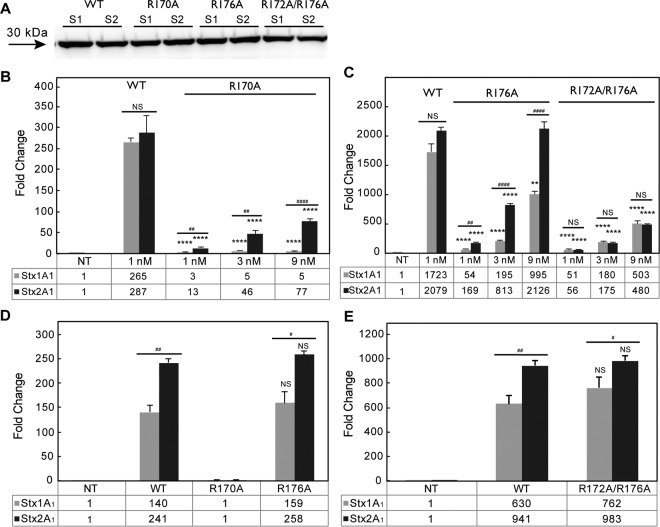
FIG 2.
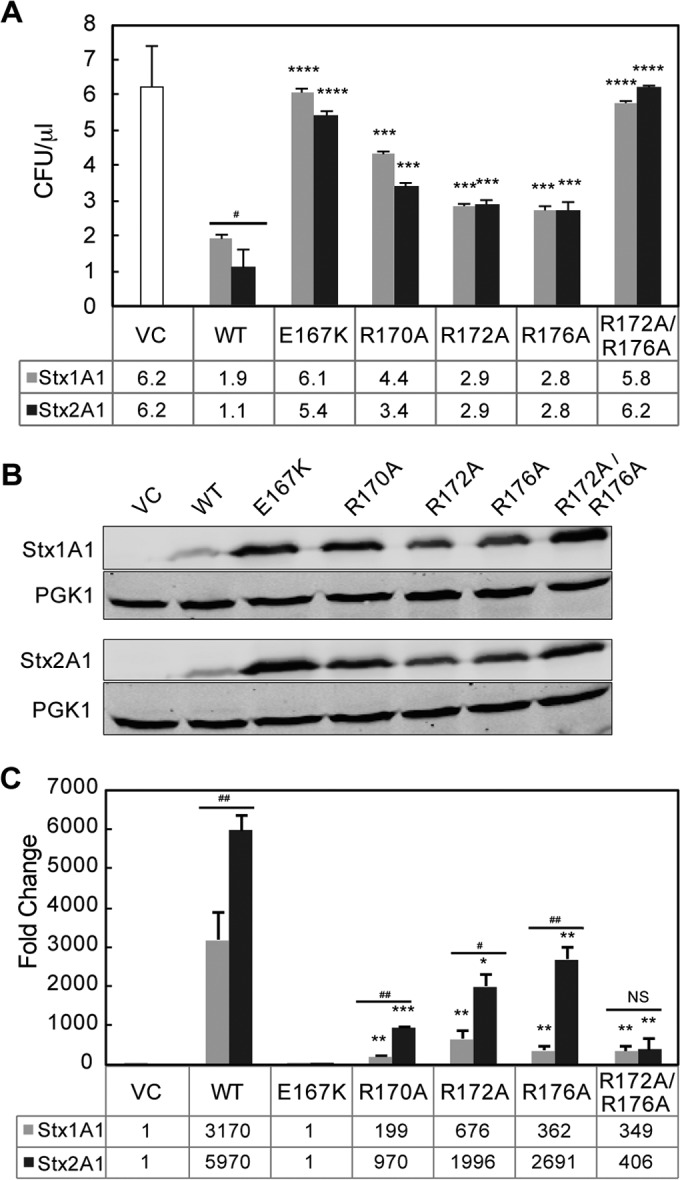
(A) Viability and ribosome depurination in yeast expressing WT or mutant Stx1A1 or Stx2A1. Yeast cells transformed with a plasmid carrying WT or mutant Stx1A1 or Stx2A1 or the empty vector (VC) were grown in SD medium supplemented with 2% glucose and then transferred to SD medium supplemented with 2% galactose. At 0 and 4 hpi, a series of 10-fold dilutions were plated on media containing 2% glucose and grown at 30°C for 1 to 2 days. The numbers of CFU per milliliter at 4 hpi were calculated from at least 3 independent transformants. The error bars represent standard errors (SE) (n = 3 independent experiments). The means of Stx1A1, Stx2A1, and their variants were significantly different using a two-sample t test (***, P < 0.001, ****, P < 0.0001 [means compared to the respective WT]; #, P < 0.05 [means compared between Stx1A1 and Stx2A1]). (B) Immunoblot analysis of yeast cells transformed with WT or mutant Stx1A1 or Stx2A1. Total protein from cells (OD600 = 5) isolated at 6 hpi was separated on an SDS-2% PAGE gel and probed with anti-V5. Anti-PGK1 was used as a loading control. (C) Depurination of ribosomes in yeast. Total RNA (375 ng) isolated from cells (OD600 = 1) expressing WT or mutant Stx1A1 or Stx2A1 collected at 1 hpi was used to quantify the relative level of depurination using qRT-PCR. The y axis shows the fold change in depurination of toxin-treated samples over the control samples without toxin (VC). The error bars represent SE (n = 3 replicates). The means of WT Stx1A1 and Stx2A1 and Stx1A1 and Stx2A1 variants were significantly different using a two-sample t test (*, P < 0.05, **, P < 0.01, ***, P < 0.001 [means compared to the respective WT]; #, P < 0.05, ##, P < 0.01 [means compared between Stx1A1 and Stx2A1]; NS, not significant).
FIG 7.
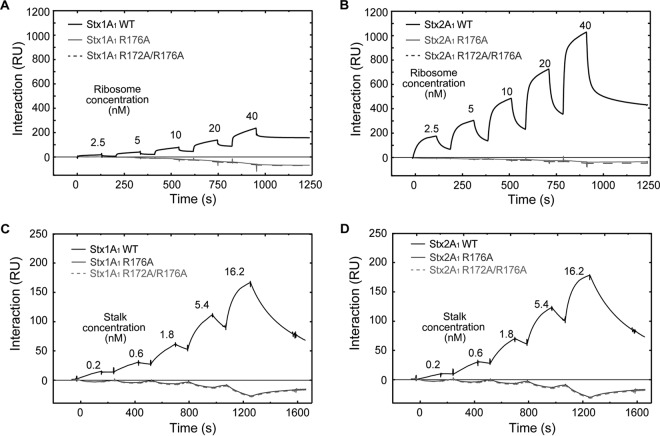
(A) Interaction of rat liver ribosomes with 10×His-tagged WT Stx1A1 and 10×His-tagged R176A and R172A/R176A variants. (B) Interaction of rat liver ribosomes with 10×His-tagged WT Stx2A1 and 10×His-tagged R176A and R172A/R176A variants. WT Stx1A1, WT Stx2A1, and R176A and R172A/R176A variants were captured on an NTA chip at 1,800 RU. Different concentrations of rat liver ribosomes were passed over the surface as analytes. (C) Interaction of rat liver ribosomes with 10×His-tagged WT Stx1A1 and 10×His-tagged R170A. (D) Interaction of rat liver ribosomes with 10×His-tagged WT Stx2A1 and 10×His-tagged R170A. 10×His-Stx1A1, 10×His-Stx2A1, and 10×His-R170A variants were captured on an NTA chip at 1,800 RU. Different concentrations of rat liver ribosomes were passed over the surface as analytes, as shown.
RESULTS
Arginines R172 and R176 are more important than R179 in the cytotoxicity and depurination activity of Stx1A1 and Stx2A1 in yeast.
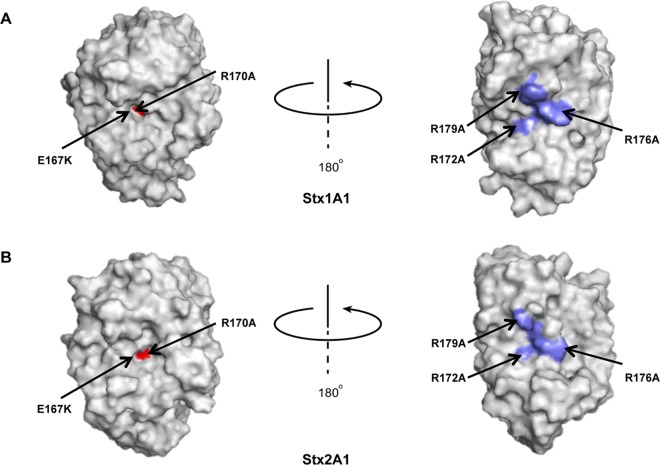
Comparison of the crystallographic structures of Stx1A1 and Stx2A1 (Fig. 1A and B) showed that R172, R176, and R179, previously shown to be involved in the interaction of Stx1A1 with P11 (24), are more exposed in Stx1A1 than in Stx2A1, suggesting that R172, R176, and R179 might play different roles in ribosome binding in Stx1A1 and Stx2A1. We therefore mutated R172, R176, and R179 to alanine in Stx1A1 and Stx2A1. We included mutations in the active-site residues Glu167 (E167K) and Arg170 (R170A) (Fig. 1A and B) as controls. The WT and mutant forms of Stx1A1 and Stx2A1 were expressed in yeast under the control of the GAL1 promoter, and viability was determined by induction on galactose for 4 h, followed by plating serial dilutions on media containing glucose. The numbers of CFU calculated based on the viability assay are shown in Fig. 2A and Fig. S1A in the supplemental material. WT Stx2A1 decreased yeast viability approximately 2-fold more than WT Stx1A1 (Fig. 2A; see S1A in the supplemental material). While there was a significant increase in the viability of R172A and R176A variants, the R179A variant did not show a significant increase in viability compared to the WT and was not analyzed further (see Fig. S1A in the supplemental material).
FIG 1.
Crystallographic structures of Stx1A1 and Stx2A1 showing the active site and the distal face of the active site. The A1 subunits of Shiga toxin and Shiga toxin 2 were modeled from the Protein Data Bank ID 1DM0 (Shiga toxin) and 1R4P (Shiga toxin 2). The active site of Stx1A1 (red) is more embedded (A) than the active site of Stx2A1 (B). Rotation about the y axis by approximately 180° reveals the conserved arginines (blue) in Stx1A1 (A) and in Stx2A1 (B).
To determine if mutation of more than one arginine would lead to a further reduction in toxicity, we mutated two or three different arginines at the distal face of the active site to alanine simultaneously and compared the viability of the mutants to that of the WT (see Fig. S1A in the supplemental material). The growth of double (R172A/R176A and R172A/R179A) and triple (R172A/R176A/R179A) arginine mutants was similar to that of the empty vector (see Fig. S1A in the supplemental material), indicating that simultaneous mutation of two arginines reduced toxicity to a minimal level in both toxins. These results showed that Arg179 is not as important as Arg172 and Arg176 but that it has an effect in reducing toxicity when combined with mutations at nearby arginines (see Fig. S1A in the supplemental material). Since there were no discernible differences between the growth rates of the double and triple mutants (see Fig. S1 in the supplemental material), R172A/R176A was selected for further analysis. The E167K mutation led to a total loss of cytotoxicity, while the R170A mutation reduced the toxicity of both Stx1A1 and Stx2A1 (Fig. 2A). Although Glu167 and Arg170 play critical roles in the catalytic activity of Shiga toxins, the complete loss of toxicity of E167K might be due to the drastic change from a negatively charged residue to a positively charged residue. In contrast, R170A retains some toxicity, possibly because the positive charge is replaced by a smaller neutral alanine (Fig. 2A).
The expression of arginine variants in yeast was analyzed by immunoblot analysis at 6 hpi with monoclonal antibodies against the V5 epitope (Fig. 2B). Monoclonal antibodies against phosphoglycerate kinase 1 (PGK1) were used as a loading control. As observed previously (26), the expression level of the arginine variants correlated inversely with their toxicity. All the mutants were expressed at higher levels than the WT, consistent with their lower toxicity (Fig. 2B).
In order to determine if there is a correlation between the reduced cytotoxicity and the depurination activity in yeast, the depurination activity of the arginine variants was examined using a previously described qRT-PCR assay (30). Total RNA was collected from yeast cells at 1 hpi, and depurination was determined relative to yeast harboring the empty vector using the ΔΔCT method. All the mutants showed a significant reduction in the depurination level compared to WT Stx1A1 and Stx2A1, except R179A (see Fig. S1B in the supplemental material). The E167K single mutation eliminated depurination, consistent with the viability assay (Fig. 2C). Stx2A1 containing the single R170A, R172A, or R176A mutation depurinated ribosomes at a significantly higher level than Stx1A1 containing the same mutations, while simultaneous mutations at Arg172 and Arg176 caused a similar reduction in the depurination level of Stx1A1 and Stx2A1 (Fig. 2C; see Fig. S1B in the supplemental material). These results showed that conserved arginines play a critical role in the toxicity and depurination activities of Stx1A1 and Stx2A1 in yeast. However, Arg170, Arg172, and Arg176 are more critical for the depurination activity and toxicity of Stx1A1 than of Stx2A1.
Arginine variants depurinate yeast ribosomes at a reduced level but depurinate yeast RNA at a level similar to that of WT Stx1A1 and Stx2A1.
To examine the depurination activities of the arginine mutants, 10×His-tagged WT and mutant forms of Stx1A1 and Stx2A1 were expressed in E. coli and purified. The dramatic change from glutamic acid to lysine likely affected folding and prevented the purification of Stx1A1 and Stx2A1 containing the E167K mutation. This mutant and Stx2A1 containing an R172A mutation could not be purified. Recombinant R170A, R176A, and R172A/R176A mutants and WT Stx1A1 and Stx2A1 were analyzed by immunoblot analysis using monoclonal antibodies against histidine (Qiagen, Valencia, CA). Each toxin migrated on SDS-PAGE as expected for its size (Fig. 3A).
FIG 3.
(A) Immunoblot analysis of purified 10×His-tagged WT Stx1A1 and Stx2A1 and 10×His-tagged R170A, R176A, and R172A/R176A variants. Equal amounts (1 μg) of purified Stx1A1 (S1) and Stx2A1 (S2) and variants were separated on a 12% SDS-polyacrylamide gel. 10×His-tagged Stx1A1 and Stx2A1 were detected by monoclonal anti-His. (B) Depurination of yeast ribosomes by 10×His-tagged WT Stx1A1 and Stx2A1 and R170A variants. (C) Depurination of yeast ribosomes by 10×His-tagged WT Stx1A1 and Stx2A1 and R176A and R172A/R176A variants. Yeast ribosomes (7 pmol) were incubated with different concentrations of 10×His-tagged WT or variant forms of Stx1A1 or Stx2A1 at 30°C for 10 min. The rRNA (375 ng) was used to quantify the relative levels of depurination by qRT-PCR. The y axis shows the fold change in depurination of toxin-treated samples over the control samples without toxin treatment (NT). The error bars represent SE (n = 3 replicates). The means of WT Stx1A1 and Stx2A1 and their variants were significantly different using a two-sample t test (**, P < 0.01, ****, P < 0.0001 [means compared to the respective WT]; ##, P < 0.01, ####, P < 0.0001 [means compared between Stx1A1 and Stx2A1]; NS, not significant). (D) Depurination of total RNA from yeast by purified Stx1A1, Stx2A1, and R170A and R176A variants. (E) Depurination of total RNA from yeast by purified Stx1A1, Stx2A1, and R172A/R176A variants. Total RNA (1 μg) was incubated with different amounts of 10×His-tagged WT Stx1A1 and Stx2A1 or 10×His-tagged R170A, R176A, and R172A/R176A variants at 37°C for 15 min. The relative levels of depurination were determined using qRT-PCR. The y axis shows the fold change in depurination of toxin-treated samples over the control samples without toxin treatment (NT). The error bars represent SE (n = 3 replicates). The means of Stx1A1, Stx2A1, and their variants were significantly different using a two-sample t test (#, P < 0.05, ##, P < 0.01 [means compared between Stx1A1 and Stx2A1]; NS, not significant).
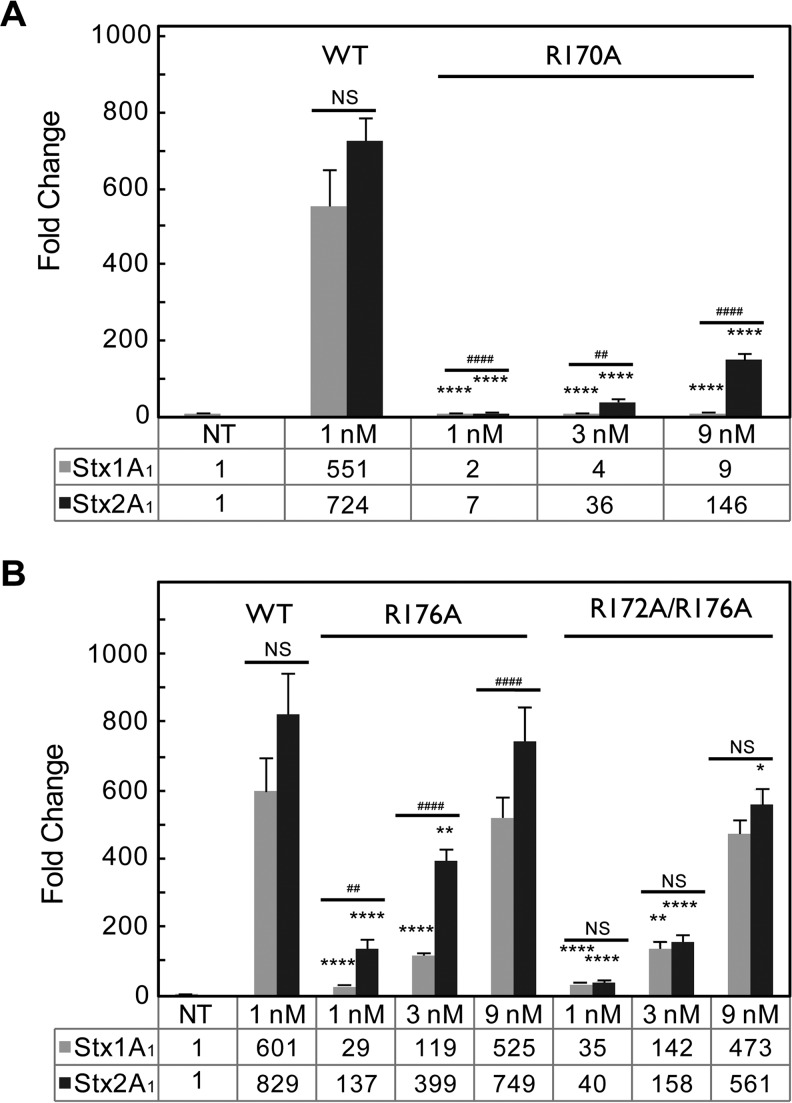
The depurination activity of recombinant Stx variants was examined using monomeric yeast ribosomes (Fig. 3B and C). The ribosomes were treated with 3-fold-higher doses of the mutant toxins in order to ascertain the linear range of increase in depurination relative to the toxin concentration. Depurination by the R170A variant was significantly reduced compared to WT Stx1A1 or Stx2A1 at all doses tested (Fig. 3B). Stx1A1 R176A and R172A/R176A had significantly lower depurination levels than the WT even at a 9 nM toxin concentration (Fig. 3C). In contrast, only Stx2A1 R172A/R176A had significantly lower depurination levels than the WT at a 9 nM toxin concentration (Fig. 3C). There was a highly significant reduction in depurination at the lower concentrations of Stx2A1 R176A, but the reduction in depurination was not observed when it was used at 9 nM (Fig. 3C). Stx2A1 variants containing single mutations depurinated ribosomes at a significantly higher level than Stx1A1 variants (Fig. 3B and C). The R172A/R176A double mutation resulted in comparable levels of depurination in Stx1A1 and Stx2A1 (Fig. 3C), indicating that mutation of both arginines is necessary to reduce the depurination activity of Stx2A1 to the same level as that of Stx1A1 (Fig. 3C).
When total yeast RNA was used as a substrate for depurination with purified toxins, Stx1A1 and Stx2A1 containing R176A and R172A/R176A mutations showed depurination activity similar to that of WT Stx1A1 and Stx2A1 (Fig. 3D and E). In contrast, the R170A mutant showed no activity on RNA, since the mutation was at the active site (Fig. 3D). WT Stx2A1 and Stx2A1 R176A and R172A/R176A depurinated RNA at a significantly higher level than WT Stx1A1 and Stx1A1 R176A and R172A/R176A, respectively, indicating that WT Stx2A1 and Stx2A1 R176A and R172A/R176A were more active. Therefore, the R176A and R172A/R176A mutations do not affect RNA binding or catalytic activity, providing evidence that the reduction in ribosome depurination is not due to a reduction in catalytic activity. In contrast, reduced catalytic activity is responsible for the reduction in ribosome depurination by R170A (Fig. 3D).
Arginine mutations affect the interaction of the A1 subunits with the ribosome and with the stalk pentamer in yeast.
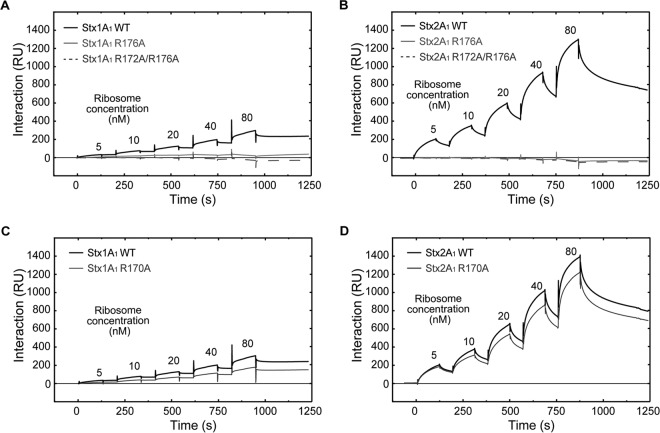
To determine if reduced depurination is due to reduced interaction with the ribosome, we examined the interaction of Stx variants with intact yeast ribosomes and with the purified stalk pentamer from yeast by surface plasmon resonance (SPR) using Biacore T200. In order to detect binding, 10×His-tagged WT Stx1A1, 10×His-tagged WT Stx2A1, and 10×His-tagged R176A and R172A/R176A variants were captured on an NTA chip at 800 RU. The same amount of 10×His-tagged EGFP was captured on the reference channel as a control (Fig. 4A and B). Since WT Stx1A1 and Stx2A1 and the variants were captured using the N-terminal 10×His, their binding sites were exposed. We could not detect binding of Stx1A1 and Stx2A1 containing R176A or R172A/R176A mutations to the ribosome at concentrations up to 40 nM (Fig. 4A and B). Binding was not detected even when R176A or R172A/R176A were captured at 2,500 RU. Our results were in agreement with a previous study, which showed that the R176A variant of Stx1A1 did not bind the P11 peptide by SPR when the biotinylated peptide was immobilized on the Biacore chip and the R176A variant was passed over the surface (24). These results highlighted the importance of the arginine residues at the distal face of the active site in the interaction of the A1 subunits with the ribosome. They also showed that Stx2A1 bound ribosomes at a higher level than Stx1A1, consistent with previous results (26).
FIG 4.
(A) Interaction of yeast ribosomes with 10×His-tagged WT Stx1A1 and 10×His-tagged R176A and R172A/R176A variants. (B) Interaction of yeast ribosomes with 10×His-tagged WT Stx2A1 and 10×His-tagged R176A and R172A/R176A variants. WT Stx1A1, WT Stx2A1, and R176A and R172A/R176A variants were captured on an NTA chip at 800 RU. Different concentrations of ribosomes were passed over the surface as analytes, as shown. (C) Interaction of the purified yeast stalk pentamer with 10×His-tagged WT Stx1A1 and 10×His-tagged R176A and R172A/R176A variants. (D) Interaction of the purified yeast stalk pentamer with 10×His-tagged WT Stx2A1 and 10×His-tagged R176A and R172A/R176A variants. WT Stx1A1 and Stx2A1 and R176A and R172A/R176A variants were captured on an NTA chip at 1,000 RU, and the same amount of EGFP was captured on the reference channel. Different concentrations of stalk pentamer were passed over the surface as analytes, as shown.
In order to examine the interaction of the Stx variants with the stalk pentamer, 10×His-tagged WT Stx1A1 and Stx2A1 and 10×His-tagged R176A and R172A/R176A variants were captured on different channels of an NTA chip at 1,000 RU, and the purified stalk pentamer from yeast was passed over the surface (Fig. 4C and D). The same amount of 10×His-tagged EGFP was captured on the reference channel as a control. R176 or R172/R176 mutations eliminated the interaction of Stx1A1 and Stx2A1 with the stalk pentamer, even at a 16.2 nM stalk pentamer concentration (Fig. 4C and D).
To determine if arginine-to-alanine conversion at the active site affected the interaction of Stx1A1 and Stx2A1 with the ribosome, we examined the interaction of the R170A variant with ribosomes and with the purified stalk pentamer. As shown in Fig. 5A and B, Stx1A1 R170A interacted with ribosomes at a 1.7-fold-lower level than the WT while Stx2A1 R170A bound ribosomes at a 1.3-fold-lower level than the WT when 40 nM yeast ribosomes were used. Since the interaction did not fit a 1:1 model, the association (kon) and dissociation (koff) rates could not be accurately determined. Instead, the binding level was used to calculate the equilibrium dissociation constant (KD) (Table 1). The apparent KD values of R170A variants were 1.2-fold higher than those of the WT for Stx1A1 and Stx2A1, and the difference was significant. These results indicate that a slight change in the overall charge of the toxin molecule due to a mutation at the active site affects its interaction with the ribosome (Fig. 5A and B). The apparent KD values for WT Stx1A1 and Stx1A1 R170A were 3-fold higher than the apparent KD values for WT Stx2A1 and Stx2A1 R170A, respectively, indicating that WT Stx2A1 and Stx2A1 R170A have higher affinity for the ribosome than WT Stx1A1 and Stx1A1 R170A, respectively.
FIG 5.
(A) Interaction of yeast ribosomes with 10×His-tagged WT Stx1A1 and 10×His-tagged R170A. (B) Interaction of yeast ribosomes with 10×His-tagged WT Stx2A1 and 10×His-tagged R170A. 10×His-Stx1A1, 10×His-Stx2A1, and 10×His-R170A variants were captured on an NTA chip at 800 RU. Different concentrations of ribosomes were passed over the surface as analytes, as shown. (C) Interaction of the purified yeast stalk pentamer with 10×His-tagged WT Stx1A1 and 10×His-tagged R170A. (D) Interaction of the purified yeast stalk pentamer with 10×His-tagged WT Stx2A1 and 10×His-tagged R170A. 10×His-Stx1A1, 10×His-Stx2A1, and 10×His-R170A variants were captured on an NTA chip at 1,000 RU, and the same amount of EGFP was captured on the reference channel. Different concentrations of the stalk pentamer were passed over the surface as analytes, as shown.
TABLE 1.
Apparent KD of the interaction of A1 subunits with ribosomes
| Toxin | Mean apparent KD (M) of ribosomesa |
|
|---|---|---|
| Yeast | Rat | |
| Stx1A1 | ||
| WT | 8.47 × 10−8 ± 0.15 × 10−8A | 9.39 × 10−8 ± 2.03 × 10−8E |
| R170A | 9.37 × 10−8 ± 0.23 × 10−8B | 11.71 × 10−8 ± 1.91 × 10−8F |
| Stx2A1 | ||
| WT | 2.89 × 10−8 ± 0.08 × 10−8C | 4.39 × 10−8 ± 0.15 × 10−8G |
| R170A | 3.45 × 10−8 ± 0.22 × 10−8D | 5.06 × 10−8 ± 0.83 × 10−8H |
The letters indicate statistical comparisons, where means were significantly different between A and B (P < 0.01), between C and D (P < 0.05), between B and D (P < 0.001), between E and F (P < 0.05), between G and H (P < 0.05), and between F and H (P < 0.001) as determined using a two-sample t test.
When the interaction of R170A variants was examined with the stalk pentamer, the interaction fit a 1:1 interaction model (Fig. 5C and D). Stx1A1 R170A showed slightly lower kon and koff than WT Stx1A1, which led to a slightly higher KD than the WT (Table 2). Stx2A1 R170A, had slightly higher kon and koff, but the KD was almost identical to that of WT Stx2A1 (Table 2). The affinity and association and dissociation rates of R170A variants for the stalk complex were not significantly different than those of the WT Stx1A1 and Stx2A1, indicating that the lower affinity of the R170A variants for the ribosome than WT Stx1A1 and Stx2A1 is not due to their interaction with the stalk pentamer.
TABLE 2.
Stalk interaction parametersa
| Toxin | kon (M−1 s−1) | koff (s−1) | KD (M) |
|---|---|---|---|
| Stx1A1 | |||
| WT | 1.53 × 106 ± 0.76 × 106A | 2.29 × 10−3 ± 0.56 × 10−3C | 1.50 × 10−9 ± 0.59 × 10−9E |
| R170A | 1.08 × 106 ± 0.55 × 106B | 2.04 × 10−3 ± 0.23 × 10−3D | 1.89 × 10−9 ± 0.52 × 10−9F |
| Stx2A1 | |||
| WT | 1.28 × 106 ± 0.23 × 106G | 2.34 × 10−3 ± 0.45 × 10−3I | 1.84 × 10−9 ± 0.21 × 10−9K |
| R170A | 1.39 × 106 ± 0.08 × 106H | 2.55 × 10−3 ± 0.28 × 10−3J | 1.83 × 10−9 ± 0.31 × 10−9L |
The letters indicate statistical comparisons, where means were not significantly different between A and B, between C and D, between E and F, between G and H, between I and J, and between K and L as determined using a two-sample t test.
Arginine mutations affect the interaction of the A1 subunits with mammalian ribosomes and depurination of mammalian ribosomes.
When purified rat liver ribosomes were treated with recombinant WT Stx1A1 and Stx2A1 and variants, all the mutants showed a significant reduction in depurination in comparison to the WT at 1 and 3 nM (Fig. 6A and B). Depurination by Stx1A1 R170 and R176A was reduced significantly more than by Stx2A1 R170A and R176A. In contrast, Stx1A1 and Stx2A1 R172A/R176A showed similar levels of reduction in depurination at the different concentrations tested. Stx2A1 R172A/R176A had significantly lower depurination than the WT even at 9 nM. These results indicate that mutation of arginines at the distal face of the active site reduce depurination of mammalian ribosomes by Stx1A1 and Stx2A1. Both arginines need to be mutated to reduce the depurination of mammalian ribosomes by Stx2A1 to the same level as that by Stx1A1.
FIG 6.
(A) Depurination of rat liver ribosomes by purified 10×His-tagged WT Stx1A1 and Stx2A1 and 10×His-tagged R170A variants. (B) Depurination of rat liver ribosomes by purified 10×His-tagged WT Stx1A1 and Stx2A1 and 10×His-tagged R176A and R172A/R176A variants. Rat liver ribosomes (7 pmol) were incubated with different concentrations of WT Stx1A1 or Stx2A1 or R170A, R176A, or R172A/R176A variants at 30°C for 10 min. Total rRNA (375 ng) was used to quantify the relative levels of depurination by qRT-PCR. The y axis shows the fold change in depurination of toxin-treated samples over the control samples without toxin treatment (NT). The error bars represent SE (n = 3 replicates). The means of WT Stx1A1 and Stx2A1 and their variants were significantly different using a two-sample t test (**, P < 0.01, ****, P < 0.0001 [means compared to the respective WT]; ##, P < 0.01, ####, P < 0.0001 [means compared between Stx1A1 and Stx2A1]; NS, not significant).
In order to determine if reduced depurination is due to reduced binding to rat liver ribosomes, the interaction of 10×His-tagged WT Stx1A1 and Stx2A1 and 10×His-tagged R176A and R172A/R176A variants with rat liver ribosomes was analyzed using Biacore T200. The A1 chains were captured on an NTA chip at 1,800 RU, and rat liver ribosomes were passed over the surface at different concentrations using single-cycle kinetics. The same amount of 10×His-tagged EGFP was captured on the reference channel as a control (Fig. 7A and B). As observed with yeast ribosomes, R176A and R172A/R176A variants failed to interact with rat liver ribosomes.
When interaction of 10×His-tagged R170A variants was examined with rat liver ribosomes, there was a 1.2-fold reduction in the binding levels of Stx1A1 and Stx2A1 R170A in comparison to WT Stx1A1 and Stx2A1 (Fig. 7C and D). This difference was significant (Table 1). These results demonstrated that conserved arginines at the distal face of the active site and Arg170 at the active site are critical for the interaction of Stx1A1 and Stx2A1 with rat liver ribosomes and highlighted the importance of the surface charge in the interaction of the A1 subunits with mammalian ribosomes.
DISCUSSION
Arginines on the distal face of the active site are critical for the depurination activity and toxicity of Stx1A1 and Stx2A1.
Our recent results indicated that there are differences in the biphasic interaction of Stx1A1 and Stx2A1 with the ribosome (26). Stx2A1 had a higher affinity for the ribosome and a higher catalytic activity than Stx1A1 in yeast and in mammalian cells. Examination of electrostatic surfaces of Stx1A1 and Stx2A1 indicated a number of differences (26). An ∼180° rotation along the y axis revealed the positively charged zone, which was composed of several arginines and was larger and more exposed in Stx1A1 than in Stx2A1 (Fig. 1) (26). To determine if these differences contribute to the higher affinity of Stx2A1 for the ribosome, we mutated Arg172, Arg176, and Arg179, previously shown to be critical for the interaction of Stx1A1 with P11 (24), and examined the effects of these mutations on ribosome binding, depurination activity, and toxicity. Point mutations at Arg179 did not show any obvious reduction in toxicity and depurination activity in either Stx1A1 or Stx2A1 in yeast (see Fig. S1A and B in the supplemental material). In contrast, R172A and R176A mutations led to a significant reduction in toxicity (Fig. 2A) and depurination activity (Fig. 2C). These results indicate that the three different arginines have different roles in the depurination activity and toxicity of Stx1A1 and Stx2A1. Point mutations at Arg172 or Arg176 showed greater reductions in Stx1A1 than in Stx2A1, indicating that they are more critical for the depurination activity and toxicity of Stx1A1 than of Stx2A1. These results demonstrate for the first time that arginines on the distal face of the active site are important for the depurination activity and toxicity of Stx1A1 and Stx2A1.
While there were significant differences in the depurination levels among the Stx1A1 and Stx2A1 mutants in yeast, this difference was not apparent in the cytotoxicity assay, indicating that the absolute level of depurination often does not correlate with cytotoxicity (32). In RTA, at least two arginines had to be mutated in order to perturb the interaction between the toxin and the stalk pentamer or the ribosome before a reduction in toxicity was observed (28). Similarly, in TCS, single point mutations reduced the interaction with the P2 protein while a triple mutation disrupted the interaction (12). The depurination levels of Stx2A1 single mutants on yeast (Fig. 3B and C) and rat liver ribosomes (Fig. 6A and B) were consistently higher than those of Stx1A1 single mutants. Unlike the single mutations, the double and triple arginine mutations had similar effects and caused a more drastic reduction in Stx2A1 than in Stx1A1 (Fig. 2; see Fig. S1 in the supplemental material).
When the effects of the mutations in conserved arginines were examined in Vero cells using an EGFP transfection assay (26), E167K and R172A/R176A variants showed significantly higher levels of EGFP fluorescence than the WT Stx1A1 and Stx2A1, indicating reduced inhibition of EGFP translation (see Fig. S2A in the supplemental material). Since protein levels of Stx1A1 and Stx2A1 were too low to detect by immunoblotting, RNA levels were measured by qRT-PCR. RNA expression levels of WT Stx1A1 and Stx2A1 and variants were similar in Vero cells (see Fig. S2B in the supplemental material). When total RNA from Vero cells was analyzed to determine the in vivo depurination levels, the depurination levels of all the Stx2A1 mutants were significantly reduced in comparison to WT Stx2A1. The R172A/R176A mutation reduced depurination by Stx2A1 to a level similar to that by Stx1A1 (see Fig. S2C in the supplemental material). These results indicate that conserved arginines on the distal face of the active site are important for the depurination activity of Stx2A1 and that mutation of more than one arginine is required to cause a substantial reduction in the toxicity and depurination activity of Stx2A1 in mammalian cells. A plausible reason might be that Stx2A1 has higher catalytic activity than Stx1A1, since it depurinates naked RNA more efficiently than Stx1A1 even in the absence of ribosomal proteins (Fig. 3D and E) (26). Further analysis is required to fully understand the basis for this variation.
Conserved arginines on the distal face of the active site are critical for the interaction of Stx1A1 and Stx2A1 with the ribosome and with the stalk pentamer.
We previously showed that the interaction of RTA with yeast ribosomes by SPR did not fit a 1:1 interaction model but rather followed a two-step binding model (27). We proposed that the non-stalk-specific electrostatic interactions with the ribosome increase the local concentration of RTA on the ribosome and promote its diffusion to the stalk. Electrostatic interactions with the P-protein stalk stimulate the depurination activity of RTA by orienting RTA toward the SRL (28). Stx1A1 and Stx2A1 interactions with yeast (Fig. 4 and 5) or rat liver ribosomes (Fig. 7) did not follow a 1:1 interaction model but instead fit a biphasic model with an initial fast association-dissociation phase followed by a slower association-dissociation phase. Stx2A1 had a faster association-dissociation pattern with ribosomes than Stx1A1 but did not show a significant difference in its interaction with the purified stalk pentamer compared to Stx1A1 (Fig. 4) (26). These results suggest that the differences in the interactions of Stx1A1 and Stx2A1 with the ribosome are not due to differences in their interactions with the stalk pentamer.
The R176A and R172A/R176A variants failed to show any interaction with the stalk pentamer or with ribosomes. Although the R176A and R172A/R176A mutations affected the depurination of ribosomes by Stx1A1 and Stx2A1 and abolished ribosome binding, they had no effect on their depurination activity when RNA was used as a substrate (Fig. 3D and E). Thus, the reduction in depurination activity of the arginine variants toward the ribosome is due to their inability to bind ribosomes and not to the loss of their catalytic activities. Although the R176A and R172A/R176A mutations caused similar reductions in the affinities of Stx1A1 and Stx2A1 for the ribosomal stalk, they had a differential effect on their depurination activities. These results indicate that conserved arginines at the distal face of the active site are critical for the interactions of Stx1A1 and Stx2A1 with the P-protein stalk, but not for the differences in their interactions with the ribosome.
Mutation of an arginine at the active site affects the binding affinity of Stx1A1 and Stx2A1 for the ribosome, but not for the stalk pentamer.
Early studies using the holotoxins identified Glu167 and Arg170 of the A1 subunits as important for catalytic activity (33–36). The corresponding Arg180 in RTA is proposed to promote cleavage of the adenine by protonating position N3 of the adenine (37–39), while the corresponding Glu177 polarizes a surrounding water molecule to produce a hydroxide ion that aids in the cleavage of adenine. In ricin, conversion of Glu177 to glutamine leads to a 180-fold loss in activity and conversion to an aspartate leads to an 80-fold reduction, while conversion to alanine leads to only a 20-fold loss (39, 40). It was proposed that the nearby Glu208 can access the SRL due to the small size of the alanine side chain (41). The conversion of Glu167 to aspartate led to only a 3-fold reduction in Stx1 (36). However, conversion to a positively charged lysine led to a total loss of toxicity. Stx1A1 and Stx2A1 containing the E167K mutation had viability and depurination patterns similar to those of the empty vector. We were unable to purify this protein, probably as a result of the drastic change in folding of the active site. Conversion of Arg170 to alanine led to a 16-fold reduction in the depurination activity of Stx1A1 and a 6-fold reduction in the depurination activity of Stx2A1. Stx1A1 R170A had 4.9-fold-lower depurination activity than Stx2A1 R170A (Fig. 2). We have reported that the region around the active site of Stx2A1 is neutral in comparison to a negatively charged region around the active site of Stx1A1 (26). This difference in the surface charge may provide better access of Stx2A1 to the SRL, leading to the observed difference in depurination activity. We examined the effect of the R170A mutation on the interaction of Stx1A1 and Stx2A1 with yeast (Fig. 5A and B) and rat liver ribosomes (Fig. 7C and D). The association and dissociation patterns of this mutant were slightly lower than those of the WT, and the apparent KD values of both toxins were significantly different than that of the WT (Table 1). We did not observe a significant effect of the R170A mutation on the interaction of either toxin with the purified stalk pentamer (Table 2) but observed differences in their interactions with the ribosome. These results indicate that a charge difference at a site away from the ribosomal stalk binding region affects the binding affinity of the toxins for the ribosome, but not the stalk pentamer, suggesting that Arg170 contributes to the accumulation of the toxins on the ribosome.
In summary, we used site-directed mutagenesis to identify the roles of the arginine residues on the distal face of the active site and at the active site in the interaction of Stx1A1 and Stx2A1 with ribosomes. Our results indicate that mutation of more than one arginine is required to reduce the toxicity and catalytic activity of Stx2A1 significantly in comparison to Stx1A1 in both yeast and mammalian cells. Arg176 and Arg172 play roles in the stalk-dependent interactions of Stx1A1 and Stx2A1 with the ribosome, while Arg170 enhances non-stalk-specific interactions with the ribosome. These results demonstrate that conserved arginines at the stalk binding site or at the active site are critical for the interaction of Stx1A1 and Stx2A1 with the ribosome and their depurination activity and toxicity, but they do not contribute to the higher affinity of Stx2A1 for the ribosome. Calculation of the electrostatic surfaces of Stx1A1 and Stx2A1 showed a number of differences (26). Stx1A1 has a pronounced negatively charged knob composed of Glu60 and Glu61, which is missing in Stx2A1, as the corresponding residues are Tyr60 and Gln61. Stx2A1 has a negatively charged region consisting of Glu215 and Asp216, while Stx1A1 has Gly215 and Gln216 forming a neutral projection. Further studies will address whether these differences in surface charge contribute to the higher affinity of Stx2A1 than Stx1A1 for the ribosome. We demonstrate here that the depurination activities and toxicity of Stx1A1 and Stx2A1 are reduced by inhibiting their interactions with the ribosome, identifying toxin-ribosome interactions as a new target for the design of inhibitors of STEC infection.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Marek Tchórzewski and Przemyslaw Grela for providing the purified stalk pentamer from yeast, Karen Chave (Northeast Biodefense Center Protein Synthesis Core, U54-AI057158-Lipkin) for purification of the Shiga toxin A1 subunits, and John McLaughlin for assistance with the statistical analysis.
The work was supported by National Institutes of Health grants AI092011 and AI072425 to Nilgun E. Tumer.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00630-16.
REFERENCES
- 1.Siegler R, Oakes R. 2005. Hemolytic uremic syndrome; pathogenesis, treatment, and outcome. Curr Opin Pediatr 17:200–204. doi: 10.1097/01.mop.0000152997.66070.e9. [DOI] [PubMed] [Google Scholar]
- 2.Boerlin P, McEwen S, Boerlin-Petzold F, Wilson J, Johnson R, Gyles C. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J Clin Microbiol 37:497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGannon CM, Fuller CA, Weiss AA. 2010. Different classes of antibiotics differentially influence Shiga toxin production. Antimicrob Agents Chemother 54:3790–3798. doi: 10.1128/AAC.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimmitt PT, Harwood CR, Barer MR. 2000. Toxin gene expression by Shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg Infect Dis 6:458. doi: 10.3201/eid0605.000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086. [DOI] [PubMed] [Google Scholar]
- 6.Bergan J, Lingelem ABD, Simm R, Skotland T, Sandvig K. 2012. Shiga toxins. Toxicon 60:1085–1107. doi: 10.1016/j.toxicon.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 7.Sandvig K, van Deurs B. 2005. Delivery into cells: lessons learned from plant and bacterial toxins. Gene Ther 12:865–872. doi: 10.1038/sj.gt.3302525. [DOI] [PubMed] [Google Scholar]
- 8.Endo Y, Tsurugi K, Yutsudo T, Takeda Y, Ogasawara T, Igarashi K. 1988. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. Eur J Biochem 171:45–50. doi: 10.1111/j.1432-1033.1988.tb13756.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Feng S, Kumar V, Ero R, Gao YG. 2013. Structure of EF-G-ribosome complex in a pretranslocation state. Nat Struct Mol Biol 20:1077–1084. doi: 10.1038/nsmb.2645. [DOI] [PubMed] [Google Scholar]
- 10.Clementi N, Chirkova A, Puffer B, Micura R, Polacek N. 2010. Atomic mutagenesis reveals A2660 of 23S ribosomal RNA as key to EF-G GTPase activation. Nat Chem Biol 6:344–351. doi: 10.1038/nchembio.341. [DOI] [PubMed] [Google Scholar]
- 11.Shi X, Khade PK, Sanbonmatsu KY, Joseph S. 2012. Functional role of the sarcin-ricin loop of the 23S rRNA in the elongation cycle of protein synthesis. J Mol Biol 419:125–138. doi: 10.1016/j.jmb.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan DS, Chu LO, Lee KM, Too PH, Ma KW, Sze KH, Zhu G, Shaw PC, Wong KB. 2007. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res 35:1660–1672. doi: 10.1093/nar/gkm065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiou JC, Li XP, Remacha M, Ballesta JP, Tumer NE. 2008. The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae. Mol Microbiol 70:1441–1452. doi: 10.1111/j.1365-2958.2008.06492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCluskey AJ, Poon GM, Bolewska-Pedyczak E, Srikumar T, Jeram SM, Raught B, Gariepy J. 2008. The catalytic subunit of Shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. J Mol Biol 378:375–386. doi: 10.1016/j.jmb.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Too PH, Ma MK, Mak AN, Wong YT, Tung CK, Zhu G, Au SW, Wong KB, Shaw PC. 2009. The C-terminal fragment of the ribosomal P protein complexed to trichosanthin reveals the interaction between the ribosome-inactivating protein and the ribosome. Nucleic Acids Res 37:602–610. doi: 10.1093/nar/gkn922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 17.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell 118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, McAlear MA, Moore PB, Noller HF, Ortega J, Panse VG, Ramakrishnan V, Spahn CM, Steitz TA, Tchorzewski M, Tollervey D, Warren AJ, Williamson JR, Wilson D, Yonath A, Yusupov M. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol 24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalo P, Reboud JP. 2003. The puzzling lateral flexible stalk of the ribosome. Biol Cell 95:179–193. doi: 10.1016/S0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 20.Krokowski D, Boguszewska A, Abramczyk D, Liljas A, Tchorzewski M, Grankowski N. 2006. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol Microbiol 60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- 21.Choi AK, Wong EC, Lee KM, Wong KB. 2015. Structures of eukaryotic ribosomal stalk proteins and its complex with trichosanthin, and their implications in recruiting ribosome-inactivating proteins to the ribosomes. Toxins 7:638–647. doi: 10.3390/toxins7030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li XP, Grela P, Krokowski D, Tchorzewski M, Tumer NE. 2010. Pentameric organization of the ribosomal stalk accelerates recruitment of ricin a chain to the ribosome for depurination. J Biol Chem 285:41463–41471. doi: 10.1074/jbc.M110.171793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grela P, Li XP, Tchorzewski M, Tumer NE. 2014. Functional divergence between the two P1-P2 stalk dimers on the ribosome in their interaction with ricin A chain. Biochem J 460:59–67. doi: 10.1042/BJ20140014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCluskey A, Bolewska-Pedyczak E, Jarvik N, Chen G, Sidhu S, Johannes L. 2012. Charged and hydrophobic surfaces on the A chain of Shiga-like toxin 1 recognize the C-terminal domain of ribosomal stalk proteins. PLoS One 7:e31191. doi: 10.1371/journal.pone.0031191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiou J-C, Li X-P, Remacha M, Ballesta JP, Tumer NE. 2011. Shiga toxin 1 is more dependent on the P proteins of the ribosomal stalk for depurination activity than Shiga toxin 2. Int J Biochem Cell Biol 43:1792–1801. doi: 10.1016/j.biocel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basu D, Li X-P, Kahn JN, May KL, Kahn PC, Tumer NE. 2016. The A1 subunit of Shiga toxin 2 has higher affinity for ribosomes and higher catalytic activity than the A1 subunit of Shiga toxin 1. Infect Immun 84:149–161. doi: 10.1128/IAI.00994-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li XP, Chiou JC, Remacha M, Ballesta JP, Tumer NE. 2009. A two-step binding model proposed for the electrostatic interactions of ricin a chain with ribosomes. Biochemistry 48:3853–3863. doi: 10.1021/bi802371h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li XP, Kahn PC, Kahn JN, Grela P, Tumer NE. 2013. Arginine residues on the opposite side of the active site stimulate the catalysis of ribosome depurination by ricin A chain by interacting with the P-protein stalk. J Biol Chem 288:30270–30284. doi: 10.1074/jbc.M113.510966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang T, Lei J, Yang H, Xu K, Wang R, Zhang Z. 2011. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast 28:795–798. doi: 10.1002/yea.1905. [DOI] [PubMed] [Google Scholar]
- 30.Pierce M, Kahn JN, Chiou J, Tumer NE. 2011. Development of a quantitative RT-PCR assay to examine the kinetics of ribosome depurination by ribosome inactivating proteins using Saccharomyces cerevisiae as a model. RNA 17:201–210. doi: 10.1261/rna.2375411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steel RG, James H, Dickey DA, James HT, David AD. 1997. Principles and procedures of statistics: a biometrical approach. McGraw-Hill series in probability and statistics. McGraw-Hill, New York, NY. [Google Scholar]
- 32.Yan Q, Li XP, Tumer NE. 2012. N-glycosylation does not affect the catalytic activity of ricin A chain but stimulates cytotoxicity by promoting its transport out of the endoplasmic reticulum. Traffic 13:1508–1521. doi: 10.1111/j.1600-0854.2012.01404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deresiewicz RL, Calderwood SB, Robertus JD, Collier RJ. 1992. Mutations affecting the activity of the Shiga-like toxin I A-chain. Biochemistry 31:3272–3280. doi: 10.1021/bi00127a032. [DOI] [PubMed] [Google Scholar]
- 34.Cao C, Kurazono H, Yamasaki S, Kashiwagi K, Igarashi K, Takeda Y. 1994. Construction of mutant genes for a non-toxic verotoxin 2 variant (VT2vp1) of Escherichia coli and characterization of purified mutant toxins. Microbiol Immunol 38:441–447. doi: 10.1111/j.1348-0421.1994.tb01805.x. [DOI] [PubMed] [Google Scholar]
- 35.Di R, Kyu E, Shete V, Saidasan H, Kahn PC, Tumer NE. 2011. Identification of amino acids critical for the cytotoxicity of Shiga toxin 1 and 2 in Saccharomyces cerevisiae. Toxicon 57:525–539. doi: 10.1016/j.toxicon.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hovde CJ, Calderwood SB, Mekalanos JJ, Collier RJ. 1988. Evidence that glutamic acid 167 is an active-site residue of Shiga-like toxin I. Proc Natl Acad Sci U S A 85:2568–2572. doi: 10.1073/pnas.85.8.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho M-C, Sturm MB, Almo SC, Schramm VL. 2009. Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins. Proc Natl Acad Sci U S A 106:20276–20281. doi: 10.1073/pnas.0911606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y, Robertus JD. 1992. Analysis of several key active site residues of ricin A chain by mutagenesis and X-ray crystallography. Protein Eng 5:775–779. doi: 10.1093/protein/5.8.775. [DOI] [PubMed] [Google Scholar]
- 39.Schlossman D, Withers D, Welsh P, Alexander A, Robertus J, Frankel A. 1989. Role of glutamic acid 177 of the ricin toxin A chain in enzymatic inactivation of ribosomes. Mol Cell Biol 9:5012–5021. doi: 10.1128/MCB.9.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ready MP, Kim Y, Robertus JD. 1991. Site-directed mutagenesis of ricin A-chain and implications for the mechanism of action. Proteins 10:270–278. doi: 10.1002/prot.340100311. [DOI] [PubMed] [Google Scholar]
- 41.Frankel A, Welsh P, Richardson J, Robertus J. 1990. Role of arginine 180 and glutamic acid 177 of ricin toxin A chain in enzymatic inactivation of ribosomes. Mol Cell Biol 10:6257–6263. doi: 10.1128/MCB.10.12.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.