Abstract
Neisseria meningitidis, the meningococcus, bears the potential to cause life-threatening invasive diseases, but it usually colonizes the nasopharynx without causing any symptoms. Within the nasopharynx, Neisseria meningitidis must face temperature changes depending on the ambient air temperature. Indeed, the nasopharyngeal temperature can be substantially lower than 37°C, the temperature commonly used in experimental settings. Here, we compared the levels of meningococcal biofilm formation, autoaggregation, and cellular adherence at 32°C and 37°C and found a clear increase in all these phenotypes at 32°C suggestive of a stronger in vivo colonization capability at this temperature. A comparative proteome analysis approach revealed differential protein expression levels between 32°C and 37°C, predominantly affecting the bacterial envelope. A total of 375 proteins were detected. Use of database annotation or the PSORTb algorithm predicted 49 of those proteins to be localized in the outer membrane, 21 in either the inner or outer membrane, 35 in the periplasm, 56 in the inner membrane, and 208 in the cytosol; for 6 proteins, no annotation or prediction was available. Temperature-dependent regulation of protein expression was seen particularly in the periplasm as well as in the outer and inner membranes. Neisserial heparin binding antigen (NHBA), NMB1030, and adhesin complex protein (ACP) showed the strongest upregulation at 32°C and were partially responsible for the observed temperature-dependent phenotypes. Screening of different global regulators of Neisseria meningitidis suggested that the extracytoplasmic sigma factor σE might be involved in temperature-dependent biofilm formation. In conclusion, subtle temperature changes trigger adaptation events promoting mucosal colonization by meningococci.
INTRODUCTION
Microorganisms constantly adapt to changing environmental conditions. These changes include oxygen and nutrient availability, osmotic conditions, and temperature. The physiology of bacterial adaptation, including the underlying regulatory mechanisms that operate in response to radical temperature changes such as classical heat shock or cold shock, has been studied intensely (1–3). The heat shock response occurs during a transient upshift of growth temperature from 37°C to approximately 50°C (4). The heat shock response is controlled by the alternative sigma factor σ32, which regulates the expression of heat shock proteins mainly involved in protein folding or processing (4). In Escherichia coli, the cold shock response is triggered by a transient downshift from 37°C to 10°C that leads to generally slower transcriptional and translational responses except for about 26 upregulated proteins (5). The functions of these so-called cold shock proteins are less well understood than those of heat shock proteins, but they are likely involved in securing proper transcription, translation, and protein folding (5). Surprisingly little is known about the influence on bacterial physiology of slight temperature differences within mammalian hosts such as temperature gradients between the core temperature and that of mucosal surfaces. This, however, might be of particular importance for human-restricted bacterial pathogens that colonize mucosal surfaces before causing systemic disease.
Neisseria meningitidis (the meningococcus) is a commensal of the human nasopharynx which in rare cases causes invasive diseases such as sepsis or meningitis (6). Since N. meningitidis exclusively colonizes humans, it needs efficient between-host transmission and host colonization strategies to avoid eradication from the human population. As with other human-specific bacteria, the biology of N. meningitidis and its interaction with the host are commonly investigated in vitro, e.g., in tissue culture infection models and biofilm assays (7–9). Traditionally, almost all aspects of the biology of N. meningitidis have been analyzed at 37°C on the basis of the assumption of thermal homogeneity throughout the body. This assumption is not entirely correct, particularly considering the primary niche of N. meningitidis in the upper respiratory tract. A steep temperature gradient in the nasal airways was found in a study by Keck et al. (10), with an actual nasopharyngeal temperature of 33°C ± 2°C at a controlled surrounding temperature of 25°C ± 0.5°C. Therefore, in order to understand the mechanisms of meningococcal colonization, it is of interest to study phenotypic changes due to adaptation to temperatures marginally below 37°C.
To address the question how the slightly reduced growth temperature (i.e., 32°C) observed in the human nasopharynx impacts the physiology of N. meningitidis, we tested N. meningitidis for early biofilm formation, autoaggregation, and adherence to human nasopharynx epithelial cells. Furthermore, we compared the proteomes of N. meningitidis grown at the two temperatures by state-of-the-art metabolic labeling with stable nitrogen isotopes and mass spectrometry (MS) (11).
MATERIALS AND METHODS
Strains and culture conditions.
Meningococcal strains and mutants used in this study are listed in Table S2 in the supplemental material. In addition to meningococci, the N. lactamica strains 020-06 (12) and Y92-1009 (13) as well as N. gonorrhoeae strain FA1090 (14) were used. All Neisseriae strains were grown using Columbia blood agar plates or GC agar (Becton Dickinson) supplemented with PolyViteX (bioMérieux) at 37°C and 5% CO2 in a water-saturated atmosphere. For selection of transformants, GC agar was appropriately supplemented with 7 μg/ml chloramphenicol, 100 μg/ml kanamycin, 7 μg/ml erythromycin, or 100 μg/ml spectinomycin. For all comparisons of temperature-induced phenotypes, N. meningitidis strains were grown at 32°C versus 37°C in anaerobic jars fitted with a CO2 generation sachet (Oxoid). For liquid growth and growth curve analyses, strains were then resuspended in modified Neisseria defined medium (MDM), consisting of Neisseria defined medium (NDM) (7) supplemented with 5 mM NaHCO3, 10% proteose–peptone medium (PPM), and PolyViteX. Preparatory cultures were started at an optical density at 600 nm (OD600) of 0.25 in MDM and grown for 2 h at 32°C or 37°C. Then, the OD600 was adjusted to 0.15 in 10 ml of fresh prewarmed MDM and the cultures were grown with shaking at 200 rpm and 32°C or 37°C. The OD600 was recorded every 30 min.
Deletion mutants of NMB1030, NMB2095 (adhesin complex protein [ACP]), NMB2132 (neisserial heparin binding antigen [NHBA]), NMB1969 (NalP), NMB2091, NMB2144 (RpoE, σE), NMB2145 (MseR), NMB0018 (PilE), and NMB0052 (PilT).
Deletion mutants in N. meningitidis strain MC58 of NMB1030, NMB2095 (ACP), NMB2132 (NHBA), NMB2091, NMB2144 (σE), and NMB2145 (anti-sigma factor E, MseR) were processed following the same principle. First, approximately 500-bp sequences upstream and downstream of the coding sequence of the targeted genes were amplified by PCR. Then, these fragments were cloned into pBluescript II SK(+) vector (Invitrogen). The resulting plasmid was restricted between the upstream and downstream sequence followed by the ligation of a restricted kanamycin or erythromycin resistance cassette. The resulting plasmid containing the kanamycin or erythromycin cassette flanked by the upstream and downstream regions of the target gene was confirmed by PCR and sequencing. Wild-type MC58 and capsule-deficient (Δcsb) MC58 were transformed with the respective knockout plasmids. Resulting clones were verified by PCR and Southern blot hybridization.
To create a deletion mutation in NMB0018 (pilE) and NMB0052 (pilT), the corresponding gene was amplified by PCR and cloned into the pTL1 vector, a derivative of the pBluescript vector (Invitrogen) containing the Neisseria uptake sequence (15). An inverse PCR was conducted with outward-facing primers introducing AvrII restriction sites. A spectinomycin resistance cassette was inserted via the AvrII sites by replacing ∼200 bp of the coding sequence. The resultant constructs were amplified in E. coli TOP10 before transformation of the plasmid into N. meningitidis. The resulting MC58ΔpilE and MC58ΔpilT mutant strains were confirmed by PCR, sequencing, and Western blot analysis.
For all mutants, the presence of Opa, Opc, and pili was verified by Western blotting; all strains expressed the opaD allele, except for the MC58ΔcsbΔNHBA mutant, which expressed the opaB allele as analyzed by reverse transcription-PCR (RT-PCR) followed by sequencing.
All primers and restriction sites and positioning of the primers corresponding to the MC58 genome are listed in Table S3 in the supplemental material. All plasmids constructed for the knockouts are listed in Table S4.
Quantification of capsule or pilin expression by whole-cell ELISA.
N. meningitidis (MC58 and MC58Δcsb) strains were grown at 32°C versus 37°C as outlined above, fixed for 30 min in phosphate-buffered saline (PBS) containing 4% paraformaldehyde (PFA), and then thoroughly washed. The suspensions were adjusted to an OD600 of 0.50 and then diluted 1:5 (OD of 0.1) or 1:50 (OD of 0.01) or 1:500 (OD of 0.001). Volumes of 50 μl per well of these dilutions, or of PBS as a blank control, were dried on 96-well enzyme-linked immunosorbent assay (ELISA) plates (Nunc) in duplicate for coating. After washing (PBS–0.05% Tween 20) and blocking (PBS–1% bovine serum albumin [BSA]), wells were probed with 8 μg/ml serogroup B capsule-specific monoclonal antibody 735 (mAb735) (16) or pilin-specific monoclonal antibody SM1 (17), followed by incubation with goat anti-mouse horseradish peroxidase (HRP) conjugate (Jackson ImmunoResearch) at 400 ng/ml in blocking buffer. To ensure equal levels of N. meningitidis coating in the wells, duplicate ELISA plates were probed with a 1:1,000 dilution of a polyclonal rabbit antiserum against N. meningitidis MC58, followed by goat anti-rabbit HRP detection. TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Pierce) was used for detection. The capsule-specific signal was normalized to the anti-MC58 signal by dividing values of blank-corrected OD reads of capsule-specific staining by those of blank-corrected OD reads of the MC58-specific staining. To avoid saturation artifacts, only those N. meningitidis coating dilutions that yielded final OD reads between 0.5 and 1.0 were taken into account.
Quantification of capsule expression by flow cytometry.
PFA-fixed N. meningitidis (MC58, MC58Δcsb mutant) suspensions were prepared as described above after overnight growth at 32°C, 37°C, or 42°C. Suspensions (50 μl) at an OD of 0.50 were blocked by addition of 1% BSA, and the capsule was probed with mAb735 (80 μg/ml) for 1 h at room temperature, followed by two wash steps and detection with goat anti-mouse Alexa 488 (Jackson ImmunoResearch) (10 μg/ml). After two wash steps, pellets were resuspended in 300 μl PBS, flow cytometry was performed using a FACSCalibur analyzer, and data were analyzed with CellQuestPro software.
Immunofluorescence microscopy for pilin expression.
N. meningitidis strains (MC58, MC58ΔpilE, and MC58ΔpilT) were grown at 32°C versus 37°C as outlined above. Culture material was carefully taken up with disposable inoculation loops, dispersed in 1 μl PBS on glass slides, and air dried. After heat fixation, samples were blocked with PBS–5% BSA for 1 h at room temperature before addition of 30 μl of a 1:100 dilution of anti-pilin antibody SM1 (17) along with a 1:1,000 dilution of rabbit antiserum against MC58 in PBS–5% BSA for 1 h. Samples were washed twice in PBS–0.05% Tween 20 and once in plain PBS. Then, samples were incubated with 30 μl goat anti-mouse Cy3 and goat anti-rabbit Alexa 488 (Jackson ImmunoResearch), each at 4 μg/ml, in PBS–5% BSA for 1 h. After washing was performed, samples were mounted with Fluoroshield (Sigma-Aldrich) and immunofluorescence microscopy was performed using a Keyence BZ-9000 digital microscope (Keyence) at 100-fold magnification.
Lipopolysaccharide (LPS) detection by silver staining following polyacrylamide gel separation.
Sample preparation was done using a method similar to that described in reference 18. MC58 was grown at 32°C versus 37°C as outlined above. A 1-ml suspension in PBS at an OD of 0.6 was pelleted at 13,000 × g for 1 min, and pellets were taken up in 50 μl lysis buffer (2% SDS, 4% β-mercaptoethanol, 10% glycerol, 1 M Tris [pH 6.8], bromophenol blue). The samples were boiled for 10 min and then cooled to 60°C. Then, 10 μl of proteinase K (Sigma-Aldrich) solution (2.5 mg/ml in lysis buffer) was added and samples were incubated at 60°C for 1 h. Equal volumes of sample and 2× Tricine sample solution (4% SDS, 12% glycerol, 50 mM Tris [pH 6.8], 2% β-mercaptoethanol, 0.01% Serva Blue G [Serva]) were mixed and boiled for 5 min, and then 3 μl per lane was loaded onto a Tricine-buffered polyacrylamide gel (16.5% acrylamide, 6% bis-acrylamide) as outlined in reference 19. After electrophoresis, gels were stained by silver staining following the protocol described in reference 20. Gels were photographed using a Bio-Rad ChemiDoc MP imaging system.
Aggregation assay.
To assess meningococcal autoaggregation, the protocol described in reference 21 was used. Bacteria were grown overnight at either 32°C or 37°C as outlined above and then thoroughly resuspended in 4 ml PBS at an OD600 of 2.0. Subsequently, the suspensions were incubated at room temperature without shaking and suspensions were sampled at 30-min intervals at the liquid-air interface to measure the OD at 600 nm.
Early biofilm formation.
Early biofilm formation was assessed in modified NDM (7) plus 10% PPM in 24-well cell culture dishes. A volume consisting of 1 ml containing 108 CFU of N. meningitidis was seeded per well to initiate biofilm formation. After 1 h, all growth medium was removed. To stain the adherent biomass, 1 ml of 0.05% crystal violet (CV) (Difco) was added for 10 min. After two wash steps were performed with 2 ml of PBS each, bound CV was dissolved in 96% ethanol and quantified by measuring optical density at 595 nm.
Infection of epithelial cells.
Human pharyngeal cell lines Detroit 562 and FaDu were used for infection assays. FaDu cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and Detroit cells in DMEM supplemented with 10% FBS, 1% nonessential amino acids, 2 mM l-glutamine, and 1 mM sodium pyruvate at 37°C and 5% CO2 in a water-saturated atmosphere. All cell culture reagents were purchased from Life Technologies.
For the infection assay, 5 × 105 Detroit 562 cells or 4 × 105 FaDu cells were seeded per 24-well plate at 24 h prior to infection, which yielded ∼90% confluent monolayers. The N. meningitidis strains were incubated overnight at 32°C or 37°C as outlined above. The resultant colonies were harvested with a sterile cotton swab and resuspended in sterile PBS, and the OD600 was measured to adjust the inoculum to 5 × 107 CFU/ml in plain DMEM. Serial dilutions of the inocula were plated and colonies enumerated to ascertain the correct bacterial density. The cell cultures were washed thrice with warm PBS, and 200 μl per well of the inoculum (i.e., 107 bacteria) was added and incubated at 37°C for 1 h. After three washes with PBS, 1 ml per well of PBS–1% saponin was added and the plates were incubated for 30 min at 37°C to lyse the cells. After thorough resuspension, serial dilutions were plated onto Columbia blood agar plates and colonies enumerated after overnight incubation.
Sample preparation for proteome analysis.
Sample preparation for proteome analysis was conducted using a method similar to that described recently (22). Strain MC58 was grown in 1 liter of liquid culture at either 32°C or 37°C for 6 h in MDM. The cultures were then immediately chilled on ice, and cells were harvested by centrifugation. In order to obtain an internal 15N standard (23), equal portions of reaction mixtures corresponding to each set of conditions were mixed with equal amounts of 15N-labeled MC58 cells grown on minimal medium for meningococci (MMM) with 15NH4Cl as the sole source of nitrogen (22). Subsequently, outer membrane (OM) preparations were prepared using the “shake-and-bake” method (24) without further purification. We selected this approach in order to enrich outer membrane proteins in relation to the cytoplasmic proteins that were dominant (with respect to relative abundance). Samples were analyzed by one-dimensional SDS-PAGE and underwent in-gel tryptic digestion (25) for subsequent mass spectrometric (MS) analysis.
Mass spectrometry measurement.
The mass spectrometric (MS) analysis was carried out as described in reference 26. In brief, peptide samples were subjected to reversed-phase C18 column chromatography on an EASY nLC Proxeon system coupled to MS with an LTQ-Orbitrap mass spectrometer (Thermo Fisher).
Data analysis and 14N/15N quantification.
Tandem MS (MS/MS) data were processed as described in detail in references 11 and 22. Database searches were conducted using Sorcerer-SEQUEST (SageN; version v.28) against the N. meningitidis strain MC58 (27) target decoy protein sequence databases (complete proteome set of N. meningitidis strain MC58 with a set of common laboratory contaminants) compiled with BioEdit. The searches were performed in two iterations, i.e., with search parameters light (enzyme type, trypsin [KR]; peptide tolerance, 10 ppm; tolerance for fragment ions, 1 amu; b- and y-ion series; variable modification, methionine [15.99 Da]; a maximum of three modifications per peptide allowed) and with search parameters heavy (parameters identical to those used for 14N/light with the mass shift of all amino acids completely labeled with 15N taken into account). Resulting *.dta and *.out files were assembled and filtered with DTASelect (revision 2.0.25) (parameters -y 2 -c 2 -C 4 -here -decoy Rev_ -p 2 -t 2 -u -MC 2 -i 0.3 -fp 0.005). The resulting protein identification data were cured with an in-house written java script to ensure a minimum of two different peptides for each protein identified.
The cured search results were then used for parsing by the use of Census software (28) to obtain the relative quantitative data of 14N peaks (sample) versus 15N peaks (pooled reference). Quantification results were exported (R2 values of >0.7; only unique peptides were included; proteins failing to be relatively quantified were checked manually in the graphical user interface for on/off proteins). Proteins relatively quantified with at least two peptides were taken into account in the subsequent analysis. The MS proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (29) (see below for the data set identifier number).
Prediction of subcellular protein localization.
Subcellular localization of proteins was allocated according to the annotation in the NeMeSys database (30) and/or UniProt (31). For all proteins without database annotation of subcellular localization, the PSORTb algorithm was used for prediction (32). The categories used were “outer membrane (OM),” “periplasm (P),” “inner membrane (IM),” and “cytoplasm (C).” There are no sorting rules established for lipoproteins in N. meningitidis. Thus, for lipoproteins (and some other proteins) without a known function or published evidence of their subcellular localization other than “membrane,” the category of “inner or outer membrane (I/OM)” was used. Proteins for which no subcellular localization was inferred by database or PSORTb query and proteins for which divergent results were obtained from all three sources were categorized as “no prediction (no).”
Total membrane vesicle preparation for Western blotting.
The N. meningitidis strains were grown overnight on Columbia blood agar at 5% CO2 and 37°C. For each tested strain, two liquid cultures (100 ml) were adjusted to an OD600 of 0.1. The bacteria were grown with shaking at 200 rpm at 32°C or 37°C until the cultures reached an OD600 between 1.0 and 2.0. Bacterial cultures were centrifuged for 10 min at 8,000 × g and 4°C, and pellets were resuspended in 50 mM Tris-HCl–5 mM EDTA (pH 8.1). Resuspended bacteria were stored overnight at −80°C. Then, bacteria were exposed to ultrasonic disintegration three times for 45 s each time. Cell debris was removed by 15 min of centrifugation at 8,000 × g and 4°C. Cell envelopes were collected from the supernatant by ultracentrifugation for 1 h at 200,000 × g and 4°C. Pellets were resuspended in distilled water (dH2O). Using a Pierce bicinchoninic acid (BCA) protein assay kit, protein concentrations of the membrane samples were determined.
PorA detection by Western blotting (nonreducing SDS-PAGE).
Membrane samples were diluted with SDS-PAGE sample buffer without β-mercaptoethanol containing 0.2% SDS. Dilutions were aimed at adjusting similar concentrations for all samples. Samples were loaded on SDS-free polyacrylamide gels (12.5%). Electrophoresis was conducted at 4°C and at 50 V or 100 V for loading or separation, respectively. A Page Ruler prestained protein marker (Thermo Scientific) was used as a molecular weight marker. After resolution on SDS-PAGE, proteins were transferred onto a nitrocellulose membrane and PorA was visualized with P1.7-specific typing antibody from NIBSC on a GS-800 calibrated densitometer (Bio-Rad) and analyzed by the use of PDQuest Advanced software (version 8.0; Bio-Rad).
qPCR analysis of bacterial gene expression.
RNA was isolated from N. meningitidis strains grown at 32°C versus 37°C in anaerobic jars fitted with a CO2 generation sachet (Oxoid) using a Qiagen RNeasy kit (Qiagen) with inclusion of a DNase treatment step. RNase quality was assessed using Agilent RNA nano chips run on a Bioanalyzer (Agilent). Reverse transcription was carried out using 2 μg of RNA with hexanucleotide mix (Roche) and Superscript II reverse transcriptase (Invitrogen). Quantitative PCR (qPCR) was performed with a StepOne Plus system, applying comparative threshold cycle (ΔΔCT) analysis. As an endogenous control for normalization of gene expression, NMB1392 (zwf) encoding glucose-6-phosphate dehydrogenase was included in the target assortment. The reaction was carried out using 5 ng/μl cDNA in Maxima SYBR green/ROX qPCR master mix in the presence of a 20 mM concentration of each primer.
Statistical analysis.
Data plotting and statistical analyses were conducted using GraphPad Prism 5.04. The statistical tests used are indicated for each experiment in the figure legends.
Accession number(s).
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (29) with the data set identifier PXD003690.
RESULTS
Growth rate and major virulence factor expression in N. meningitidis grown at 32°C versus 37°C.
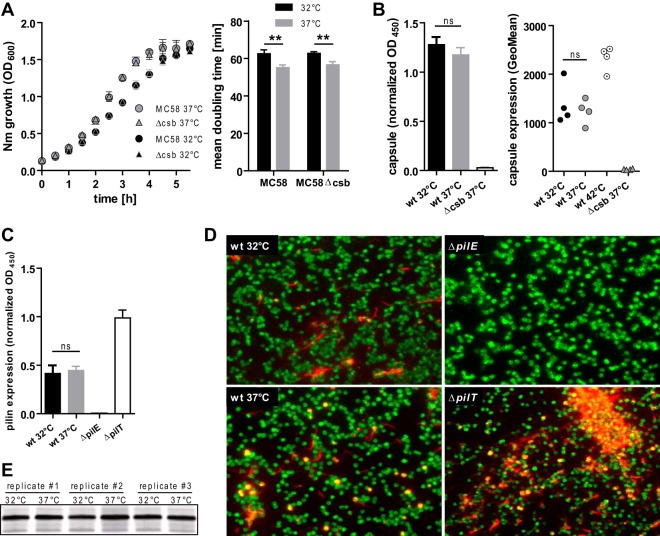
In order to compare general phenotypic changes in N. meningitidis strains due to growth at 32°C versus 37°C, we determined the growth kinetics of the bacteria as well as the critical virulence determinants capsule, pilus, and lipooligosaccharide (LOS). Growth kinetics of N. meningitidis MC58 and its capsule-deficient derivative MC58Δcsb mutant were assayed in liquid culture at 32°C versus 37°C, with the results showing only a slight reduction in the growth rate of both strains at 32°C in the log phase (Fig. 1A). It has recently been demonstrated that N. meningitidis virulence factors such as the capsule can be upregulated at 42°C (33). We therefore compared the levels of capsule expression of MC58 grown at 32°C versus 37°C by ELISA as well as by flow cytometry (Fig. 1B) and did not detect a difference in capsulation. As a control, we included MC58 grown at 42°C, which displayed increased capsule expression (Fig. 1B). In addition, we assayed the levels of piliation of MC58 grown at 32°C versus 37°C, as the pilus governs several aspects of N. meningitidis virulence such as natural competence, aggregation, biofilm formation, and cellular adherence (34). Using whole-cell ELISA and immunofluorescence microscopy, we found similar levels of pilin expression in N. meningitidis grown at 32°C and 37°C, whereas pilin expression was absent in a pilE mutant and was abundant in a hyperpiliated pilT mutant (Fig. 1C and D). Furthermore, we did not observe any molecular weight changes in LOS between 32°C and 37°C, as analyzed by polyacrylamide gel electrophoresis and subsequent silver staining (Fig. 1E). We therefore conclude that the levels of capsule expression, pilus expression, and LOS composition are not different in N. meningitidis grown at either 32°C or 37°C.
FIG 1.
Phenotypic characterization of N. meningitidis grown at 32°C or 37°C. (A) Growth curves of capsulated (MC58) or capsule-deficient (MC58Δcsb) N. meningitidis (Nm) were obtained as the OD600 of agitated liquid cultures (left panel). Mean doubling time was calculated from the logarithmic-growth phase (first 2 h) (right panel). Data are presented as means ± standard deviations of results of three independent measurements. (B) Determination of temperature-dependent capsule expression by whole-cell ELISA (left panel) or flow cytometry (right panel). For whole-cell ELISA, PFA-fixed N. meningitidis cells were coated onto ELISA plates and probed with capsule-specific mAb735. OD reads were normalized to those obtained with MC58-specific rabbit immune sera on an identical control plate as a coating control. Data represent means ± standard deviations of results of four independent experiments. Flow cytometry was done using mAb735 to probe capsule, followed by goat anti-mouse Alexa 488 detection. Geometric means (GeoMean) of results of four independent experiments are plotted. wt, wild type. (C) Pilus expression assessed by whole-cell ELISA (left panel) in MC58 grown at 32°C or 37°C with the MC58ΔpilE strain as a pilin-deficient negative control and the MC58ΔpilT strain as a hyperpiliated positive control. Whole-cell ELISA was conducted as described for panel B, using pilin-specific monoclonal antibody SM1. Means ± standard deviations of results of five independent experiments are plotted. (D) Immunofluorescence staining of pilin with SM1 (red) and rabbit antiserum against N. meningitidis MC58 and whole bacteria (green). Representative images from three independent experiments are shown. (E) Silver staining of lipooligosaccharide from MC58 grown at either 32°C or 37°C (three independent replicates), with separation performed by the use of Tricine-buffered polyacrylamide gel electrophoresis. For panels A to C, ** and ns denote P < 0.01 and P > 0.05, respectively, for results from unpaired Student's t tests.
Increased aggregation, biofilm formation, and cellular adherence in N. meningitidis grown at 32°C versus 37°C.
We sought to determine whether N. meningitidis displays differences in its ability to form aggregates and biofilms as well as in cellular adhesion at 32°C (representing a temperature encountered during asymptomatic colonization) versus 37°C, which N. meningitidis might rather face during invasion. Since the capsule can be a significant negative modulator of outer membrane protein interactions through steric hindrance, we examined capsulated as well as acapsulate N. meningitidis of strain MC58 in these analyses.
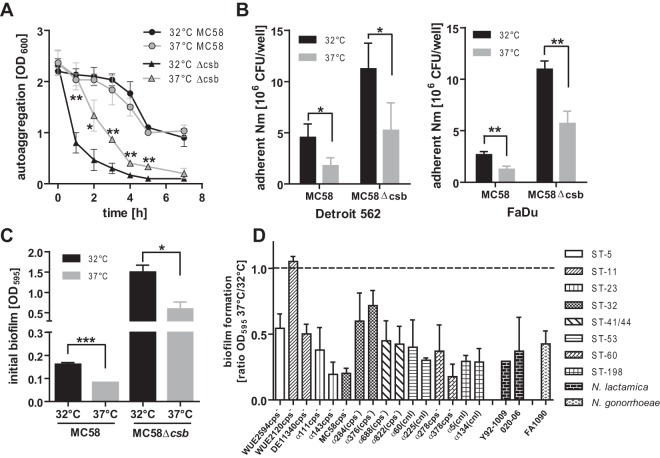
Acapsulate N. meningitidis grown at 32°C showed significantly stronger autoaggregation than N. meningitidis grown at 37°C (Fig. 2A). However, the temperature dependency of autoaggregation was absent in capsulated N. meningitidis, which showed an overall weak autoaggregation phenotype. This decrease in aggregation in capsulated N. meningitidis would be consistent with steric hindrance of interactions between outer membrane proteins other than pili. Capsulated as well as acapsulate N. meningitidis grown at 32°C displayed significantly stronger adhesion to the Detroit 562 and FaDu pharyngeal cell lines than inocula grown at 37°C (Fig. 2B). As expected, acapsulate N. meningitidis displayed stronger overall adherence; however, this did not have an impact on the temperature dependency of the adhesion phenotype. Furthermore, early biofilm formation of capsulated as well as acapsulate N. meningitidis was strongly increased in cells grown at 32°C (Fig. 2C). Since N. meningitidis is a genetically diverse species (35), the temperature-dependent early biofilm formation of 16 different meningococcal strains belonging to eight different multilocus sequence types was tested. With one exception, all tested meningococcal strains exhibited increased early biofilm formation at the lower temperature (Fig. 2D). Since capsule expression strongly reduces N. meningitidis biofilm formation (7) (Fig. 2C), we assayed only naturally acapsulate strains or capsule-deficient derivatives in the experiments represented in Fig. 2D for better interstrain comparability. Similar phenotypic changes at lower temperature were observed in other Neisseria species such as Neisseria lactamica and Neisseria gonorrhoeae (Fig. 1D). Thus, increased biofilm formation at lower temperature is an inherent property of pathogenic Neisseriae as well as in N. lactamica.
FIG 2.
Temperature dependency of N. meningitidis autoaggregation, adherence, and biofilm formation. (A) Determination of N. meningitidis autoaggregation by measuring meniscal absorbance at 600 nm over time in nonagitated liquid cultures. Data presented are means ± standard deviations of results of three independent experiments. (B) Adherence of N. meningitidis to Detroit and FaDu pharyngeal cell lines was tested by infecting monolayers with 107 meningococci (MC58) per 24-well plate. Adherent bacteria were enumerated after 1 h of incubation by plating serial dilutions. Means ± standard deviations of results of three to four independent experiments are shown. (C) Early biofilm formation of capsulated and acapsulate N. meningitidis (strain MC58) in 24-well plates after 1 h was tested by crystal violet staining. Data presented are means ± standard deviations of results of three independent experiments. In panels A to C, ***, **, *, and ns denote P < 0.001, P < 0.01, P < 0.05, and P > 0.05, respectively, for results from unpaired Student's t tests. (D) Early biofilm formation assessed as described for panel C for a collection of meningococcal strains belonging to different sequence types (ST, as indicated on the right) as well as for other Neisseria species. All meningococci were either capsule-deficient mutants [cps−] or naturally acapsulate due to natural mutations in single capsule genes [(cps−)] or due to natural replacement of the entire locus by the capsule null locus [(cnl)] (see Table S2 in the supplemental material). Data are presented as the mean ratios ± standard deviations of results of biofilm formation at 37°C and 32°C determined in three independent experiments.
Taking the data together, N. meningitidis grown at 32°C display enhanced aggregation, adhesion, and biofilm formation, and the temperature dependency of these phenotypes does not appear to be influenced by the capsule.
Differential N. meningitidis proteomes at 32°C versus 37°C.
Since the aggregation and adhesion properties of N. meningitidis were significantly temperature dependent, we used global proteome analysis to identify differences in protein expression at 32°C and 37°C. This approach entails the use of 15N-labeled proteins as an internal standard, which allows very precise and reliable proteomic comparisons (11). By addition of equal amounts of the internal 15N standard to the 32°C and 37°C samples compared, any possible bias inferred during sample preparation was kept to a minimum.
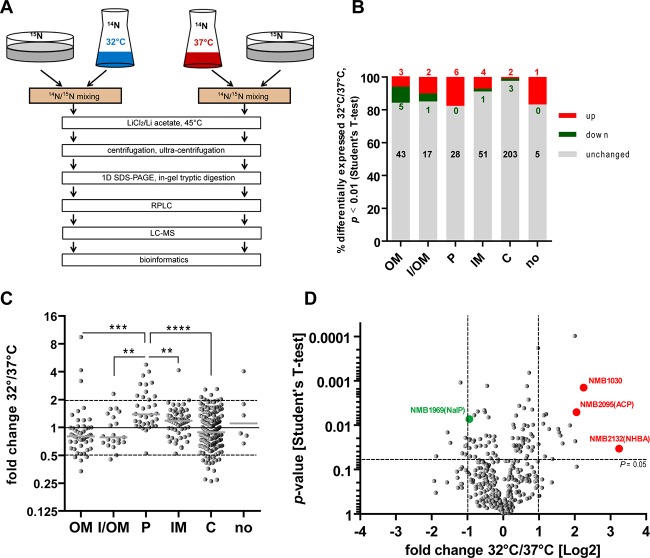
Since aggregation and adhesion properties of bacteria are governed by the protein composition of the outer membrane, we used outer membrane protein-enriched samples for a differential proteomics approach using highly accurate mass spectrometry (Fig. 3A). Using this method, a total of 375 distinct proteins were quantified from both the 32°C and 37°C conditions (see Table S1 in the supplemental material). All quantified proteins were classified according to their known or predicted subcellular localization using the NeMeSys database, the UniProt database, and/or the PSORTb algorithm (30–32). Among the 375 identified proteins, 49 were predicted to be localized in the outer membrane (OM), 21 (predominantly lipoproteins) in either the inner or outer membrane (I/OM), 35 in the periplasm (P), 56 in the inner membrane (IM), and 208 in the cytosol (C), whereas the location of 6 proteins could not be inferred (no) by either database annotation or computational prediction (Fig. 3B). Judged by the data corresponding to the significance level (P ≤ 0.01) of the 32°C/37°C ratio of the regulated proteins in three independent experiments, about 7.5% of all detected proteins showed temperature-dependent regulation. The biggest relative proportion of regulated proteins was found among I/OM proteins (4 of 21, 19%), followed by P (6 of 34, 18%) and OM (7 of 49, 14%), while the smallest relative proportion of regulated proteins was in C (5 of 209, 2.4%) (Fig. 3B). These different cellular compartments showed significant differences in the proportion of temperature-regulated proteins (χ2 = 20.82; P = 0.0003). Judged by the averaged fold expression, the modulation of temperature-guided proteome changes was most pronounced in the periplasm, where almost all detected proteins were upregulated (Fig. 3C). In conclusion, the moderate temperature change of 5°C had the highest impact on protein regulation of the bacterial envelope comprised of outer and inner membrane plus periplasm, while little change was seen in the cytoplasm (Fig. 3B and C). A function was assigned to most of the regulated OM proteins, while many of the regulated periplasmic proteins are of as-yet-unknown function. The three most strongly upregulated proteins were the neisserial heparin binding antigen (NHBA, NMB2132) (36), the conserved hypothetical protein NMB1030 (37), and the adhesin complex protein ACP (NMB2095) (38). A role in meningococcal biofilm formation has been described for the NHBA outer membrane protein (39), and ACP is known to facilitate N. meningitidis adherence to different human cell types (38). NMB1030 was recently described as a periplasmic ubiquinone-8 binding protein, and yet its function(s) in oxidative stress responses or electron chain transport was not apparent (40).
FIG 3.
N. meningitidis proteome comparison between growth levels at 32°C and 37°C using 15N-labeled N. meningitidis as an internal control. (A) Meningococci grown at 32°C versus 37°C in liquid culture (samples) were mixed with equal amounts of meningococci grown on agar plates containing 15NH4Cl as the sole source of nitrogen (internal control to minimize sample preparation artifacts). OMs were extracted with lithium chloride/lithium acetate buffer at 45°C and subjected to one-dimensional (1D) SDS gel electrophoresis. Separated proteins were subjected to in-gel digestion and then to reverse-phase liquid chromatography (RPLC). LC-MS/MS was performed with high resolution and high mass accuracy. (B) Protein expression of N. meningitidis at 32°C versus 37°C was quantified and referenced against an internal 15N-labeled standard. Quantified proteins were sorted according to the predicted subcellular localization (outer membrane, OM; inner or outer membrane, I/OM; periplasm, P; inner membrane, IM; cytoplasm, C; subcellular localization unclear, no). The proportions of up- or downregulated proteins (P < 0.01 in Student's t tests of three independent experiments) as well as of nonregulated proteins are plotted, with numbers of identified proteins indicated in the graph. (C) Fold change of protein expression between 32°C and 37°C by cellular localization. The mean regulation factor between 32°C and 37°C calculated from three independent experiments is plotted. ****, ***, and ** refer to P < 0.0001, P < 0.001, and P < 0.01, respectively, in one-way analysis of variance (ANOVA) applying Bonferroni's post hoc test for comparing OM, I/OM, P, IM, and C (excluding “no”). (D) Proteome analysis visualized as a volcano plot. Fold change of differential regulation is presented as log2 ratio of 32°C/37°C on the x axis, and the P values of results of unpaired t tests of differential expression between 32°C and 37°C are inversely plotted on the y axis. The genes of the proteins marked in red (highest expression at 32°C) or green (NMB1969, NalP) were chosen for further analyses.
Role of NMB2132, NMB1030, and NMB2095 in temperature-dependent N. meningitidis adhesion phenotypes.
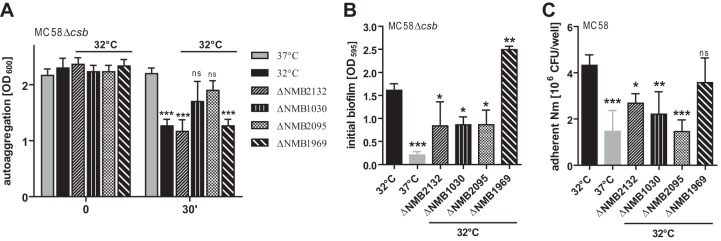
Since temperature reduction increased the strength of the adhesion and aggregation properties in N. meningitidis, we tested whether the three proteins that were most strongly upregulated (as shown by the data corresponding to the 32°C/37°C ratio) at 32°C, NMB2132 (NHBA, 9.47-fold upregulated, P = 0.0337), NMB1030 (4.759-fold upregulated, P = 0.0014), and NMB2095 (ACP, 4.15-fold regulated, P = 0.0051), are involved in adhesion and autoaggregation. Therefore, the respective deletion mutants were constructed; furthermore, we included a deletion mutant of the downregulated autotransporter protease NalP (NMB1969). Although NalP downregulation (fold expression, 0.52; P = 0.0072) was not as strong as for some other proteins, we pursued its influence, since NalP has been shown to cleave several N. meningitidis outer membrane proteins and thereby acts as a negative regulator of meningococcal biofilm formation (39). Acapsulate meningococci and their corresponding NMB2132, NMB1030, NMB2095, and NMB1969 deletion mutants grown overnight at 32°C were assayed for autoaggregation, early biofilm formation, and adherence to nasopharyngeal epithelial cells (Fig. 4). Acapsulate N. meningitidis grown overnight at 37°C served as a control. NMB1030 and NMB2095 deletion mutants showed aggregation similar to that seen with the parental strain grown at 37°C, whereas the NMB2132 (NHBA) and NMB1969 (NalP) mutants strongly aggregated similarly to the parental strain grown at 32°C (Fig. 4A). Thus, NMB1030 and NMB2095 (ACP) seem to be involved in temperature-dependent regulation of autoaggregation. In agreement with a previous report in which the roles of NMB2132 and NMB1969 in biofilm formation (at 37°C) were analyzed (39), our assays performed with the 32°C-grown NMB2132 (NHBA) mutant revealed reduced biofilm formation, while the NMB1969 (NalP) mutant showed increased biofilm formation (Fig. 4B). In alignment with the autoaggregation phenotype, the NMB1030 and NMB2095 (ACP) mutants, but also the NMB2132 (NHBA) mutant, displayed reduced early biofilm formation (Fig. 4B).
FIG 4.
Influence of NMB2132 (NHBA), NMB1030, NMB2095 (ACP), and NMB1969 (NalP) on temperature-dependent N. meningitidis phenotypes. Deletion mutants of the indicated genes were generated in N. meningitidis MC58 or its capsule-deficient derivative MC58Δcsb as indicated above each plot. (A) Autoaggregation was determined as decreases in the OD600 of meniscal samples taken at 0 min and 30 min from nonagitated liquid cultures. (B) Early biofilm formation of parental N. meningitidis MC58Δcsb and corresponding mutants grown at either 32°C or 37°C as indicated below the x axis. (C) Adherence of N. meningitidis MC58 and corresponding mutants to Detroit nasopharyngeal epithelial cells was tested by infecting 5 × 105 Detroit cells with 107 CFU N. meningitidis per well. Adherent bacteria were enumerated after 1 h of incubation by plating serial dilutions on Columbia blood agar. (A to C) Mean values ± standard deviations (SD) from at least three independent experiments are shown. ***, **, and * refer to P < 0.001, P < 0.01, and P < 0.05, respectively, in one-way ANOVA applying Dunnett's post hoc test for comparing all groups to the corresponding parental strain at 37°C at 30 min (A) or to the parental strain at 32°C (B and C).
The NMB2132 (NHBA), NMB1030, and NMB2095 (ACP) mutants showed decreased adherence to pharyngeal epithelial cells (Fig. 4C). In particular, the reduction of cellular adhesion of the NMB2095 is in good agreement with its description as an adhesin (38). The NMB1969 (NalP) mutant did not show a significant change in the level of cell adhesion. Taking the data together, the three most strongly upregulated proteins at 32°C convincingly contribute to the elevated adhesion and aggregation properties of N. meningitidis seen at 32°C.
Regulation of phenotypic changes induced by temperature reduction.
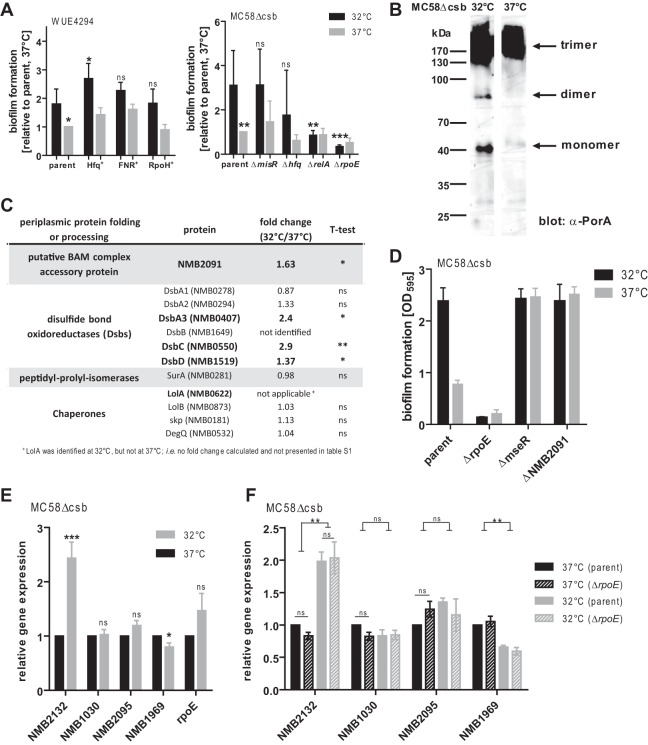
In order to elucidate the underlying regulatory mechanisms for the altered proteome and phenotypes at lower temperature, we tested the influence of known global regulators of bacterial gene expression. We took advantage of a collection of global regulator mutants that were available in two different N. meningitidis genetic backgrounds (WUE4294 and MC58), which were screened for early biofilm formation. Overexpression of the RNA chaperone Hfq (41), the oxygen-responsive transcriptional regulator FNR (42), or the heat shock-associated sigma factor RpoH (43) in strain WUE4294 did not change the phenotype of increased N. meningitidis biofilm production at 32°C. And yet, the overall biofilm production appeared to be increased in the Hfq-overexpressing mutant (Fig. 5A, left panel). Analyzing global regulators in acapsulate MC58 (Fig. 5A, right panel), no altered phenotype of the temperature dependence of biofilm formation was seen in deletion mutants of misR, which is part of a two-component regulatory system (MisR/MisS) (44), or in a deletion mutant of hfq. In contrast, there was a significant loss of biofilm formation in a relA mutant and a rpoE mutant at both temperatures. While relA as a regulator in the stringent response appears to be essential in overall biofilm formation across different species (45–47), we were surprised to find an even stronger phenotype in the rpoE mutant lacking σE. In E. coli, the classical σE response is activated by outer membrane stress or misfolded proteins in the periplasm (48), leading to the decay of the anti-σE factor and subsequently to the release of functional σE into the cytosol. σE then stimulates the expression of—among others—proteins involved in periplasmic protein folding or processing (48). In contrast to E. coli, previous studies did not suggest a σE response to periplasmic stress in N. meningitidis (49, 50). Since those studies were conducted using log-phase bacteria, we speculated that a σE response to outer membrane stress may still occur in mature N. meningitidis cultures such as those used in our system. In order to test whether misfolded proteins might accumulate in the periplasm, which in turn might trigger expression of σE, we analyzed the major outer membrane porin PorA as an indicator for membrane stress. PorA porins are inserted into the OM as functional trimers, whereas monomers and dimers in outer membrane preparations indicate dysfunctional assembly of integral outer membrane proteins, thereby indicating conditions of membrane stress (51). In agreement with our hypothesis, immunoblotting after semi-native SDS-PAGE revealed a higher proportion of monomeric/dimeric PorA porins in membrane preparations at 32°C than at 37°C (Fig. 5B). Further evidence for the presence of periplasmic protein folding or processing stress at lower temperature came from our proteome analysis: Among all 12 quantified meningococcal proteins likely to be involved in periplasmic protein folding or processing, the four proteins NMB2091, DsbA3, DsbC, and DsbD were upregulated at 32°C whereas none of the proteins was downregulated (Fig. 5C). Furthermore, the chaperone LolA was detected in our proteome analysis only at 32°C and not at 37°C (and the corresponding data are therefore not shown in table S1 in the supplemental material), likely indicating increased expression at 32°C. In agreement with a functional role of σE in the temperature dependence of biofilm formation, a knockout mutant of the MseR anti-σE factor exhibited elevated levels of biofilm formation at 32°C as well as 37°C (Fig. 5D). Similarly, deletion of NMB2091, an important factor for the assembly of outer membrane proteins (52) which was also upregulated at 32°C (see Fig. 5C), yielded the same temperature-independent biofilm phenotype as that seen with the anti-σE factor mutant (Fig. 5D). Thus, we conclude that the σE/anti-σE factor system might be a potent switch for the regulation of early biofilm formation in N. meningitidis, apparently in response to outer membrane stress that occurs at temperatures found in the nasopharynx.
FIG 5.
Possible involvement of σE in temperature-dependent early biofilm formation. (A) Influence of global regulator knockout mutants on early biofilm formation. (Left panel) hfq, fnr, or rpoH overexpression mutants in comparison to the corresponding parental strain (“parent”) WUE4294. (Right panel) ΔmisR, Δhfq, ΔrelA, and ΔrpoE (encoding σE) deletion mutants in comparison to the corresponding parental MC58Δcsb strain. In both panels, N. meningitidis was grown overnight at 32°C or 37°C, and early biofilm formation was assayed after 1 h. Means of results of at least three independent experiments in which the values were normalized to the corresponding parent strain at 37°C were plotted. ***, **, and * refer to P < 0.001, P < 0.01, and P < 0.05, respectively, in one-way ANOVA applying Dunnett's post hoc test for comparing all groups to the corresponding parental strain at 32°C. (B) Outer membrane preparations of planktonic N. meningitidis cultures grown at 32°C or 37°C were analyzed on nonreducing SDS-PAGE and immunoblotted using a PorA-specific monoclonal antibody. Positions of monomeric, dimeric, and trimeric PorA complexes are indicated. (C) List of all identified proteins in the proteomic comparison between 32°C and 37°C resembling proteins involved in periplasmic protein folding or processing (proven in N. meningitidis or homologs to E. coli proteins). Significantly regulated proteins are indicated in bold. (D) Parental strain MC58Δcsb and corresponding mutants of σE (ΔrpoE), anti-σE factor (ΔmseR), and the periplasmic protein NMB2091 (ΔNMB2091), which is involved in OM protein folding, were assayed for early biofilm formation after growth at 32°C or 37°C. (E) qPCR analysis of expression of target genes (indicated on the x axis) in the MC58Δcsb mutant at 32°C versus 37°C. The relative abundance of each transcript normalized to 37°C is plotted as the mean ± standard error of the mean (SEM) of results of 8 to 11 independent experiments carried out in triplicate. ***, *, and ns denote P < 0.001, P < 0.05, and P > 0.05, respectively, in two-tailed paired Student's t tests. (F) qPCR analysis of expression of target genes (indicated on the x axis) in the MC58Δcsb mutant versus the MC58ΔcsbΔrpoE mutant at 32°C versus 37°C. The relative abundance of each transcript normalized to 37°C is plotted as the mean ± SEM of results of 4 independent experiments carried out in triplicate. ** and ns denote P < 0.01 and P > 0.05, respectively, in repeated-measure ANOVA applying Bonferroni's post hoc test.
In order to elucidate whether de novo gene transcription is involved in the temperature-dependent proteomic changes, we conducted qPCR analysis of our target genes NMB2132 (NHBA), NMB1030, NMB2095 (ACP), and NMB1969 (NalP) as well as of rpoE. NMB2132 displayed robust and significant transcriptional upregulation at 32°C, whereas NMB1969 was downregulated (Fig. 5E). The results seen with the other genes were mostly unchanged, with NMB2095 (ACP) and rpoE showing a slight trend toward higher expression at 32°C. Hence, transcriptional control is indeed one mechanism—but not the only mechanism—by which temperature-dependent proteomic changes are invoked. To elucidate whether σE might be a transcriptional regulator of NMB2132 (NHBA) transcription, we compared its levels of transcription at 32°C versus 37°C in the MC58 and MC58ΔrpoE strains (Fig. 5F). NMB2132 transcription was not at all affected by σE, indicating that the link with the role in σE-dependent temperature-affected biofilm formation is not a direct one or does not depend on NHBA. The power of our analysis was not sufficient to allow us to state that the—admittedly marginal—differences in transcription of NMB1030 and NMB2095 between the parental strain and the rpoE mutant shown in Fig. 5F were in fact due to the mutation in rpoE.
DISCUSSION
In vitro experiments to investigate pathogen-host interactions are commonly conducted at 37°C, as this is the approximate temperature which the bacteria encounter upon invasion of the human host. However, despite strict thermal regulation, the human body temperature is neither spatially nor temporally perfectly constant. In fact, the nasopharynx temperature is coupled to that of the inhaled air (10). Therefore, bacteria such as N. meningitidis are subjected to temperature changes within this niche. Here, we have demonstrated that even slight temperature differences—and not only the stark changes encountered during classical heat shock or cold shock—have a significant impact on the phenotype and proteome of N. meningitidis, a strictly human-adapted bacterium.
From a technical perspective, our work suggests that 32°C appears to be a physiologically more meaningful temperature at which to monitor the interactions of N. meningitidis with the host's mucosal epithelium in in vitro settings: the adhesion properties of N. meningitidis appeared to be stronger at 32°C than at 37°C (Fig. 2). Likely, N. meningitidis is subjected to a temperature of 37°C only upon initial invasion into the bloodstream. However, systemic dissemination of N. meningitidis induces pyrexia and it has recently been shown that at 42°C, a N. meningitidis transcriptional response is triggered by a temperature-sensing noncoding RNA, which results in the upregulation of virulence factors (33). Biologically, this can be interpreted as the bacterium sensing the host's immune response; however, the overcoming of the host immune system by N. meningitidis is fatal for the host and poses a dead end for N. meningitidis itself. In contrast, increased expression of N. meningitidis factors that allow initial host colonization at the site of the nasopharyngeal mucosa at a temperature that is indicative of a steady-state host (i.e., 32°C) fit well into our view of the biology of N. meningitidis as a commensal. This is further substantiated by the fact that the major determinants of N. meningitidis virulence, namely, capsule, type IV pilus, and LOS, are not affected by the slight temperature difference. N. meningitidis capsule expression must be considered in context, since virtually all isolates from invasive disease are capsulated, whereas very few carriage isolates possess a capsule (53). During the colder seasons, the nasopharyngeal temperature is likely even lower than 33°C, as observed by Keck et al. at 25°C ambient temperature, possibly enhancing the effects on N. meningitidis adhesion phenotypes. In winter months, the rate of meningococcal disease cases rises quickly, and this has been linked to the pattern of seasonal flu (54). In light of our findings, it seems plausible that acquisition of N. meningitidis during winter, when ambient and nasopharyngeal temperatures are low and when N. meningitidis is therefore best equipped for host colonization based on its expressional profile, might be more efficient.
By comparing the proteomes of N. meningitidis strains grown at 32°C and 37°C (Fig. 2), we gained insight into the adaptation of N. meningitidis as a commensal of the human nasopharynx to the slightly differing temperatures which can be observed in this ecological niche. To our knowledge, this is the first comprehensive proteomic comparison of the effects of a temperature shift as small as 5°C on the physiology of a bacterial commensal. Although there have already been a few other studies addressing the impact of bigger temperature alterations on bacterial transcriptomes (55–57), our study was unique in that it focused on the global impact of a small temperature variation, such as is frequently seen within a mammalian host, on the proteome and thereon coupled phenotypes that impact the colonization capabilities of a human-adapted bacterium.
In E. coli, 4 to 5°C temperature downshifts from 37°C to 33°C and from 33°C to 28°C altered the expression of genes over a wide range of functional classes, with energy metabolism genes being particularly affected (55). For instance, genes involved in glycolysis, the tricarboxylic acid (TCA) cycle, and amino acid synthesis and transport were regulated, but so were high percentages of hypothetical and unclassified genes. In that study, a marked growth speed reduction of about 40% accompanied the 4°C-to-5°C temperature shifts (55). In our study, the 5°C temperature downshift caused only a 10% reduction of growth speed in N. meningitidis, based on the mean doubling time. Based on our proteome data, significant alterations occurred predominantly in the meningococcal envelope, while cytoplasmic proteins (including those involved in energy metabolism) showed very little change. This finding must be interpreted carefully, since the samples in our analysis were enriched for outer membrane proteins.
NMB2132 (NHBA), NMB1030, and NMB2095 (ACP) showed the greatest degree of temperature-dependent regulation and were identified as factors contributing to the temperature dependence of autoaggregation, biofilm formation, and cellular adherence (Fig. 4). N. meningitidis adhesion properties are usually governed by outer membrane proteins. Indeed, NMB2132 (NHBA) is a lipoprotein of the outer membrane and is known to contribute to biofilm formation (39). NMB2095 (ACP) is described as outer membrane adhesin (38) and was thus categorized as an outer membrane protein in our analysis. Outer membrane proteins in Gram-negative bacteria either are lipoproteins or form beta-barrels (58, 59). NMB2095 (ACP) is not a lipoprotein, and, with ∼100 amino acids after signal peptide cleavage (deduced from reference 27), it is too small to form a minimal beta-barrel of 142 amino acids (60). Therefore, its nature as a true outer membrane protein is not certain. Our data also demonstrate a contribution of NMB1030 to aggregation, biofilm formation, and cellular adherence. This function, whether exerted in a direct or an indirect fashion, appears distinct from its postulated role as a ubiquinone-8 binding protein (40), which would also imply a periplasmic instead of an outer membrane localization, but its distinct subcellular localization has not been directly shown. Further studies are required in order to gain better insight into the true subcellular location as well as the function and possible interaction partners of NMB1030 and might aid in revealing its mechanistically unclear contribution to biofilm formation and adhesion. In our study, we focused on the three proteins with the strongest degree of regulation, but it stands to reason that other regulated proteins may have an impact on the observed temperature-dependent adhesion properties as well. Of note, we have dissected the effect of NMB2132 (NHBA), NMB1030, and NMB2095 (ACP) using deletion mutants but we have not investigated overexpression or complementation mutants to further underscore the obtained results.
By qPCR, we found that the temperature-dependent abundance of NMB2132 (NHBA) and NMB1969 (NalP) is controlled at least in part at the transcript level (Fig. 5E and F), i.e., through transcription or mRNA stability; on the other hand, NMB1030 and NMB2095 (ACP) appear to be regulated by other mechanisms, since we did not observe changes in their transcript profiles with temperature. Others have investigated the temperature-dependent expression of NMB2132 (NHBA) and came to the conclusion that a posttranslational regulatory switch in the regulatory upstream region is responsible for enhanced expression at 30°C (61). We do not think that our findings are in conflict with that report, since transcriptional control does not exclude additional levels of protein expression control. We conclude that both transcriptional regulation and translational and/or posttranslational regulation occur to regulate the envelope proteome content at different temperatures.
In our experiments, we determined a significant contribution of the extracytoplasmic sigma factor σE to the temperature dependence of N. meningitidis biofilm formation (Fig. 5). It appears that σE triggers biofilm formation at 32°C, since in a mutant with a deletion of its coding gene, rpoE, biofilm formation was basically abrogated, whereas a mutant with a deletion of the gene encoding the corresponding anti-sigma factor MseR displayed intense biofilm formation at both temperatures (Fig. 5A and D). σE activation is possibly due to the relative abundance of outer membrane proteins that have not fully adopted their functional tertiary structure upon slight temperature reduction, as monitored with PorA in an exemplary manner in our study (Fig. 5B). Outer membrane proteins require the assistance of the beta-barrel assembly machinery (BAM) for their proper folding and membrane insertion. According to our proteomics data, the BAM components BamA, BamC, BamD, and BamE display lower expression at 32°C (see Table S1 in the supplemental material), which may be the underlying cause of the accumulation of misfolded proteins. Therefore, periplasmic proteins involved in proper protein folding appear upregulated (Fig. 5C), possibly to counteract this accumulation of misfolded proteins. A possible link between σE and misfolded envelope proteins was found by use of a NMB2091 mutant, which is known to have an impact on the BAM (52) and the gene homolog of which in E. coli (yraP) is part of the σE regulon (62). And yet, our finding that N. meningitidis seems to possess a functional σE response to membrane stress is in conflict with reports that suggested a function of this sigma factor/anti-sigma factor system outside the periplasm (49, 50). The σE response in N. gonorrhoeae was suggested to be triggered by oxidative stress (63), and the levels of expression of the proteins MsrAB, AniA, and NspA, which are known to be regulated by σE (49, 63), were unchanged in our proteomic analysis. However, the latter studies (49, 50, 63) were all conducted using N. meningitidis grown to log phase, whereas we used mature overnight cultures for our analyses here, which might explain potential differences in the outcomes with respect to apparent σE-dependent effects during distinct stages of N. meningitidis growth. Although we could show NMB2132 (NHBA) and NMB1969 (NalP) to be regulated on the transcript level, there was no effect of σE in their regulation (Fig. 5E and F). Hence, the possible contribution of σE to temperature-dependent phenotypes does not involve the direct upregulation of the target genes in our analyses but appears to be rather obscure and indirect.
Overall, our study showed that N. meningitidis displays an optimal phenotype for mucosal colonization at 32°C and that a temperature upshift of only 5°C already leads to significant proteomic changes. At 32°C, outer membrane proteins such as NMB2132 (NHBA), NMB1030, and NMB2095 (ACP) seem to govern autoaggregation, biofilm formation, and cellular adherence. First data presented here suggest that the σE response to outer membrane stress at this temperature seems to play a crucial role in temperature-dependent biofilm formation. Future studies are needed to further unravel the signaling events driving the σE response and how this contributes to biofilm formation, aggregation, and cellular adherence.
Supplementary Material
ACKNOWLEDGMENTS
This project was funded by a grant to U.V. from the German Research Foundation, project no. VO 718/6-1. Furthermore, this study was supported by Bundesministerium für Bildung und Forschung grant Medizinische Infektionsgenomik: Proteomics von Meningokokken und Pneumokokken, Teilprojekt Würzburg, Foerderkennzeichen 0315828D, to U.V. and D.B.
We thank Alexandra Schubert-Unkmeir (Institute for Hygiene and Microbiology, University of Würzburg) for kindly providing MC58ΔpilE and MC58ΔpilT mutant strains. We furthermore acknowledge the excellent technical assistance of Daniela Stanke and Lea Strobel.
We declare that we have no conflict of interest.
M.L., A.O., U.V., D.B., and K.J. conceived and designed the study; M.L., M.B., A.O., and K.J. did data acquisition and analysis; M.L., A.O., U.V., D.B., and K.J. interpreted data; M.L., U.V., and K.J. wrote the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00584-16.
REFERENCES
- 1.Gophna U, Ron EZ. 2003. Virulence and the heat shock response. Int J Med Microbiol 292:453–461. doi: 10.1078/1438-4221-00230. [DOI] [PubMed] [Google Scholar]
- 2.Weber MH, Marahiel MA. 2003. Bacterial cold shock responses. Sci Prog 86:9–75. doi: 10.3184/003685003783238707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arsène F, Tomoyasu T, Bukau B. 2000. The heat shock response of Escherichia coli. Int J Food Microbiol 55:3–9. doi: 10.1016/S0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- 4.Rosen R, Ron EZ. 2002. Proteome analysis in the study of the bacterial heat-shock response. Mass Spectrom Rev 21:244–265. doi: 10.1002/mas.10031. [DOI] [PubMed] [Google Scholar]
- 5.Gualerzi CO, Giuliodori AM, Pon CL. 2003. Transcriptional and post-transcriptional control of cold-shock genes. J Mol Biol 331:527–539. doi: 10.1016/S0022-2836(03)00732-0. [DOI] [PubMed] [Google Scholar]
- 6.Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. 2001. Meningococcal disease. N Engl J Med 344:1378–1388. doi: 10.1056/NEJM200105033441807. [DOI] [PubMed] [Google Scholar]
- 7.Lappann M, Haagensen JA, Claus H, Vogel U, Molin S. 2006. Meningococcal biofilm formation: structure, development and phenotypes in a standardized continuous flow system. Mol Microbiol 62:1292–1309. doi: 10.1111/j.1365-2958.2006.05448.x. [DOI] [PubMed] [Google Scholar]
- 8.Lappann M, Claus H, van Alen T, Harmsen M, Elias J, Molin S, Vogel U. 2010. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol Microbiol 75:1355–1371. doi: 10.1111/j.1365-2958.2010.07054.x. [DOI] [PubMed] [Google Scholar]
- 9.Lappann M, Vogel U. 2010. Biofilm formation by the human pathogen Neisseria meningitidis. Med Microbiol Immunol 199:173–183. doi: 10.1007/s00430-010-0149-y. [DOI] [PubMed] [Google Scholar]
- 10.Keck T, Leiacker R, Riechelmann H, Rettinger G. 2000. Temperature profile in the nasal cavity. Laryngoscope 110:651–654. doi: 10.1097/00005537-200004000-00021. [DOI] [PubMed] [Google Scholar]
- 11.Otto A, Bernhardt J, Meyer H, Schaffer M, Herbst FA, Siebourg J, Mäder U, Lalk M, Hecker M, Becher D. 2010. Systems-wide temporal proteomic profiling in glucose-starved Bacillus subtilis. Nat Commun 1:137. doi: 10.1038/ncomms1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett JS, Griffiths DT, McCarthy ND, Sleeman KL, Jolley KA, Crook DW, Maiden MC. 2005. Genetic diversity and carriage dynamics of Neisseria lactamica in infants. Infect Immun 73:2424–2432. doi: 10.1128/IAI.73.4.2424-2432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver KJ, Reddin KM, Bracegirdle P, Hudson MJ, Borrow R, Feavers IM, Robinson A, Cartwright K, Gorringe AR. 2002. Neisseria lactamica protects against experimental meningococcal infection. Infect Immun 70:3621–3626. doi: 10.1128/IAI.70.7.3621-3626.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nachamkin I, Cannon JG, Mittler RS. 1981. Monoclonal antibodies against Neisseria gonorrhoeae: production of antibodies directed against a strain-specific cell surface antigen. Infect Immun 32:641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unkmeir A, Kammerer U, Stade A, Hubner C, Haller S, Kolb-Maurer A, Frosch M, Dietrich G. 2002. Lipooligosaccharide and polysaccharide capsule: virulence factors of Neisseria meningitidis that determine meningococcal interaction with human dendritic cells. Infect Immun 70:2454–2462. doi: 10.1128/IAI.70.5.2454-2462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frosch M, Gorgen I, Boulnois GJ, Timmis KN, Bitter-Suermann D. 1985. NZB mouse system for production of monoclonal antibodies to weak bacterial antigens: isolation of an IgG antibody to the polysaccharide capsules of Escherichia coli K1 and group B meningococci. Proc Natl Acad Sci U S A 82:1194–1198. doi: 10.1073/pnas.82.4.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Virji M, Heckels JE, Watt PJ. 1983. Monoclonal antibodies to gonococcal pili: studies on antigenic determinants on pili from variants of strain P9. J Gen Microbiol 129:1965–1973. [DOI] [PubMed] [Google Scholar]
- 18.Hitchcock PJ, Brown TM. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol 154:269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 20.Tsai CM, Frasch CE. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem 119:115–119. doi: 10.1016/0003-2697(82)90673-X. [DOI] [PubMed] [Google Scholar]
- 21.Hubert K, Pawlik MC, Claus H, Jarva H, Meri S, Vogel U. 2012. Opc expression, LPS immunotype switch and pilin conversion contribute to serum resistance of unencapsulated meningococci. PLoS One 7:e45132. doi: 10.1371/journal.pone.0045132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lappann M, Otto A, Becher D, Vogel U. 2013. Comparative Proteome analysis of spontaneous outer membrane vesicles and purified outer membranes of Neisseria meningitidis. J Bacteriol 195:4425–4435. doi: 10.1128/JB.00625-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacCoss MJ, Wu CC, Liu H, Sadygov R, Yates JR III. 2003. A correlation algorithm for the automated quantitative analysis of shotgun proteomics data. Anal Chem 75:6912–6921. doi: 10.1021/ac034790h. [DOI] [PubMed] [Google Scholar]
- 24.Heckels JE. 1977. The surface of Neisseria gonorrhoeae: isolation of the major components of the outer membrane. J Gen Microbiol 99:333–341. doi: 10.1099/00221287-99-2-333. [DOI] [PubMed] [Google Scholar]
- 25.Dreisbach A, Otto A, Becher D, Hammer E, Teumer A, Gouw JW, Hecker M, Völker U. 2008. Monitoring of changes in the membrane proteome during stationary phase adaptation of Bacillus subtilis using in vivo labeling techniques. Proteomics 8:2062–2076. doi: 10.1002/pmic.200701081. [DOI] [PubMed] [Google Scholar]
- 26.Bonn F, Bartel J, Büttner K, Hecker M, Otto A, Becher D. 2014. Picking vanished proteins from the void: how to collect and ship/share extremely dilute proteins in a reproducible and highly efficient manner. Anal Chem 86:7421–7427. doi: 10.1021/ac501189j. [DOI] [PubMed] [Google Scholar]
- 27.Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, Dodson RJ, Nelson WC, Gwinn ML, DeBoy R, Peterson JD, Hickey EK, Haft DH, Salzberg SL, White O, Fleischmann RD, Dougherty BA, Mason T, Ciecko A, Parksey DS, Blair E, Cittone H, Clark EB, Cotton MD, Utterback TR, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith HO, Fraser CM, Moxon ER, Rappuoli R, Venter JC. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 28.Park SK, Venable JD, Xu T, Yates JR III. 2008. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods 5:319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vizcaíno JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rusniok C, Vallenet D, Floquet S, Ewles H, Mouze-Soulama C, Brown D, Lajus A, Buchrieser C, Medigue C, Glaser P, Pelicic V. 2009. NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol 10:R110. doi: 10.1186/gb-2009-10-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res 43(Database issue):D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FS. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh E, Kugelberg E, Tracy A, Zhang Q, Gollan B, Ewles H, Chalmers R, Pelicic V, Tang CM. 2013. Temperature triggers immune evasion by Neisseria meningitidis. Nature 502:237–240. doi: 10.1038/nature12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 35.Claus H, Maiden MC, Wilson DJ, McCarthy ND, Jolley KA, Urwin R, Hessler F, Frosch M, Vogel U. 2005. Genetic analysis of meningococci carried by children and young adults. J Infect Dis 191:1263–1271. doi: 10.1086/428590. [DOI] [PubMed] [Google Scholar]
- 36.Serruto D, Spadafina T, Ciucchi L, Lewis LA, Ram S, Tontini M, Santini L, Biolchi A, Seib KL, Giuliani MM, Donnelly JJ, Berti F, Savino S, Scarselli M, Costantino P, Kroll JS, O'Dwyer C, Qiu J, Plaut AG, Moxon R, Rappuoli R, Pizza M, Arico B. 2010. Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci U S A 107:3770–3775. doi: 10.1073/pnas.0915162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giuliani MM, Adu-Bobie J, Comanducci M, Arico B, Savino S, Santini L, Brunelli B, Bambini S, Biolchi A, Capecchi B, Cartocci E, Ciucchi L, Di Marcello F, Ferlicca F, Galli B, Luzzi E, Masignani V, Serruto D, Veggi D, Contorni M, Morandi M, Bartalesi A, Cinotti V, Mannucci D, Titta F, Ovidi E, Welsch JA, Granoff D, Rappuoli R, Pizza M. 2006. A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci U S A 103:10834–10839. doi: 10.1073/pnas.0603940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hung MC, Heckels JE, Christodoulides M. 2013. The adhesin complex protein (ACP) of Neisseria meningitidis is a new adhesin with vaccine potential. mBio 4:e00041-13. doi: 10.1128/mBio.00041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arenas J, Nijland R, Rodriguez FJ, Bosma TN, Tommassen J. 2013. Involvement of three meningococcal surface-exposed proteins, the heparin-binding protein NhbA, the alpha-peptide of IgA protease and the autotransporter protease NalP, in initiation of biofilm formation. Mol Microbiol 87:254–268. doi: 10.1111/mmi.12097. [DOI] [PubMed] [Google Scholar]
- 40.Donnarumma D, Golfieri G, Brier S, Castagnini M, Veggi D, Bottomley MJ, Delany I, Norais N. 2015. Neisseria meningitis GNA1030 is a ubiquinone-8 binding protein. FASEB J 29:2260–2267. doi: 10.1096/fj.14-263954. [DOI] [PubMed] [Google Scholar]
- 41.Moll I, Leitsch D, Steinhauser T, Bläsi U. 2003. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep 4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones HM, Gunsalus RP. 1987. Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J Bacteriol 169:3340–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grossman AD, Zhou YN, Gross C, Heilig J, Christie GE, Calendar R. 1985. Mutations in the rpoH (htpR) gene of Escherichia coli K-12 phenotypically suppress a temperature-sensitive mutant defective in the sigma 70 subunit of RNA polymerase. J Bacteriol 161:939–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tzeng YL, Kahler CM, Zhang X, Stephens DS. 2008. MisR/MisS two-component regulon in Neisseria meningitidis. Infect Immun 76:704–716. doi: 10.1128/IAI.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balzer GJ, McLean RJ. 2002. The stringent response genes relA and spoT are important for Escherichia coli biofilms under slow-growth conditions. Can J Microbiol 48:675–680. doi: 10.1139/w02-060. [DOI] [PubMed] [Google Scholar]
- 46.Lemos JA, Brown TA Jr, Burne RA. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect Immun 72:1431–1440. doi: 10.1128/IAI.72.3.1431-1440.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor CM, Beresford M, Epton HA, Sigee DC, Shama G, Andrew PW, Roberts IS. 2002. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J Bacteriol 184:621–628. doi: 10.1128/JB.184.3.621-628.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duguay AR, Silhavy TJ. 2004. Quality control in the bacterial periplasm. Biochim Biophys Acta 1694:121–134. doi: 10.1016/j.bbamcr.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 49.Huis in 't Veld RA, Willemsen AM, van Kampen AH, Bradley EJ, Baas F, Pannekoek Y, van der Ende A. 2011. Deep sequencing whole transcriptome exploration of the sigmaE regulon in Neisseria meningitidis. PLoS One 6:e29002. doi: 10.1371/journal.pone.0029002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hopman CT, Speijer D, van der Ende A, Pannekoek Y. 2010. Identification of a novel anti-sigmaE factor in Neisseria meningitidis. BMC Microbiol 10:164. doi: 10.1186/1471-2180-10-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volokhina EB, Beckers F, Tommassen J, Bos MP. 2009. The beta-barrel outer membrane protein assembly complex of Neisseria meningitidis. J Bacteriol 191:7074–7085. doi: 10.1128/JB.00737-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bos MP, Grijpstra J, Tommassen-van Boxtel R, Tommassen J. 2014. Involvement of Neisseria meningitidis lipoprotein GNA2091 in the assembly of a subset of outer membrane proteins. J Biol Chem 289:15602–15610. doi: 10.1074/jbc.M113.539510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Claus H, Maiden MC, Maag R, Frosch M, Vogel U. 2002. Many carried meningococci lack the genes required for capsule synthesis and transport. Microbiology 148:1813–1819. doi: 10.1099/00221287-148-6-1813. [DOI] [PubMed] [Google Scholar]
- 54.Rameix-Welti MA, Zarantonelli ML, Giorgini D, Ruckly C, Marasescu M, van der Werf S, Alonso JM, Naffakh N, Taha MK. 2009. Influenza A virus neuraminidase enhances meningococcal adhesion to epithelial cells through interaction with sialic acid-containing meningococcal capsules. Infect Immun 77:3588–3595. doi: 10.1128/IAI.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gadgil M, Kapur V, Hu WS. 2005. Transcriptional response of Escherichia coli to temperature shift. Biotechnol Prog 21:689–699. [DOI] [PubMed] [Google Scholar]
- 56.Galletti MF, Fujita A, Nishiyama MY Jr, Malossi CD, Pinter A, Soares JF, Daffre S, Labruna MB, Fogaca AC. 2013. Natural blood feeding and temperature shift modulate the global transcriptional profile of Rickettsia rickettsii infecting its tick vector. PLoS One 8:e77388. doi: 10.1371/journal.pone.0077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White-Ziegler CA, Um S, Perez NM, Berns AL, Malhowski AJ, Young S. 2008. Low temperature (23 degrees C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154:148–166. doi: 10.1099/mic.0.2007/012021-0. [DOI] [PubMed] [Google Scholar]
- 58.Koebnik R, Locher KP, Van Gelder P. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol 37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 59.Calmettes C, Judd A, Moraes TF. 2015. Structural aspects of bacterial outer membrane protein assembly. Adv Exp Med Biol 883:255–270. doi: 10.1007/978-3-319-23603-2_14. [DOI] [PubMed] [Google Scholar]
- 60.Johansson MU, Alioth S, Hu K, Walser R, Koebnik R, Pervushin K. 2007. A minimal transmembrane beta-barrel platform protein studied by nuclear magnetic resonance. Biochemistry 46:1128–1140. doi: 10.1021/bi061265e. [DOI] [PubMed] [Google Scholar]
- 61.Delany I, Antunes A, Haag A, Boccadifuoco G, Biolchi A, Burnelli B. 2012. A novel regulatory switch for increased expression of Neisseria meningitidis NHBA at physiological temperatures found in the human nasopharynx. Abstr 18th Int Pathog Neisseria Conf (IPNC) Meet, 9 to 14 September, Würzburg, Germany, poster 013. [Google Scholar]
- 62.Onufryk C, Crouch ML, Fang FC, Gross CA. 2005. Characterization of six lipoproteins in the sigmaE regulon. J Bacteriol 187:4552–4561. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gunesekere IC, Kahler CM, Ryan CS, Snyder LA, Saunders NJ, Rood JI, Davies JK. 2006. Ecf, an alternative sigma factor from Neisseria gonorrhoeae, controls expression of msrAB, which encodes methionine sulfoxide reductase. J Bacteriol 188:3463–3469. doi: 10.1128/JB.188.10.3463-3469.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.