Abstract
A reference 535 bp barcode sequence from a fragment of the mitochondrial gene cytochrome oxidase I (COI), acquired from specimens of An. neivai Howard, Dyar & Knab, 1913 from its type locality in Panama, was used as a tool for distinguishing this species from others in the subgenus Kerteszia. Comparisons with corresponding regions of COI between An. neivai and other species in the subgenus (An. bellator Dyar & Knab 1906, An. homunculus Komp 1937, An cruzii Dyar & Knab, 1908 and An. laneanus Corrêa & Cerqueira, 1944) produced K2P genetic distances of 8.3–12.6%, values well above those associated with intraspecific variation. In contrast, genetic distances among 55 specimens from five municipalities in the Colombian Pacific coastal state of Chocó were all within the range of 0–2.5%, with an optimized barcode threshold of 1.3%, the limit for unambiguous differentiation of An. neivai. Among specimens from the Chocó region, 18 haplotypes were detected, two of which were widely distributed over the municipalities sampled. The barcode sequence permits discrimination of An. neivai from sympatric species and indicates genetic variability within the species; aspects key to malaria surveillance and control as well as defining geographic distribution and dispersion patterns.
Keywords: DNA Barcode, Malaria, Kerteszia, COI
Introduction
In the Neotropical region, Colombia is second only to Brazil in number of malaria cases each year (Chaparro & Padilla 2012). Among malaria vectors in Colombia, Anopheles (Kerteszia) neivai Howard, Dyar & Knab, 1913 is often considered to be a secondary vector due to localized distribution, apparent natural infection rates and its larval habitat (Gutiérrez et al. 2008; Sinka et al. 2010). However, in the Pacific Coast of Colombia it may be considered a primary vector of malaria, especially in isolated municipalities where medical access is still limited such as Litoral de San Juan (Charambirá) and Santa Bárbara-Iscuandé (Astaiza et al. 1988; Murillo et al. 1988). Those municipalities are located within mangrove environments, which are ideally suited to An. neivai, a species that undergoes larval development in bromeliads. Within this region, in Buenaventura, Valle del Cauca, 4.76% of this species has been found to be naturally infected with Plasmodium falciparum (Gutiérrez et al. 2008). In the mangrove environment of this region, the relative abundance of An. neivai surpasses other known malaria vectors, such as An. albimanus Wiedemann, 1820, and its biting activity has been shown to peak at dawn and dusk, times of increased fishing activity by locals (Escovar et al. 2013).
From a phylogenetic perspective, An. neivai occurs within the subgenus Kerteszia Theobald, which includes several species associated with malaria transmission and whose larvae undergo development primarily in bromeliads located in forests (Zavortink 1973). While one of the earliest recognized species of the subgenus Kerteszia, it has an extensive geographic distribution extending from Chiapas, Mexico to Peru and Bolivia (Stone et al. 1959; Zavortink 1973). Distinguishing An. neivai often has been difficult; questionable records of its distribution exist, especially from topographic areas of relatively high elevation (González & Carrejo 2009).
Evidence from molecular variability in mitochondrial DNA fragments and variation in morphological characters suggests that the current concept of An. neivai may represent a complex of closely related species (Linton 2009; Montoya-Lerma et al. 1987). Several Neotropical species of Anopheles have been shown to consist of one or more sibling (cryptic) species (Rosa-Freitas et al. 1998). Even though closely related, such species may exhibit ecological and behavioral differences that may affect their malaria transmission potential or susceptibility to insecticides (Collins & Paskewitz 1996). Molecular markers are now being used to elucidate the presence of cryptic species within long established malaria vectors such as An. cruzii, An. albitarsis Lynch Arribálzaga, 1878, and An. triannulatus (Neiva & Pinto, 1922) (Gómez et al. 2013; Gutiérrez et al. 2010; Lehr et al. 2005; Rona et al. 2009, 2012; Rosero et al. 2012; Silva-do-Nascimento et al. 2011).
In the current study we evaluated a fragment of mitochondrial Cytochrome Oxidase I (COI), which represents the barcode region as described by Hebert et al. (2003, 2004). Standardized mitochondrial fragments from COI (DNA barcodes) have been used to identify mosquitoes (Cywinska et al. 2006; Harrison et al. 2012; Kumar et al. 2007; Ruiz-Lopez et al. 2012), including species from the Colombian Andes (Rozo-Lopez & Mengual 2015). Also, in Colombia, DNA barcodes were used to distinguish An. calderoni Wilkerson, 1991, An. punctimacula Dyar & Knab, 1906 and An, malefactor Dyar & Knab, 1906; part of a closely related species group with high morphological similarities and of medical importance, especially with regard to An. calderoni (González et al. 2010). DNA barcodes also have been used to evaluate intraspecific variability, especially in some Neotropical Anopheles (Gutierrez et al. 2010; Jaramillo et al. 2011; Mirabello & Conn 2006). Thus, when haplotype diversity in the malaria vector An. darlingi Root, 1926 was assessed among several municipalities in two western states of Colombia, many shared haplotypes were encountered, inferring a high level of gene flow (Gutierrez et al. 2010). Similarly, in the same region of Colombia, low genetic differentiation was noted among populations of the malaria vector An. nuneztovari Gabaldon, 1940, where previous evidence suggests the existence of a species complex (Jaramillo et al. 2011).
The present study was designed to obtain a reference barcode sequence from specimens of An. neivai from its type locality in Panama and to evaluate the efficacy of this barcode region to differentiate An. neivai from other species in the subgenus Kerteszia. Also, haplotype diversity in An. neivai was evaluated over a similar size area as that described for An. darlingi and An. nuneztovari (Gutierrez et al. 2010; Jaramillo et al. 2011) but for the distinctive Pacific coastal region of Colombia. Haplotype structure in An. neivai was further contrasted between the Colombian municipalities and the species’ type locality in Panama.
Materials and methods
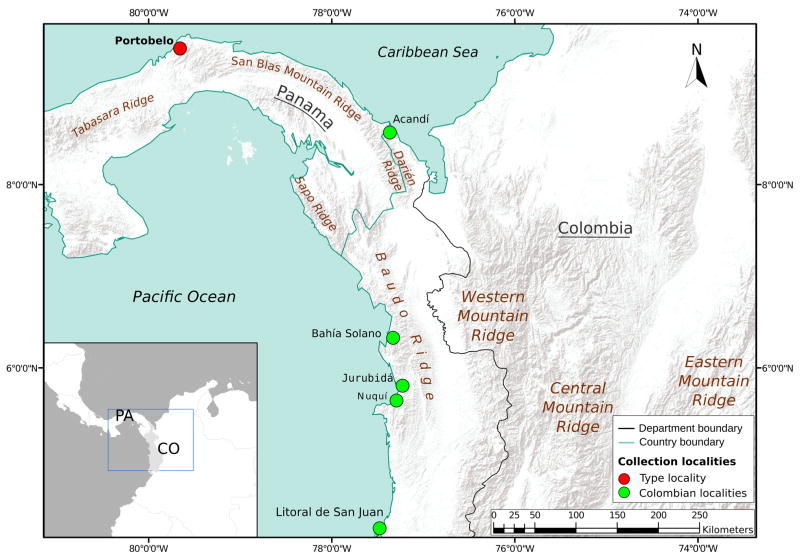
We collected specimens from the type locality at Portobelo, Colon Panama; as well as five municipalities within the Colombian department of Chocó: Acandí, Jurubidá, Nuquí, Bahía Solano, and Litoral de San Juan (Fig. 1, Table 1). The Colombian localities were chosen on the basis of epidemiological indications of malaria transmission (SEM 1957, Astaiza et al. 1988, INS 2015). While a few adult specimens were obtained using aspirators, most were collected as larvae and pupae in tank bromeliads and reared to adults under laboratory conditions similar to those described by Pecor & Gaffigan (1997). Fourth-stage larval and pupal exuviae, as well as male genitalia, associated with some of the reared specimens were mounted in Euparal following procedures presented by Pecor & Gaffigan (1997). Identification of many specimens (n = 34) was based on morphological characters as presented in Zavortink (1973), González & Carrejo (2009) and Harrison et al. (2012). Specimens were deposited at Museo Entomológico Francisco Luis Gallego, at Universidad Nacional de Colombia sede Medellin (MELFG). Additionally, thirty-one specimens were later identified on the basis of the COI-barcode sequences obtained, and added to the genetic pool for analysis.
FIGURE 1.
Collection municipalities map for Anopheles neivai in Colombia (CO) and Panama (PA).
TABLE 1.
Collection information for Anopheles neivai in Panama and Colombia and its associated haplotypes (based on a 535 bp COI alignment).
| Country, Dept. | Municipality | n | Haplotype (n) | Capture site: [NCBI register/Museum Catalog No (life stage*)] |
|---|---|---|---|---|
| Panama, Colón | Portobelo (Pbelo)a | 10 | 17 (5) | cs1: [KM234373/NC_27056 (L), † KM234396 (L), † KM234409 (L), KM234433/NC_27057 (L)] |
| cs2: [KM234435/NC_27059(A)] | ||||
| 18 (3) | cs1: [† KM234385(L), † KM234400(L), † KM234429(L)] | |||
| 19 (2) | cs1: [KM234402/NC_27058 (L), † KM234415(L)] | |||
| Colombia, Chocó | Acandí (Aca) | 2 | ‡1 | cs3: [† KM234371(F)] |
| 2 | cs3: [† KM229741(F)] | |||
| Jurubidá (Juru) | 10 | ‡1 (6) | cs4: [KM234376/NC_27055 (F)] | |
| cs5: [† KM234401(A), † KM234404(A), † KM234405(A), † KM234406(A), † KM234419(A)] | ||||
| ‡8 (2) | cs5: [† KM234398(A), † KM234425(A)] | |||
| 12 | cs5: [† KM234411(A)] | |||
| 20 | cs5: [† KM234387(A)] | |||
| Nuquí (Nq) | 3 | ‡8 (3) | cs6: [† KM234403/ NC 26137 (M), † KM234432(F)] | |
| cs7: [† KM234413/ NC 26136 (M)] | ||||
| Bahía Solano (BS) | 21 | 3 (6) | cs8: [KM234375(A), † KM234395(F), KM234397(A), KM234408(A), † KM234434(F)] | |
| cs9: [† KM234412(F)] | ||||
| 4 | cs8: [KM234389(A)] | |||
| 5 | cs10: [† KM234420(M)] | |||
| 6 (2) | cs11: [† KM234379(M)] | |||
| cs8: [KM234416(A)] | ||||
| 7 | cs8: [† KM234381(A)] | |||
| ‡8 (2) | cs8: [KM234374(A), KM234378(A)] | |||
| 10 | cs8: [† KM234388(F)] | |||
| 13 (2) | cs8: [† KM234372(M), KM234424(A)] | |||
| 14 | cs8: [ KM234417(A)] | |||
| 15 | cs8: [KM234384(A)] | |||
| 16 (3) | cs8: [KM234380(A), † KM234382(F), KM234391(A)] | |||
| Litoral de San Juan (LSJ) | 19 | ‡1 (9) | cs12: [† KM234383(F), KM234386(A), KM234394(A), KM234399(A), KM234410(A), KM234426(A), KM234428(A), KM234430(A), † KM234431/NC 26433(F)] | |
| ‡8 (7) | cs12: [KM234390(A), KM234393/NC_26391 (A), KM234414(A), † KM234418(F), † KM234421(F), KM234423(A), † KM234427(F)] | |||
| 9 | cs12: [KM234422(A)] | |||
| 11 | cs12: [KM234407(A)] | |||
| 21 | cs12: [KM234392(A)] |
[L: larvae, A: adult, M: adult Male, F: adult Female].
Type locality for An. neivai.
cs: capture site
species previously identified with taxonomy based in morphology,
haplotypes found in several municipalities.
Total DNA was extracted from each sample using the macerate method of Collins et al. (1987), as adapted by Uribe et al. (1998) and (Uribe et al. 2001) in a final elution volume of 50 μL. A 650–700 bp fragment of the COI-barcode region was amplified with the primers described in Kumar et al. (2007) because the primers provided in Folmer (1994) failed to yield consistent positive amplicons. PCR amplification was carried out in a final volume of 30 μL, including 6 μL of GoTaq® 5X buffer (Promega), 1.2 μL of dNTP (2.5 mM), 0.48 μL of MgCl2 (25 mM), 0.6 μL of each primer, 0.18 μL of GoTaq® DNA polymerase and 2 μL of purified DNA. All PCR amplifications were verified using electrophoresis on a 1% agarose gel with Gelstar® (Lonza), only positive PCR products were purified for cycle sequencing using a MultiScreen®HTS Vacuum Manifold (EMD Millipore). DNA sequencing was performed on both strands for each positive PCR product using BigDye 3.1® sequencing kit (ThermoFisher Scientific), in a 10 μL reaction including 2 μL ABI 5X dilution buffer, 0.5 μL Big Dye, 1 μL of primer and 2 μL of PCR product. Both strands were sequenced. Each sequencing reaction were purified using BigDye XTerminator® purification kit (ThermoFisher Scientific) using 45 μL of SAM solution, 10 μL XTerminator and 2 μL cycle sequencing for each sample. Finally, all samples were analyzed in an ABI 3500XL® (Applied Biosystems) automated capillary sequencer.
Sequence quality was assessed with Sequence Scanner 1.0® (Applied Biosystems 2011) based on the quality estimators CRL and QV20. Consensus sequences were obtained with Bioedit 7.25 (Hall 1999) and Geneious ® 6 (Kearse et al. 2012). A multiple alignment was constructed using Muscle (Edgar 2004). The final aligned data matrix was comprised of 535 base pairs. The presence of nuclear copies of mitochondrial origin (NUMTs) was evaluated as in Hlaing et al. (2009). To verify the identity of the amplified region, the sequences were aligned with the whole mitochondrial genome of An. albitarsis (NCBI accession number NC020662). Sequences from the type locality were compared to deposited sequences of species in the subgenus Kerteszia in GenBank using the Blast algorithm implemented by Altschul et al. (1990) and, also, by using the Barcoding of Life (BOLD) Identification System (IDS) described by Ratnasingham & Hebert (2007). The sequences included were: An. cruzii (KU551285), An. homunculus (KU551283); An. laneanus (KU551288) as part of recent Kerteszia mitochondrial genome sequences from Oliveira et al. (2016), as well as those of An. bellator (KU551287), An. lepidotus Zavortink, 1973 (JQ041286) and An. pholidotus Zavortink, 1973 (JQ041288-87). In addition, sequence from a collected specimen of An. marajoara Galvao & Damasceno, 1942 and a COI sequence of An. albitarsis (NC020662) were included, since the subgenus Nyssorhynchus Blanchard is closely related to Kerteszia (Harbach 2013). No barcode sequences of An. neivai from the type locality were recovered from GenBank. From the BOLD database, 51 records of sequences described as An. neivai were retrieved, 33 of which originated from Colombia. However, none of the sequences have been published nor are they available from the BOLD database.
Polymorphism and haplotype diversity (Hd) in the COI barcoding region of An. neivai was evaluated using DNASP 5 (Librado & Rozas 2009). Intra- and inter-species variation (barcoding gap) was estimated from genetic distances based on the K2P model that distinguishes between transitions and transversions (Kimura 1980), available in MEGA 6 (Tamura et al. 2013). A threshold for barcode gap at the species level was calculated for detecting the possible presence of cryptic species using the SPIDER package (Brown et al. 2012), available in R 3.2 (R Developement Core Team 2015). To further explore the existence of cryptic species associated to An. neivai, a statistical parsimony network was obtained using TCS 1 (Clement et al. 2000), based on previously identified haplotypes. Comparisons with other species in the subgenus Kerteszia and An. albitarsis were derived from K2P distances using criteria presented by Hebert et al. (2003) and Kumar et al. (2007). A K2P based dendrogram with bootstrap support was estimated using the APE package (Paradis 2012), also available in R.
Results
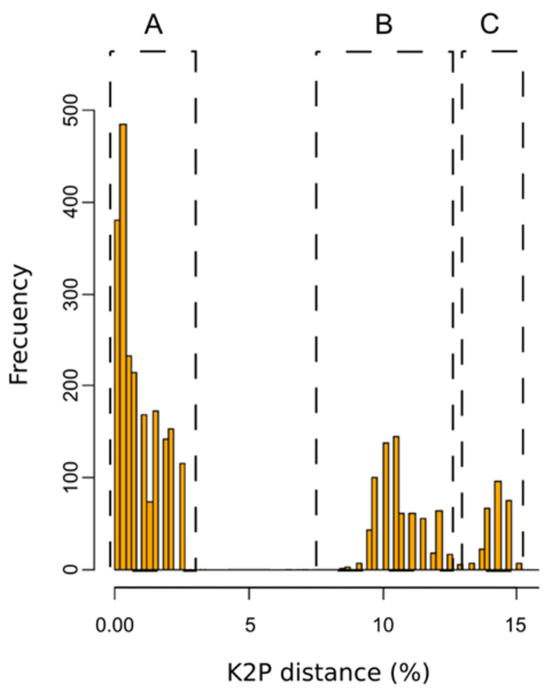
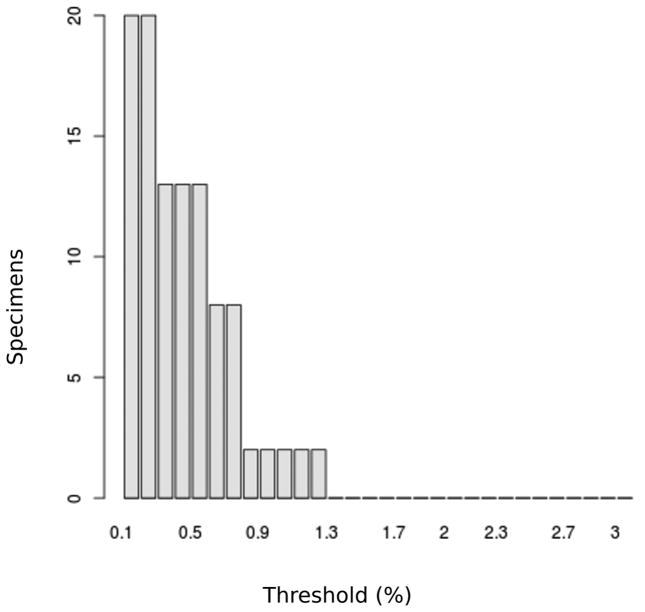
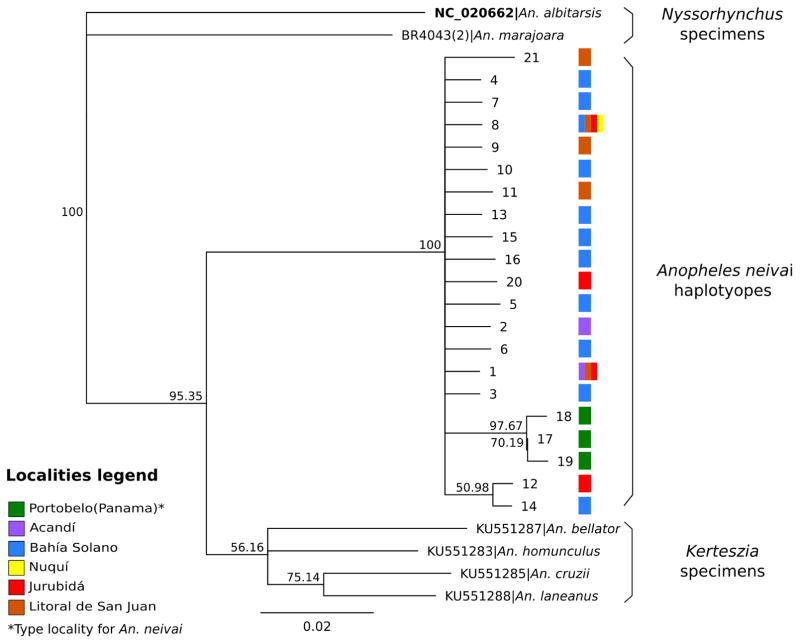
Genetic variability within the 535 base pair barcode region of COI was examined for 65 individuals of An. neivai. Within the type locality (n=10), three haplotypes were encountered, which were divergent by four substitutions (Tables 1 and 2). These haplotypes were compared with sequences available in GenBank for other species of the subgenus Kerteszia. The pairwise genetic similarity between An. neivai and other species of Kerteszia ranged between 88–92%: An. pholidotus (90–91%), An. lepidotus (88–90%), An. cruzii (90%), and An. homunculus (92%). Differences between the sequences of An. neivai, other species of Kerteszia and An. albitarsis are presented in terms of genetic distance (K2P distances) in Fig. 2. In this figure the first dotted box (A) of the three distinct ranges presented represents intraspecies distances for An. neivai, which varied between 032.5%. The second range (B) represents the differences between An. neivai and other species in Kerteszia (An. pholidotus, An. lepidotus, An. cruzii and An. homunculus), which varied between 8.3 to 12.6 %. The last range (C) represents differences between specimens of Kerteszia and An. albitarsis, which varied from 13 to 15.2%. For An. neivai, a barcode threshold of 1.3% (Fig. 3) was calculated, indicating the limit at which accumulative errors are minimized and no concurrence occurs with the distance rank of specimens assigned to another taxonomic identity (species) nor are nonspecific DNA sequences present (false and positive negatives) (Brown et al. 2012). The neighbor joining dendrogram in Fig. 4 presents a summary of K2P distances and infers that all An. neivai sequences conform to a single cluster with 100% bootstrap support. Considered together, these results indicate a clear separation of An. neivai from other species in the subgenus Kerteszia.
TABLE 2.
Variable sites in An. neivai haplotypes (based in a 535 pb COI alignment).
| Haplotype | n | Municipality | Variable sites (nucleotide position)
|
Ndb | K2P %c | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 4 | 4 | 4 | 5 | |||||||||
| 2 | 5 | 7 | 0 | 0 | 1 | 5 | 6 | 7 | 9 | 0 | 0 | 1 | 4 | 5 | 6 | 6 | 6 | 7 | 9 | 9 | 1 | 5 | 6 | 8 | 4 | 7 | 8 | 1 | ||||||
| 5*+ | 3* | 0 | 7 | 1 | 7 | 0+ | 5 | 7 | 9 | 1 | 0 | 6 | 9 | 5 | 7 | 6 | 7 | 9 | 3 | 3 | 4 | 4* | 6* | 8 | 0 | 3* | 6 | 3 | 2 | |||||
| 17 | 5 | Pbeloa | A | G | G | T | C | C | A | T | A | C | T | A | T | T | A | T | C | T | A | T | A | G | C | C | C | T | G | A | G | C | ||
| 18 | 3 | Pbeloa | . | A | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 2 | 0.37 |
| 19 | 2 | Pbeloa | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | C | . | . | . | . | 2 | 0.37 |
| 1 | 16 | Aca, Juru,LSJ | T | . | A | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | T | T | . | A | . | . | . | 7 | 1.31 |
| 2 | 1 | Aca | T | . | A | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | T | T | . | A | . | A | . | 8 | 1.50 |
| 3 | 6 | BS | T | . | A | . | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | T | T | . | A | . | . | T | 8 | 1.50 |
| 4 | 1 | BS | T | . | A | . | . | T | . | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | T | T | T | . | A | . | . | T | 9 | 1.68 |
| 5 | 1 | BS | T | . | A | . | . | T | . | . | . | . | . | . | . | . | G | . | . | . | . | C | . | . | T | T | T | . | A | . | . | T | 10 | 1.87 |
| 6 | 2 | BS | T | . | A | . | . | T | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | T | T | T | . | A | . | . | T | 9 | 1.68 |
| 7 | 1 | BS | C | . | . | . | . | T | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | T | T | . | A | . | . | . | 7 | 1.31 |
| 8 | 14 | BS, Nq, Juru, LSJ | T | . | A | . | . | T | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | T | T | . | A | . | . | . | 8 | 1.50 |
| 9 | 1 | LSJ | T | . | A | . | . | T | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | T | T | . | A | . | . | T | 9 | 1.68 |
| 10 | 1 | BS | T | . | A | . | . | T | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | T | T | C | A | . | . | T | 10 | 1.87 |
| 11 | 1 | LSJ | T | . | A | C | . | T | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | T | T | . | A | . | . | . | 9 | 1.68 |
| 12 | 1 | Juru | T | . | A | . | . | T | G | . | G | . | C | . | C | . | . | . | . | . | . | . | . | . | T | T | T | . | A | . | . | . | 11 | 2.06 |
| 13 | 2 | BS | T | . | A | . | . | .T | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | T | . | . | A | . | . | . | 7 | 1.31 |
| 14 | 1 | BS | T | . | A | . | T | T | G | . | G | . | . | . | . | . | . | . | . | . | . | . | G | . | T | T | . | . | A | . | . | . | 10 | 1.87 |
| 15 | 1 | BS | T | . | A | . | . | T | G | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | T | T | . | . | A | . | . | . | 8 | 1.50 |
| 16 | 3 | BS | C | . | A | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | T | . | . | A | . | . | . | 6 | 1.12 |
| 20 | 1 | Juru | T | . | A | . | . | T | . | . | . | . | . | G | . | . | . | . | . | . | G | . | . | . | T | T | . | . | A | . | . | . | 8 | 1.50 |
| 21 | 1 | LSJ | T | . | A | . | . | T | . | C | . | T | . | G | . | . | . | . | T | . | . | . | . | A | T | T | . | . | A | G | . | . | 12 | 2.24 |
Municipality acronyms: Pbelo: Portobelo (Panama, Colon); Aca: Acandí, Bahía Solano (BS), Nq: Nuquí, Juru: Jurubidá, LSJ: Litoral de San Juan (Colombia, Chocó).
Sites that separate Panama from Colombia.
Transversion
Type locality for An. neivai.
Nucleotide differences against Haplotype 17.
K2P % distance (%) against Haplotype 17.
Average K2P distance (%) between samples from Colombia and Panama: 1.62; between samples from Colombia: 0.56.
FIGURE 2.
Genetic differences (K2P) among An. neivai collected specimens (A) and against other species from Kerteszia (B) and Nyssorhynchus from NCBI (C).
FIGURE 3.
DNA barcode threshold optimization for An. neivai.
FIGURE 4.
Dendrogram (K2P) for An. neivai specimens from Colombia, Panama, with other Kerteszia and Nyssorhynchus species (based on a 535 bp COI alignment. Branch support was provided by bootstrap resampling (10000 replicates).
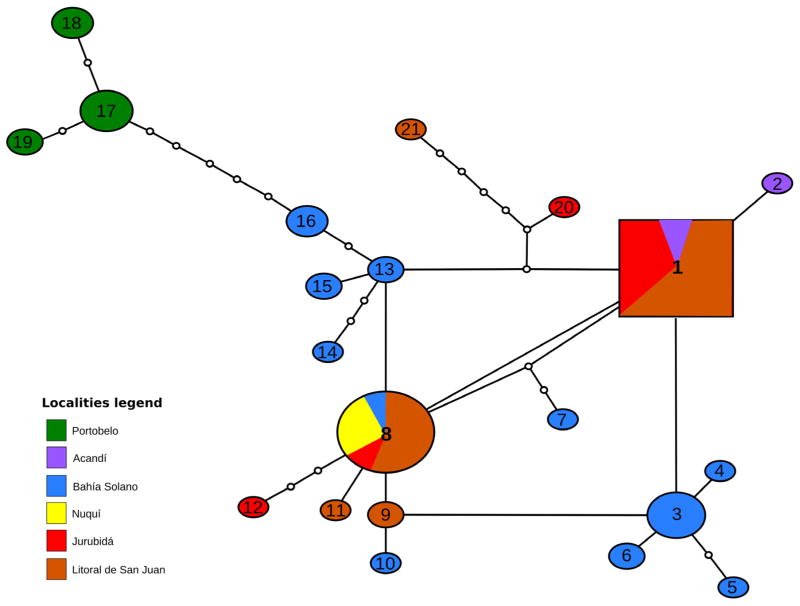
Among the 55 specimens of An. neivai from five municipalities in the Chocó region of Colombia, 18 COI haplotypes were detected in association with 26 polymorphic sites. All variation among these sites were transitions with the exception of a single transversion. The Colombian haplotypes varied from the Panamanian ones by 6312 nucleotide differences (Table 2). A majority of the Colombian specimens were from the municipalities of Bahía Solano (21) and Litoral de San Juan (19). While 11 haplotypes were detected from Bahía Solano only 5 were observed from Litoral de San Juan. Two Colombian haplotypes (1 and 8), differing by a single transition, were the most frequently encountered with similar distributions spanning approximately 500 km. In addition, they exhibited a close relationship to most of the other Colombian haplotypes (Table 1, Fig. 5). Haplotype diversity (Hd), Nei (1987) ranged from 0 at Nuquí (3 specimens) to 0.99 at Bahía Solano (21 specimens), while in Litoral de San Juan (19 specimens) and Portobelo (10 specimens) Hd values were similar (0.67–0.69 respectively).
FIGURE 5.
Haplotype network for An. neivai collected in Colombia and Panama (based on a 535 bp COI alignment).
Discussion
With an extensive geographic distribution and indications of both morphologic and molecular variability, emphasis was placed on developing a molecular marker for An. neivai based on specimens originating from its type locality in Panama (Linton 2009; Montoya-Lerma et al. 1987; Zavortink 1973). Despite of its usefulness for most metazoans, the barcode universal primers (Folmer 1994) do not always produce consistent PCR products for specimens preserved under differing conditions. Furthermore, presence of mutations at nucleotide positions where primers anneal require alternative primers sets in order to produce COI DNA barcodes (Hajibabaei et al. 2006; Kumar et al. 2007; Park et al. 2010). The 535 bp COI barcode region utilized in this study appears to clearly distinguish An. neivai from other species in Kerteszia (An. bellator, An. homunculus, An. cruzii, and An. laneanus) as revealed by K2P genetic distance divergences of 8.3312.6%. This range is similar to the interspecific divergence (mean 8.2%, range 7.638.7%) derived from COI sequences by Harrison et al. (2012) for comparisons between An. pholidotus and An. lepidotus. Also, using the COI barcoding region, Linton (2009) observed interspecific differences averaging 7.2% among four species of Kerteszia. Furthermore, comparisons between Panamanian specimens of An. neivai and those from the Pacific coast of Colombia revealed only small differences in K2P distances, all less than 2.5%, and, thus, below the barcode limit of 4% (0–3.9 %) for defining a species [single taxonomic unit] (Cywinska et al. 2006; Escovar et al. 2012; Foster et al. 2013; Kumar et al. 2007; Ruiz-Lopez et al. 2010). In addition, Arregui et al. (2015) report differences from 3–7% between species complexes, such as An. albitarsis and An. oswaldoi, Peryassú, 1922. Considered together, these results indicate that the 535 bp COI barcoding region appears to be an effective tool for differentiating An. neivai.
With the relative ease in which COI barcode sequences are now produced and their unique specificity, the COI barcode region provides a means to evaluate the extensive geographic distribution of An. neivai and may be helpful in evaluating the existence of sibling or cryptic species from different environments and geographic regions (Hebert et al. 2003). Indications of morphological variation in An. neivai have been noted by Montoya-Lerma et al. (1987), and differences exceeding 3% in the COI barcode region have been reported by Linton (2009) among specimens from Colombia, Ecuador and Venezuela. This level of variability could indicate the presence of cryptic species, especially when compared to local distributions in Colombia and the species type locality. When comparing mutation rates of the same sequence region in An. fluviatilis James 1902, differences reached only 0.8 % including those between forms S and T (Kumar et al. 2013).
The presence of two highly frequent haplotypes (1 and 8), which differed by a single transition, among specimens from Colombia (Fig. 2) suggests they may represent hypothetical ancestors from which many of the other haplotypes were derived as was suggested for An. pseudopunctipennis Theobald, 1901 (Dantur Juri et al. 2014). Both of these haplotypes were most abundant at the southernmost Colombian municipality sampled, and while also present near the Panamanian border, they were not encountered at the type locality in Panamá. The minimal number of mutations between specimens of An. neivai from the type locality and those from Colombia was six. Based on a larger fragment of the COI gene, 978 bp, Mirabello & Conn (2006) found differences of seven mutational steps between specimens of An. darlingi from Central America and Colombia. Comparisons of haplotype divergence among Kerteszia species by Lorenz et al. (2015) detected a magnitude of difference between An. cruzii and An. homunculus of 23 mutations.
While An. neivai is primarily associated with low elevation coastal regions, several records of its presence at relatively high elevations occur in the literature including municipalities in the Colombian Departments of Antioquia, Cundinamarca, and Boyacá (González & Carrejo 2009; SEM 1957), the Venezuelan Andes (Rubio-Palis 1991) and Ecuador (Arregui et al. 2015). The purported presence of An. neivai in atypical environments may be evaluated on the basis of barcode sequences. Such analyses would be appropriate for delimiting An. neivai’s distribution, from the perspectives of both elevation and geographic range, and also for detecting potential cryptic species. In turn, this knowledge should be beneficial for understanding An. neivai’s role in malaria transmission and for undertaking control measures.
Acknowledgments
Partial funding for this work was provided by the Amazon Malaria Initiative under the auspice of the US Agency for International Development (USAID). Special thanks are given to Robert A. Wirtz and Audrey Lenhart (both at the Centers for Disease Control and Prevention) for support throughout this endeavor. We are grateful for partial funding from Programa de Estudio y Control de Enfermedades Tropicales (PECET) of the Universidad de Antioquia, COLCIENCIAS-COLFUTURO grant for PhD studies (Convocatoria 567) and also for logistic support from Fundación Salud para el Trópico, Santa Marta, Colombia. We also express our appreciation to Rafael Vivero, Diego Puerta and David Gallo (members of Grupo de Investigación en Sistemática Molecular - Universidad Nacional de Colombia sede Medellín and PECET) for their participation in the fieldwork. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. http://dx.doi.org/10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Applied Biosystems. [accessed 1 July 2015];Sequence Scanner Software v1.0 Sequence Trace Viewer and Editor. 2011 Available from: https://www3.appliedbiosystems.com/cms/groups/mcb_marketing/documents/generaldocuments/cms_042188.pdf.
- Arregui G, Enríquez S, Benítez-Ortiz W, Navarro JC. Molecular Taxonomy of Anopheles from Ecuador, using mitochondrial DNA (Cytochrome c Oxidase I) and Maximum Parsimony optimization. Boletín de Malariología y Salud Ambiental. 2015;55(2):128–136. [Google Scholar]
- Astaiza R, Murillo C, Fajardo P. Biología de Anopheles (Kerteszia) neivai H., D. & K., 1913 (Diptera: Culicidae) en la costa pacífica de Colombia. II Fluctuación de la población adulta. Revista de Saúde Pública. 1988;22(2):101–108. doi: 10.1590/s0034-89101988000200005. http://dx.doi.org/10.1590/S0034-89101988000200005. [DOI] [PubMed] [Google Scholar]
- Brown SDJ, Collins RA, Boyer S, Lefort MC, Malumbres-Olarte J, Vink CJ, Cruickshank RH. Spider: An R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Molecular Ecology Resources. 2012;12:562–565. doi: 10.1111/j.1755-0998.2011.03108.x. http://dx.doi.org/10.1111/j.1755-0998.2011.03108.x. [DOI] [PubMed] [Google Scholar]
- Chaparro P, Padilla J. Mortalidad por paludismo en Colombia, 1979–2008. Biomédica. 2012;32(Supplement 1):95–105. doi: 10.1590/S0120-41572012000500011. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall K. TCS: a computer program to estimate gene genealogies. Molecular Ecology. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. http://dx.doi.org/10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Collins F, Mendez M, Rasmussen M, Mehaffey P, Besansky N, Finnerty V. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae Complex. The American Journal of Tropical Medicine and Hygiene. 1987;37(1):37–41. doi: 10.4269/ajtmh.1987.37.37. [DOI] [PubMed] [Google Scholar]
- Collins FH, Paskewitz SM. A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Molecular Biology. 1996;5(1):1–9. doi: 10.1111/j.1365-2583.1996.tb00034.x. http://dx.doi.org/10.1111/j.1365-2583.1996.tb00034.x. [DOI] [PubMed] [Google Scholar]
- Cywinska A, Hunter FF, Hebert PDN. Identifying Canadian mosquito species through DNA barcodes. Medical and Veterinary Entomology. 2006;20(4):413–424. doi: 10.1111/j.1365-2915.2006.00653.x. http://dx.doi.org/10.1111/j.1365-2915.2006.00653.x. [DOI] [PubMed] [Google Scholar]
- Dantur Juri MJ, Moreno M, Prado Izaguirre MJ, Navarro JC, Zaidenberg MO, Almirón WR, Claps GL, Conn JE. Demographic history and population structure of Anopheles pseudopunctipennis in Argentina based on the mitochondrial COI gene. Parasites & Vectors. 2014;7(1):423. doi: 10.1186/1756-3305-7-423. http://dx.doi.org/10.1186/1756-3305-7-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. http://dx.doi.org/10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escovar J, González R, Quiñones ML. Anthropophilic biting behaviour of Anopheles (Kerteszia) neivai Howard, Dyar & Knab associated with Fishermen’s activities in a malaria-endemic area in the Colombian Pacific. Memórias do Instituto Oswaldo Cruz. 2013;108(8):1057–1064. doi: 10.1590/0074-0276130256. http://dx.doi.org/10.1590/0074-0276130256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escovar J, González R, Quiñones ML, Wilkerson RC, Harrison B, Ruiz-lopez F. Morphological and molecular identification of Anopheles (Kerteszia) present in two endemic foci of malaria in Colombia. Journal of the American Mosquito Control Association. 2012;28(2):106–107. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology. 1994;3:294–299. [PubMed] [Google Scholar]
- Foster PG, Bergo ES, Bourke BP, Oliveira TMP, Nagaki SS, Sant’Ana DC, Sallum MAM. Phylogenetic analysis and DNA-based species confirmation in Anopheles (Nyssorhynchus) Plos ONE. 2013;8(2):e54063. doi: 10.1371/journal.pone.0054063. http://dx.doi.org/10.1371/journal.pone.0054063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez G, Jaramillo L, Correa MM. Wing geometric morphometrics and molecular assessment of members in the Albitarsis Complex from Colombia. Molecular Ecology Resources. 2013;2:1082–1092. doi: 10.1111/1755-0998.12126. http://dx.doi.org/10.1111/1755-0998.12126. [DOI] [PubMed] [Google Scholar]
- González R, Carrejo N, Wilkerson RC, Alarcon J, Alarcon-Ormasa J, Ruiz F, Bhatia R, Loaiza J, Linton YM. Confirmation of Anopheles (Anopheles) calderoni Wilkerson, 1991 (Diptera: Culicidae) in Colombia and Ecuador through molecular and morphological correlation with topotypic material. Memórias do Instituto Oswaldo Cruz. 2010;105(8):1001–1009. doi: 10.1590/s0074-02762010000800009. http://dx.doi.org/10.1590/S0074-02762010000800009. [DOI] [PubMed] [Google Scholar]
- González R, Carrejo N. Introducción al estudio taxonómico de Anopheles de Colombia: Claves y notas de distribución. Programa Editorial Universidad del Valle; Cali: 2009. p. 260. [Google Scholar]
- Gutiérrez L, Orrego LM, Gómez GF, López A, Luckhart S, Conn JE, Correa MM. A new mtDNA COI gene lineage closely related to Anopheles janconnae of the Albitarsis complex in the Caribbean region of Colombia. Memórias do Instituto Oswaldo Cruz. 2010;105(8):1019–1025. doi: 10.1590/s0074-02762010000800011. http://dx.doi.org/10.1590/S0074-02762010000800011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez LA, Gomez GF, Gonzalez JJ, Castro MI, Luckhart S, Conn JE, Correa MM. Microgeographic Genetic Variation of the Malaria Vector Anopheles darlingi Root (Diptera: Culicidae) from Cordoba and Antioquia, Colombia. American Journal of Tropical Medicine and Hygiene. 2010;83(1):38–47. doi: 10.4269/ajtmh.2010.09-0381. http://dx.doi.org/10.4269/ajtmh.2010.09-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez LA, Naranjo N, Jaramillo LM, Muskus C, Luckhart S, Conn JE, Correa MM. Natural infectivity of Anopheles species from the Pacific and Atlantic Regions of Colombia. Acta Tropica. 2008;107:99–105. doi: 10.1016/j.actatropica.2008.04.019. http://dx.doi.org/10.1016/j.actatropica.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN. DNA barcodes distinguish species of tropical Lepidoptera. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:968–971. doi: 10.1073/pnas.0510466103. http://dx.doi.org/10.1073/pnas.0510466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Harbach RE. The Phylogeny and classification of Anopheles. In: Manguin S, editor. Anopheles mosquitoes - New insights into malaria vectors. InTech; Rijeka: 2013. p. 55. [Google Scholar]
- Harrison BA, Ruiz-Lopez F, Falero GC, Savage HM, Pecor JE, Wilkerson RC. Anopheles (Kerteszia) lepidotus (Diptera: Culicidae), not the malaria vector we thought it was: Revised male and female morphology; larva, pupa,and male genitalia characters; and molecular verification. Zootaxa. 2012;3218:1–17. [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, Jeremy R, DeWaard JR. Biological identifications through DNA barcodes. Proceedings of the Royal Society B Biological Sciences. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. http://dx.doi.org/10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of Birds through DNA Barcodes. PLoS Biology. 2004;2(10):1657–1663. doi: 10.1371/journal.pbio.0020312. http://dx.doi.org/10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlaing T, Tun-Lin W, Somboon P, Socheat D, Setha T, Min S, Chang MS, Walton C. Mitochondrial pseudogenes in the nuclear genome of Aedes aegypti mosquitoes: implications for past and future population genetic studies. BMC Genetics. 2009;10:11. doi: 10.1186/1471-2156-10-11. http://dx.doi.org/10.1186/1471-2156-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INS (Instituto Nacional de Salud) Boletín Epidemiológico Semanal. [accessed 10 January 2016];Estadísticas del sistema de vigilancia en salud pública- SIVIGILA. 2015 Available from: http://www.ins.gov.co/boletin-epidemiologico/BoletnEpidemiolgico/2015BoletinepidemiologicoSemana52.pdf.
- Jaramillo LM, Gutiérrez LA, Luckhart S, Conn JE, Correa MM. Molecular evidence for a single taxon, Anopheles nuneztovari s.l., from two endemic malaria regions in Colombia. Memórias do Instituto Oswaldo Cruz. 2011;106(8):1017–23. doi: 10.1590/s0074-02762011000800020. http://dx.doi.org/10.1590/S0074-02762011000800020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. http://dx.doi.org/10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16(2):111–120. doi: 10.1007/BF01731581. http://dx.doi.org/10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar NP, Krishnamoorthy N, Sahu SS, Rajavel AR, Sabesan S, Jambulingam P. DNA Barcodes indicate members of the Anopheles fluviatilis (Diptera: Culicidae) species complex to be conspecific in India. Molecular Ecology Resources. 2013;13(3):354–361. doi: 10.1111/1755-0998.12076. http://dx.doi.org/10.1111/1755-0998.12076. [DOI] [PubMed] [Google Scholar]
- Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae) Journal of Medical Entomology. 2007;44(1):1–7. doi: 10.1603/0022-2585(2007)44[1:dbcdso]2.0.co;2. http://dx.doi.org/10.1603/0022-2585(2007)44[1:DBCDSO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lehr MA, Kilpatrick CW, Wilkerson RC, Conn JE. Cryptic species in the Anopheles (Nyssorhynchus) albitarsis (Diptera: Culicidae) complex: incongruence between random amplified polymorphic DNA-polymerase chain reaction identification and analysis of mitochondrial DNA COI gene sequences. Annals of the Entomological Society of America. 2005;98(6):908–917. doi: 10.1603/0013-8746(2005)098[0908:CSITAN]2.0.CO;2. http://dx.doi.org/10.1603/0013-8746(2005)098[0908:CSITAN]2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. http://dx.doi.org/10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Linton Y-MM. Mosquito Barcoding Initiative. The first barcode release paper. [accessed 11 September 2015];Third International Barcode of Life Conference. 2009 Available from: https://vimeo.com/8996184.
- Lorenz C, Patané JSL, Suesdek L. Morphogenetic characterisation, date of divergence, and evolutionary relationships of malaria vectors Anopheles cruzii and Anopheles homunculus. Infection, Genetics and Evolution. 2015;35:144–152. doi: 10.1016/j.meegid.2015.08.011. http://dx.doi.org/10.1016/j.meegid.2015.08.011. [DOI] [PubMed] [Google Scholar]
- Mirabello L, Conn JE. Molecular population genetics of the malaria vector Anopheles darlingi in Central and South America. Heredity. 2006;96:1–11. doi: 10.1038/sj.hdy.6800805. http://dx.doi.org/10.1038/sj.hdy.6800805. [DOI] [PubMed] [Google Scholar]
- Montoya-Lerma J, Murillo C, Solarte Y. Variación fenotípica de Anopheles (K) neivai (Diptera: Culicidae) en la costa Pacífica de Colombia. Colombia Médica. 1987;8(1):25–27. [Google Scholar]
- Murillo C, Astaiza R, Fajardo P. Biología de Anopheles (Kerteszia) neivai h., d. & k., 1913 (Diptera: Culicidae) en la costa pacífica de Colombia. I fluctuación de la población larval y características de sus criaderos. Revista de Saúde Pública. 1988;22(2):94–100. doi: 10.1590/s0034-89101988000200004. http://dx.doi.org/10.1590/S0034-89101988000200004. [DOI] [PubMed] [Google Scholar]
- Oliveira TMP, Foster PG, Bergo ES, Nagaki SS, Sanabani SS, Marinotti O, Marinotti PN, Sallum MAM. Mitochondrial genomes of Anopheles (Kerteszia) (Diptera: Culicidae) from the Atlantic Forest, Brazil. Journal of Medical Entomology. 2016:1–8. doi: 10.1093/jme/tjw001. published online. [DOI] [PubMed] [Google Scholar]
- Park DS, Suh SJ, Oh HW, Hebert PDN. Recovery of the mitochondrial COI barcode region in diverse Hexapoda through tRNA-based primers. BMC Genomics. 2010;11:423. doi: 10.1186/1471-2164-11-423. http://dx.doi.org/10.1186/1471-2164-11-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E. Analysis of Phylogenetics and Evolution with R. Springer; New York: 2012. p. 386. [Google Scholar]
- Pecor J, Gaffigan T. [accessed 12 August 2015];Collecting, rearing, preserving, mounting and shipping techniques for mosquitoes. 1997 Available from: http://wrbu.si.edu/Techniques.html.
- R Developement Core Team. [accessed 5 October 2015];R: A Language and Environment for Statistical Computing. 2015 Available from: http://www.r-project.org.
- Ratnasingham S, Hebert PDN. BOLD: The Barcode of Life Data System ( http://www.barcodinglife.org) Molecular Ecology Notes. 2007;7(3):355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rona LDP, Carvalho-Pinto CJ, de Peixoto A. Speciation in brazilian Atlantic Forest mosquitoes: A mini-review of the Anopheles cruzii species complex. In: Fusté C, editor. Studies in population genetics. INTECH Open Access Publisher; Rijeka: 2012. pp. 105–116. http://dx.doi.org/10.5772/35693. [Google Scholar]
- Rona LDP, Carvalho-Pinto CJ, Gentile C, Grisard EC, Peixoto A. Assessing the molecular divergence between Anopheles (Kerteszia) cruzii populations from Brazil using the timeless gene: further evidence of a species complex. Malaria Journal. 2009;8:60. doi: 10.1186/1475-2875-8-60. http://dx.doi.org/10.1186%2F1475-2875-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Freitas MG, Lourenço-de-Oliveira R, de Carvalho-Pinto CJ, Flores-Mendoza C, Silva-do-Nascimento TF. Anopheline species complexes in Brazil. Current knowledge of those related to malaria transmission. Memórias do Instituto Oswaldo Cruz. 1998;93(5):651–655. doi: 10.1590/s0074-02761998000500016. http://dx.doi.org/10.1590/S0074-02761998000500016. [DOI] [PubMed] [Google Scholar]
- Rosero DA, Jaramillo LM, Gutiérrez La, Conn JE, Correa MM. Genetic diversity of Anopheles triannulatus s.l. (Diptera: Culicidae) from Northwestern and Southeastern Colombia. American Journal of Tropical Medicine and Hygiene. 2012;87(5):910–920. doi: 10.4269/ajtmh.2012.12-0285. http://dx.doi.org/10.4269/ajtmh.2012.12-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozo-Lopez P, Mengual X. Mosquito species (Diptera, Culicidae) in three ecosystems from the Colombian Andes: identification through DNA barcoding and adult morphology. ZooKeys. 2015;513:39–64. doi: 10.3897/zookeys.513.9561. http://dx.doi.org/10.3897/zookeys.513.9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Palis Y. Vector biology and malaria transmission in western Venezuela. University of London; London: 1991. p. 261. [Google Scholar]
- Ruiz-Lopez F, Linton YM, Ponsonby DJ, Conn JE, Herrera M, Quiñones ML, Vélez ID, Wilkerson RC. Molecular comparison of topotypic specimens confirms Anopheles (Nyssorhynchus) dunhami Causey (Diptera: Culicidae) in the Colombian Amazon. Memórias do Instituto Oswaldo Cruz. 2010;105(7):899–903. doi: 10.1590/s0074-02762010000700010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Lopez F, Wilkerson RC, Conn JE, McKeon SN, Levin DM, Quiñones ML, Póvoa MM, Linton YMM. DNA barcoding reveals both known and novel taxa in the Albitarsis Group (Anopheles: Nyssorhynchus) of Neotropical malaria vectors. Parasites & Vectors. 2012;5(1):44. doi: 10.1186/1756-3305-5-44. http://dx.doi.org/10.1186/1756-3305-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEM (Servicio de Erradicación de la Malaria. Ministerio de Salud Pública. República de Colombia) Plan de erradicación de la Malaria en Colombia. II. Ministerio. de Salud. Pública; Bogotá: 1957. p. 635. [Google Scholar]
- Silva-do-Nascimento TF, Pitaluga LDR, Peixoto AA, Lourenço-de-Oliveira R. Molecular divergence in the timeless and cpr genes among three sympatric cryptic species of the Anopheles triannulatus complex. Memórias do Instituto Oswaldo Cruz. 2011;106(Supplement 1):218–22. doi: 10.1590/s0074-02762011000900027. http://dx.doi.org/10.1590/S0074-02762011000900027. [DOI] [PubMed] [Google Scholar]
- Sinka ME, Rubio-Palis Y, Manguin S, Patil AP, Temperley WH, Gething PW, Van Boeckel T, Kabaria CW, Harbach RE, Hay SI. The dominant Anopheles vectors of human malaria in the Americas: occurrence data, distribution maps and bionomic précis. Parasites & Vectors. 2010;3(4):72. doi: 10.1186/1756-3305-3-72. http://dx.doi.org/10.1186/1756-3305-3-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone A, Knight KL, Starcke H. A Synoptic Catalog of the mosquitoes of the World (Diptera: Culicidae) The Thomas Say Foundation, Entomological Society of America; Washington D.C: 1959. p. 358. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. http://dx.doi.org/10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe SI, Lehmann T, Rowton ED, Vélez BID, Porter CH. Speciation and population structure in the morphospecies Lutzomyia longipalpis (Lutz & Neiva) as derived from the mitochondrial ND4 gene. Molecular Phylogenetics and Evolution. 2001;18(1):84–93. doi: 10.1006/mpev.2000.0863. http://dx.doi.org/10.1006/mpev.2000.0863. [DOI] [PubMed] [Google Scholar]
- Uribe SI, Porter CH, Vélez ID. Amplificación y obtención de secuencias de rRNA mitocondrial en Lutzomyia spp (Diptera: Psychodidae) vectores de Leishmaniosis. Revista Colombiana de Entomología. 1998;23(3–4):177–185. [Google Scholar]
- Zavortink TJ. Mosquito studies (Diptera, Culicidae) XXIX. A review of the subgenus Kerteszia of Anopheles. Contributions of the American Entomological Institute. 1973;9(3):1–54. [Google Scholar]