ABSTRACT
One of the distinct reproductive programs in orchid species is pollination-triggered ovule development and megasporogenesis. During sexual reproduction, fertilization occurs days to months after pollination. The molecular mechanisms evolved to carry out this strategic reproductive program remain unclear. In the August issue of Plant Physiology1, we report comprehensive studies of comparative genome-wide gene expression in various reproductive tissues and the molecular events associated with developmental transitions unique to sexual reproduction of Phalaenopsis aphrodite. Transcriptional factors and signaling components whose expression is specifically enriched in interior ovary tissues when fertilization occurs and embryos start to develop have been identified. Here, we report verification of additional fertilization-associated genes, DOMAINS REARRANGED METHYLTRANSFERASE 1 (PaDRM1), CHROMOMETHYLTRANSFERASE 1 (PaCMT1), SU(VAR)3-9 RELATED PROTEIN 1 (PaSUVR1), INDOLE-3-ACETIC ACID inducible 30-like 1 (PaIAA30L1), and ETHYLENE INSENSITIVE 3-like 1 (PaEIN3L1), and discuss their potential roles in gametophyte development, epigenetic reprogramming, and hormone regulation during fertilization and establishment of embryo development in Phalaenopsis orchids.
KEYWORDS: Fertilization; gene expression, orchid, Phalaenopsis aphrodite, sexual reproduction
Fertilization and embryo development starts by fusion of the sperm cell and egg from the female gametophyte. Successful fertilization and seed development requires coordinated development of male and female gametophytes and intensive communication between male and female reproductive cells. Tremendous progress has been made recently in understanding the molecular mechanism of fertilization and embryo development in plants. Particularly, studies on regulation of gametophyte development, prevention of polyspermy, and epigenetic reprogramming after fertilization have provided new insights into hormone signaling and the gene regulatory paradigm for plant sexual reproduction.2-4
In our previous study, we reported that pollen tubes enter the ovule at approximately 65 d after pollination (DAP) followed by expression of an egg-specific marker EGG CELL 1 Like 1 (PaEC1L1) in P. aphrodite.5 These data provide cellular and molecular evidence that fertilization occurs at approximately 70 d after pollination (DAP) in P. aphrodite. In addition to the egg-specific marker, several transcription factors, methyltransferases, hormone signaling components, and cell cycle regulators6 have also been shown to display fertilization-associated expression patterns.1 Isolation of theses genes provides a novel opportunity to study the unique sexual reproduction of Phalaenopsis orchids.
Development of reproductive cells and establishment of embryogenesis
Among the over-represented transcription factors enriched during fertilization and early embryo development (70-80 DAP) are type I MADS domain transcription factors. Plant MADS domain transcription factors play important regulatory roles at various stages in plant development.7 Type I MADS domain transcription factors are involved in regulation of plant reproduction, particularly female gametophyte, embryo, and endosperm development. For example, Arabidopsis AGAMOUS-LIKE 23 (AGL23), which is regulated by the epigenetic regulator PRC2-type polycomb group complex, is expressed in developing ovules, embryo, and endosperm. AGL23 is required for successful development of female gametophyte and embryogenesis.8 Expression of AGL62, another Type I MADS domain transcription factor, is detected in antipodal cells of the female gametophyte and in developing endosperm. AGL62 regulates cellularization of developing endosperm during seed development.9 In addition to Type I MADS domain transcription factors, the divergent MADS domain transcription factors (MIKC*) have been shown to regulate male reproductive cells. Members of the Arabidopsis and rice MIKC* subgroup are predominantly expressed during the late stage of pollen development.10,11 In line with the expression patterns, mutations in MIKC*-type transcription factors cause reduction in pollen viability and defects in pollen tube growth.11,12
Our previous study reported that expression of PaMADS39, PaMADS51, and PaMADS63 are tightly associated with the stage (70-80 d after pollination) when the female and male gametophytes are established and fertilization occurs (Table 1).1 PaMADS39 and PaMADS51 belong to the Mα-subclass of type I MADS-box genes and are closely related to Arabidopsis AGL23 and AGL62.1 Similar to temporal expression patterns of AGL23 and AGL62, PaMADS39 and PaMADS51 are expressed in reproductive tissues when fertilization occurs and embryo development initiates. It will be of future interest to investigate the tissue specific localization and molecular functions of PaMADS39 and PaMADS51 during sexual reproduction of Phalaenopsis orchids.
Table 1.
Transcript abundances of the indicated genes enriched in interior ovary tissues collected at 70/80 DAP of P. aphrodite by RNA-seq analysis. Expression pattern of the transcripts was confirmed by qRT-PCR (Fig. 1).
| FPKM values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Transcript ID |
Annotation |
30/40 DAP |
50/60 DAP |
70/80 DAP |
90/100/120 DAP |
140/160 DAP |
180/200 DAP |
PLB |
Protocorm |
Young leaves |
Stalk buds |
| orchid.id141182.tr411193 | PaDRM1 | 0.0 | 0.0 | 7.5 | 0.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| orchid.id175307.tr208591 | PaCMT1 | 13.9 | 30.7 | 103.7 | 15.3 | 7.8 | 4.6 | 7.1 | 9.0 | 11.4 | 8.5 |
| orchid.id159191.tr200827 | PaSUVR1 | 0.7 | 4.6 | 13.5 | 3.9 | 1.9 | 1.0 | 0.2 | 0.7 | 1.3 | 0.5 |
| orchid.id153148.tr220461 | PaIAA30L1 | 0.0 | 0.0 | 8.9 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.2 |
| orchid.id123237.tr401072 | PaEIN3L1 | 2.8 | 0.8 | 8.4 | 1.9 | 0.0 | 0.0 | 0.1 | 0.1 | 0.4 | 0.0 |
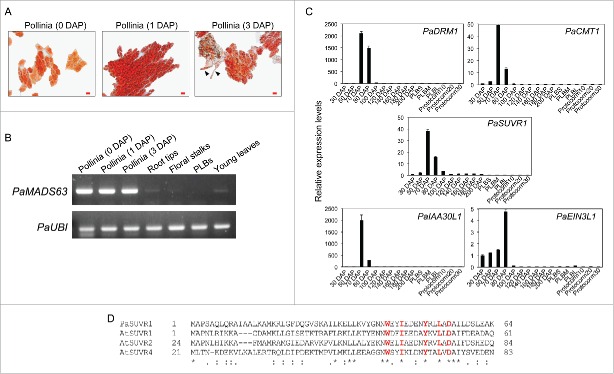
Phylogenetic analysis puts PaMADS63 in the MIKC* subclass of type II MADS domain transcription factors.1 The MIKC* type MADS domain proteins belong to a divergent class of type II MADS domain proteins and are expressed specifically in mature pollen of Arabidopsis13 and rice,11 and in gametophytic generation of moss Funaria hygrometrica.14 Functional characterization of MIKC* heterodimers has shown that they function as master regulators of pollen maturation programs.11,12 Similarly, PaMADS63 mRNA was preferentially expressed in pollinia, and pollinia in hydrated (one day after pollination) and germinated (3 d after pollination) conditions (Fig. 1A and Fig. 1B). PaMADS63 mRNA was hardly detectable in root tips, floral stalks, PLBs, and young leaves (Fig. 1B). This suggests that PaMADS63 may play an important role during pollen development in Phalaenopsis orchids.
Figure 1.
(A) Acetocarmine staining images showing pollinia at 0, 1, and 3 d after pollination (DAP). Arrowheads point to the pollen tubes. Scale bar = 20 μm. (B) Semi-quantitative RT-PCR showing expression of PaMADS63 mRNA in the indicated tissues. PaUBI1 was used as an internal control. PLB, Protocorm-like-body. (C) Quantitative RT-PCR showing expression patterns of PaDRM1, PaCMT1, PaSUVR1, PaIAA30L1, and PaEIN3L1 in interior tissues of developing ovaries from 30 to 200 d after pollination (DAP). Small-sized PLB (PLBS), medium-sized PLB (PLBM), large-sized PLB (PLBL), 10-day-old protocorms (protocorm10), 20-day-old protocorms (protocorm20), and 30-day-old protocorms (protocorm30). (D) Alignment of the WIYLD domain of Arabidopsis SUVR proteins and PaSUVR1. AtSUVR1, AT1G04050; AtSUVR2, AT5G43990; AtSUVR4, AT3G04380.
DNA methylation and fertilization
Dynamic regulation of DNA methylation is important for plant sexual reproduction.15 In plants, de novo and maintenance of DNA methylation are important to regulate genome imprinting and gene expression. Data indicate an overall reduction of DNA methylation in the germ line during gametogenesis.16 The partially demethylated genome of the zygote undergoes methylation reprogramming through de novo DNA methylation after fertilization.17 In Arabidopsis, resetting the methylation of the early-stage of embryos requires the DOMAINS REARRANGED METHYLTRNSFERASES 2 (DRM2) and CHROMOMETHYLASE 3 (CMT3), which methylate cytosine residues in the CHH, CHG, and CG context,18 and in CHG context19 respectively. It is hypothesized that DNA methylation reprogramming in the zygotes and early stage of embryos after fertilization contributes to the maintenance of genome integrity in the embryo.2 Interestingly, our RNA-seq dataset identified PaDRM1 and PaCMT1 as overrepresented genes expressing at 70-80 DAP when fertilization occurs (Table 1). These temporal expression patterns were verified by qRT-PCR (Fig. 1C). This data suggests that PaDRM1 and PaCMT1 may be important in resetting methylation status after fertilization in Phalaenopsis orchids. In addition to DRM and CMT genes, the SET-domain histone methyltransferase has been shown to facilitate DNA methylation by enabling the propagation of CHG methylation through methylation of the lysine residue 9 of histone 3 (H3K9). Specifically, the methylated CHG DNA recruits the histone H3K9 methyltransferase SU(VAR)3-9 homolog 4 (SUVH4) that deposits H3K9met2 marks, which promote binding to CMT3.20 We have identified a Phalaenopsis histone H3K9 methyltransferase SU(VAR)3-9 related gene, PaSUVR1, whose expression was specifically enriched during fertilization and early establishment of embryogenesis (Table 1 and Fig. 1C). The SUVR proteins are plant-specific SET-domain proteins that belong to a subgroup of SU(VAR)3-9-like proteins and are related to SUVH proteins.21 The SUVR proteins contain a conserved WIYLD domain (Fig. 1D) that has been shown to bind ubiquitin and regulate methylation of H3K9.22 It is possible that PaDRM1, PaCMT1, and PaSUVR1 are coordinated to regulate epigenetic reprogramming after fertilization of Phalaenopsis orchids.
Hormone regulation and fertilization
Ethylene and auxin play essential roles in regulating fertilization in plants.23-25 Successful fertilization requires termination of subsequent pollen tube attraction to prevent polyspermy. The pollen tube block is mediated by ETHYLENE INSENSITIVE 3 (EIN3)-dependent ethylene response signaling, which leads to programmed cell death of synergid cells that guide pollen tubes to the ovules.24 Degeneration of the synergid cells ensures one-on-one pairing of male and female gametes and prevents fertilization of an egg by more than one sperm. Fertilization triggers auxin response that facilitates GA biosynthesis specifically in the ovules.23 GA signaling is important to coordinate growth and development of fruits. The increasing auxin levels after fertilization is also important to drive central cell division and endosperm development via the AGL62-dependent pathway.25
Ethylene and auxin have been shown to regulate ovary and ovule development after pollination in orchids.26,27 However, their roles during fertilization have yet to be determined. Based on our RNA-seq data set, expression of Phalaenopsis INDOLE-3-ACETIC ACID inducible 30 like (PaIAA30L1) and EIN3-like 1 (PaEIN3L1) genes is specifically enriched at 70–80 DAP when fertilization occurs and embryogenesis initiates (Table 1).1 PaIAA30L1 encodes an auxin-induced AUX/IAA transcriptional repressor. PaEIN3L1 encodes an EIN3 domain transcriptional activator.28 The fertilization-associated expression pattern of PaIAA30L1 and PaEIN3L1 was validated by qRT-PCR analysis (Fig. 1C). Given the temporal expression pattern of PaIAA30L1 and PaEIN3L1 and their annotated functions, we hypothesize that auxin and ethylene may play regulatory roles during fertilization and/or early embryo development in Phalaenopsis orchids. Similar to the expression pattern of PaIAA30L1, Arabidopsis IAA30 is expressed in seeds containing zygotes. In addition, Arabidopsis IAA30 is directly regulated by 2 embryogenesis regulators, LEAFY COTYLEDON 2 (LEC2, a B3 domain transcription factor) and AGL15. These experiments suggest that IAA30 plays roles at an earlier stage of embryo development.29,30 Consistently, IAA30 induced auxin signaling has been reported to be required for the AGL15-induced somatic embryogenesis program.30 Despite the absence of AGL15 in the Phalaenopsis genome, it is speculated that the 2 orchid B3 transcription factors (PaABI3L1 and PaABI3L2) may coordinate with PaIAA30L1-induced auxin signaling to regulate orchid embryogenesis.
Our comparative genome-wide transcriptome analysis in reproductive tissues has allowed us to identify fertilization/embryogenesis-associated genes in P. aphrodite. Further characterization of these genes will help understanding the molecular mechanisms governing fertilization and establishment of embryogenesis in Phalaenopsis orchids.
Methods
Plant materials and growth conditions
Phalaenopsis aphrodite Subsp. formosana (m1663) plants in 2.5- or 3-inch pots were purchased from Chain Port Orchid Nursery (Ping Tung, Taiwan). Plants were grown in a growth chamber with alternating 12 h light (23°C)/12 h dark (18°C) cycles.
Sample collection and acetocarmine staining
Orchid flowers were hand pollinated and developing ovaries were harvested as described previously.1 To visualize pollen and pollen tubes, samples were stained with acetocarmine as described previously.5 Protocorms and PLBs were categorized and harvested for RNA extraction as described previously.1 Pollen, root tips with sizes smaller than 2 cm in length, 5-10 cm long floral stalks, and young leaves were collected for RNA extraction. The tissues samples were flash frozen in liquid nitrogen and stored in a freezer at -80°C.
RNA isolation and RACE-PCR
RNA was isolated as described previously.1 DNA-free RNA was reverse transcribed in the presence of a mixture of oligo dT and random primers (9:1 ratio) using the GoScript Reverse Transcription System (Promega) according to the manufacturer's instructions.
RACE-PCR was carried out using a SMARTer RACE cDNA amplification kit according to the manufacturer's instructions (Clontech). The amplified PaCMT1, PaDRM1, PaSUVR1, and PaEIN3L1 cDNAs were verified by sequencing. The Genbank accession numbers of PaCMT1, PaDRM1, PaEIN3L1, PaIAA30L1, and PaSUVR1 are KX759037, KX759038, KX759039, KX759040, and KX759041 respectively.
Quantitative RT-PCR and semi-quantitative RT-PCR
The RNA samples used for quantitative RT-PCR were independent from samples used for RNA-seq. For quantitative and semi-quantitative RT-PCR, 10 μl of PCR reaction containing 2.5 μl of 1/20 diluted cDNA, 0.2 μM of primers, and 5 μl of 2× KAPA SYBR FAST master mix (KAPA Biosystems). PCR was programmed as follows: 95°C for 1 min, 40 cycles at 95°C for 5 s and 55 to 62°C for 20 s. Each reaction was performed in triplicate. Primer pairs and the annealing temperature used for quantitative PCR are listed in Table 2. Ubiquitin (PaUBI1)6 was used as an internal control.
Table 2.
List of primer pairs and annealing temperature used for semi-quantitative RT-PCR and qRT-PCR.
| Gene name | Forward primer | Reverse primer | Amplicon size (bp) | TM(°C) |
|---|---|---|---|---|
| PaDRM1 | 5′-ACATCAAGCCGGAGTTTGTC-3′ | 5′-ATGCCTCTTGGATGGTTTTG-3′ | 132 | 58 |
| PaCMT1 | 5′-TATGACCATCGGCCTCTACC-3′ | 5′-GCTTCATGGATGGGTCAAGT-3′ | 145 | 58 |
| PaSUVR1 | 5′-TGGGAGGTCTTTCAATGGTT-3′ | 5′-TTGAGGACTCTGAAGCACCA-3′ | 134 | 62 |
| PaIAA30L1 | 5′-GGAAAAAGAAGCAGGCCTTT-3′ | 5′-TCCAAATGAAGTTCCATCCA-3′ | 159 | 58 |
| PaEIN3L1 | 5′-CAAGGACCTCTCCGATACCA-3′ | 5′-GCGGAGGTACATTCTTCGTC-3′ | 170 | 60 |
| PaUBI1 | 5-AACTCCATCGCCTTCCTCTT-3′ | 5′-TGAAGCATGGCATCAATTTC-3′ | 101 | 55 to 62 |
| PaMADS63 | 5′-AGGGAGGGAAAACAGCAAGT-3′ | 5′-ATGCCCTGAGAGTGTGTTCC-3′ | 248 | 59 |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We express our appreciation to Ms. Miranda Loney for English editing and the AS-BCST Greenhouse Core Facility for greenhouse service.
Funding
This work was supported by the Development Program of Industrialization for Agricultural Biotechnology grants (to SCF); and in part by a grant (to SCF) from the Biotechnology Center in Southern Taiwan, Academia Sinica.
Author Contributions
SCF conceived and designed the experiments. JCC, and MJW performed the experiments and analyzed the data. SCF wrote the paper.
ORCID
Su-Chiung Fang http://orcid.org/0000-0001-6894-5025
References
- 1.Fang SC, Chen JC, Wei MJ. Protocorms and protocorm-like bodies are molecularly distinct from zygotic embryonic tissues in Phalaenopsis aphrodite. Plant Physiol 2016; 171:2682-700; PMID:27338813; http://dx.doi.org/ 10.1104/pp.16.00841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawashima T, Berger F. Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet 2014; 15:613-24; PMID:25048170; http://dx.doi.org/ 10.1038/nrg3685 [DOI] [PubMed] [Google Scholar]
- 3.Rutley N, Twell D. A decade of pollen transcriptomics. Plant Reprod 2015; 28:73-89; PMID:25761645; http://dx.doi.org/ 10.1007/s00497-015-0261-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dresselhaus T, Sprunck S, Wessel GM. Fertilization mechanisms in flowering plants. Curr Biol 2016; 26:R125-39; PMID:26859271; http://dx.doi.org/ 10.1016/j.cub.2015.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen JC, Fang SC. The long pollen tube journey and in vitro pollen germination of Phalaenopsis orchids. Plant Reprod 2016; 29:179-88; PMID:27016359; http://dx.doi.org/ 10.1007/s00497-016-0280-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin HY, Chen JC, Wei MJ, Lien YC, Li HH, Ko SS, Liu ZH, Fang SC. Genome-wide annotation, expression profiling, and protein interaction studies of the core cell-cycle genes in Phalaenopsis aphrodite. Plant Mol Biol 2014; 84:203-26; PMID:24222213; http://dx.doi.org/22872082 10.1007/s11103-013-0128-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smaczniak C, Immink RG, Angenent GC, Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 2012; 139:3081-98; PMID:22872082; http://dx.doi.org/ 10.1242/dev.074674 [DOI] [PubMed] [Google Scholar]
- 8.Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM, Colombo L. AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J 2008; 54:1037-48; PMID:18346189; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03485.x [DOI] [PubMed] [Google Scholar]
- 9.Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 2008; 20:635-47; PMID:18334668; http://dx.doi.org/ 10.1105/tpc.107.055137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verelst W, Saedler H, Munster T. MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol 2007; 143:447-60; PMID:17071640; http://dx.doi.org/ 10.1104/pp.106.089805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Cui S, Wu F, Yan S, Lin X, Du X, Chonga K, Schilling S, Theißenc G, Meng Z. Functional conservation of MIKC*-Type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell 2013; 25:1288-303; PMID:23613199; http://dx.doi.org/ 10.1105/tpc.113.110049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adamczyk BJ, Fernandez DE. MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol 2009; 149:1713-23; PMID:19211705; http://dx.doi.org/ 10.1104/pp.109.135806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verelst W, Twell D, de Folter S, Immink R, Saedler H, Munster T. MADS-complexes regulate transcriptome dynamics during pollen maturation. Genome Biol 2007; 8:R249; PMID:18034896; http://dx.doi.org/ 10.1186/gb-2007-8-11-r249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zobell O, Faigl W, Saedler H, Munster T. MIKC* MADS-box proteins: conserved regulators of the gametophytic generation of land plants. Mol Biol Evol 2010; 27:1201-11; PMID:20080864; http://dx.doi.org/ 10.1093/molbev/msq005 [DOI] [PubMed] [Google Scholar]
- 15.Jullien PE, Berger F. DNA methylation reprogramming during plant sexual reproduction? Trends Genet 2010; 26:394-9; PMID:20609490; http://dx.doi.org/ 10.1016/j.tig.2010.06.001 [DOI] [PubMed] [Google Scholar]
- 16.Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 2006; 124:495-506; PMID:16469697; http://dx.doi.org/ 10.1016/j.cell.2005.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calarco JP, Borges F, Donoghue MT, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijó J, Becker J, et al.. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 2012; 151:194-205; PMID:23000270; http://dx.doi.org/ 10.1016/j.cell.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, Vashisht AA, Chory J, Wohlschlegel JA, Patel DJ, Jacobsen SE. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 2014; 157:1050-60; PMID:24855943; http://dx.doi.org/ 10.1016/j.cell.2014.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 2001; 292:2077-80; PMID:11349138; http://dx.doi.org/ 10.1126/science.1059745 [DOI] [PubMed] [Google Scholar]
- 20.Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 2007; 17:379-84; PMID:17239600; http://dx.doi.org/ 10.1016/j.cub.2007.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorstensen T, Fischer A, Sandvik SV, Johnsen SS, Grini PE, Reuter G, Aalen RB. The Arabidopsis SUVR4 protein is a nucleolar histone methyltransferase with preference for monomethylated H3K9. Nucleic Acids Res 2006; 34:5461-70; PMID:17020925; http://dx.doi.org/ 10.1093/nar/gkl687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veiseth SV, Rahman MA, Yap KL, Fischer A, Egge-Jacobsen W, Reuter G, Zhou MM, Aalen RB, Thorstensen T. The SUVR4 histone lysine methyltransferase binds ubiquitin and converts H3K9me1 to H3K9me3 on transposon chromatin in Arabidopsis. PLoS Genet 2011; 7:e1001325; PMID:21423664; http://dx.doi.org/ 10.1371/journal.pgen.1001325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorcey E, Urbez C, Blazquez MA, Carbonell J, Perez-Amador MA. Fertilization-dependent auxin response in ovules triggers fruit development through the modulation of gibberellin metabolism in Arabidopsis. Plant J 2009; 58:318-32; PMID:19207215; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03781.x [DOI] [PubMed] [Google Scholar]
- 24.Völz R, Heydlauff J, Ripper D, von Lyncker L, Gross-Hardt R. Ethylene signaling is required for synergid degeneration and the establishment of a pollen tube block. Dev Cell 2013; 25:310-6; PMID:23673332; http://dx.doi.org/ 10.1016/j.devcel.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 25.Figueiredo DD, Batista RA, Roszak PJ, Köhler C. Auxin production couples endosperm development to fertilization. Nature Plants 2015; 1:15184; PMID:27251719; http://dx.doi.org/ 10.1038/nplants.2015.184 [DOI] [PubMed] [Google Scholar]
- 26.Zhang XS, O'Neill SD. Ovary and gametophyte development are coordinately regulated by auxin and ethylene following pollination. Plant Cell 1993; 5:403-18; PMID:12271070; http://dx.doi.org/ 10.1105/tpc.5.4.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai WC, Hsiao YY, Pan ZJ, Kuoh CS, Chen WH, Chen HH. The role of ethylene in orchid ovule development. Plant Science 2008; 175:98-105; http://dx.doi.org/ 10.1016/j.plantsci.2008.02.011 [DOI] [Google Scholar]
- 28.Yamasaki K, Kigawa T, Inoue M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, Terada T, et al.. Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. J Mol Biol 2005; 348:253-64; PMID:15811366; http://dx.doi.org/ 10.1016/j.jmb.2005.02.065 [DOI] [PubMed] [Google Scholar]
- 29.Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci U S A 2006; 103:3468-73; PMID:16492731; http://dx.doi.org/ 10.1073/pnas.0511331103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 2009; 21:2563-77; PMID:19767455; http://dx.doi.org/ 10.1105/tpc.109.068890 [DOI] [PMC free article] [PubMed] [Google Scholar]