Abstract
Here, we present a natural product discovery approach whereby structures are bioinformatically predicted from primary sequence and produced by chemical synthesis (synthetic-bioinformatic natural products, syn-BNPs), circumventing the need for bacterial culture and gene expression. When applied to nonribosomal peptide synthetase gene clusters from human-associated bacteria we identified the humimycins. These antibiotics inhibit lipid II flippase and potentiate β-lactam activity against methicillin-resistant Staphylococcus aureus in mice, potentially providing a new treatment regimen.
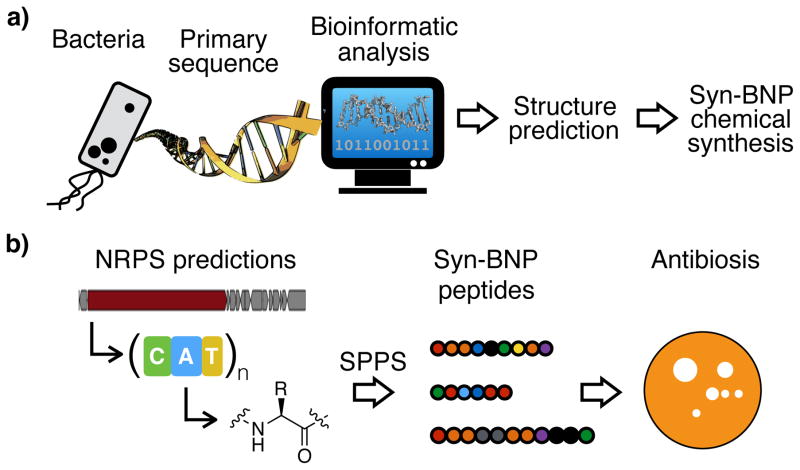
The characterization of small molecules produced by bacteria in laboratory culture has been a key step to understanding bacterial physiology and developing small molecule therapeutics.1 As successful as this approach has been for identifying novel bioactive small molecules, extensive sequencing of bacterial genomes and metagenomes has revealed that the bacterial biosynthetic diversity traditionally accessed in the laboratory represents only a small fraction of what is predicted to exist in nature.2,3 This shortcoming arises from our inability to culture most bacteria in the laboratory and from the fact that most biosynthetic gene clusters remain silent under laboratory fermentation conditions.4 Here, we present a bioactive small molecule discovery pipeline that circumvents the requirement for either bacterial culture or gene cluster expression. In our approach, natural product structures are bioinformatically predicted from primary sequence data and produced by chemical synthesis. Since natural products often appear in nature as families of related structures with the same biological activity, we reasoned that even if our structural predictions were not perfect, many syn-BNPs would be sufficiently accurate representations of nature to elicit the intended bioactivities. We have called these bioinformatically inspired compounds syn-BNPs for Synthetic Bioinformatic Natural Products (Fig. 1a).
Figure 1. Overview of the Syn-BNP approach.
a) Advances in our understanding of natural product biosynthesis have enabled the prediction of natural product structures from primary sequence data alone. In a syn-BNP approach these structures are accessed through chemical synthesis instead of biosynthesis. b) Here we apply a syn-BNP approach to NRPs predicted from human microbiome sequence data and assay these new molecules for antibiosis activities.
The human microbiome is an exemplary test case for a syn-BNP discovery approach. Tremendous resources have been allocated to the sequencing and bioinformatic analysis of the human microbiome.5,6 Nevertheless, the functional characterization of this data, including commensal bacteria-encoded natural product biosynthetic gene clusters, remains rare. Our interest in exploring syn-BNPs encoded by the human microbiota stems from the potential use of these metabolites as therapeutics and as tools for improving our understanding of human microbiome functions. Because antibiotics can serve as medicines and as modulators of the composition of the human microbiome, we chose to screen syn-BNPs predicted from the human microbiome for antibacterial activity against human associated commensal and pathogenic bacteria.
Systematic bioinformatic analysis of sequenced bacterial genomes indicate that nonribosomal peptides (NRPs) are one of the most common and diverse families of complex secondary metabolites produced by bacteria.7,8 Over the past two decades, a number of models have been developed for predicting the identity, order, and modification of the amino acids comprising an NRP, based solely on the primary sequence of NRP megasynthetases.9–12 Concurrently, solid phase peptide synthesis (SPPS) of structurally diverse peptides has become rapid and economical, making NRP gene clusters an ideal test case for a syn-BNP approach (Fig. 1b).
Genomic sequence data from human (commensal and pathogenic) associated bacteria were bioinformatically queried for gene clusters predicted to encode large NRPs (≥5 residues), as short NRPs are often highly modified and are therefore not easily accessible using SPPS alone. This analysis led to the identification of 57 unique nonribosomal peptide synthetase (NRPS) gene clusters, from which we removed those that appeared to be incomplete in the existing sequence data and those containing more than one PKS module, a thioreductase domain, or any heterocyclization domains. The chemical outputs of the remaining 25 gene clusters, which we believed to be amenable to SPPS, were predicted using three published NRPS prediction algorithms (Stachelhaus, Minowa, and NRPSPredictor2) to produce syn-BNP targets.9–12 In instances where bioinformatic predictions for human microbiome associated gene clusters diverged strongly between algorithms (e.g., side-chains were predicted to carry opposite charges), multiple syn-BNP peptides were designed and synthesized. In all cases where NRPS gene clusters were bioinformatically predicted to encode an N-terminally acylated peptide, we elected to design the syn-BNP to be N-acylated with β-hydroxymyristic acid (HMA), a fatty acid commonly observed in NRPs.13 In total, 30 syn-BNPs targets were designed based on the gene clusters found in human commensal bacterial sequence data. After two rounds of SPPS using standard Fmoc chemistry, we obtained pure samples for 25 of the 30 targeted syn-BNPs (Supplementary Results, Supplementary Table 1).
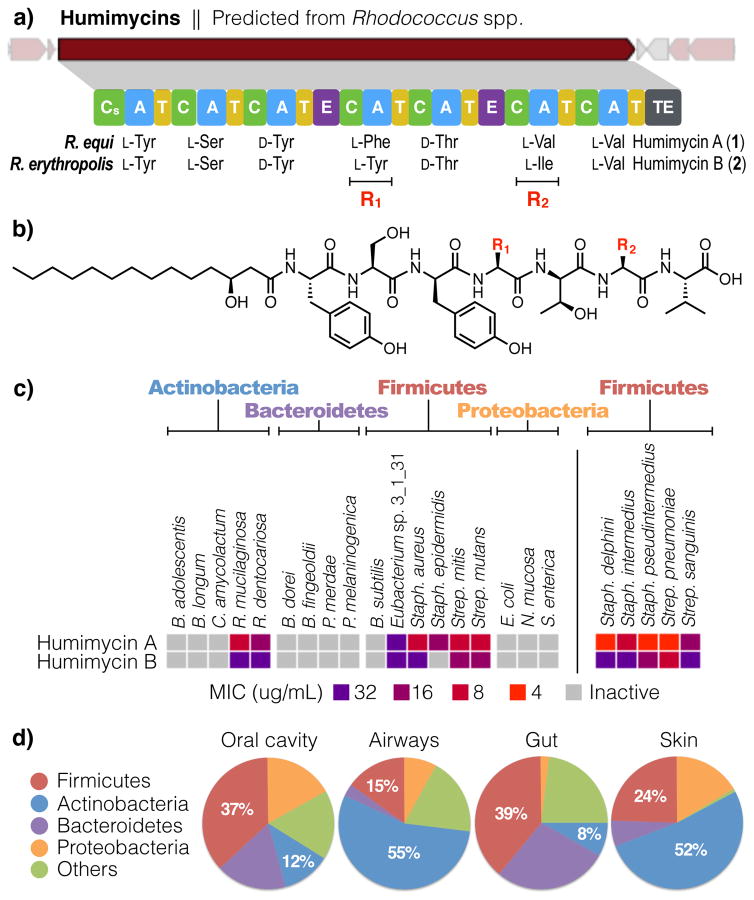
To identify novel antibiotic scaffolds with potential in vivo roles in shaping the ecology of the human microbiome, we assayed this collection of syn-BNPs for antibacterial activity against a panel of common human commensal and pathogenic bacteria. This led to the identification of two antibiotics we have trivially named humimycin A (1) and B (2) (human microbiome mycin, Fig. 2a). The humimycins were predicted from closely related NRPS gene clusters found in the genomes of Rhodococcus equi and Rhodococcus erythropolis, respectively. Bioinformatic analyses of these two NRPS gene clusters indicated that they encoded hepta-peptides that differed at only the fourth and sixth residues (F/Y and V/I, respectively, Fig. 2b). Both syn-BNPs were synthesized with N-terminal HMA modifications, due to the presence of starter condensation domains (Cs) in the gene clusters, which are associated with acylation of the first amino acid of an NRP.13
Figure 2. Discovery and screening of the humimycins.
a) The humimycins were predicted from closely related gene clusters found in two Rhodococcus spp. cultured from human subjects. b) Chemical structures of humimycin A (1) and B (2). The two antibiotics differ only at the fourth (F/Y) and sixth (V/I) residues. c) MIC values for the humimycins against a panel of human commensal and pathogenic bacteria were determined (n = 2). The right panel shows that the humimycins are particularly active against bacteria in the Staphylococcus and Streptococcus genus (n = 3).
Rhodococcus species have been extensively studied for natural product production using traditional fermentation-based discovery methods. None of these studies report the identification of a metabolite resembling the humimycins.14 Likewise, our extensive analysis of Rhodococcus species culture broth extracts by both LCUV and LCMS analysis did not reveal any metabolites related to the humimycins, suggesting that the humimycin gene cluster is silent under laboratory fermentation conditions.
The humimycins were found to be broadly active against Firmicutes and to show some activity against Actinobacteria, when screened for antibiosis against commensal and pathogenic bacteria (Fig. 2c). The humimycins are particularly active against Staphylococcus and Streptococcus species, including common members of the normal human flora such as S. aureus (minimum inhibitory concentration (MIC) 8 μg/mL) and S. pneumoniae (MIC 4 μg/mL). This spectrum of activity is interesting in light of the fact that Firmicutes and Actinobacteria dominate the human microbiota of the gut (Fig. 2d).15 In a structure-activity relationship study, we found that no residue in 1 could be replaced with alanine without dramatically impacting the potency of the antibiotic (Supplementary Table 3).
Humimycin A exhibits MICs ranging from 8–128 μg/mL against methicillin-resistant S. aureus (MRSA) clinical isolates (Supplementary Table 4). To study the antibacterial mode of action of the humimycins, we selected S. aureus USA300 mutants that could survive on 2.5 times the MIC (20 μg/mL) and sequenced the genomes of 23 resistant mutants. Upon comparison to the parent strain, we found that all 23 mutants contained one non-synonymous mutation in SAV1754, an essential gene in S. aureus (Supplementary Figure 3 and Table 5). Fifteen of these strains contained no other detectable mutations. Overexpression of SAV1754 in S. aureus confers resistance to humimycin A (MIC >128 μg/mL), further supporting inhibition of SAV1754 as a likely mode of action of the humimycins (Supplementary Table 6). The gene product of SAV1754 is believed to be a homolog of MurJ, a flippase responsible for the translocation of peptidoglycan precursors from the inside to the outside of the cell.16
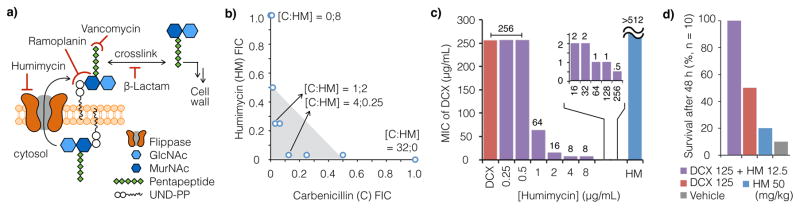
While MurJ is essential in many bacteria, including many important pathogens, it remains an underexplored antibacterial target.17 The ability of SAV1754 inhibitors to potentiate β-lactam antibiosis is thought to arise from the fact that both antibiotics target the same essential pathway, peptidoglycan biosynthesis (Fig. 3a). In a high throughput screen for molecules that could potentiate β-lactam antibiosis against otherwise resistant strains Merck & Co. identified synthetic small molecule inhibitors of SAV1754.18,19 Humimycin A exhibits a similar ability to restore β-lactam sensitivity to β-lactam resistant bacteria. For example, the MIC of carbenicillin (carboxypenicillin) was reduced from 32 to 1 μg/mL in the presence of 2 μg/mL humimycin A (0.25× MIC) against MRSA USA300 (Fig. 3b), one of the most predominant community-associated MRSA strains in the U.S.
Figure 3. Humimycin A and β-lactam act in synergy.
a) SAV1754 is the S. aureus homolog of MurJ, which is a flippase responsible for the transportation of peptidoglycan precursors across the cytoplasmic membrane. b) Carbenicillin (C) and humimycin A (HM) act synergistically to inhibit the growth of MRSA USA300 (n = 2). Fraction inhibitory concentration (FIC) values ≤0.5 defines synergy between two agents (shaded in light gray); [C:HM] denotes the respective inhibitory concentrations at each data point (μg/mL). c) The minimum inhibitory concentration (MIC) of humimycin A with dicloxacillin (DCX) alone and at various humimycin A concentrations against MRSA COL (n = 2) are shown in red and purple, respectively. Humimycin A alone does not inhibit MRSA COL growth (MIC >512 μg/mL, blue). d) Survival data for mice treated with humimycin A or dicloxacillin either alone or together using a MRSA COL peritonitis model (n = 10 mice per cohort) are shown. In this model humimycin potentiates β-lactam activity in vivo.
Humimycin A’s ability to potentiate β-lactam activity is also seen with strains where it alone shows no detectable antibacterial activity. For example, the MRSA COL strain, while not susceptible to humimycin A (MIC >512 μg/ml) and exhibiting a very high MIC for the β-lactam dicloxacillin (MIC 256 μg/mL), is sensitive to dicloxacillin at 8 μg/mL in the presence of as little as 4 μg/mL of humimycin A (Fig. 3c). The ability of humimycin A to potentiate β-lactam activity in vitro led us to explore the possibility that it might do the same in vivo. In murine tolerability studies humimycin A is tolerated at concentrations (>50 mg/kg), far exceeding those expected to be necessary for β-lactam potentiation. In a murine peritonitis-sepsis model treatment of a MRSA COL infection with dicloxacillin and humimycin together dramatically increases survival compared to treatment with either humimycin or dicloxacillin alone (Fig. 3d), potentially providing a novel MRSA treatment regimen.
While R. equi has historically been regarded as an opportunistic pathogen seen in animals and immune-compromised patients,20 R. erythropolis is found as a part of the normal human nasal, mouth and eye microbiota.21,22 Interestingly, the occurrence of Rhodococcus species in the gut increases dramatically to a median of 30% in some patients diagnosed with ulcerative colitis (UC).23 The production of an antibiotic with activity against Firmicutes and Actinobacteria could play a role in establishing the overpopulation of R. erythropolis in the UC gut as Firmicutes and Actinobacteria normally represent nearly half of the gut microbiota.15 In addition to the potential to provide new small molecule therapeutics, characterization of molecules inspired by commensal bacteria biosynthetic gene clusters can provide a means for developing hypotheses about how commensal bacteria affect human physiology. For example, the discovery of the humimycins provides a testable mechanistic hypothesis for how dysbiosis of gut microbiota might evolve in UC.24
Our identification of the humimycins using a syn-BNP approach validates this as a strategy for identifying bioactive metabolites and highlights the unique state of the field of natural product chemistry today. Extensive biosynthetic studies have culminated in our emerging ability to predict the structures of many natural products from primary sequence alone. While in this study we focused on the synthesis of linear peptides because of the ease with which they can be generated by SPPS, there are many ways to expand this approach to more topologically and functional complex NRPs. For example, the construction of cyclic peptides using purified thioesterase domains is compatible with SPPS.25 Based on our analysis of high-quality sequenced bacterial genomes in GenBank not associated with the human microbiome, there are currently more than 1,500 unique large NRPS gene clusters (encoding ≥5 amino acids) amenable to a SPPS-based syn-BNP approach. As the sequencing of microbial genomes is still in an early exponential growth phase, this number should only continue to grow for the foreseeable future (Supplementary Figure 2). With the development of improved bioinformatic prediction algorithms for biosynthetic gene cluster families beyond NRPSs and the incorporation of more sophisticated chemical and chemo-enzymatic synthesis steps into the production of syn-BNPs, we believe this approach will enable broad and rapid access to diverse bioactive compounds that are inspired by gene clusters found within the ever-growing assemblage of microbial sequence data.
Online Methods
Bioinformatic prediction of NRPs
Genome sequences of the human microbiota were downloaded from the NIH Human Microbiome Project (HMP, (ftp://ftp.ncbi.nlm.nih.gov/genomes/HUMAN_MICROBIOM/Bacteria)26 and the Human Oral Microbiome Database (HOMD, (ftp://ftp.homd.org/HOMD_annotated_genomes).27 The software package Antibiotics and Secondary Metabolite Analysis Shell (antiSMASH) v2.0 was used for the identification and prediction of NRP biosynthetic gene clusters encoded by these genomes.28 Syn-NRPs originating from the HMP and HOMD databases were named serially as [Human.N] and [Oral.N]. All syn-NRPs discussed in this manuscript are listed in Supplementary Table 1. AntiSMASH consults three prediction algorithms to call the amino acid substrate specificity of an adenylation domain (NRPSPredictor2, Stachelhaus code, and Minowa). A consensus prediction refers to the situation wherein two (or all three) algorithms make consistent substrate predictions for a given adenylation domain. In this case the predicted amino acid was used in the synthesis of the syn-BNP. In case of a minor conflict between prediction algorithms we opted for the amino acid with the smaller side-chain, e.g., Val/Leu/Ile and Ser/Thr. In case of major conflicts (e.g., where side-chains were predicted to carry opposite charges), both peptides were synthesized. Tyrosine and phenylalanine prediction made by NRPSPredictor2 or Stachelhaus code were chosen over tryptophan (Trp) predictions made Minowa, as we noticed that Trp is overrepresented in Minowa predictions. Lastly, tyrosine (Tyr) was used at the first residue in place of p-hydroxyphenylglycine (Hpg) in Human.8v1 and v2. To check the robustness of these NRPS prediction algorithms we carried out a similar analysis of NRPS gene clusters deposited in the MiBIG database and found that the core peptide encoded by the vast majority NRPS gene clusters was predicted correctly (Supplementary Figure 2).
Peptide synthesis
Resins for peptide synthesis were purchased from AnaSpec. Coupling reagents (PyBOP) and Nα-Fmoc/side-chain protected amino acids were purchased from P3BioSystems. 3-Hydroxymyristic acids were purchased from TCI America (racemic mixture) and Santa Cruz Biotechnology (pure enantiomers). All other chemical reagents and solvents were purchased from Sigma Aldrich. Reaction vessels were custom made by the Scientific Glassblowing Laboratory at the Department of Chemistry of Yale University.
Pure samples were obtained for 25 of the 30 syn-BNP peptides targeted for chemical synthesis (Supplementary Table 1). 20 of these peptides were purchased through the custom peptide synthesis service of GenScript Biotech Corporation and five were synthesized in-house. Peptides from GenScript were delivered as lyophilized materials that had been HPLC-purified and MS-verified (MALDI). All pure peptides were dissolved in DMSO at 12.8 mg/mL as stock solutions and stored at −20 °C. In-house peptide syntheses, including humimycin A and B, were built on Wang resin29 following standard Fmoc/tBu SPPS methods. The first amino acid (6 equiv.) was activated using DIC (3 equiv.) in 10% DMF/DCM (0°C), added to the resins in the presence of DMAP as a catalyst (0.1 equiv.) and shaken under nitrogen (4 h at 0°C). Unreacted resins were capped using acetic anhydride in pyridine (1 h). Fmoc removal was accomplished using three rounds of treatment with 20% piperidine in DMF (15, 10, and 5 min. each). All ensuing amino acids were coupled twice. In each coupling an Nα-Fmoc and side-chain protected amino acid was activated using a mixture of PyBOP (4 equiv.) and DIEA (8 equiv.), followed by reaction with the peptide on-resin (1 h). Peptides were cleaved by 95% TFA supplemented with TIS and H2O (2.5% of each, v/v) for 2 h, concentrated to approximately 10% of the original volume, diluted with aqueous MeCN (75%, v/v), passed through a 0.45 μm filter and HPLC-purified. All purified peptides were examined by LC/MS (ESI).
Characterization of the humimycins
A racemic mixture of 3-hydroxymyristic acid was used for N-terminal modification in our initial syntheses of all syn-BNPs. Humimycin A diastereomers showed different MIC values when tested against MRSA USA300 (Supplementary Table 3). The absolute stereochemistry of the most active diastereomer was determined by comparing HPLC-purified peptides from the bulk synthesis to independent batches of small-scale syntheses using enantiopure (R) and (S)-3-hydroxymyristic acid (Supplementary Figure 1). The more potent (S)-isomer is referred to as humimycin A (1). In the case of humimycin B the analogous (S)-isomer was purified and is referred to as compound 2. Humimycin A (1) HRMS: m/z calculated for [M − H]− (C58H84N7O14): 1102.6076, found: 1102.6075. Humimycin B (2) HRMS: m/z calculated for [M − H]− (C59H86N7O15): 1132.6182, found: 1132.6194.
Syn-BNP screening
Syn-BNPs were screened against a panel of commensal and pathogenic bacteria covering the four major phyla associated with the human microbiome. This included five Actinobacteria, four Bacteroidetes, six Firmicutes and three Proteobacteria species. All peptides were tested in duplicate for antibiosis activity. Assays were performed in microtiter plates, wherein each well contained growth media (see Supplementary Table 2 for a list of growth media) (100 μL), syn-BNP (32 μg/mL) and bacteria diluted 1,000-fold from a stationary phase culture. Binary antibiosis results for most bacteria were determined by visual inspection after static incubation at 37 °C for 18 h. P. melaninogenica and Eubacterium sp. 3_1_31 were grown for 36 h, and C. amycolactum was grown for 60 h. Specific MICs were determined for syn-BNPs that inhibited bacterial growth in this initial screen (see Susceptibility assays, part a). Bacteria species associated with the human flora were obtained from BEI Resources.
Susceptibility assays
a) Standard assays
MIC assays were performed in duplicate in 96-well microtiter plates based on the protocol recommended by Clinical and Laboratory Standards Institute.30 DMSO stock solutions of syn-BNPs (12.8 mg/mL) were added to the first well in a row and serially diluted (2 fold per transfer) across the microtiter plate. The last well was reserved for a peptide-free control. Overnight cultures of bacteria were diluted 5,000-fold and 50 μL was used as an inoculum in each well. MIC values were determined by visual inspection after 18 h incubation (37 °C, static growth).
b) Synergy assays
Synergistic β-lactam-humimycin activities were assessed through a two-dimensional (2D) susceptibility assay. Two fold serial dilutions were carried out as described above. Carbenicillin (a β-lactam antibiotic) was diluted serially from left to right, and humimycin was diluted serially from top to bottom. The highest concentration tested for both antibiotics was 32 μg/mL. Fractional inhibitory concentration (FIC) is defined as the ratio of the apparent synergistic MIC divided by the MIC of the antibiotic measured alone.29
Selection of humimycin A resistant mutants
A single S. aureus USA300 colony (the parent) from a freshly struck plate was inoculated into LB medium and grown overnight at 37 °C. Part of the overnight culture (4 mL) was spun down and kept frozen at −20 °C. The rest of the overnight culture was diluted 100-fold, supplemented with humimycin A at 20 μg/mL (2.5X MIC) and 100 μL aliquots was distributed into 200 unique microtiter plate wells. Growth was observed in 50 wells after overnight incubation, indicating the presence of bacteria with mutation(s) conferring humimycin A resistance. Approximately 2 μL of culture from each of these wells was used to inoculate freshly prepared 100 μL aliquots of LB media supplemented with humimycin A (20 μg/mL). The resulting cultures after overnight incubation were struck out for single colonies on LB/agar plates supplemented with humimycin A (20 μg/mL) for single colonies.
Genome sequencing
Single colonies of 23 humimycin A resistant mutants as well as the USA300 parent were individually inoculated into 4 mL of LB media free of any antibiotics. After overnight incubation cells were collected by centrifugation. DNA extractions were performed using a MasterPure Purification Kit (EpiCentre Biotechnologies). Multiplex sequencing libraries were prepared from the resulting genomic DNA using a Nextera XT DNA Sample Preparation Kit (FC-131-1024) with Nextera XT Index kit (FC-131-1001) based on protocols provided by the manufacturer (Illumina). Briefly, the genomic DNA was treated with RNase and quantified using the Qubit dsDNA HS Assay System (Q32854, ThermoFisher Scientific). Tagmentation and PCR amplification proceeded according to the manufacturer’s protocol, after which the quality and size of the libraries were verified using HS D1000 ScreenTape (TapeStation 2200, Agilent Technologies). Libraries were pooled at equimolar concentrations and column purified by NucleoSpin Gel and PCR Cleanup (MN-750609-250, Macherey-Nagel). The resulting tagged DNA library was size-selected by E-Gel (Life Technologies) and the 450 bp band was excised. The final library pool was checked for molarity on TapeStation and sequenced using MiSeq Reagent Kit v3 (MS-102-3003, Illumina).
Mutation (SNP) identification
De-barcoded MiSeq reads were assessed for mutations by comparing each read against the reference genome of Staphylococcus aureus USA300_FPR3757 (RefSeq assembly accession: GCF_000013465.1). All reads were mapped to the reference genome using SNIPPY (https://github.com/tseemann/snippy) for the identification of variants. SNIPPY is a wrapper of several programs including freebayes (https://github.com/ekg/freebayes).32 Single-nucleotide polymorphisms (SNP) observed in the parent strain were then subtracted from those observed in the humimycin A resistant strains, resulting in a final list of SNPs (Supplementary Table 5).
Cloning and overexpression of SAV1754
The SAV1754 gene was PCR amplified from wild type S. aureus USA300 and a mutant resistant to humimycin A (mutant no. 8, Supplementary Table 5). PCR products and the pRMC233 vector were digested (SacI/KpnI) and ligated, followed by transformation into S. aureus RN4220 and selection on BHI agar plates containing chloramphenicol (10 μg/mL). The recombinant plasmids were verified by DNA sequencing. Overnight cultures of the resulting S. aureus strains were used to inoculate LB containing chloramphenicol (10 μg/mL). Late log-phase cultures (OD600 ~0.8) were induced by anhydrotetracycline (50 ng/mL) for 5 h and then tested in the presence of anhydrotetracycline (5 ng/mL) for susceptibilities against humimycin A. Primer sequences, PCR conditions, and susceptibility data are listed in Supplementary Table 6.
Murine peritonitis-sepsis model
Female outbred Swiss Webster mice were used for this study. MRSA COL was grown in Mueller-Hinton broth at 37°C overnight and diluted with 5% hog mucin and 0.9% NaCl to provide challenge inoculum of approximately 5 × 108 CFU per mouse in a volume of 0.5 mL via intraperitoneal injection. Forty mice were randomly grouped into 10 per cohort, and each group was given single doses of vehicle (20% DMA, 40% PEG, 40% D5W), humimycin (HM) at 50 mg/kg, dicloxacillin (DCX) at 125 mg/kg, and HM:DCX combination at 12.5 mg/kg HM:125 mg/kg DCX 1 h post-infection via IV injection. Mice were maintained in accordance with American Association for Accreditation of Laboratory Care criteria. The Rutgers University Institutional Animal Care and Use Committee approved all animal procedures.
Supplementary Material
Acknowledgments
We thank the Fischetti (MRSA), Tomasz (MRSA) and Marraffini (S. aureus, S. delphini, S. intermedius, and S. pseudo-intermedius) laboratories at the Rockefeller University for providing strains. This work was support by the Rainin Foundation, NIH grants U19AI109713 (D.S.P) and F32 29 AI110029 (Z.C.P.).
Footnotes
Author Contributions
SFB conceived of the project. JC and XVF carried out antibiosis assays, spectrum of activity screening, and resistant mutant selection. DI, HAZ, RGM, MJ, SS, and JSF carried out peptide synthesis on large scale. MAT carried out genome sequencing. LJC and EAG screened anaerobic bacteria. BVBR and ZCP carried out bioinformatic analysis. SP and DSP carried out mice studies.
Financial Interest Statement
The authors declare no competing financial interests.
References
- 1.Newman DJ, Cragg GM. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 2.Charlop-Powers Z, Milshteyn A, Brady SF. Curr Opin Microbiol. 2014;19:70–75. doi: 10.1016/j.mib.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piel J. Annu Rev Microbiol. 2011;65:431–453. doi: 10.1146/annurev-micro-090110-102805. [DOI] [PubMed] [Google Scholar]
- 4.Rutledge PJ, Challis GL. Nat Rev Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 5.Qin J, et al. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, et al. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlop-Powers Z, et al. eLife. 2015;4 doi: 10.7554/eLife.05048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doroghazi JR, et al. Nat Chem Biol. 2014;10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stachelhaus T, Mootz HD, Marahiel MA. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 10.Minowa Y, Araki M, Kanehisa M. J Mol Biol. 2007;368:1500–1517. doi: 10.1016/j.jmb.2007.02.099. [DOI] [PubMed] [Google Scholar]
- 11.Rottig M, et al. Nucleic Acids Res. 2011;39:W362–367. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber T, et al. Nucleic Acids Res. 2015;43:W237–243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH. BMC Evol Biol. 2007;7:78. doi: 10.1186/1471-2148-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitagawa W, Tamura T. Microbes Environ. 2008;23:167–171. doi: 10.1264/jsme2.23.167. [DOI] [PubMed] [Google Scholar]
- 15.D’Argenio V, Salvatore F. Clin Chim Acta. 2015;451:97–102. doi: 10.1016/j.cca.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Sham LT, et al. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sewell EW, Brown ED. J Antibiot. 2014;67:43–51. doi: 10.1038/ja.2013.100. [DOI] [PubMed] [Google Scholar]
- 18.Lee SH, et al. Sci Transl Med. 2016;8:329ra332. doi: 10.1126/scitranslmed.aad7364. [DOI] [PubMed] [Google Scholar]
- 19.Huber J, et al. Chem Biol. 2009;16:837–848. doi: 10.1016/j.chembiol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Kraal L, Abubucker S, Kota K, Fischbach MA, Mitreva M. PLoS One. 2014;9:e97279. doi: 10.1371/journal.pone.0097279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen TT, Kirkeby LP, Poulsen K, Reinholdt J, Kilian M. APMIS. 2000;108:663–675. doi: 10.1034/j.1600-0463.2000.d01-13.x. [DOI] [PubMed] [Google Scholar]
- 22.Graham JE, et al. Invest Ophthalmol Vis Sci. 2007;48:5616–5623. doi: 10.1167/iovs.07-0588. [DOI] [PubMed] [Google Scholar]
- 23.Lepage P, et al. Gastroenterology. 2011;141:227–236. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Jostins L, et al. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohli RM, Walsh CT, Burkart MD. Nature. 2002;418:658–661. doi: 10.1038/nature00907. [DOI] [PubMed] [Google Scholar]
- 26.Human Microbiome Project Consortium. Nature. 2012;486:215. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen T, et al. Database (Oxford) 2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blin K, et al. Nucleic Acids Res. 2013;41:W204. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang SS. J Am Chem Soc. 1973;95:1328. doi: 10.1021/ja00785a602. [DOI] [PubMed] [Google Scholar]
- 30.Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard. 9. Clinical and Laboratory Standards Institute; Wayne, PA: 2012. [Google Scholar]
- 31.Hall MJ, Middleton RF, Westmacott D. J Antimicrob Chemother. 1983;11:427. doi: 10.1093/jac/11.5.427. [DOI] [PubMed] [Google Scholar]
- 32.Garrison E, Marth G. 2012 arXiv. [Google Scholar]
- 33.Corrigan RM, Foster TJ. Plasmid. 2009;61:126. doi: 10.1016/j.plasmid.2008.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.