Abstract
Elicitation of broadly neutralizing antibody (bNAb) responses to the conserved elements of the HIV-1 envelope glycoproteins (Env), including the primary receptor CD4 binding site (CD4bs), is a major focus of vaccine development yet to be accomplished. However, a large number of CD4bs-directed bNAbs have been isolated from HIV-1 infected individuals. Comparison of the the routes of binding used by the CD4bs-directed bNAbs from patients and the vaccine-elicited CD4bs-directed monoclonal antibodies (MAbs) indicates that the latter fail to neutralize primary virus isolates since they approach the Env spike with a vertical angle and contact the specific surface residues occluded in the native spike, including the bridging sheet on gp120. To preferentially expose the CD4bs and direct the immune response away from the bridging sheet, resulting in an altered angle of approach, we engineered an immunogen consisting of gp120 core in complex with the prototypic CD4i antibody, 17b. This mAb directly contacts the bridging sheet but not the CD4bs. The complex was further stabilized by chemical crosslinking to prevent dissociation. Rabbits immunized with the crosslinked complex displayed earlier affinity maturation, achieving tier 1 virus neutralization compared to animals immunized with gp120 core alone. Immunization with the crosslinked complex induced transient antibody responses with binding specificity similar to the CD4bs-directed bNAbs. MAbs derived from complex-immunized rabbits displayed footprints on gp120 more distal from the bridging sheet as compared to previous vaccine-elicited CD4bs antibodies, indicating that Env-antibody complexes effectively dampen immune responses to undesired immunodominant bridging sheet determinants.
Keywords: HIV-1, Env, neutralizing antibody, CD4 binding site, CD4i, gp120 core, complex, cross linking, immunization, VRC01
INTRODUCTION
The human immunodeficiency viruses type 1 (HIV-1) cause the destruction of CD4+ lymphocytes in their host, resulting in the development of acquired immunodeficiency syndrome (AIDS) (1, 2). The entry of HIV into host cells is mediated by the viral envelope glycoproteins (Env), a trimeric complex consisting of the exterior surface protein, gp120 and the transmembrane protein, gp41. Gp120 first contacts the host target cell primary receptor, CD4, which creates a high-affinity binding site for the coreceptor CCR5, subsequently leading to membrane fusion and virus entry (3). Thus the CD4 binding site (CD4bs) on gp120 is a conserved and accessible structural element critical for Env function and virus entry.
Besides its crucial role in mediating virus entry, HIV Env is the sole viral target of host neutralizing antibodies. It has become increasingly clear that a protective Env-specific antibody response is likely the most critical component of an effective HIV-1 vaccine (4). Due to the extremely high genetic and antigenic variability of HIV Env, elicitation of broadly reactive antibody responses to the conserved Env elements, including the CD4bs, is a logical focus of HIV vaccine development.
Recent progress in isolating monoclonal antibodies (MAbs) including CD4bs-directed broadly neutralizing antibodies (bNAbs) (5–10) such as VRC01 from HIV-infected individuals allows detailed characterization of bNAb cognate epitopes on Env. These high-resolution functional and structural epitope analyses revealed that CD4bs-directed bNAbs include antibodies with two distinct modes of recognition: 1) VRC01-class bNAbs with either VH1-2 or VH1-46 immunoglobulin (Ig) heavy chain gene segment using CDRH1 &2 to contact gp120 CD4bs; and 2) CDRH3-dominant recognition (11). Structural and mutagenic studies suggest that VRC01 targets the viral Env functional spike by partially mimicking the viral receptor CD4 and approaching the CD4bs at a more favorable angle than other CD4bs-directed antibodies or even the soluble form of the receptor CD4 (sCD4) (6, 12). A unique feature of VRC01-class CD4bs bNAbs is that they display a high affinity for modified gp120 core proteins with conformational constraints, which is distinct from CD4bs-directed non-broadly neutralizing MAbs (non-bNAbs). For example, VRC01-class bNAbs bind a stabilized gp120 core (13, 14) engineered with additional intra-domain disulfide bonds and a few pocket-filling mutations to stabilize gp120 core into a CD4-bound conformation. VRC01-like bNAbs also bind a further modified stabilized gp120 core, the resurfaced core (RSC3) (7), with nanomolar affinities. In fact, VRC01 was isolated by FACS-based sorting with RSC3 and CD4bs knockout (K/O) mutant RSC3 Δ371 as sorting probes. In contrast, CD4bs non-bNAbs poorly bind these modified gp120 core (7). Furthermore, VRC01-class bNAbs share similar footprints on the gp120 surface (12). Besides contacting the CD4 binding loop region (residues 365-373) on gp120, which is shared by all CD4bs MAbs, VRC01-class bNAbs also contact loop D, V5, and β24-α5 connections. Mutations in these regions dramatically affect VRC01-class antibody binding to gp120 (6). Due to their superior neutralization breadth, VRC01-class antibodies are considered as our vaccine template to elicit CD4bs bNAb responses. Such unique structural and functional properties of VRC01-class bNAbs offer us rationales to elicit and assess VRC01-class neutralizing antibody response in vaccine setting.
CD4bs bNAb responses are infrequently elicited during natural infections, whereas CD4bs non-bNAb responses are elicited at relatively high frequency by both natural infections and Env-based vaccines (15–18). Recent advancements in antigen-specific memory B cell repertoire analysis and structural definition of antigen-antibody recognition interface revealed the mechanism underlying the limited neutralization breadth elicited by current vaccines (16, 19, 20). Analysis of CD4bs-directed MAbs isolated from non-human primates (NHP) immunized with the current prototypic vaccine candidate, Env trimer YU2gp140-F revealed that the epitopes of these vaccine-elicited MAbs only partially overlap with that of bNAb VRC01, while more closely aligning with the human infection-elicited non-bNAb, F105 (16, 20, 21). Previous studies suggest that the vaccine-elicited NHP MAbs sample sub-optimal approaching angles and footprints on gp120 surface to access the CD4bs on the Env trimer. Therefore, these vaccine-elicited NHP CD4bs MAbs subsequently failed to overcome the steric hindrance to the CD4bs on the primary isolate (tier 2 virus) Env trimer spikes and displayed poor inhibition of the viral entry of the more neutralization-resistant tier 2 virus isolates (16, 20). In particular, recognition by the NHP CD4bs MAbs of gp120 is focused primarily on the α5 and bridging sheet on gp120, but not on loop D and V5 of gp120, which are extensively contacted by VRC01-class CD4bs bNAbs. Moreover, the bridging sheet structural elements have been previously reported as immunodominant sites as part of the epitopes of the human infection-elicited CD4bs non-bNAbs, F105 and b13 (22). Therefore, a modified Env immunogen with constrained access to these less accessible elements on Env functional spike might lower the frequency of activating the cognate subset of dominant B cells, thereby leading to a more focused CD4bs-directed neutralizing response with improved neutralization capacity.
In this study, we sought to use Env-CD4 induced (CD4i) MAb complex as an immunogen to direct the immune response away from the immunodominant determinant, and thus elicit more focused neutralizing antibody responses to the conserved and accessible epitopes including that of bNAb VRC01. CD4i antibodies, a unique subset of antibodies whose binding affinity for gp120 can be induced by the presence of sCD4, recognize the conserved coreceptor binding sites including the bridging sheet (23). Interestingly, similar to sCD4, VRC01 and CD4i antibodies can enhance binding to gp120 in a mutual manner (7). Thus, utilizing an Env-CD4i MAb complex as an immunogen is anticipated to have minimal bridging sheet access and augmented affinity for VRC01-class CD4bs bNAbs. We selected Env components consisting of HXBc2 gp120 Core (V3S) (13), derived from the HIV virus isolate HXBc2 gp120 Env with truncated V1, V2, V3 loop regions, as well as the N-/C- termini to minimize dominant immune responses toward the variable loops. 17b, one of the prototypic CD4i MAbs, whose presence enhances VRC01 binding to gp120 and whose epitope overlaps with the four stranded bridging sheets (23), is used to form a complex with HXBc2 gp120 core in this study. To stabilize the internal core-17b association within the complex, chemical crosslinkers such as glutaraldehyde (GLA) and dimethyl pimelimidate (DMP), were used to crosslink the complex. The gp120-MAb complexes have been previously tested for immunogenicity with moderate improvements (24–26). However, their impact on the immune response can vary depending on the nature of the immune complex and the targeted epitope. In this study, with carefully selected loop-deleted gp120, which can accommodate binding MAbs better than full-length gp120 due to the loop removal (27), we anticipated that this novel complex would lead to improved immunogenicity outcome.
We immunized rabbits with the gp120 core-17b complex four times. To further augment the immune response, we continued to immunize the rabbits with membrane-bound form of gp160 Env (pSVIII JRFL(+)ΔCT) plasmid DNA and boosted with a soluble form of trimeric JRFLgp140-F, a regimen that elicited robust neutralizing antibody responses in NHP in a previous study (28). We found that the gp120 core-17b complex immunization elicited potent neutralization against tier 1 virus, and induced transient antibody responses of similar binding specificity of CD4bs-directed bNAbs such as VRC01, suggesting that VRC01-like bNAb precursors were activated. To gain insight into the specificity of the complex-elicited antibody responses, we isolated MAbs from one of the immunized rabbits screened with CD4bs antibody binding phenotypes. Characterization of the epitopes of these MAbs elicited by the complex immunogen revealed that their footprints on gp120 are distal from the bridging sheet, compared to the prototypic Env immunogen elicited CD4bs MAbs in NHP and the non-bNAbs isolated from natural infections. Thus, this Env-antibody complex serves as an effective priming immunogen to direct immune response away from the undesired immunodominant determinants, which can be applied in future immunogenicity studies to focus the immune response. Our study is a clear addition to a number of recent immunogenicity studies using the next generation of well-ordered soluble Env SOSIP trimer (29, 30) or trimers in virus-like particle form (31), in which there was no evidence suggesting that VRC01-like bNAb precursors were efficiently activated and expanded.
Materials and Methods
Molecular modeling
The ClusPro server 2.0 (32) was used in the Ab mode to produce structural models of the vaccine-elicited NHP CD4bs MAbs GE148 and GE136 (16, 20) in complex with previously determined crystal structures of gp120 core (PDB ID codes 3HI1 and 3IDX). The structures of the two CD4bs non-bNAbs, F105 and b13, were used since these MAbs displayed a similar binding and neutralizing profile as the NHP MAbs. The structure of gp120 core was extracted from PDB files, 3HI1 and 3IDX, respectively, with the removal of F105 and b13, respectively to present the CD4bs on the core molecular surface. The resulting gp120 molecule coordinates were superimposed on the 17b-liganded gp120 core structure (PDB ID 1C9M) using the gp120 cores as superimposable units to deduce the relative angle of approach of the NHP antibodies to 17b, the CD4i MAb. The same procedure was carried out with the crystal structure of the VRC01-liganded core complex (PDB ID 3NGB). The use of the gp120 core and F105/b13 complex structural information as template to build the structural models for gp120 core:NHP MAbs GE136/GE148 complexes was validated in previously study (20). Validation included specific paratope mutagenesis analysis of GE136 and GE148 followed by binding and EM negative stain images of the GE136 and GE148 in complex with gp120.
Expression and purification of HIV-1 glycosylation protein
Env ligands, gp120 (derived from isolate YU2), gp120 core (previously referred to as V3S core, derived from isolate HXBc2), soluble gp140-F trimers (derived from JRFL and YU2) (33), resurfaced stabilized core (RSC3), RSC3 Δ371I (with the deletion of I371 residue of RSC3) (7), RSC3/G367R and RSC3 Δ371I/P363N (34) were produced by transient transfection of expression plasmid into FreeStyle 293-F suspension cells (Invitrogen) as previously described (28, 35). Soluble gp140-F trimer, RSC3, RSC3 Δ371I, RSC3/G367R and RSC3 Δ371I/P363N were purified by lentil lectin affinity chromatography followed by chelating chromatography over an Ni2+ column (GE Health Care, Piscataway, NJ) as described previously (36). Others were purified by a MAb 17b-coupled protein A-Sepharose column (37). All the proteins were subjected to size exclusion chromatography to remove undesired oligomeric forms when applicable.
Gp120 core-17 complex preparation and crosslinking
To form complexes, gp120 cores and 17b were mixed at molar ratio of 2:1 in PBS buffer and incubated at room temperature for 1 hour with gentle rocking. Excess non-complexed gp120 core or 17b was removed by size-exclusive chromatography using a Superdex 200 column (GE Health Care, Piscataway, NJ). The gp120 core-17b complexes were collected and stored at −80°C prior to further application. The gp120 core-17b complexes at a concentration of 100 μg/ml (presented as gp120 concentration) in 200 mM sodium borate buffer at pH 9.3, was crosslinked with DMP (Sigma) or GLA (Sigma), at room temperature and various concentrations for selected periods of time to optimize the crosslinking efficiency that was monitored by SDS PAGE. 2M Tris-HCl buffer (pH 7.4) was added to quench the crosslinking reaction at 4°C overnight with final concentration of 250 mM Tris-HCl, followed by buffer exchange to PBS pH 7.4. Under the optimized crosslinking condition, gp120 core-17b complexes crosslinked with DMP at 20 mM for 20 minutes and GLA at 20 mM for 5 minutes, respectively, were used in immunogenicity study.
HIV-1 gp160 DNA vaccine
The non-codon-optimized (non-CO) gene segment coding for the cleaved gp160 region of HIV-1 isolate JRFL, namely JRFL(+)ΔCT, was cloned in the vector pSVIII as described previously (28). Tat expression was carried out in pCTat vector (38).
Rabbit immunization and rabbit hybridoma production
New Zealand White rabbits (6 to 8 weeks of age) were divided into four groups with core, complex, complex-GLA and complex-DMP as immunogen, respectively, with 4 animals in each group. All procedures were carried out by following animal research guidelines and approved by institutional IACUC. Each animal received a 50 μg (week 0), and 25 μg (weeks 4, 12, 20 and 41) dose of protein immunogen, respectively, in 0.5 mL of PBS containing 100 μl of Adjuplex (ADVANCED BIOADJUVANTS, LLC) as adjuvant, by intramuscular injection of two hind legs. At weeks 28 and 33, rabbits received two separate DNA immunizations with each consisting of a total of 500 μg non-CO JRFL(+)ΔCT and pCTat (at a ratio of 20:1) plasmid DNA by intramascular route followed by electroporation as previously described (28). Blood was withdrawn from animals 2 weeks after each immunization for serum preparation. Rabbit #906 from complex-immunized group received 50 μg of JRFLgp140-F trimer protein intravenous immunization at week 45, and its spleen was harvested 4 days later for hybridoma production at Epitomics (Burlingame, CA), according to the previously established procedures (39–41).
ELISA Assay
ELISA assays were performed in 96-well MaxiSorp plates (Nalgene Nunc International). Plates were coated with envelope protein ligands at 2 μg/ml with 100 μl/well overnight at 4°C. After blocking the plates with blocking buffer consisting of phosphate buffered saline buffer (PBS), 5% fetal bovine serum (FBS), and 2% non-fat milk, the plates were incubated at 37°C for 1 hr with the immune sera, human or rabbit MAbs, respectively in five-fold serial dilutions in blocking buffer starting at 1:50 dilution for sera and 10 μg/ml for MAbs, followed by wash and incubation with HRP-conjugated goat anti-rabbit or anti-human IgG Fc secondary antibody (Jackson ImmunoResearch, West Grove, PA) at 1:10, 000 dilution in PBS/0.05% TWEEN 20.
For sandwich ELISA, 17b IgG at 5 μg/ml was coated on Reacti-Bind (Pierce) ELISA plates, in PBS overnight at 4°C. After plate blocking with blocking buffer, monomeric gp120 core was added at 2 μg/ml and incubated for 1h at 37°C. The human MAbs, biotin-labeled with Pierce EZ-Link NHS Biotin reagent (Thermo Scientific, Rockford, IL) per the manufacturer’s instruction, was added at 10 μg/ml followed by 5-fold serial dilutions. Biotin-labeled protein binding was detected by secondary streptavidin HRP polymer (SA-HRP, Sigma-Aldrich) at 1:1000 dilutions in PBS/0.05% TWEEN 20/10% blocking buffer for 1 hr at room temperature.
Competition ELISA was performed as previously described (7). Gp120 was captured in the ELISA wells pre-coated with the sheep anti-gp120 C5 region-specific Ab, D7324 (Aalto Bio Reagents). The unlabeled competitors, human MAbs or CD4-Ig, were diluted in blocking buffer and added at 5-fold serial dilution, starting at 50 μg/ml. After 30-min incubation at 37°C, biotin-labeled human MAbs or rabbit MAbs at a single concentration were added to the wells. This concentration was determined by previous titration experiments to give an OD 450 nm value in the range of 1 to 2. Biotin-labeled protein binding was detected as described earlier.
Between each incubation step, the plates were washed four times with PBS containing 0.05% TWEEN 20. The HRP-conjugate signal was developed by addition of 100 μl of TMB single solution (Invitrogen, Camarillo, CA) to each well. The reaction was stopped by adding 100 μl of 3% sulfuric acid and optical absorbance was measured at 450 nm.
Analysis of rabbit MAb heavy-chain and light-chain sequences
The rabbit MAb IgG heavy / light chain encoding genes were amplified by RT-PCR protocols with total mRNA from the hybridoma cell as template. The products were sequenced at Epitomics with rabbit IgG heavy/light chain sequence specific primers. After verification by sequencing, the heavy/light chain encoding gene nucleotide sequence data were analyzed by IMGT V-QUEST using rabbit Ig germline reference database (42) available at http://www.imgt.org/IMGT_vquest/share/textes/.
Neutralization assays and competition neutralization assay
Neutralization assays were performed with an HIV-1 Env pseudovirus assay with TZM-bl as target cells (43). To determine the serum/MAb dilution that resulted in a 50% reduction in relative luciferase signal units that reflect virus entry level, we performed serial dilutions of sera or antibodies and fitted the neutralization dose-response curves by nonlinear regression with a four-parameter Hill slope equation programmed into JMP statistical software (JMP 5.1, SAS Institute Inc.). The results are reported as the serum neutralization ID50 or MAb neutralization IC50, which is the reciprocal of the serum dilution or MAb concentration producing 50% virus neutralization. Diverse HIV-1 virus isolates, including viruses from clades A, B, and C, were used in the neutralization assays. The sources of the Env-encoding plasmids were described previously (44). To verify the virus neutralization activity of the sera, sera IgGs were further purified for virus neutralization assay. Serum IgG was purified by diluting 5 ml sera at 10-fold in PBS and passing through an affinity column packed with 5 ml of Protein A sepharose bead (GE Healthcare, Piscataway, NJ). The column was washed with PBS and the column-bound IgG was eluted by 100 mM Glycine-HCl buffer, pH 2.7, neutralized by addition of 2M Tris-HCl pH 7.4 buffer. The eluted IgG was subjected to dialysis against PBS, pH 7.4. The flow-through fraction was checked for possible remaining IgG, which was further purified to assure the complete IgG yield.
For the competition neutralization assay, RSC3 variants at 25 μg/ml were mixed with the rabbit sera in serial dilutions and incubated for 30 minutes at 37 °C to allow the formation of IgG-RSC3 variants complex prior to the addition of the pseudovirus to the assay (7), followed by the subsequent neutralization assay procedures. The working concentration of the RSC3 variants was optimized by initially titrating the RSC3 variants against a panel of selected HIV-1 infected individual plasma or sera with potent neutralizing activities as well as broadly neutralizing monoclonal antibodies to achieve saturating inhibition effects in previous study (7). In each experiment, we used fixed amounts of the CD4bs-directed bNAb VRC01 and non-CD4bs bNAb 2G12 as positive and negative control for the competition neutralization assay.
Ab binding kinetics analysis
The antibody and gp120 variants binding kinetics analyses were performed as previously described (16). Briefly, an Octet RED96 system (ForteBio) was used to assess the kinetics of rabbit MAb and human MAb binding to gp120 core or RSC3 with Bio-Layer Interferometry (BLI). Prior to the assay, both gp120 core and RSC3 were subjected to size exclusion chromatography to remove undesired oligomeric forms when applicable. Human MAbs at 10 μg/ml in PBS/0.2% TWEEN 20 were captured on the surface of the anti-human IgG Fc capture biosensors (ForteBio) for 1 min. SA-sensor (ForteBio) was used to initially capture biotin-labeled mouse anti-rabbit IgG Fc (Sigma-Aldrich) MAb at 10 μg/ml, followed by loading the rabbit MAbs. The biosensor tip was then immersed in wells containing gp120 core or RSC3 in PBS/0.2% TWEEN 20. The samples were two-fold serially diluted from an initial starting concentration of 250 nM. KD, the affinity constant value in nanomolars was calculated as off-rate/on-rate. The sensograms were corrected with the blank reference and fit with the software ForteBio Data Analysis 6.4 using a 1:1 binding model with the global fitting function (grouped by color, Rmax).
Human and NHP MAbs
The HIV-specific human MAb 2G12 was purchased from Polymun Scientific Inc. The CD4bs MAb F105 was provided by M. Posner (Dana-Farber Cancer Institute), and the CD4bs MAbs b6, b12, b13 and PGV04 were provided by D. Burton (The Scripps Research Institute), VRC01 and CH103 were previously described (7, 10). NHP CD4bs MAbs GE125, GE136 and GE140 were previously described (16). The CD4-Ig plasmid expression construct was provided by J. Sodroski (Dana-Farber Cancer Institute). The CD4i MAb 17b was provided by J. Robinson (Tulane University). HIV-Ig was obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. V3-specific MAb, 447, was provided by Susan Zolla-Pazner (New York University CFAR).
Ala scanning
A panel of 27 gp120 Ala mutants containing alterations known to affect CD4 binding or recognition by a set of CD4bs-directed MAbs was selected for analysis (16). The Ala mutations were generated previously in the context of the full-length JRCSF gp160 expression plasmid (45). The gp160 plasmids were individually co-transfected into 293T cells together with a plasmid containing the remaining HIV structural genes to produce Env pseudoviruses as described previously (43). For binding analysis, the gp120 was released from the pseudovirus by detergent lysis and captured by sheep anti-gp120 C5 Ab, D7324 (Aalto Bio Reagents) pre-coated into wells of a 96-well ELISA plate. After washing, the gp120 binding avidity of the human and rabbit MAbs was assessed with an anti-human or rabbit Fc HRP secondary Ab and the TMB single solution as described earlier. The level of binding avidity of the human MAb 2G12 was used to normalize the variant gp120 expression levels. The effect of a given Ala mutation on Ab binding was represented by apparent affinity (avidity) relative to the binding level to wildtype (WT) gp120, calculated with the formula [(EC50_WT/EC50_mutant)/(EC50_WT for 2G12/EC50_mutant for 2G12)] × 100, where EC50 is the median effective concentration, as previously described (6).
Avidity index
Avidity index was evaluated as described previously (46, 47) with some modifications. ELISA plates were coated overnight at 4°C with 200 ng/well of gp120 core. Rabbit serum was added in dilution series and incubated for 1.5 h before exposure to 1.5 M NaSCN or PBS for 15 min at room temperature. ED50 titers, the half-maximal efficient dilution value for each sample/treatment were determined. The avidity index was calculated with the formula, (ED50 titer_NaSCN-treated/ED50 titer _PBS-treated) × 100).
Statistical analysis
Statistical analyses were performed using the GraphPad Prism software version 5.04. ANOVA test was used to assess differences in immune functions in vaccinated animal groups, with P-values <0.05 considered significant. Pearson correlation assay was used for correlation analysis, with P-value <0.05 considered significant.
RESULTS
CD4i MAb 17b inhibits CD4bs non-bNAbs binding to gp120 while augmenting bNAb VRC01 binding to gp120
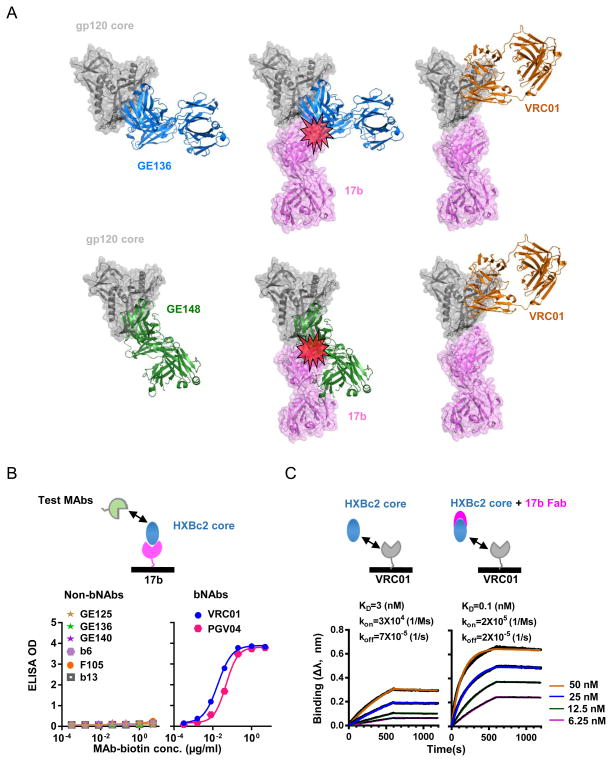
17b, a prototypic CD4i MAb recognizes a gp120 region that overlaps with the chemokine receptor-binding site including the bridging sheet (23). Most of the CD4bs non-bNAbs such as F105 and b13 elicited from natural infections also contact the bridging sheet (22). It has been observed that CD4i MAbs often compete with the natural infection-elicited CD4bs MAbs binding to gp120 (48), presumably due to their overlapping contact with bridging sheet epitopes. Using computer modeling, we had similar prediction supporting this observation. This modeling suggested that 17b would clash with most of the vaccine-elicited CD4bs non-bNAbs, including those that were isolated in our previous studies (Fig. 1a). We sought to determine if 17b could inhibit the vaccine-elicited CD4bs non-bNAbs binding to gp120. The gp120 core was captured on an ELISA plate pre-coated with 17b to form a gp120 core-17b complex. The binding of the complex to the CD4bs non-bNAbs, including the vaccine-elicited MAbs GE125, GE136, and GE140 as well as the natural infection-elicited MAbs b6, F105, and b13, was abolished as expected (Fig. 1b, left). In contrast, the complex is efficiently recognized by the CD4bs bNAbs including VRC01 and PGV04 (Fig. 1b, right). Therefore, the gp120 core-17b complex has modified antigenicity resulting in reduced CD4bs non-bNAbs recognition.
Fig. 1. CD4i MAb, 17b, enhances CD4bs bNAbs while inhibits non-bNAbs binding to gp120 core.
(A) Schematic presentation of the CD4i MAb 17b providing steric clash against CD4bs non-bNAb recognition. Upper, (left) A model of gp120 core (PDB 3IDX) complex with NHP CD4bs MAb GE136 Fab (in blue) which was isolated and whose crystal structure was solved in our previous study; (middle), 17b Fab (PDB 1C9M), in purple, clashes with GE136 on gp120 core when superimposed onto the left model; (right) VRC01 (PDB 3NGB), in orange, is compatible with 17b binding to gp120 core. Lower, A model of gp120 core (PDB 3HI1) complex with NHP CD4bs MAb GE148 Fab (in green). The legends are the same as in the upper panel. (B) Effect of 17b on bNAb and non-bNAb binding to monomeric HXBc2 gp120 core by Sandwich ELISA assay. Saturating amount of 17b was coated on ELISA plate and HXBc2 gp120 core was added to form complex with 17b, followed by the addition of biotinylated human MAbs (VRC01, PGV04, F105, b6, b13) and NHP MAbs (GE125, GE136 and GE140). Binding was detected with a streptavidin-HRP conjugate and the absorbance was read at 450 nm. OD, optical density. (C) The kinetics of binding of VRC01 to HXBc2 gp120 core (left) or HXBc2 gp120 core pre-mixed with 17b Fab (right) revealed by Bio-Layer Interferometry. The binding affinity was analyzed using an Octet system with VRC01 immobilized on the sensor surface, and the HXBc2 core (left) or HXBc2 core pre-mixed with 17b Fab (right) as analyte. The concentration of HXBc2 core or HXBc2 core +17b Fab mixture ranged from 6.25–50 nM. 17b enhances VRC01 binding affinity (KD) to gp120 core over an order of magnitude.
A previous study found that the binding of CD4i MAbs, including 17b to YU2gp120, was markedly enhanced in the presence of VRC01 or sCD4 (7), which is consistent with the notion that both sCD4 and VRC01 can stabilize monomeric gp120 in a CD4i-preferred binding conformation (6). To examine if 17b has a reciprocal effect on VRC01 binding to the gp120 core, we determined the kinetics of VRC01 binding to the gp120 core, in the absence and presence of 17b Fab, respectively, by Bio-Layer Interferometry (BLI). We found that VRC01 binding affinity for the gp120 core, assessed by dissociation constant KD, is approximately 3 nM (Fig. 1c, left). The KD changed to 0.1 nM in the presence of 17b Fab, which forms complex with gp120 core prior to VRC01 binding (Fig. 1c, right). Therefore, the gp120 core-17b complex has increased affinity for VRC01 by more than an order of magnitude compared to the unbound gp120 core. Taken together, the gp120 core-17b complex has attenuated binding to CD4bs non-bNAbs and augmented affinity for bNAb VRC01, compared to the free gp120 core. Subsequently, this complex might selectively activate bNAb B cell precursors and dampen the CD4bs non-bNAb response. We then sought to constitute gp120 core-17b complex for immunogenicity study in small animals.
The constitution and stabilization of gp120 core-17b complex
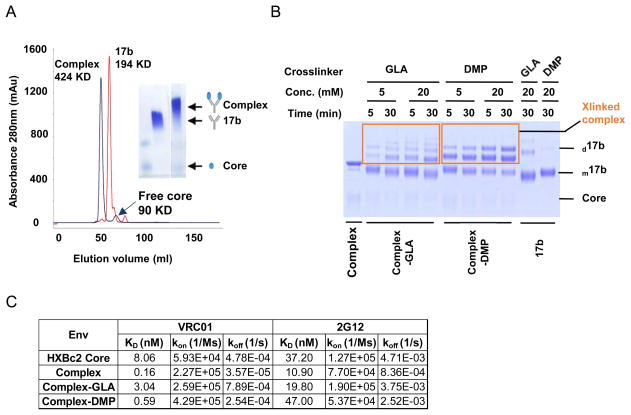
As the first step to prepare gp120 core-17b complex as immunogen, we titrated the ratio of gp120 core to 17b IgG to form a complex. We found that when 17b IgG (kept constantly at 20 μM) and gp120 core were mixed at 1:2 molar ratio, the gp120 core-17b complex formed well, with virtually no free form of either 17b IgG or gp120 core (Fig. 2a, right), as visualized by a blue-native PAGE gel. We then used this molecular ratio to perform further complex preparation. In a larger scale of complex preparation, we conducted size-exclusive gel-filtration chromatography to purify the complex (Fig. 2a, left). In the gel-filtration elution fraction, the complex appeared as a major peak of molecular weight (MW) around 424 kDa as expected, consisting of one 17b IgG (MW=194 kDa) and two gp120 core (MW=90 kDa) molecules, which was collected and subsequently confirmed by ELISA and SDS-PAGE gel analysis.
Fig. 2. Biophysical characterization of the HXBc2 core-17b complex.
(A) Size-exclusion chromatography (SEC) (left) and blue native gel (right) of complex. The complex, which was resolved as the peak of molecular weight of 424 kDa (in black color), consisting of one 17b IgG and two gp120 core molecules, was collected for subsequent applications. The SEC profile of 17b IgG (in red color) was overlaid for comparison. (B) Optimization of complex chemical crosslinking conditions, verified by non-reducing SDS PAGE gel of complex-GLA and complex-DMP crosslinking reaction products. The reaction condition at 20 mM of GLA and DMP, with complex crosslinking for 5 mins and 30 mins, respectively, was selected for large scale preparation of these two complexes. d17b and m17b, dimeric and monomeric form of 17b antibody, respectively. (C) The binding kinetics of complex with VRC01 and 2G12. Biotinylated VRC01 or 2G12 was captured on streptavidin-coated sensor in BLI system, which was immersed into wells containing Env immunogen variants to detect Ab-immunogen binding. The dissociation constants (KD), association rates (kon), and dissociation rates (koff) values of each Ab-immunogen pair were determined and displayed.
To further stabilize the complex, by preventing complex dissociation, we used chemical crosslinkers with differently sized spacer arms such as glutaraldehyde (GLA, 5 Å spacer arm) and Dimethyl pimelimidate (DMP, 9.5 Å spacer arm) to crosslink the gp120 core and 17b IgG molecules within the complex. We tested various conditions including crosslinker concentrations, duration of crosslinking reactions, and permutations, to optimize the crosslinking efficiency, as assessed by non-reducing SDS PAGE (Fig. 2b). The complex, resolved on the SDS PAGE gel, had the highest crosslinked fraction when crosslinked with 20 mM GLA for 5 minutes or 20 mM DMP for 30 minutes (Fig. 2b). We then chose these crosslinking conditions for complex stabilization. Note that there were minor oligomer forms of the complex after crosslinking by both GLA and DMP (Fig. 2b), which were referred to as complex-GLA and complex-DMP, respectively.
To determine if crosslinked complexes retain their native antigenicity, we tested their binding affinity for conformation-dependent bNAbs such as CD4bs MAb VRC01, and the glycan-specific MAb 2G12. As expected, the affinities of all the crosslinked complexes for bNAbs were similar to the uncrosslinked complex (Fig. 2c), except that for Complex-DMP, the affinity (KD) for 2G12 was slightly lower than the free HXBc2 Core and Complex-GLA. These results suggest that the crosslinking process allowed the complex to retain its native conformation and antigenicity to great extent. We then investigated the immunogenicity of these various forms of the complex in small animals.
Gp120 complex-GLA elicited neutralizing antibody response of earlier kinetics and higher degree of antibody affinity maturation
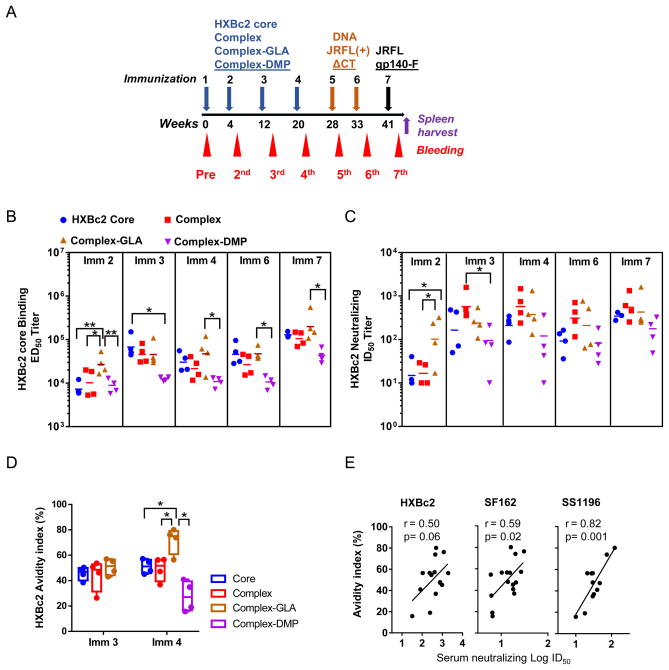
Rabbits divided into 4 groups, with 4 animals in each group, received a total of 4 immunizations with immunogens including the gp120 core, complex, complex-GLA, or complex-DMP, respectively, with adjuplex utilized as adjuvant, followed by 2 injections and electroporations of JRFL(+)ΔCT DNA, and a final boost with JRFLgp140-F trimer/adjuplex (Fig. 3a & Fig. S1). To study the kinetics of the antibody response during immunization, serum samples collected from animals 2 weeks after each immunization were tested for the presence of antibodies recognizing HXBc2 gp120 core by ELISA, presented as half-maximum binding titers (ED50 value, Fig. 3b). After the 2nd immunization, sera from all of the immunized animals displayed ED50 binding titers above 104-fold reciprocal dilution, while the complex-GLA immunized group animals had 3-fold higher ED50 titers than all of the other groups (Fig. 3b, Imm 2, P<0.05). This is also consistent with the notable virus neutralization titers (mean ID50> 100) observed with the sera from the complex-GLA immunized animals (Fig. 3c, Imm 2, P<0.05), while the sera from other groups barely displayed virus neutralization capacity (Fig. 3c). This data suggests that complex-GLA immunization sped up the antibody response kinetics compared to the free gp120 core immunization.
Fig. 3. gp120 core-17b complex immunized rabbit antibody response specificity and affinity maturation.
(A) Schematic presentation of the rabbit immunization and sampling schedule. Rabbits were immunized with Env protein (core or complex) in adjuplex 4 times (indicated by blue arrow) followed by 2 times of DNA immunizations (indicated by orange arrow), further boosted with JRFLgp140-F trimer protein in adjuplex (indicated by black arrow). The pre-immunization sera were harvested on week 0, and post-immunization sera were harvested 2 weeks after each immunization (indicated by red arrowheads). To generate hybridoma cell lines, the spleen of rabbit #906, was harvested 4 days after one additional JRFLgp140-F trimer hyper immunization on week 45 (indicated by purple arrow). (B) Serum of each rabbit from individual groups at different immunization time points was assessed for HXBc2 gp120 core binding titer. The half-maximal effective dilution (ED50) value of each animal was plotted, with the ED50 geometric mean of each group indicated as a line. (C) Serum virus neutralization activity was assessed against HIV-1 HXBc2 pseudovirus. The half-maximal inhibitory dilution (ID50) of each animal was plotted; ID50 geometric mean of each group was indicated as a line. (D) Rabbit serum antigen-specific response affinity maturation assessed by binding avidity index. Serum samples (n=4 for each group) were collected 2 weeks after the 3rd (Imm 3) and 4th immunization (Imm 4). Binding avidity index of serum samples for HXBc2 core was determined. Significance was evaluated using ANOVA. *, P≤0.05; **, P≤0.01. (E) Correlation of rabbit serum affinity maturation (avidity index) with neutralization capacity (Log ID50 titers) against HXBc2, SF162 and SS1196 of Imm 4 sera. Correlation was determined through Pearson’s correlation analysis, where P< 0.05 was considered as significant.
The sera antibody ELISA binding titers for gp120 core and neutralization titers against HXBc2 virus were all greatly elevated by the 3rd immunization in the core and complex immunized animal groups, whereas the binding and neutralization titers from complex-GLA immunized group increased moderately following the 3rd immunization (Fig. 3b & 3c). No significant change for the antibody response of the complex-DMP immunized animals was observed (Fig. 3b, 3c) following the 3rd immunization. The 4th immunization did not cause notable changes to the binding and neutralization titers of all animal sera (Fig. 3b, 3c). However, the 7th immunization with JRFLgp140-F as a heterologous protein boost elevated the gp120 core ED50 binding titers of most animals above 105 (Fig. 3b), while the neutralization titers against HXBc2 increased slightly (Fig. 3c). The animal group immunized with complex-DMP constantly displayed lower binding and neutralization titers, suggesting that crosslinking with DMP impeded the immunogenicity of the gp120 core.
The finding that the complex-GLA immunization elicited neutralization antibody responses of earlier kinetics suggested that the antibodies elicited by complex-GLA may possess higher levels of affinity maturation than the response elicited by other immunogens. To evaluate the affinity maturation of the Env immunogen elicited antibody response, which is often referred to as avidity, we measured the serum antibody binding avidity to the gp120 core under assay conditions of different stringency (in the presence and absence of chaotropic agent, e.g. 1.5M NaSCN). We found that the avidity indices of the serum gp120 core binding antibodies from the complex-GLA immunized animal group sera were higher than those from other animal groups following the 4th immunization (Fig. 3d, *P<0.5). The elevated antibody avidity indices were demonstrated to correlate with neutralization potency against a couple of viruses (SF162 and SS1196) of heterologous Env (Fig. 3e). This indicates that Env-specific antibody responses elicited by the complex-GLA immunogen underwent a higher degree of affinity maturation leading to increased cross-reactive neutralizing capacity, in addition to the observed earlier kinetics.
The transient elicitation of CD4bs bNAb precursors
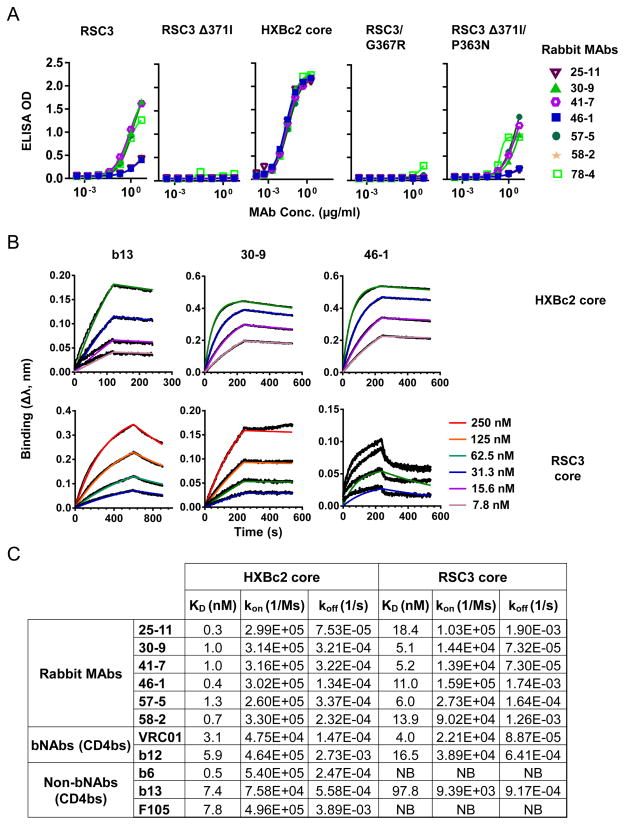
To gain insight into the binding specificity of the immunogen elicited antibody response we took advantage of a set of well-established protein probes that specifically differentiates the CD4bs-directed antibody response from the other subset of responses. Previous works demonstrated that CD4bs MAbs bind recombinant monomeric gp120 well, but only a small subset of CD4bs MAbs are able to access the CD4bs on the native viral Env functional spike and subsequently neutralize primary HIV isolates (tier 2 viruses). The resurfaced core RSC3, one of the differentiating protein probes, binds CD4bs bNAbs such as VRC01 well, but poorly binds most of the CD4bs non-bNAbs such as F105 and b6 (7), with the exception of b13 (Fig. S2a). RSC3 Δ371I, a CD4bs knockout (K/O) mutant, greatly diminished VRC01 binding (Fig. S2a). Using this pair of Env resurfaced core probes, we were able to detect a CD4bs-directed antibody response in most of the immunized animal sera (Fig. S2b & c). We found that sera from the complex and complex-GLA groups had slightly higher CD4bs-directed neutralizing titers than those from the core-immunized group (Fig. S2b & c). However, this difference was not statistically significant. Of note, such antibody responses revealed by RSC3/RSC3 Δ371I probing includes specificity of CD4bs non-bNAbs such as b13.
Additional mutant versions of RSC3 have been designed to further differentiate CD4bs bNAbs from non-bNAbs by their distinctive binding phenotypes (34). One such mutant, RSC3/G367R, which has abolished binding affinity for prototypic CD4bs non-bNAbs such as b13, b6, and F105 and modest binding to bNAbs such as VRC01 (34) (Fig. 4a, left), was used as a more selective probe for CD4bs bNAb response in this study. To examine if there were precursors of CD4bs bNAbs elicited in the immunized animals, we assessed the sera ELISA binding and neutralization specificity by using RSC3/G367R and the corresponding CD4bs K/O mutant, RSC3 Δ371I/P363N (34) as probes. We detected the different binding patterns of the Imm 3 sera from animals immunized with complex-GLA. We found that at this time point antibodies with binding specificity of CD4bs bNAbs were likely elicited in some of the animals (Fig. 4a, right), with higher binding titers for RSC3/G367R than for the CD4bs K/O mutant RSC3 Δ371I/P363N (with an ED50 value of 3-fold difference). To examine the evolving CD4bs bNAb response kinetics, we tested sera from different immunization time points and immunization regimens. We found that a CD4bs bNAb response was detectable in multiple animals (3 out of 4), but only in the complex-GLA group after the 3rd immunization (Imm 3, Fig. 4b). This suggested that CD4bs bNAb B cell precursors were activated in a relatively smaller population than other Env-specific precursors, given the observed low ED50 binding titer (Imm 3, Fig. 4b). After the 4th immunization, there was only one animal serum from each group possessing CD4bs bNAb antibody binding profile, which waned following the 7th immunization (Imm 7). Therefore, the antibody response of the CD4bs bNAb B cell precursors was elicited transiently and at a higher frequency in complex-GLA immunized animal group than in other groups.
Fig. 4. CD4bs bNAb response transiently elicited by complex-GLA immunization.
(A) ELISA binding profiles of human CD4bs and non-CD4bs MAbs (left), and selected sera of Imm 3 and Imm 7 (right) from complex-GLA immunized rabbit group tested with CD4bs bNAb probes, RSC3/G367R and RSC3 Δ371I/P363N. (B) Immune sera CD4bs bNAb ELISA binding titers at different immunization time points. The value after subtracting ED50 of RSC3 Δ371I/P363N from RSC3/G367R was deduced as CD4bs bNAb binding ED50 titer and the ED50 geometric mean of each group was indicated as a line. (C) CD4bs bNAb neutralizing activity of Imm 3 sera was assessed against HXBc2 pseudovirus in the presence of RSC3/G367R and RSC3 Δ371I/P363N. The value after subtracting ID50 of RSC3/G367R from RSC3 Δ371I/P363N was deduced as CD4bs bNAb neutralizing ID50 and ID50 geometric mean of each group was indicated as a line. The legends are identical to those in (B).
To test if the transient CD4bs bNAb precursor response inferred from the RSC3/G367R and RSC3 Δ371I/P363N binding specificity assessment following the Imm 3 time point had virus neutralizing capacity, we performed an HXBc2 virus neutralization assay with the sera from the Imm 3 time point, in the presence and absence of the probes. The CD4bs bNAb ID50 titer was deduced by subtracting the serum ID50 in the presence of the probe RSC3/G673R from that in the presence of CD4bs K/O mutant probe RSC3 Δ371I/P363N. We found that only two sera from the complex-GLA immunized animals after the 3rd immunization (Imm 3) displayed HXBc2 virus neutralizing activity (Fig. 4c), and diminished following the 7th immunization (not shown). These results confirmed that the transiently elicited CD4bs bNAb precursor response after the 3rd immunization in complex-GLA animal group possessed VRC01-like footprints on gp120 and was capable of neutralizing tier 1 virus.
Neutralizing antibody response breadth of the rabbit immune sera
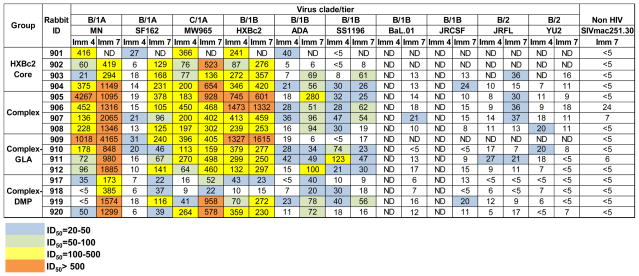
To examine the overall virus neutralization breadth of the immunized animals, animal sera 2 weeks after the 4th and 7th immunization, namely Imm 4 and Imm 7, respectively, were analyzed for their capacity to neutralize a panel of viruses by TZM-bl assay (43). The Imm 4 sera from most animals immunized by core, complex, or complex-GLA, respectively, demonstrated a range of neutralization capacity from weak (ID50 >20), moderate (ID50 >100) to potent (ID50 >500) neutralization against neutralization sensitive tier 1A viruses from clade B and C, such as MN, SF162 and MW965, and the autologous tier 1B virus HXBc2 (Fig. 5). When tested against other neutralization moderately-resistant tier 1B or resistant tier 2 viruses, sera from all groups displayed weak, moderate or no neutralization capacity (Fig. 5). Tier 1 virus neutralization titers in complex and complex-GLA immunized animals were slightly higher than those immunized by core (Fig. 5; Fig. 3c, Imm 2, P<0.05) and complex-DMP (Fig. 5; Fig. 3c, Imm 2 &3, P<0.05). One animal in the core primed group, #901, died after the 4th immunization, whose death was not related to vaccination.
Fig. 5. The overall outcome of the gp120 core-17b complex immunogenicity study in rabbits.
Neutralization capacity of antisera at 2 weeks post the 4th (Imm 4) and 7th immunization (Imm 7), respectively, against a panel of HIV-1 isolates was assessed. Values of the reciprocal dilution of each animal serum sample to achieve 50% neutralization (ID50) were reported. ND, not determined.
The virus neutralization titers of the Imm 7 sera samples were modestly higher than the Imm 4 time point (Fig. 5), suggesting that the 2 times of DNA immunizations followed by JRFLgp140-F trimer boost elevated the neutralizing antibody response of animals from all groups. The observed serum virus neutralization capacity was verified by neutralization assays using purified IgG from selected sera from the Imm 7 time point, which displayed neutralizing profiles similar to the sera samples (Fig. S3). Weak and sporadic neutralization (ID50> 20) against tier 2 viruses, the autologous isolate JRFL and heterologous isolate YU2 was only detected in complex/complex-GLA immunized animals (Figs. 5 & S3).
Boost immunizations (Imm 5–7) with membrane-bound Env trimer DNA JRFL(+)ΔCT followed by soluble Env trimer protein JRFLgp140-F injection increased the potency of the neutralizing response with limited breadth against tier 2 viruses including the cognate tier 2 virus JRFL. Therefore, the observed weak tier 2 virus neutralization was unlikely mediated by CD4bs bNAb responses. The overall outcome of our immunization is similar to previously published HIV Env trimer immunogenicity studies in small animals (18, 28, 49, 50), despite that we observed early transient elicitation of CD4bs bNAb precursor response in animals primed with complex-GLA. Apparently, the boost immunizations did not elevate the CD4bs bNAb precursor response substantially as evidenced by the steady serum neutralization titers of the majority of the animals at Imm 4 and Imm 7 against HXBc2, a sensitive indicator for CD4bs-directed neutralization specificity (Figs. 3c & 5). These data suggest that complex-GLA only elicited CD4bs bNAb B cell precursors in a temporary manner, which failed to expand in subsequent immunizations. In contrast, the CD4bs non-bNAb response in the immunized animal sera from the other groups was relatively robust and durable (Fig. S2c). Alternatively, the bNAb response elicited by the immunization may be too low to be detected by tier 2 virus neutralization assays.
On the whole, the majority of the CD4bs antibody response reactive with RSC3 in most of the animal sera (Fig. S2b & C) is distinct from the transient CD4bs bNAb response mainly detected in the complex-GLA immunized animals (Fig. S4a). We then sought to characterize the predominant RSC3-reactive CD4bs antibody response by isolating MAbs from one of the representative animals, which is critical to delineate the factors that limit the neutralization breadth and potency of the predominant antibody subsets elicited by our vaccination in order to inform vaccine re-design.
Isolation of RSC3 reactive MAbs from selected immunized rabbit
To isolate the predominant RSC3-reactive antibodies detected by binding and competitive neutralization assay (Fig. S2b, c), we undertook a recently published platform of dissecting the HIV-1 Env-specific B cell repertoire in immunized rabbits (51). We chose the representing animal #906 from the complex-immunized group to receive an antigen hyper-immunization 4 weeks after the 7th immunization, followed by termination and spleen harvest. The splenocytes of animal #906 were used to produce proximately 4000 hybridoma cells, from which a total of 78 multi-clonal stage hybridoma cells were positively screened for their supernatant reactivity against the YU2gp140-F trimer WT. These 78 hybridoma cells were subsequently screened for binding affinity with the probe RSC3 and RSC3 Δ371I (Fig. S4a). Among them, those having RSC3 high/ RSC3 Δ371I low binding profiles were selected for further production of stable hybridoma cells (monoclonal stage). A total of 9 cell lines from clones #15, 25, 30, 41, 46, 49, 57, 58 and 78 were chosen to produce MAbs, with 8 MAbs expressed sufficiently. 4 MAbs, 30-9, 41-7, 57-5 and 78-4 showed moderate binding with RSC3 but abolished binding with RSC3 Δ371I (Fig. 6a); 3 MAbs, 25-11, 46-1 and 58-2 showed weak binding with RSC3. Clone 15-6 was identical to clone 25-11 by gene segment analysis (next), and therefore was not included in the functional analysis. Some clones lost the RSC3 binding activity after in vitro expansion, which resulted in their exclusion from further analysis. Consistent with the serum binding profile of animal #906, these RSC3-reactive MAbs displayed no binding affinity for probe RSC3/G367R (Fig 6a, right), distinctive from that of the CD4bs bNAbs (Fig. 4a).
Fig. 6. The affinity of rabbit MAbs for HXBc2 core and RSC3 core variants.
(A) ELISA binding profiles of rabbit MAbs with HXBc2 core and RSC3 variants. (B) The binding kinetics of CD4bs MAb b13 and representative rabbit MAbs 30-9 and 46-1 to HXBc2 core and RSC3 core. Different concentrations of the core were shown. (C) The binding kinetic parameters of the rabbit MAbs to HXBc2 core and RSC3 core. Selected human MAbs were examined for comparison. The dissociation constants KD, between the different MAbs and ligands were shown in nanomolar (nM). NB, not binding.
The binding kinetics of these rabbit MAbs with HXBc2 and RSC3 gp120 core, respectively, was revealed by BLI (Fig. 6b). All rabbit MAbs exhibited remarkably high binding affinity to HXBc2 core, with KD ranging from 0.3 nM to 2.5 nM, comparable to that of the known human CD4bs MAbs (Fig. 6c). Consistent with the ELISA binding results, the binding affinity of rabbit MAbs to RSC3 varied, with KD ranging from 0.8 nM to 18.4 nM, similar to the bNAbs VRC01 and b12, and higher than non-bNAb b13. Two control non-bNAbs, b6 and F105 displayed no binding with RSC3 (Fig. 6c) (16), as expected.
To verify the binding specificity of these MAbs on gp120, a competition ELISA was performed with CD4bs ligands, including CD4-Ig and VRC01, and CD4i MAb17b as competitor, respectively. As seen in Table 1 & Fig. S4b, all of the rabbit MAbs compete well with CD4bs ligands including both CD4-Ig and VRC01 as competitor, indicating that the epitope of the rabbit MAbs may overlap with CD4bs. Interestingly, the rabbit MAbs did not compete well with the CD4i MAb, 17b, when used as competitor (Table 1 & Fig. S4b). In contrast, 17b competed well with most CD4bs MAbs such as b12, F105, and previously isolated vaccine-elicited NHP MAbs (GE125, 136, and 140) for binding to gp120 (Table 1 & Fig. S4b). Conversely, 17b enhanced the binding of VRC01 (Table 1, Figs. 1a & S4b). Our data is consistent with the notion that 17b prefers to bind gp120 in a CD4-bound conformation, which is not compatible with most other CD4bs MAb binding conformations except VRC01 (6, 7, 16). This result suggested that these rabbit MAbs elicited by the gp120 core-17b complex bind gp120 in a conformation compatible with 17b binding, a unique feature distinct from that of the conventional CD4bs non-bNAbs isolated from either natural infections or vaccinations.
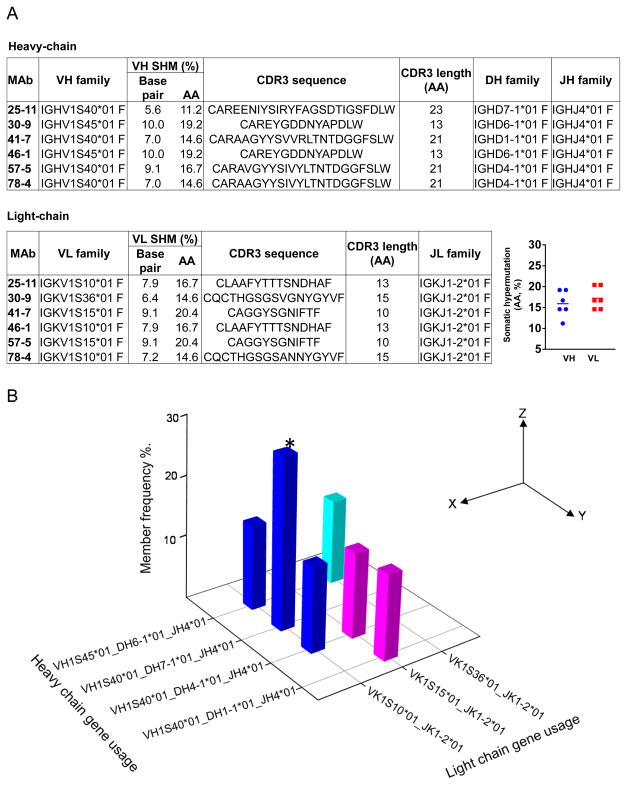
To investigate the genetic composition and clonal relationship of these rabbit MAbs, we amplified and sequenced the full length IgG heavy-chain and light-chain genes of these individual hybridoma clones. By searching the IMGT rabbit IgG gene database using program V-QUEST, the germline family usage, mutation frequency, and CDR3 features of the heavy-chain and light-chain genes were determined (Fig. 7). We found that these RSC3-reactive rabbit MAbs have skewed gene family usages with both heavy and light chain genes. Sharing exactly the same heavy and light chain sequences, clones 25-11 and 15-6 were identical clones, suggesting that they were derived from a well expanded memory B cell precursor. Most of the isolated rabbit MAb heavy-chain genes preferentially utilized germline gene family IGHV1S40*01, except for clones 30-9 and 46-1 that used IGHV1S45*01 (Fig. 7b). For the light-chain genes, 3 different germline families were utilized by these 8 clones. The genetically highly related gene family usages between these complex-elicited RSC3-reactive MAbs have been observed previously with RSC3-reactive CD4bs bNAbs (7, 9, 52).
Fig. 7. The Ig heavy and light chain gene family usages of the complex induced rabbit MAbs.
(A) Left, VDJ (VJ) gene family detail of heavy (light) chains of rabbit MAbs. Right, somatic hypermutation of each MAb, with mean value of VH and VL indicated as a line (B) The frequency of heavy and light chain gene family usage combination of rabbit MAbs. *, MAb 15-6, with sequence identical to 25-11, was also included in this analysis.
Since somatic hypermutation (SHM) is a key parameter in evaluating antibody affinity maturation, we sought to assess the divergence of the heavy/light chain genes from their germline counterpart as an indicator of SHM and affinity maturation. We found that heavy chain V genes of the rabbit MAbs exhibited an average SHM (amino acid sequence) level of 15.9±3.1%, while light-chain V genes had an average of 17.2±2.62%. These data indicated that the immunization regimen used in this study was effective in stimulating a modestly high SHM level for antigen-specific Abs (Fig. 7a). Examination of the heavy-chain CDR3 region revealed an average length of 18.6 amino acid residues, which is similar to typical CD4bs-directed antibody CDR3 length (53, 54).
The RSC3-reactive MAbs displayed modest virus neutralization breadth and potency
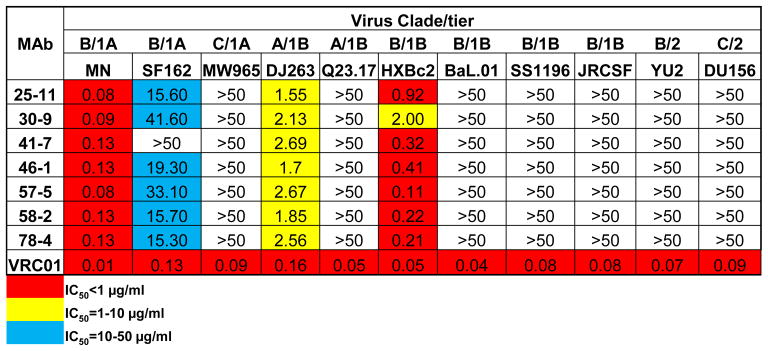
We next examined the neutralizing capacity of the rabbit MAbs using a standardized pseudotype virus neutralization assay (43). The virus panel included isolates from clades A, B and C consisting of tier 1 and tier 2 viruses. All rabbit MAbs displayed neutralizing activity against several tier 1A and tier 1B viruses, including clade A virus DJ263 and clade B viruses MN and SF162 (Fig. 8). This result demonstrated that the rabbit MAbs were capable of recognizing the functionally conserved antigenic surface of Env across HIV-1 clades, but were limited to those of the neutralization-sensitive tier 1 viruses. Additionally, the neutralization breadth of these MAbs was similar to that of the poly-clonal sera (Fig. 5 & Fig. S3).
Fig. 8. Virus neutralization capacity of rabbit MAbs against a panel of HIV-1 isolates, from both tiers 1 and 2.
Values represent the MAb concentration (μg/ml) to achieve 50% neutralization (IC50).
Fine mapping of rabbit MAbs revealed footprints on gp120 distinct from the known prototypic CD4bs antibodies
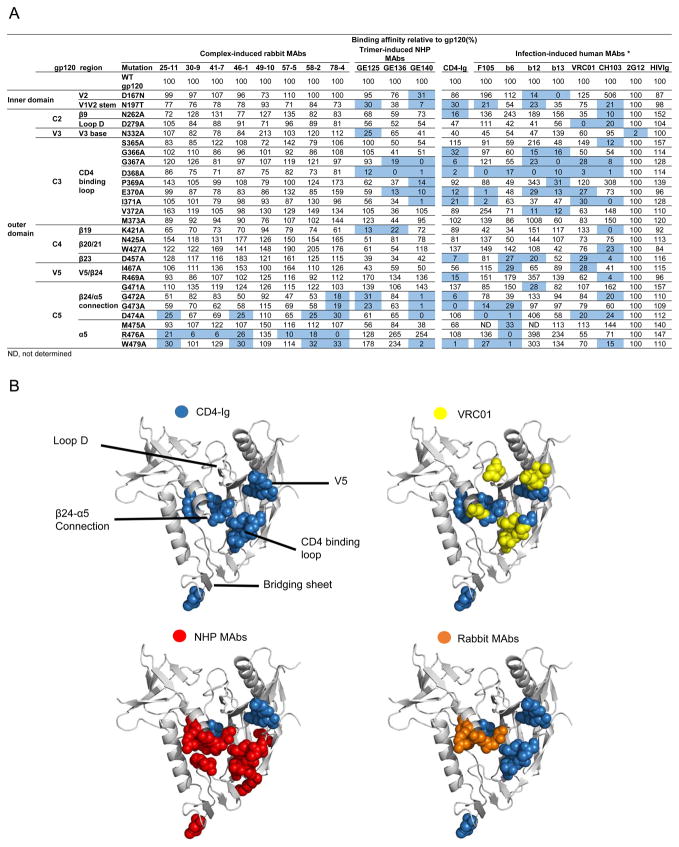
Since these RSC3-reactive rabbit MAbs competed well with CD4bs ligands, including most of the known CD4bs MAbs and CD4-Ig, it is likely that the epitopes recognized by these rabbit MAbs overlap with these CD4bs ligands. To more precisely define the sub-specificities of rabbit MAbs on gp120, we selected 27 JRCSF gp120 mutants containing single point mutation, mostly mutated to alanine (45), on selected residues which were contact residues for CD4, or CD4bs-directed MAbs (6, 14, 16, 22, 23). This gp120 mutation panel has been used in previous studies to define sub-specificities of known CD4bs MAbs (6, 14, 16, 22, 23) including 1) bNAbs such as VRC01, CH103, and b12; and 2) non-bNAbs including those elicited during natural infections (b6, b13, and F105) and by Env vaccination in NHP animals (GE125, GE136, and GE140) (Fig. 9a).
Fig. 9. Epitope mapping of complex-elicited rabbit MAbs by Ala scanning of JRCSF gp120.
(A) A panel of JRCSF Env mutants containing single Ala point mutations in gp120 was used for limited Ala scanning of the CD4bs region for the rabbit MAbs. Well-characterized CD4bs MAbs elicited either by trimeric gp140-F in NHP (GE125, 136, and 140), or by HIV-1 infections, were used as comparison. MAb 2G12 and HIV-Ig binding signals were used for normalization. The effect of a given Ala mutation on ligand binding relative to wild-type (WT) gp120 is shown. Mutations resulting in a three-fold reduction in binding relative to WT gp120 are highlighted in blue. * indicated that the data for CD4-Ig, F105, b6, b12, b13 and VRC01 was from reference (16). (B) The gp120 core molecular surface was used to highlight the residues affecting recognition by CD4-Ig, previously known MAbs, and complex-elicited rabbit MAbs. The critical residues inferred from the binding for CD4-Ig are highlighted in blue, VRC01 in yellow, NHP MAbs in red, and rabbit MAbs in orange. The structure of gp120 core was obtained from the Protein Data Bank (PDB ID: 2NY3).
We found that the regions targeted by rabbit MAbs significantly overlap with the epitopes of CD4bs ligands. Interestingly, recognition by all of the rabbit MAbs was markedly changed by mutations in the conserved elements, β24/α5 connection and α5 region (residues 474-479), in a manner similar to most CD4bs ligands including CD4-Ig, CD4bs bNAbs such as VRC01 as well as non-bNAbs (Fig. 9a). Additionally, MAb 78-4 was affected by the mutation of residues 472 and 473 in the β24/α5 connection, which is similar to CD4-Ig and CD4bs non-bNAbs including the vaccine-elicited MAbs previously isolated from NHP animals immunized with trimeric gp140-F proteins (Fig. 9a). The β24/α5 connection and α5 region, consisting of the residues D474, M475, and R476 at the outer domain/inner domain junction of gp120 were targeted by the rabbit MAbs. These regions have been shown to be essential for viral fitness and thus are frequent targets of HIV-neutralizing antibodies (55).
In contrast, mutations in the CD4 binding loop that affect recognition by both CD4bs bNAbs and non-bNAbs have no effect on rabbit MAb binding (Fig. 9a). Mutations in regions including the bridging sheet (V1V2 stem, β19, β20/21), V2, and V5/ β24 affected most of the non-bNAbs such as b6, b13 and F105, and NHP MAbs but not rabbit MAbs (Fig. 9a). Finally, mutations in loop D and the V5 regions affecting bNAb VRC01 binding have no effect on rabbit MAb binding. These results indicate that vaccine-induced rabbit MAbs target epitopes that only partially overlap with the CD4bs ligands in certain gp120 regions and these epitopes are very distinct from those of the previously known CD4bs ligands, including the vaccine-elicited NHP MAbs.
The unique distribution of the rabbit RSC3-reactive MAb epitopes compared to the CD4bs MAbs elicited during natural infection and the prototypical Env vaccination could be better appreciated when we modeled the epitopes inferred from their gp120 mutant binding profiles (Fig. 9b). The RSC3-reactive rabbit MAbs displayed minor contact to the bridging sheet, which is one of the major targets of the uncleaved trimeric gp140-F protein elicited NHP MAbs (16). Of note, the RSC3-reactive rabbit MAbs were elicited by the gp120 core-17b complex, in which the immunodominant bridging sheet region was masked by the bridging sheet-interacting CD4i MAb 17b (23). Therefore, the composition of the gp120 core complex immunogen likely changed the immune-response distribution on the gp120 core and shifted the dominant immune response away from the bridging sheet.
On the other hand, the gp120 core complex-elicited rabbit MAbs and the uncleaved trimeric gp140-F protein elicited NHP MAbs all contact the α5 region extensively (Fig 9b), unlike VRC01, which contacts loop D and V5 regions. The α5 region is closer to the axis of the Env trimer and is thus occluded on the well-packed Env functional spikes of primary virus isolates, which accounts for the restricted access of the vaccine-elicited MAbs to the Env functional spikes and the neutralization breadth limited to only tier 1 viruses (16). Therefore, future work should focus on further dampening the undesired antibody response toward the α5 region of gp120 to improve immunogenicity outcome.
DISCUSSION
In this study, we developed complex immunogens composed of gp120 core and CD4i MAb and used priming/boosting immunization regimens in an effort to focus antibody responses to conserved and accessible epitopes, including the CD4bs region of gp120.
We observed that complex-GLA immunization led to a modestly improved antibody response compared to immunization with gp120 core, including higher antigen binding and neutralization titers and an elevated avidity index associated with virus neutralization potency (Fig. 3). Our data suggest that crosslinked gp120 core-antibody complex is a superior early priming immunization regimen to monomeric gp120 core. It has been reported that GLA-treated gp140-F elicited higher tier 1 virus neutralization titers than untreated gp140-F in immunized rabbits (50). The observed modest immunogenicity improvement may be caused by higher quality antigen-presentation due to the presence of higher-ordered complexes formed by GLA crosslinking (Fig. 2b). These large crosslinked molecular complexes include dimers, trimers or tetramers, which may contribute to the earlier and more potent response induced by complex-GLA. Consistent with this notion, such large trimer or tetramer complexes did not exist in the complex-DMP crosslinking product (Fig. 2b), which elicited the lowest immune response in animals (Fig. 3). Previous studies have shown that crosslinking tumor antigens using the crosslinker DTSSP, which formed a panel of higher-ordered antigen complexes as well, can generate cellular immune responses that produce anti-tumor effects similar to those seen with non-covalent complexes of heat shock protein and tumor antigen (56). Furthermore, oligomeric hen egg lysozyme (HEL) could more efficiently promote both B cell receptor activation and internalization than monovalent HEL (57). These results are consistent with the theory that particulate antigens are better activators of the B-cell response than their monovalent forms (57, 58). The overall principle of higher-ordered antigen or antigen-antibody complex as immunogen is to present the candidate protein firmly and repetitively to stimulate the crosslinking of B-cell receptors and more effectively initiate antigen specific B-cell response (57). This notion suggests that the fusion of Env antigen and antibody in one complex, or biological nanostructures, such as exosomes, virus-like particles, and attenuated viruses (59, 60) may help improve Env immunogenicity.
Complex-DMP immunized animals showed the lowest immunogenicity compared with core, complex and complex-GLA (Fig. 4). Previous DMP crosslinking of monomeric gp120 or complex consisting of gp120 and MAb A32 also displayed weak antibody responses (26), suggesting that DMP crosslinking of gp120 has a deleterious effect on its immunogenicity. The DMP crosslinking may disrupt the native conformation of gp120. Alternatively, the DMP-crosslinked gp120 may lose the flexibility to expose certain moieties required for in vivo antigen-presentation (61–63) during immune response elicitation, although the DMP-crosslinked gp120 core complex displays unchanged binding affinity for the conformation-dependent antibodies VRC01 and 2G12 in vitro (Fig. 2c). In contrast, the complex crosslinked by GLA displayed augmented immunogenicity, suggesting that the nature of the crosslinkers could significantly impact target protein immunogenicity. We suspect that the differently sized spacer arms between crosslinked chemical groups on gp120 and antibody 17b may lead to differential effects on immunogenicity. GLA, has spacer arm of 5 Å, while DMP has 9.5 Å spacer arm. Crosslinking by DMP with the longer spacer arm may cause a more global surface change of the immunogen than GLA with the shorter spacer arm, which could subsequently affect the immunogenicity of gp120 such as impeding the antigen-presentation process or through other unknown mechanisms. For DMP crosslinked Complex, the affinity (KD) for 2G12 is slightly lower than that of the free HXBc2 Core and GLA-crosslinked Complex. These relatively subtle differences could be related to the different immunogenicity outcomes.
As confirmed by the binding sub-specificity of the MAbs isolated from the complex-immunized animal group, the complex vaccination-elicited neutralizing antibody response displayed a different epitope distribution on gp120 compared to the non-bNAb response elicited by conventional Env immunogens as well as natural infections (Fig. 9). The isolated rabbit MAbs had binding footprints shifted away from the bridging sheet region, which is predominantly targeted by the previously isolated CD4bs non-bNAbs. One of the components of the gp120 core complex is the CD4i MAb, 17b, whose epitope overlaps with the bridging sheet region. The occupancy of 17b on the bridging sheet region could block the Ab response toward this immunodominant surface and focus the antibody response on other conserved and accessible elements. Consistent with the improved immunogenicity outcome of a previous study using a gp120-specific antibody as a component of an immunogen to enhance V3-specfic neutralizing antibody responses, the gp120-antibody complex efficiently alters and focuses the immune response by attenuating off-target responses (24).
Lastly, while the CD4bs bNAb response was weakly and transiently elicited, mostly in the complex-GLA immunized animal group at early immunization time points, there is still predominant antibody response in the sera of complex-immunized animals targeting the α5 region adjacent to the inner domain of gp120, which is more occluded on the virus Env functional trimer (Fig. 9). Perhaps the predominant non-bNAb B cells, which recognize epitopes including the α5 region, eventually out-compete the CD4bs bNAb precursor B cells in the germinal center during the immunization course. It is conceivable that efforts aiming to dampen antibody response toward the α5 region could further improve the quality of the neutralizing antibody response. Engineering additional glycosylation sites in the gp120 inner domain surrounding the α5 region to dampen the cognate predominant immune response could be pursued (64).
In summary, we examined the immunogenicity of the HIV envelope glycoprotein, gp120, in complex with a CD4i MAb to focus the antibody immune response towards the receptor binding site, CD4bs. We found that the crosslinked complex elicited antibodies with a similar binding specificity as the bNAb VRC01. However, such CD4bs bNAb response was transient and only displayed a modest neutralizing capacity against neutralization-sensitive tier 1 viruses. Higher levels of affinity maturation and clonal expansion of such bNAb clones will likely be required to achieve expanded neutralization breadth and potency. Future effort should focus on improving the durability and potency of such a response. It would be intriguing to test immunization regimens using Env-MAb fusion proteins as immunogens presented in particulate formats such as virus-like-particles, immunogen modifications to initially dampen the immunodominant response toward the α5 region, followed by further boost immunizations with the next generation of well-ordered Env trimer mimetic reported recently (31, 65–68), which provides a more stringent selection for bNAb B cell precursors with VRC01 binding specificity. Since it was speculated that not much CDbs bNAb would be raised in rabbits due to the lack of an optimal germline VH segment (69), NHP or transgenic mice with the human IgG repertoire could also be tested with such regimens.
Supplementary Material
Table I.
Competition of rabbit MAbs with CD4Ig, VRC01 and 17b
| MAbs | Competitors
|
|||
|---|---|---|---|---|
| CD4Ig | VRC01 | 17b | ||
| Rabbit MAbs | 25-11 | ++ | +++ | − |
| 30-9 | ++ | +++ | − | |
| 41-7 | ++ | ++ | − | |
| 46-1 | ++ | +++ | − | |
| 57-5 | ++ | ++ | − | |
| 58-2 | ++ | ++ | − | |
| 78-4 | ++ | +++ | − | |
|
| ||||
| CD4bs MAbs | VRC01 | +++ | +++ | Enhance* |
| b12 | +++ | +++ | + | |
| F105 | +++ | +++ | +++ | |
| GE125 | +++ | +++ | ++ | |
| GE136 | +++ | +++ | ++ | |
| GE140 | +++ | +++ | ++ | |
|
| ||||
| V3 MAb | 447 | − | − | − |
+++, 75–100% competition; ++, 50–75% competition; +, 25–50 competition; −, < 25% competiti on.
17b enhanced VRC01 binding more than 30%.
Acknowledgments
The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: anti-HIV-1 gp120 antibodies including 2G12 from Dr. Hermann Katinger, 17b and 39F from Dr. James Robinson (Tulane University), F105 from Dr. Marshall Posner (Dana Farber Cancer Institute), 447 from Dr. Susan Zolla-Pazner and Dr. Miroslaw K Gorny (New York University School of Medicine), and HIV immune globulin (HIVIG). We are grateful to Dr. Joseph Sodroski (Dana Farber Cancer Institute) for providing the plasmid encoding CD4-Ig, Dr. Dennis Burton (The Scripps Research Institute) for providing JRCSF gp120 alanine scanning mutant panel plasmids, and Advanced Bioadjuvants, LLC for providing Adjuplex through an material transfer agreement. We thank Amy Xiao and James Steinhardt for proofreading this manuscript.
This study was supported by NIH/NIAID grant R01AI102766 (Y.L.), NIAID Development Grant P30AI36214 (Y.L.), Center for AIDS Research (CFAR), University of California, San Diego, P01 AI104722 (R.T.W., Y.L.), UM1 AI100663 (R.T.W.) and the Intramural Research Program of the Vaccine Research Center (J.R.M), National Institute of Allergy and Infectious Diseases, National Institutes of Health. This work was also partially funded by the International AIDS Vaccine Initiative (IAVI) (R.T.W.) with the generous support of United States Agency for International Development (USAID), Ministry of Foreign Affairs of the Netherlands, and the Bill & Melinda Gates Foundation; a full list of IAVI donors is available at www.iavi.org. The contents of this manuscript are the responsibility of the authors and do not necessarily reflect the views of USAID or the US Government.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
CONFLICT OF INTEREST: The authors declare that they have no conflicts of interest with the contents of this article.
AUTHOR CONTRIBUTIONS: Project designed by Y.C., Y.L., R.T.W. Experiment performed by Y.C., R.W., S.O’D. Y.F., K.T., C.C., H.E.A., J.D., Y.L. Data analyzed by Y.C., R.W., S.O’D., Y.F., K.T., J.G., C.C., J.R.M., Y.L. Manuscript written by Y.C., Y.L., R.T.W., J.R.M. All authors were asked to comment on the manuscript.
References
- 1.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, Haynes BF, Palker TJ, Redfield R, Oleske J, Safai B, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 3.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 4.Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol. 2010;28:413–444. doi: 10.1146/annurev-immunol-030409-101256. [DOI] [PubMed] [Google Scholar]
- 5.Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, Bjorkman PJ. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, O’Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, Graham BS, Haynes BF, Burton DR, Wyatt RT, Mascola JR. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. J Virol. 2011;85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, O’Dell S, Perfetto S, Schmidt SD, Shi W, Wu L, Yang Y, Yang ZY, Yang Z, Zhang Z, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Simek M, Burton DR, Koff WC, Doria-Rose NA, Connors M, Program NCS, Mullikin JC, Nabel GJ, Roederer M, Shapiro L, Kwong PD, Mascola JR. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, Hurley A, Myung S, Boulad F, Poignard P, Burton DR, Pereyra F, Ho DD, Walker BD, Seaman MS, Bjorkman PJ, Chait BT, Nussenzweig MC. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Program NCS, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, Ernandes MJ, Kong R, Longo NS, Louder MK, McKee K, O’Dell S, Schmidt SD, Tran L, Yang Z, Druz A, Luongo TS, Moquin S, Srivatsan S, Yang Y, Zhang B, Zheng A, Pancera M, Kirys T, Georgiev IS, Gindin T, Peng HP, Yang AS, Program NCS, Mullikin JC, Gray MD, Stamatatos L, Burton DR, Koff WC, Cohen MS, Haynes BF, Casazza JP, Connors M, Corti D, Lanzavecchia A, Sattentau QJ, Weiss RA, West AP, Jr, Bjorkman PJ, Scheid JF, Nussenzweig MC, Shapiro L, Mascola JR, Kwong PD. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey B, Svehla K, Xu L, Wycuff D, Zhou T, Voss G, Phogat A, Chakrabarti BK, Li Y, Shaw G, Kwong PD, Nabel GJ, Mascola JR, Wyatt RT. Structure-based stabilization of HIV-1 gp120 enhances humoral immune responses to the induced co-receptor binding site. PLoS Pathog. 2009;5:e1000445. doi: 10.1371/journal.ppat.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, Burton DR, Dimitrov DS, Sodroski J, Wyatt R, Nabel GJ, Kwong PD. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature. 2007;445:732–737. doi: 10.1038/nature05580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morner A, Douagi I, Forsell MN, Sundling C, Dosenovic P, O’Dell S, Dey B, Kwong PD, Voss G, Thorstensson R, Mascola JR, Wyatt RT, Karlsson Hedestam GB. Human immunodeficiency virus type 1 env trimer immunization of macaques and impact of priming with viral vector or stabilized core protein. J Virol. 2009;83:540–551. doi: 10.1128/JVI.01102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O’Dell S, Feng Y, Mascola JR, Wyatt RT, Karlsson Hedestam GB. High-Resolution Definition of Vaccine-Elicited B Cell Responses Against the HIV Primary Receptor Binding Site. Sci Transl Med. 2012;4:142ra196. doi: 10.1126/scitranslmed.3003752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douagi I, Forsell MN, Sundling C, O’Dell S, Feng Y, Dosenovic P, Li Y, Seder R, Lore K, Mascola JR, Wyatt RT, Karlsson Hedestam GB. Influence of novel CD4 binding-defective HIV-1 envelope glycoprotein immunogens on neutralizing antibody and T-cell responses in nonhuman primates. J Virol. 2010;84:1683–1695. doi: 10.1128/JVI.01896-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Y, McKee K, Tran K, O’Dell S, Schmidt SD, Phogat A, Forsell MN, Karlsson Hedestam GB, Mascola JR, Wyatt RT. Biochemically defined HIV-1 envelope glycoprotein variant immunogens display differential binding and neutralizing specificities to the CD4-binding site. J Biol Chem. 2012;287:5673–5686. doi: 10.1074/jbc.M111.317776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navis M, Tran K, Bale S, Phad GE, Guenaga J, Wilson R, Soldemo M, McKee K, Sundling C, Mascola J, Li Y, Wyatt RT, Karlsson Hedestam GB. HIV-1 receptor binding site-directed antibodies using a VH1-2 gene segment orthologue are activated by Env trimer immunization. PLoS Pathog. 2014;10:e1004337. doi: 10.1371/journal.ppat.1004337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran K, Poulsen C, Guenaga J, de Val N, Wilson R, Sundling C, Li Y, Stanfield RL, Wilson IA, Ward AB, Karlsson Hedestam GB, Wyatt RT. Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign. Proc Natl Acad Sci U S A. 2014;111:E738–747. doi: 10.1073/pnas.1319512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posner MR, Cavacini LA, Emes CL, Power J, Byrn R. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J Acquir Immune Defic Syndr. 1993;6:7–14. [PubMed] [Google Scholar]
- 22.Chen L, Kwon YD, Zhou T, Wu X, O’Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, Yang ZY, Zhang MY, Arthos J, Burton DR, Dimitrov DS, Nabel GJ, Posner MR, Sodroski J, Wyatt R, Mascola JR, Kwong PD. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hioe CE, Visciano ML, Kumar R, Liu J, Mack EA, Simon RE, Levy DN, Tuen M. The use of immune complex vaccines to enhance antibody responses against neutralizing epitopes on HIV-1 envelope gp120. Vaccine. 2009;28:352–360. doi: 10.1016/j.vaccine.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Visciano ML, Tuen M, Gorny MK, Hioe CE. In vivo alteration of humoral responses to HIV-1 envelope glycoprotein gp120 by antibodies to the CD4-binding site of gp120. Virology. 2008;372:409–420. doi: 10.1016/j.virol.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao HX, Alam SM, Mascola JR, Robinson J, Ma B, Montefiori DC, Rhein M, Sutherland LL, Scearce R, Haynes BF. Immunogenicity of constrained monoclonal antibody A32-human immunodeficiency virus (HIV) Env gp120 complexes compared to that of recombinant HIV type 1 gp120 envelope glycoproteins. J Virol. 2004;78:5270–5278. doi: 10.1128/JVI.78.10.5270-5278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, Godillot AP, Wyatt R, Sodroski J, Chaiken I. Antibody 17b binding at the coreceptor site weakens the kinetics of the interaction of envelope glycoprotein gp120 with CD4. Biochemistry. 2001;40:1662–1670. doi: 10.1021/bi001397m. [DOI] [PubMed] [Google Scholar]
- 28.Chakrabarti BK, Feng Y, Sharma SK, McKee K, Karlsson Hedestam GB, Labranche CC, Montefiori DC, Mascola JR, Wyatt RT. Robust neutralizing antibodies elicited by HIV-1 JRFL envelope glycoprotein trimers in nonhuman primates. J Virol. 2013;87:13239–13251. doi: 10.1128/JVI.01247-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Taeye SW, Ozorowski G, Torrents de la Pena A, Guttman M, Julien JP, van den Kerkhof TL, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, LaBranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramirez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crooks ET, Tong T, Chakrabarti B, Narayan K, Georgiev IS, Menis S, Huang X, Kulp D, Osawa K, Muranaka J, Stewart-Jones G, Destefano J, O’Dell S, LaBranche C, Robinson JE, Montefiori DC, McKee K, Du SX, Doria-Rose N, Kwong PD, Mascola JR, Zhu P, Schief WR, Wyatt RT, Whalen RG, Binley JM. Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site. PLoS Pathog. 2015;11:e1004932. doi: 10.1371/journal.ppat.1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- 33.Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol. 2002;76:4634–4642. doi: 10.1128/JVI.76.9.4634-4642.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch RM, Tran L, Louder MK, Schmidt SD, Cohen M, Members CCT, Dersimonian R, Euler Z, Gray ES, Abdool Karim S, Kirchherr J, Montefiori DC, Sibeko S, Soderberg K, Tomaras G, Yang ZY, Nabel GJ, Schuitemaker H, Morris L, Haynes BF, Mascola JR. The development of CD4 binding site antibodies during HIV-1 infection. J Virol. 2012;86:7588–7595. doi: 10.1128/JVI.00734-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forsell MN, Dey B, Morner A, Svehla K, O’Dell S, Hogerkorp CM, Voss G, Thorstensson R, Shaw GM, Mascola JR, Karlsson Hedestam GB, Wyatt RT. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 2008;4:e1000171. doi: 10.1371/journal.ppat.1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Svehla K, Louder MK, Wycuff D, Phogat S, Tang M, Migueles SA, Wu X, Phogat A, Shaw GM, Connors M, Hoxie J, Mascola JR, Wyatt R. Analysis of neutralization specificities in polyclonal sera derived from human immunodeficiency virus type 1-infected individuals. J Virol. 2009;83:1045–1059. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dey B, Pancera M, Svehla K, Shu Y, Xiang SH, Vainshtein J, Li Y, Sodroski J, Kwong PD, Mascola JR, Wyatt R. Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J Virol. 2007;81:5579–5593. doi: 10.1128/JVI.02500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sodroski JG, Rosen CA, Haseltine WA. Trans-acting transcriptional activation of the long terminal repeat of human T lymphotropic viruses in infected cells. Science. 1984;225:381–385. doi: 10.1126/science.6330891. [DOI] [PubMed] [Google Scholar]
- 39.Spieker-Polet H, Sethupathi P, Yam PC, Knight KL. Rabbit monoclonal antibodies: generating a fusion partner to produce rabbit-rabbit hybridomas. Proc Natl Acad Sci U S A. 1995;92:9348–9352. doi: 10.1073/pnas.92.20.9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.GLY . Cell fusion method. Aug, 2009. [Google Scholar]
- 41.Pytela R, ZW, Ke Y, Qian R, Au HC. Fusion partner for production of monoclonal rabbit antibodies. Jun, 2010. [Google Scholar]
- 42.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sundling C, Forsell MN, O’Dell S, Feng Y, Chakrabarti B, Rao SS, Lore K, Mascola JR, Wyatt RT, Douagi I, Karlsson Hedestam GB. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J Exp Med. 2010;207:2003–2017. doi: 10.1084/jem.20100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vermont CL, van Dijken HH, van Limpt CJ, de Groot R, van Alphen L, van Den Dobbelsteen GP. Antibody avidity and immunoglobulin G isotype distribution following immunization with a monovalent meningococcal B outer membrane vesicle vaccine. Infect Immun. 2002;70:584–590. doi: 10.1128/IAI.70.2.584-590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai L, Vodros D, Kozlowski PA, Montefiori DC, Wilson RL, Akerstrom VL, Chennareddi L, Yu T, Kannanganat S, Ofielu L, Villinger F, Wyatt LS, Moss B, Amara RR, Robinson HL. GM-CSF DNA: an adjuvant for higher avidity IgG, rectal IgA, and increased protection against the acute phase of a SHIV-89.6P challenge by a DNA/MVA immunodeficiency virus vaccine. Virology. 2007;369:153–167. doi: 10.1016/j.virol.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moore JP, Sodroski J. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bower JF, Li Y, Wyatt R, Ross TM. HIV-1 Envgp140 trimers elicit neutralizing antibodies without efficient induction of conformational antibodies. Vaccine. 2006;24:5442–5451. doi: 10.1016/j.vaccine.2006.03.063. [DOI] [PubMed] [Google Scholar]
- 50.Schiffner T, Kong L, Duncan CJ, Back JW, Benschop JJ, Shen X, Huang PS, Stewart-Jones GB, DeStefano J, Seaman MS, Tomaras GD, Montefiori DC, Schief WR, Sattentau QJ. Immune focusing and enhanced neutralization induced by HIV-1 gp140 chemical cross-linking. J Virol. 2013;87:10163–10172. doi: 10.1128/JVI.01161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Vaine M, Wallace A, Han D, Wan S, Seaman MS, Montefiori D, Wang S, Lu S. A novel rabbit monoclonal antibody platform to dissect the diverse repertoire of antibody epitopes for HIV-1 Env immunogen design. J Virol. 2013;87:10232–10243. doi: 10.1128/JVI.00837-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, O’Dell S, Wilson R, Wu X, Schmidt SD, Hogerkorp CM, Louder MK, Longo NS, Poulsen C, Guenaga J, Chakrabarti BK, Doria-Rose N, Roederer M, Connors M, Mascola JR, Wyatt RT. HIV-1 neutralizing antibodies display dual recognition of the primary and coreceptor binding sites and preferential binding to fully cleaved envelope glycoproteins. J Virol. 2012;86:11231–11241. doi: 10.1128/JVI.01543-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Breden F, Lepik C, Longo NS, Montero M, Lipsky PE, Scott JK. Comparison of antibody repertoires produced by HIV-1 infection, other chronic and acute infections, and systemic autoimmune disease. PLoS One. 2011;6:e16857. doi: 10.1371/journal.pone.0016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Sundling C, Wilson R, O’Dell S, Chen Y, Dai K, Phad GE, Zhu J, Xiao Y, Mascola JR, Karlsson Hedestam GB, Wyatt RT, Li Y. High-Resolution Longitudinal Study of HIV-1 Env Vaccine-Elicited B Cell Responses to the Virus Primary Receptor Binding Site Reveals Affinity Maturation and Clonal Persistence. J Immunol. 2016 doi: 10.4049/jimmunol.1502543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pietzsch J, Scheid JF, Mouquet H, Klein F, Seaman MS, Jankovic M, Corti D, Lanzavecchia A, Nussenzweig MC. Human anti-HIV-neutralizing antibodies frequently target a conserved epitope essential for viral fitness. J Exp Med. 2010;207:1995–2002. doi: 10.1084/jem.20101176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Wang XY, Subjeck JR, Kim HL. Covalent crosslinking of tumor antigens stimulates an antitumor immune response. Vaccine. 2010;28:6613–6620. doi: 10.1016/j.vaccine.2010.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim YM, Pan JY, Korbel GA, Peperzak V, Boes M, Ploegh HL. Monovalent ligation of the B cell receptor induces receptor activation but fails to promote antigen presentation. Proc Natl Acad Sci U S A. 2006;103:3327–3332. doi: 10.1073/pnas.0511315103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bachmann MF, Rohrer UH, Kundig TM, Burki K, Hengartner H, Zinkernagel RM. The influence of antigen organization on B cell responsiveness. Science. 1993;262:1448–1451. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- 59.Storni T, Kundig TM, Senti G, Johansen P. Immunity in response to particulate antigen-delivery systems. Adv Drug Deliv Rev. 2005;57:333–355. doi: 10.1016/j.addr.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 60.Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, Nieusma T, Mathison J, Baker D, Ward AB, Burton DR, Stamatatos L, Nemazee D, Wilson IA, Schief WR. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fabris D, Yu ET. Elucidating the higher-order structure of biopolymers by structural probing and mass spectrometry: MS3D. J Mass Spectrom. 2010;45:841–860. doi: 10.1002/jms.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vasilescu J, Guo X, Kast J. Identification of protein-protein interactions using in vivo cross-linking and mass spectrometry. Proteomics. 2004;4:3845–3854. doi: 10.1002/pmic.200400856. [DOI] [PubMed] [Google Scholar]
- 63.Seong SY, Matzinger P. Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nat Rev Immunol. 2004;4:469–478. doi: 10.1038/nri1372. [DOI] [PubMed] [Google Scholar]
- 64.Ingale J, Tran K, Kong L, Dey B, McKee K, Schief W, Kwong PD, Mascola JR, Wyatt RT. Hyperglycosylated stable core immunogens designed to present the CD4 binding site are preferentially recognized by broadly neutralizing antibodies. J Virol. 2014;88:14002–14016. doi: 10.1128/JVI.02614-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guenaga J, de Val N, Tran K, Feng Y, Satchwell K, Ward AB, Wyatt RT. Well-ordered trimeric HIV-1 subtype B and C soluble spike mimetics generated by negative selection display native-like properties. PLoS Pathog. 2015;11:e1004570. doi: 10.1371/journal.ppat.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, Wyatt RT. Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 2015;11:539–550. doi: 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, van Gils MJ, King CR, Wilson IA, Ward AB, Klasse PJ, Moore JP. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guenaga J, Dubrovskaya V, de Val N, Sharma SK, Carrette B, Ward AB, Wyatt RT. Structure-Guided Redesign Increases the Propensity of HIV Env To Generate Highly Stable Soluble Trimers. J Virol. 2015;90:2806–2817. doi: 10.1128/JVI.02652-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci U S A. 2012;109:E2083–2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.