Abstract
Salmonella enterica serovar Pullorum/Gallinarum is an important infectious pathogen that has caused widespread problems for chicken industry. Traditional Salmonella serotyping is an expensive and time-consuming process. In this study, we developed a rapid one-step polymerase chain reaction (PCR) method to identify S. Pullorum/Gallinarum. The PCR-based assay focuses on flhB, which shows a deficient region only in S. Pullorum/Gallinarum, compared with that of other serovars. The specificity and sensitivity of the PCR system were evaluated. The developed PCR method could identify S. Pullorum/Gallinarum from 27 different Salmonella serovars and eight non-Salmonella pathogens. The minimum limit of DNA and the lowest number of cells of S. Pullorum for the PCR detection were no less than 5.85 pg/μL and 10 CFU, respectively. The method was applied to the analysis of Salmonella strains isolated from the chicken farm. The PCR-based testing results of the farm isolates were in concordance with those obtained using traditional serotyping method. This newly developed PCR-based system could be used to accurately screen for the presence of S. Pullorum/Gallinarum, and support traditional serotyping methods, especially in high-throughput screening situations.
Keywords: Salmonella Pullorum/Gallinarum, flhB, PCR detection, chicken farm, one-step
Introduction
Although there are more than 2,600 Salmonella serovars (Ranieri et al., 2013), most animal infections are caused by relatively few serovars (Nielsen, 2013; Saeki et al., 2013; Zhu et al., 2015). Fowl are the specific host of S. Gallinarum biovars Pullorum and Gallinarum, which cause “white diarrhea” (pullorum disease) and fowl typhoid, respectively (Soria et al., 2012). S. Gallinarum can spread to reproductive organs, resulting in vertical transmission of the pathogen, as well as egg-related salmonellosis (Keller et al., 1997). Thus, timely detection of S. Pullorum/Gallinarum is very important. Because food animals and poultry are important reservoirs of Salmonella (Henson, 1997; Lynch et al., 2006), the United States Department of Agriculture and Food Safety Inspection Service carry out an “in plant” Hazard Analysis and Critical Control Point program to reduce the prevalence of food-borne pathogen contamination in meats, eggs, and milk (Hong et al., 2008).
Traditional Salmonella serotyping is conducted according to the White–Kauffmann–Le Minor scheme, which identifies the somatic (O) and flagellar (H) antigens based on the agglutination of bacteria with specific antisera (Majchrzak et al., 2014). Serotyping allows comparison with historical data, because it has been used for almost 70 years. Identifying the causative S. enterica serovars is a necessary first step in any epidemiological investigation of food-borne outbreaks. Despite its widespread use, traditional serotyping has a number of drawbacks. It takes at least 3 days to complete, is labor-intensive and expensive, requires the maintenance of 250 typing antisera and 350 different antigens, and is unable to differentiate between rough or mucoid strains (Ranieri et al., 2013). Recently, polymerase chain reaction (PCR) has shown great potential as a tool for pathogen detection, as it is a high-throughput approach with a high degree of sensitivity and specificity (Abdissa et al., 2006; Moyo et al., 2007). PCR-based molecular serotyping is a simple and rapid technique for identifying Salmonella enterica isolates (Karns et al., 2015).
The bacterial flagellum is a large, complex molecular machine made up of more than 30 different proteins. The membrane protein FlhB is a highly conserved component of the flagellar secretion system (Meshcheryakov et al., 2013), and it plays an important role in the determination of flagellar hook length and regulation of protein export (Hirano et al., 1994). Most Salmonella species possess flagella and exhibit motility. However, S. Pullorum and S. Gallinarum are two notable exceptions, having been shown lack of motility and flagella (Holt and Chaubal, 1997). Thus, the flhB gene of S. Pullorum/Gallinarum may own some special features that are different from other serovar.
In the present study, we developed a rapid one-step PCR system to identify S. enterica serovar Pullorum/Gallinarum. The approach used one pair of primers targeting flhB, which in silico analysis showed a deficient region in Salmonella Pullorum/Gallinarum, compared with that of other serovars. The specificity and sensitivity of the PCR system were evaluated, and the assay was applied to Salmonella strains isolated from one chicken farm.
Materials and Methods
Bacterial Strains
A mix of commercially available and previously isolated environmental Salmonella and non-Salmonella isolates, including S. Enteritidis, S. Pullorum, S. Gallinarum, S. Dublin, S. Uganda, S. Meleagridis, S. Anatis, S. London, S. Rissen, S. Derby, S. Typhimurium, S. Choleraesuis, S. Indiana, S. Sinstorf, S. Newlands, S. Muenster, S. Yoruba, S. Dumfries, S. Kentucky, S. Agona, S. Thompson, S. Senftenberg, S. Blockley, S. Inchpark, S. Virchow, S. Farsta, S. Dabou, Mycobacterium tuberculosis, Campylobacter jejuni, Brucella abortus, Listeria monocytogenes, and Escherichia coli, were used in this study (Table 1). These strains were used for testing the specificity and sensitivity of the PCR system.
Table 1.
Salmonella and non-Salmonella strains used to evaluate the specificity and sensitivity of the developed PCR system.
| Strain | Serovar/Species | Source | flhB-PCR result (182 bp/379 bp) | |
|---|---|---|---|---|
| Salmonella | C50041 | Enteritidis | Laboratory stock | –/+ |
| C50336 | Enteritidis | Laboratory stock | –/+ | |
| S06004 | Pullorum | Laboratory stock | +/– | |
| 6508 | Pullorum | Isolate from chicken | +/– | |
| SG9 | Gallinarum | Wigley et al., 2005 | +/– | |
| SL5928 | Dublin | Laboratory stock | –/+ | |
| T3 | Uganda | Cai et al., 2016 | –/+ | |
| T9 | Meleagridis | Li et al., 2016 | –/+ | |
| T8 | Anatis | Li et al., 2016 | –/+ | |
| G2 | London | Cai et al., 2016 | –/+ | |
| ZX | Rissen | Cai et al., 2016 | –/+ | |
| Y7 | Derby | Cai et al., 2016 | –/+ | |
| Y8 | Typhimurium | Li et al., 2016 | –/+ | |
| C500 | Choleraesuis | Laboratory stock | –/+ | |
| ZH65 | Indiana | Cai et al., 2016 | –/+ | |
| ZH5 | Sinstorf | Laboratory stock | –/+ | |
| ZH10 | Newlands | Isolate from cattle | –/+ | |
| ZH24 | Muenster | Laboratory stock | –/+ | |
| ZH82 | Yoruba | Isolate from pig | –/+ | |
| G449 | Dumfries | Laboratory stock | –/+ | |
| G241 | Kentucky | Laboratory stock | –/+ | |
| G382 | Agona | Laboratory stock | –/+ | |
| ZH35 | Thompson | Cai et al., 2016 | –/+ | |
| P192 | Senftenberg | Laboratory stock | –/+ | |
| G439 | Blockley | Laboratory stock | –/+ | |
| G86 | Inchpark | Laboratory stock | –/+ | |
| P122 | Virchow | Laboratory stock | –/+ | |
| P74 | Farsta | Laboratory stock | –/+ | |
| G85 | Dabou | Laboratory stock | –/+ | |
| Non-Salmonella | H37Rv | Mycobacterium tuberculosis | ATCC 27294 | –/– |
| 11168 | Campylobacter jejuni | ATCC 700819 | –/– | |
| 110 | Campylobacter jejuni | Isolate from chicken | –/– | |
| S19 | Brucella abortus | Laboratory stock | –/– | |
| EGDe | Listeria monocytogenes | ATCC BAA-679 | –/– | |
| JS15 | Listeria monocytogenes | Isolate from sheep | –/– | |
| 1314 | Escherichia coli | Isolate from chicken | –/– | |
| 1352 | Escherichia coli | Isolate from chicken | –/– |
Isolation and Serotyping of Salmonella
Additional Salmonella isolates of unknown serovars were isolated from naturally contaminated samples from one chicken farm in Jiangsu, China. Salmonella were isolated from floors and feces, and characterized as described elsewhere (Cai et al., 2016; Li et al., 2016). The pre-enrichment step was performed by suspending each sample in 50 mL buffered peptone water (BPW; Difco, BD, Sparks, MD, USA), and incubating samples at 37°C for 16–18 h. Then, 0.1 mL of the broth culture was subcultured in 10 mL subpackaged Rappaport–Vassiliadis (RV) enrichment broth (Difco, BD) at 42°C for 24 h. One loopful of each RV broth culture was streaked onto xylose lysine tergitol 4 (Difco, BD) agar plates, which were incubated at 37°C for 24–48 h. The presumptive Salmonella colony was picked from each plate and biochemically confirmed using an API-20E test kit (bioMérieux, Marcy l’Etoile, France). All isolated Salmonella strains were serotyped by slide agglutination using specific antisera (Tianrun Bio-Pharmaceutical, Ningbo, China) according to the White–Kauffmann–LeMinor scheme (Grimont and Weill, 2007).
Bacterial Growth and Genomic DNA Isolation
All of the bacterial strains used in this study were grown in Luria-Bertani broth (Oxoid, Basingstoke, Hampshire, England) or Brain Heart Infusion broth (Becton, Dickinson and Company, Sparks, MD, USA) at 37°C and 180 rpm overnight. Genomic DNA was extracted using a TIANamp Bacterial DNA kit (TianGen, Beijing, China) according to the manufacturer’s instructions. The concentration and purity of the isolated genomic DNA were measured using a NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE, USA), and DNA was subsequently stored at -20°C until use.
In silico Analysis
To develop a PCR- and sequence-based serotyping approach for identifying S. Pullorum/Gallinarum, the difference of flhB gene between S. Pullorum/Gallinarum and non-S. Pullorum/Gallinarum was analyzed. S. Typhimurium flhB nucleotide sequence (GenBank accession no. NC_003197.1 segment 2010283-2011434) was used to search the NCBI non-redundant nucleotide database using the basic local alignment search tool (BLAST) algorithm. The maximum number of aligned sequences to display was set to the maximum value of 20,000, and other parameters were set to default values. Primers to amplify flhB were designed and checked using Primer Premier 5 (Premier, Palo Alto, CA, USA).
PCR Procedure
Polymerase chain reactions were performed in a final volume of 25 μL, containing 100 ng of the isolated genomic DNA, 1 U of Taq polymerase (Takara Biotechnology Co., Dalian, China), 1x polymerase buffer, 200 μM each deoxynucleoside triphosphate, and 0.4 μM flhB primers. PCR amplifications were performed using a T100 Thermal Cycler (Bio-Rad, Hercules, CA, USA), with an initial denaturation step of 95°C for 5 min, 30 cycles of 95°C for 45 s, 59°C for 45 s, and 72°C for 1 min, followed by a final extension step of 72°C for 10 min. The resulting amplicons were resolved by horizontal electrophoresis on a 1% agarose gel in 1x TAE buffer.
Specificity of the PCR Primers
The specificity of the flhB primers was assessed using genomic DNA from 29 Salmonella strains, including 27 different serovars, and eight non-Salmonella pathogens as negative control strains (Table 1).
Sensitivity of the PCR Assay
The genomic DNA from S. Pullorum strain S06004 was serially diluted from 58.5 ng/μL to 5.85 fg/μL in sterile water. Aliquots (2 μL) of each dilution were used as templates for PCR. The objective of the sensitivity analysis was to define the lowest concentration of genomic DNA that could be detected using the PCR assay.
Overnight culture of S. Pullorum strain S06004 was consecutively diluted 10-fold in phosphate buffered saline (PBS), and the colony forming unit (CFU) of each dilution was determined by the plate count method. The pure culture was washed with PBS twice, adjusted to the desired bacterial concentrations 2 × 106–2 × 10 CFU/mL, and boiled in a water bath for 10 min to harvest the bacterial genomic DNA, respectively. Finally, 5 μL of each dilution was used for the PCR method to define the least cells of S. Pullorum that could be detected using the PCR assay.
Application of the PCR Assay for Naturally Contaminated Samples
The PCR assay was evaluated using genomic DNA from Salmonella isolates collected from the chicken farm (24 Salmonella isolates). The results obtained from the assays were compared with the results of the traditional serotyping.
Results and Discussion
Primer Design for S. Pullorum/Gallinarum-Specific Detection
As the BLAST program is further improved and genomic data continues to be supplemented with newly published Salmonella sequences, using comparative genomic analysis to exploit novel serovar-specific genes is becoming more common (Zhai et al., 2014). To develop a PCR- and sequence-based serotyping approach for identifying S. Pullorum/Gallinarum, the difference of flhB gene between S. Pullorum/Gallinarum and non-S. Pullorum/Gallinarum was analyzed. S. Typhimurium flhB nucleotide sequence was used to search the NCBI non-redundant nucleotide database using the BLAST algorithm. The results showed that flhB gene of S. Pullorum/Gallinarum is 955 bp, covering 83% of other serovars in length (Supplementary Figure S1). Thus, the deficient region of flhB could be exploited to distinguish S. Pullorum/Gallinarum from other serovars. One pair of oligonucleotide primers covering the deficient region of the flhB gene was designed. The sequences of the forward (flhB-F) and reverse (flhB-R) primers were: 5′-TTC GCG ACG AAT TTA AAG AGA GCG AAG-3′ and 5′-CAG CGT TTA AGC TGC CAG ACC CAG GCC-3′, respectively. These primers amplified a 182-bp fragment of flhB of S. Pullorum/Gallinarum and a 379-bp fragment of flhB of non-S. Pullorum/Gallinarum (Figure 1). This allowed development of a rapid and reliable one-step PCR assay targeting flhB to specifically screen for S. Pullorum/Gallinarum. To the best of our knowledge, this is the first single PCR assay to detect S. Pullorum/Gallinarum based on the flhB gene.
FIGURE 1.
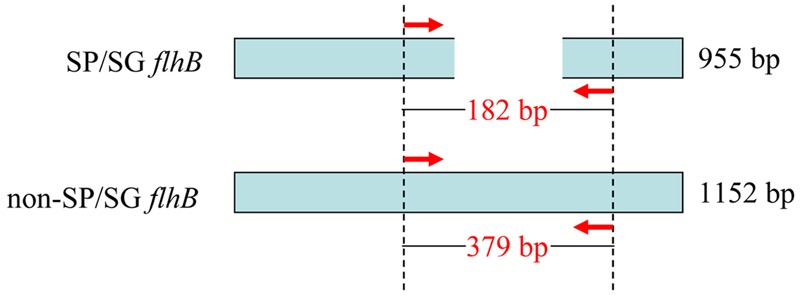
Schematic for the primer design of flhB to distinguish Salmonella Pullorum/Gallinarum from other serovars. flhB gene of S. Pullorum/Gallinarum has a deficient region compared with that of other serovars, which was exploited to design the primers. The red arrows indicate the positions of the designed primers. The PCR amplifies a product of 182 bp of S. Pullorum/Gallinarum and 379 bp of non-S. Pullorum/Gallinarum. SP/SG was referred to S. Pullorum/Gallinarum.
Specificity of the flhB-Based PCR Method for S. Pullorum/Gallinarum Detection
The specificity of the PCR system was evaluated using 29 Salmonella strains including 27 different serovars and eight non-Salmonella strains (Table 1). The results revealed that only S. Pullorum/Gallinarum generated the specific 182-bp target band. In contrast, the other 25 Salmonella serovars generated the specific 379-bp band, and eight non-Salmonella strains showed no amplification products using this primer pair (Figure 2). Previous studies have shown that S. Pullorum and S. Gallinarum are lack of motility and flagella (Holt and Chaubal, 1997), which may be related to the difference of flhB gene between S. Pullorum/Gallinarum and non-S. Pullorum/Gallinarum.
FIGURE 2.
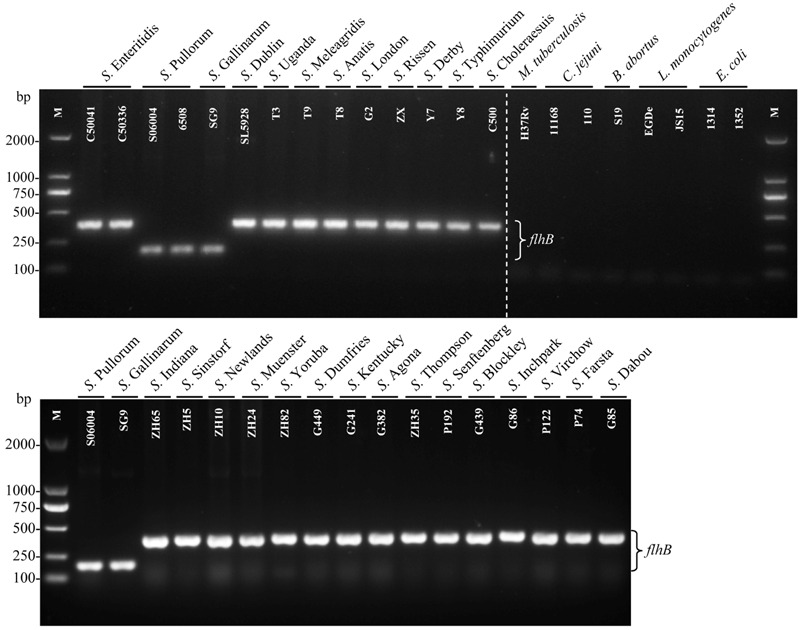
Specificity of the one-step PCR for the detection of Salmonella Pullorum/Gallinarum. The single PCR assays, using genomic DNA from various Salmonella and non-Salmonella strains, were conducted using the designed primers targeting flhB. The PCR amplifies a product of 182 bp of S. Pullorum/Gallinarum. Lane M: DL2000 DNA marker (Takara Biotechnology Co., Dalian, China). Detailed strain information is given in Table 1.
Sensitivity of the flhB-Based PCR Method for S. Pullorum/Gallinarum Detection
To determine the sensitivity of the PCR assay, genomic DNA from S. Pullorum strain S06004 was serially diluted from 58.5 ng/μL to 5.85 fg/μL and used as the template for the assay. The target fragment was amplified at concentrations of 58.5 ng/μL to 5.85 pg/μL DNA (Figure 3A). The results suggested that at least 5.85 pg/μL of genomic DNA was required for detection of S. Pullorum using this assay, which was slightly lower than previous studies (10 pg/μL) using Salmonella genomic DNA (Nithya et al., 2015). On the other hand, a 10-fold serial dilution of S. Pullorum cells that ranged from 104 CFU to 10-1 CFU per PCR system was evaluated. Using the developed PCR assay on different concentrations of S. Pullorum, we validated that the limit of detection was 10 CFU (Figure 3B).
FIGURE 3.
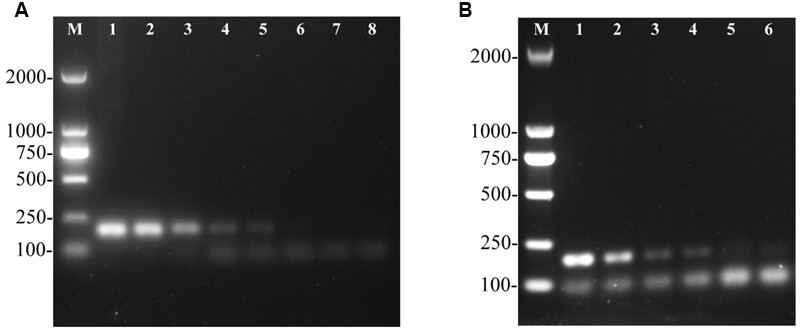
Sensitivity of the one-step PCR assay for detection of genomic DNA and cells from Salmonella Pullorum (strain S06004). The PCR amplifies a product of 182 bp. Lane M: DL2000 DNA marker (Takara Biotechnology Co., Dalian, China). (A) The PCR for the detection of genomic DNA, lanes 1–8, S. Pullorum genomic DNA used as template at the following concentrations, respectively: 58.5 ng/μL, 5.85 ng/μL, 585 pg/μL, 58.5 pg/μL, 5.85 pg/μL, 585 fg/μL, 58.5 fg/μL, 5.85 fg/μL; (B) The PCR for the detection of S. Pullorum cells, lanes 1 to 6, the number of cells per PCR assay, respectively: 104 CFU, 103 CFU, 102 CFU, 101 CFU, 100 and 10-1 CFU.
Application of the S. Pullorum/Gallinarum-Specific PCR Method
To evaluate the effectiveness of the established PCR assay, additional Salmonella isolates of unknown serovars were isolated from naturally contaminated samples from one chicken farm. The isolated Salmonella strains were examined by the developed PCR system. The PCR results showed that 10 samples from the chicken farm contained the specific 182-bp target band of flhB, suggesting that these 182-bp flhB-positive strains were S. Pullorum/Gallinarum (Figure 4). PCR results were confirmed by the traditional serotyping, the results of which showed concordance between the two methods for all samples (Table 2).
FIGURE 4.
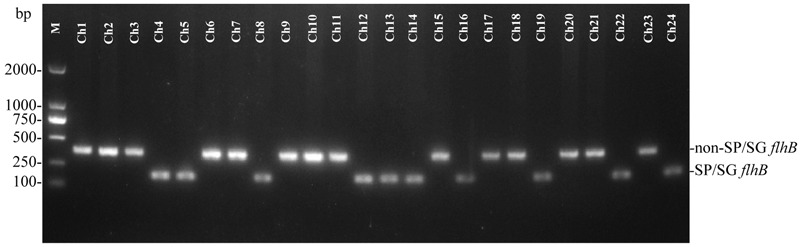
One-step PCR for the detection of Salmonella Pullorum/Gallinarum from Salmonella isolates from one chicken farm. The PCR assay produces a target amplicon of 182 bp of S. Pullorum/Gallinarum. Lane M: DL2000 DNA marker (Takara Biotechnology Co., Dalian, China). SP/SG was referred to S. Pullorum/Gallinarum. Detailed information on the Salmonella isolates is given in Table 2.
Table 2.
Salmonella strains isolated from one chicken farm to examine the application of the developed PCR system.
| Serovar (no. of isolates) | Isolate no. | flhB-PCR result (182 bp/379 bp) | Serovar (no. of isolates) | Isolate no. | flhB-PCR result (182 bp/379 bp) |
|---|---|---|---|---|---|
| Pullorum (10) | Ch4 | +/– | Enteritidis (4) | Ch2 | –/+ |
| Ch5 | +/– | Ch7 | –/+ | ||
| Ch8 | +/– | Ch11 | –/+ | ||
| Ch12 | +/– | Ch23 | –/+ | ||
| Ch13 | +/– | Indiana (7) | Ch1 | –/+ | |
| Ch14 | +/– | Ch3 | –/+ | ||
| Ch16 | +/– | Ch9 | –/+ | ||
| Ch19 | +/– | Ch10 | –/+ | ||
| Ch22 | +/– | Ch15 | –/+ | ||
| Ch24 | +/– | Ch17 | –/+ | ||
| Thompson (3) | Ch6 | –/+ | Ch21 | –/+ | |
| Ch18 | –/+ | ||||
| Ch20 | –/+ |
The serotyping of the Salmonella isolates was determined based on the traditional serotyping tests according to the White–Kauffmann–Le Minor scheme.
This PCR assay was also very rapid, taking less than 3 h to complete. The obtained experimental results were in agreement with the comparative genomic analysis used for primer design, and the proposed application was validated by screening for S. Pullorum/Gallinarum in samples isolated from the chicken farm. Although traditional serotyping should still be performed, a rapid screen using this flhB-based PCR assay may greatly reduce the need for antisera, and may assist in further investigation of Salmonella strains. In addition, the combination of PCR-based serotyping and traditional serotyping approaches will allow improved serovar classification of Salmonella isolates.
Conclusion
flhB, a gene found a deficient region only in S. Pullorum/Gallinarum to be exploited to distinguish this serovar from others, was identified in this study. The difference of flhB sequence between S. Pullorum/Gallinarum and non-S. Pullorum/Gallinarum was used to design a one-step PCR assay specific for S. Pullorum/Gallinarum. The assay was used to examine an extensive library of Salmonella isolates from one farm, thereby validating the specificity and effectiveness of the method. Our results suggest that this simple and economical PCR system could be used as a rapid diagnostic method for detection of S. Pullorum/Gallinarum accurately, especially in a high-throughput screen.
Author Contributions
ZP and XJ designed the experiments; DX and LS performed the PCR assays; DX, SG, and JT isolated the samples from the chicken farm; SA participated in the data analysis and interpretation; ZP, XJ, and DX wrote the paper. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Special Fund for Agroscientific Research in the Public Interest (201403054), the National Key Research and Development Program Special Project (2016YFD0501607), the National Natural Science Foundation of China (nos. 31320103907 and 31230070), the Project for Agricultural Products Quality and Safety Supervision (Risk Assessment) (18162130109236), the Program for New Century Excellent Talents in University (NCET-12-0745), the “Six Talent Peaks Program” of Jiangsu Province (NY-028), the Yangzhou University Science and Technology Innovation Team, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01863/full#supplementary-material
References
- Abdissa A., Asrat D., Kronvall G., Shittu B., Achiko D., Zeidan M., et al. (2006). High diversity of group A streptococcal emm types among healthy schoolchildren in Ethiopia. Clin. Infect. Dis. 42 1362–1367. 10.1086/503422 [DOI] [PubMed] [Google Scholar]
- Cai Y., Tao J., Jiao Y., Fei X., Zhou L., Wang Y., et al. (2016). Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int. J. Food Microbiol. 222 56–64. 10.1016/j.ijfoodmicro.2016.01.020 [DOI] [PubMed] [Google Scholar]
- Grimont P. A. D., Weill F.-X. (2007). Antigenic Formulae of the Salmonella serovars 9th Edn. Paris: WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur. [Google Scholar]
- Henson S. (1997). Estimating the incidence of food-borne Salmonella and the effectiveness of alternative control measures using the Delphi method. Int. J. Food Microbiol. 35 195–204. 10.1016/S0168-1605(96)01235-4 [DOI] [PubMed] [Google Scholar]
- Hirano T., Yamaguchi S., Oosawa K., Aizawa S. (1994). Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J. Bacteriol. 176 5439–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt P. S., Chaubal L. H. (1997). Detection of motility and putative synthesis of flagellar proteins in Salmonella pullorum cultures. J. Clin. Microbiol. 35 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Liu T., Lee M. D., Hofacre C. L., Maier M., White D. G., et al. (2008). Rapid screening of Salmonella enterica serovars enteritidis, hadar, heidelberg and typhimurium using a serologically-correlative allelotyping PCR targeting the O and H antigen alleles. BMC Microbiol. 8:178 10.1186/1471-2180-8-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karns J. S., Haley B. J., Van Kessel J. A. (2015). Improvements to a PCR-based serogrouping scheme for Salmonella enterica from dairy farm samples. J. Food Prot. 78 1182–1185. 10.4315/0362-028X.JFP-14-475 [DOI] [PubMed] [Google Scholar]
- Keller L. H., Schifferli D. M., Benson C. E., Aslam S., Eckroade R. J. (1997). Invasion of chicken reproductive tissues and forming eggs is not unique to Salmonella enteritidis. Avian Dis. 41 535–539. 10.2307/1592142 [DOI] [PubMed] [Google Scholar]
- Li Y., Cai Y., Tao J., Kang X., Jiao Y., Guo R., et al. (2016). Salmonella isolated from the slaughterhouses and correlation with pork contamination in free market. Food Control 59 591–600. 10.1016/j.foodcont.2015.06.040 [DOI] [Google Scholar]
- Lynch M., Painter J., Woodruff R., Braden C., Centers for Disease Control and Prevention. (2006). Surveillance for foodborne-disease outbreaks–United States, 1998-2002. MMWR Surveill. Summ. 55 1–42. [PubMed] [Google Scholar]
- Majchrzak M., Krzyzanowska A., Kubiak A. B., Wojtasik A., Wolkowicz T., Szych J., et al. (2014). TRS-based PCR as a potential tool for inter-serovar discrimination of Salmonella Enteritidis, S. Typhimurium, S. Infantis, S. Virchow, S. Hadar, S. Newport and S. Anatum. Mol. Biol. Rep. 41 7121–7132. 10.1007/s11033-014-3592-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshcheryakov V. A., Barker C. S., Kostyukova A. S., Samatey F. A. (2013). Function of FlhB, a membrane protein implicated in the bacterial flagellar type III secretion system. PLoS ONE 8:e68384 10.1371/journal.pone.0068384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyo S. J., Maselle S. Y., Matee M. I., Langeland N., Mylvaganam H. (2007). Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect. Dis. 7:92 10.1186/1471-2334-7-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen L. R. (2013). Review of pathogenesis and diagnostic methods of immediate relevance for epidemiology and control of Salmonella Dublin in cattle. Vet. Microbiol. 162 1–9. 10.1016/j.vetmic.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Nithya R., Ahmed S. A., Hoe C. H., Gopinath S. C., Citartan M., Chinni S. V., et al. (2015). Non-protein coding RNA genes as the novel diagnostic markers for the discrimination of Salmonella species using PCR. PLoS ONE 10:e0118668 10.1371/journal.pone.0118668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranieri M. L., Shi C., Moreno Switt A. I., den Bakker H. C., Wiedmann M. (2013). Comparison of typing methods with a new procedure based on sequence characterization for Salmonella serovar prediction. J. Clin. Microbiol. 51 1786–1797. 10.1128/JCM.03201-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki E. K., Alves J., Bonfante R. C., Hirooka E. Y., Oliveira T. C. R. M. (2013). Multiplex PCR (mPCR) for the detection of Salmonella spp. and the differentiation of the Typhimurium and Enteritidis serovars in chicken meat. J. Food Saf. 33 25–29. 10.1111/jfs.12019 [DOI] [Google Scholar]
- Soria M. C., Soria M. A., Bueno D. J. (2012). Comparison of 2 culture methods and PCR assays for Salmonella detection in poultry feces. Poult. Sci. 91 616–626. 10.3382/ps.2011-01831 [DOI] [PubMed] [Google Scholar]
- Wigley P., Hulme S., Powers C., Beal R., Smith A., Barrow P. (2005). Oral infection with the Salmonella enterica serovar gallinarum 9R attenuated live vaccine as a model to characterise immunity to fowl typhoid in the chicken. BMC Vet. Res. 1:2 10.1186/1746-6148-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L., Kong X., Lu Z., Lv F., Zhang C., Bie X. (2014). Detection of Salmonella enterica serovar Dublin by polymerase chain reaction in multiplex format. J. Microbiol. Methods 100 52–57. 10.1016/j.mimet.2014.02.014 [DOI] [PubMed] [Google Scholar]
- Zhu C., Yue M., Rankin S., Weill F. X., Frey J., Schifferli D. M. (2015). One-step identification of five prominent chicken Salmonella serovars and biotypes. J. Clin. Microbiol. 53 3881–3883. 10.1128/JCM.01976-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


