ABSTRACT
In 2014, the identification of stone fruits contaminated with Listeria monocytogenes led to the subsequent identification of a multistate outbreak. Simultaneous detection and enumeration of L. monocytogenes were performed on 105 fruits, each weighing 127 to 145 g, collected from 7 contaminated lots. The results showed that 53.3% of the fruits yielded L. monocytogenes (lower limit of detection, 5 CFU/fruit), and the levels ranged from 5 to 2,850 CFU/fruit, with a geometric mean of 11.3 CFU/fruit (0.1 CFU/g of fruit). Two serotypes, IVb-v1 and 1/2b, were identified by a combination of PCR- and antiserum-based serotyping among isolates from fruits and their packing environment; certain fruits contained a mixture of both serotypes. Single nucleotide polymorphism (SNP)-based whole-genome sequencing (WGS) analysis clustered isolates from two case-patients with the serotype IVb-v1 isolates and distinguished outbreak-associated isolates from pulsed-field gel electrophoresis (PFGE)-matched, but epidemiologically unrelated, clinical isolates. The outbreak-associated isolates differed by up to 42 SNPs. All but one serotype 1/2b isolate formed another WGS cluster and differed by up to 17 SNPs. Fully closed genomes of isolates from the stone fruits were used as references to maximize the resolution and to increase our confidence in prophage analysis. Putative prophages were conserved among isolates of each WGS cluster. All serotype IVb-v1 isolates belonged to singleton sequence type 382 (ST382); all but one serotype 1/2b isolate belonged to clonal complex 5.
IMPORTANCE WGS proved to be an excellent tool to assist in the epidemiologic investigation of listeriosis outbreaks. The comparison at the genome level contributed to our understanding of the genetic diversity and variations among isolates involved in an outbreak or isolates associated with food and environmental samples from one facility. Fully closed genomes increased our confidence in the identification and comparison of accessory genomes. The diversity among the outbreak-associated isolates and the inclusion of PFGE-matched, but epidemiologically unrelated, isolates demonstrate the high resolution of WGS. The prevalence and enumeration data could contribute to our further understanding of the risk associated with Listeria monocytogenes contamination, especially among high-risk populations.
INTRODUCTION
Listeria monocytogenes has been associated with foodborne outbreaks linked to contaminated ice cream (1), meat (2), caramel apples (3), and cheese (4). Recently, whole-genome sequencing has been employed to assist in listeriosis outbreak investigations (5–9). To do this, bioinformatics tools have been developed to target different genomic variations of L. monocytogenes (single nucleotide polymorphisms, allelic differences, k-mer, etc.) in either the core or whole genome (10–13). In the United States, nationwide real-time whole-genome sequencing (WGS) was implemented using the GenomeTrakr and PulseNet network to enhance listeriosis outbreak detection and investigation (14). In several outbreak investigations, the U.S. Centers for Disease Control and Prevention (CDC) had employed a whole-genome multilocus sequence typing (wgMLST) tool that targets the allelic differences in genome-wide coding regions (14), and the U.S. Food and Drug Administration (FDA) had employed a reference-based Center for Food Safety and Applied Nutrition (CFSAN) SNP Pipeline that identifies single nucleotide polymorphisms (SNPs) in the entire genome, including core genes, accessory genes, and intergenic regions (8, 11, 15).
In July 2014, certain lots of fresh stone fruits, including whole peaches, nectarines, plums, and pluots, were recalled by a packing company (company A) due to contamination with L. monocytogenes (16). Subsequently, two clinical cases were linked to these recalled fruits, which were the first reported human listeriosis cases caused by the consumption of contaminated stone fruit (14, 16). Initially, pulsed-field gel electrophoresis (PFGE) comparisons were performed on nationwide clinical cases reported in PulseNet, with isolation dates between 1 May and 31 August 2014, indicating possible exposure to the recalled fruits. This identified isolates from 4 patients in 4 states (Illinois, South Carolina, Minnesota, and Massachusetts) having PFGE profiles indistinguishable from those of isolates collected from the stone fruits and their packing environment (16). Subsequent epidemiological investigation and WGS analysis suggested that only patients in Minnesota and Massachusetts were likely linked to the contaminated stone fruits (14, 16). The wgMLST phylogeny was reported on isolates from stone fruits and their packing environment obtained in the present study and clinical isolates obtained during the epidemiological investigation (14).
Enumerating L. monocytogenes in food products linked to human listeriosis can provide a data set for our further understanding of the risk associated with L. monocytogenes contamination. Therefore, when isolating L. monocytogenes from fruits in contaminated lots, we performed simultaneous detection and enumeration. To explore whether WGS analyses of outbreak-associated isolates using different tools could lead to the same conclusions as wgMLST, an SNP-based WGS method was also performed on isolates from stone fruits, their packing environment, and patients during the outbreak investigation. This study describes the enumeration and WGS of L. monocytogenes associated with this outbreak.
MATERIALS AND METHODS
Fruits.
We randomly collected 105 unwounded stone fruits from seven lots (15 fruits/lot) available at the company's facility during the time of sampling. These lots were on the company's recall list, but the 105 fruits were stored in the packing facility and had never been distributed to commerce. These fruits were physiologically mature, but not fully ripe, at the time of analysis. The seven lots of fruits were composed of organic yellow nectarines, two cultivars of conventional white nectarines, conventional yellow nectarines, organic white peaches, conventional white peaches, and organic yellow peaches. Each fruit weighed between 127 and 145 g, with an average of 132 g.
Enumeration.
Preliminary analysis did not recover L. monocytogenes from the fruit pulp; therefore, we performed rinsing and direct plating enumeration of L. monocytogenes on the surface of each individual fruit. Briefly, each fruit was submerged in a Whirl-Pak bag (Nasco, Inc., Fort Atkinson, WI) with 80 ml of Butterfield's phosphate buffer (BPB), and the sealed bags were subsequently hand-massaged for 1 min. These bagged fruits were then put in a shaking incubator (Innova 44; Eppendorf, Inc., Hauppauge, NY) at 250 rpm for 5 min at room temperature. The resulting BPB rinsate was centrifuged for 10 min at 3,500 × g, and cell pellets were then resuspended in 1 ml of BPB. One hundred microliters of suspended cells was plated onto each of the two ALOA (Agar Listeria according to Ottavani and Agosti) plates (catalog no. AEB520080; bioMérieux, Inc., St. Louis, MO) and two RAPID'L. mono agar plates (catalog no. 3563694; Bio-Rad Laboratories, Inc., Hercules, CA), followed by incubation for up to 48 h at 37°C. A subset of typical colonies were confirmed using the real-time PCR scheme as described in the L. monocytogenes chapter of the FDA's Bacteriological Analytical Manual (17). This enumeration scheme had a lower limit of detection (LOD) of 5 CFU/fruit, and the rinsing generated an average of 75% recovery rate for peaches and 85% recovery rate for nectarines (our unpublished data).
Statistical analysis.
In order to perform statistics, we assumed the concentration of L. monocytogenes to be half of the LOD (2.5 CFU/fruit) for the fruit that did not yield L. monocytogenes. Medians, geometric means, and arithmetic means were calculated for each lot. Comparisons of medians among different lots were performed using a Mann-Whitney test with Holm-Bonferroni correction (18, 19). Log-transformed values of geometric means were compared with one-way analysis of variance (ANOVA) (20).
Serotyping and PFGE.
We performed serotyping on the following isolates: one isolate from each of the 56 fruits that yielded L. monocytogenes, and an additional isolate from 38 out of these 56 fruits, as well as 2 fruit isolates and 17 environmental isolates obtained from company A. Serotyping was performed using either a combination of antiserum agglutination and multiplex PCR (21) before WGS was obtained or the in silico serotyping tool in the Pasteur MLST L. monocytogenes database (http://bigsdb.web.pasteur.fr/listeria/listeria.html) after WGS was obtained. We performed PFGE on a subset of isolates representing all fruit cultivars, lots, and environmental samples using the standard PulseNet protocol (22). We obtained the PFGE profiles from PulseNet on the 4 clinical isolates identified during the initial epidemiological investigation.
WGS.
We performed WGS on a subset of isolates representing all fruit cultivars, lots, and environment samples.
DNA was isolated from pure cultures using the Qiagen DNeasy blood and tissue kit (catalog no. 69582; Qiagen, Inc., Valencia, CA). Sequencing libraries were prepared using the Nextera XT sample preparation kit (catalog no. FC-131-1024; Illumina, Inc.), and WGS was performed using a MiSeq (Illumina, Inc., San Diego, CA) with the version 2 kit (2 × 250 bp), according to the manufacturer's instructions (23).
A serotype IVb-v1 isolate (CFSAN023463) and a serotype 1/2b isolate (CFSAN023459), both collected from fruits, were selected to be fully sequenced using PacBio RSII (Pacific Biosciences, Menlo Park, CA, USA), as previously described (24, 25). Briefly, a single 10-kb library was prepared and sequenced using C2 chemistry on 8 single-molecule real-time (SMRT) cells with a 90-min collection protocol on the PacBio RS. These 10-kb continuous-long-read data were then de novo assembled using the PacBio Hierarchical Genome Assembly Process (HGAP2.0)/Quiver software package, followed by Minimus 2 to yield a single chromosomal contig and a plasmid contig. Raw reads were subsequently mapped to the contigs using Quiver for error correction. These two complete genomes were annotated with the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html). During sequencing, epigenetic modifications at each nucleotide position were measured as kinetic variations (KV) in the nucleotide incorporation rates, and methylase activities were deduced from the KV data (26, 27). The methylomes of these two complete genomes were analyzed and deposited in REBASE (http://tools.neb.com/genomes/view.php?list=0&view_id=35417 and http://tools.neb.com/genomes/view.php?view_id=36969).
Reference-based SNP analyses.
WGS analyses were performed using the CFSAN SNP pipeline 0.6.1 (8, 11). Briefly, raw reads from each serotype IVb-v1 isolate were mapped to the complete genome of CFSAN023463, and raw reads from each serotype 1/2b isolate were mapped to the complete genome of CFSAN023459, using default settings within Bowtie 2 version 2.2.2 (28). For each SNP analysis, the resulting BAM file was sorted using SAMtools version 1.3.1 (29), and a pileup file for each isolate was produced. These files were then processed using VarScan2 version 2.3.9 (30) to identify high-quality variant sites, using the mpileup2snp option. An in-house Python script was used to parse the .vcf files and construct an initial SNP matrix. We then removed high-density variant sites (≥3 within 1,000 bp in the reference), including 5 among outbreak isolates, 8 between the South Carolina isolate and the outbreak cluster, and 3 among serotype 1/2b isolates. Additional information about these procedures, e.g., codes and instructions, is available at https://github.com/CFSAN-Biostatistics/snp-pipeline. The Genetic Algorithm for Rapid Likelihood Inference (GARLI) (31) was used to infer phylogenies based on each of the SNP matrices. WGS analyses were performed separately on (i) serotype IVb-v1 isolates, including the two clinical isolates not associated with the outbreak; and (ii) serotype 1/2b isolates, with and without an outgroup (CFSAN010068 [NCBI SRA ID: SRR1181548]) of multilocus sequence typing (MLST) sequence type 5 (ST5) from a 2013 U.S. Hispanic-style cheese outbreak (32).
The draft genomes were de novo assembled using the CLC Genomics Workbench 8.0.3 (Qiagen, Waltham, MA), and genes containing the SNPs were extracted by BLAST, translated to protein sequences, and then aligned using the global alignment program MUSCLE (33), with default settings. This enabled the identification of SNPs as being synonymous or nonsynonymous and of the amino acid changes for nonsynonymous SNPs. We searched against NCBI, the protein families database (Pfam) (34), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (35) to further identify the putative functions of the genes containing the SNPs.
Prophage, clonal group, and inlA analyses.
We identified putative prophages from the complete genomes of CFSAN023463 and CFSAN023459 using PHAST (36). We then used BLAST to align these putative prophages with the draft genomes and determined the percentage of query coverage (percentage of the query sequence that overlaps the subject sequence) and sequence identity. In silico MLST analysis was performed using the CLC Genomics Workbench 8.0.3 (Qiagen, Waltham, MA) and tools in the Pasteur MLST L. monocytogenes database (http://bigsdb.pasteur.fr/listeria/listeria.html). The alignment and mapping tools from the Qiagen CLC Genomics Workbench were used to extract inlA sequences to determine whether there were premature stop codons in inlA.
Accession number(s).
The WGS sequences were deposited under their GenBank accession numbers for complete genomes and Sequence Read Archive (SRA) identifiers for draft genomes. We obtained the WGS data from GenomeTrakr on the 4 clinical isolates previously submitted by the CDC, seen in Table 2.
TABLE 2.
L. monocytogenes isolates from food, environment, and patients
| Source | Sample identifier | Serotype | PFGE profile (AscI/ApaI)a | NCBI accession no. or SRA identifier |
|---|---|---|---|---|
| Illinois patient | PNUSAL000957 | IVb-v1 | AscP0/ApaP0 | SRR1562157 |
| South Carolina patient | PNUSAL000730 | IVb-v1 | AscP0/ApaP0 | SRR1393979 |
| Massachusetts patient | PNUSAL000870 | IVb-v1 | AscP0/ApaP0 | SRR1534987 |
| Minnesota patient | PNUSAL001024 | IVb-v1 | AscP0/ApaP0 | SRR1597487 |
| Nectarineb | CFSAN023085 | IVb-v1 | AscP0/ApaP0 | SRR1554259 |
| CFSAN023086 | IVb-v1 | AscP0/ApaP0 | SRR1554256 | |
| Yellow nectarine, organic | CFSAN023490 | 1/2b | NA | SRR1553725 |
| CFSAN023476 | 1/2b | AscP1/ApaP1 | SRR1553871 | |
| CFSAN023477 | 1/2b | AscP2/ApaP2 | SRR1553850 | |
| CFSAN023489 | IVb-v1 | AscP0/ApaP0 | SRR1553739 | |
| CFSAN023491 | IVb-v1 | AscP0/ApaP0 | SRR1553882 | |
| CFSAN023506 | IVb-v1 | AscP0/ApaP0 | SRR1553851 | |
| CFSAN023475 | IVb-v1 | AscP0/ApaP0 | SRR1553866 | |
| CFSAN023478 | IVb-v1 | AscP0/ApaP0 | SRR1553750 | |
| Yellow nectarine | CFSAN023457 | 1/2b | AscP2/ApaP2 | SRR4090659 |
| White nectarine cv. 1 | CFSAN023473 | IVb-v1 | AscP0/ApaP0 | SRR1553779 |
| CFSAN023472 | IVb-v1 | AscP0/ApaP0 | SRR1553855 | |
| White nectarine cv. 2 | CFSAN023459 | 1/2b | AscP1/ApaP6 | NZ_CP014252.1 |
| CFSAN023488 | 1/2b | NA | SRR1553796 | |
| CFSAN023879 | 1/2b | AscP1/ApaP6 | SRR1574290 | |
| CFSAN023474 | IVb-v1 | AscP0/ApaP0 | SRR1553906 | |
| CFSAN023458 | IVb-v1 | AscP0/ApaP0 | SRR1556285 | |
| CFSAN023460 | 1/2b | AscP2/ApaP2 | SRR1556287 | |
| Yellow peach, organic | CFSAN023880 | 1/2b | AscP1/ApaP6 | SRR1574296 |
| CFSAN023462 | IVb-v1 | AscP0/ApaP0 | SRR1556292 | |
| CFSAN023463 | IVb-v1 | AscP0/ApaP0 | NZ_CP012021.1 | |
| CFSAN023464 | IVb-v1 | AscP0/ApaP0 | SRR1556294 | |
| CFSAN023465 | IVb-v1 | AscP0/ApaP0 | SRR1556293 | |
| CFSAN023466 | IVb-v1 | AscP0/ApaP0 | SRR1556289 | |
| White peach | CFSAN023492 | IVb-v1 | NA | SRR1553856 |
| CFSAN023505 | 1/2b | NA | SRR1553784 | |
| CFSAN023484 | IVb-v1 | AscP0/ApaP0 | SRR1553821 | |
| CFSAN023495 | IVb-v1 | NA | SRR1553791 | |
| CFSAN023496 | IVb-v1 | NA | SRR1553816 | |
| CFSAN023498 | IVb-v1 | NA | SRR1553788 | |
| CFSAN023500 | IVb-v1 | NA | SRR1553907 | |
| CFSAN023483 | IVb-v1 | AscP0/ApaP0 | SRR1553773 | |
| CFSAN023503 | IVb-v1 | NA | SRR1553792 | |
| CFSAN023507 | IVb-v1 | NA | SRR1553827 | |
| CFSAN023508 | IVb-v1 | NA | SRR1553774 | |
| CFSAN023479 | IVb-v1 | AscP0/ApaP0 | SRR1553797 | |
| CFSAN023480 | IVb-v1 | AscP0/ApaP0 | SRR1553820 | |
| CFSAN023481 | IVb-v1 | AscP0/ApaP0 | SRR1553798 | |
| CFSAN023482 | IVb-v1 | AscP0/ApaP0 | SRR1553826 | |
| CFSAN023497 | IVb-v1 | NA | SRR1553756 | |
| CFSAN023499 | IVb-v1 | NA | SRR1553804 | |
| CFSAN023501 | IVb-v1 | NA | SRR1566225 | |
| CFSAN023502 | IVb-v1 | NA | SRR1553840 | |
| CFSAN023504 | IVb-v1 | NA | SRR1553867 | |
| CFSAN023493 | IVb-v1 | NA | SRR1553764 | |
| CFSAN023494 | IVb-v1 | NA | SRR1553740 | |
| White peach organic | CFSAN023469 | IVb-v1 | AscP0/ApaP0 | SRR1556291 |
| CFSAN023470 | IVb-v1 | AscP0/ApaP0 | SRR1556297 | |
| CFSAN023471 | IVb-v1 | AscP0/ApaP0 | SRR1556295 | |
| CFSAN023882 | 1/2b | AscP2/ApaP2 | SRR1574321 | |
| CFSAN023881 | 1/2b | AscP2/ApaP2 | SRR1574272 | |
| CFSAN023468 | IVb-v1 | AscP0/ApaP0 | SRR1556290 | |
| CFSAN023467 | IVb-v1 | AscP0/ApaP0 | SRR1556296 | |
| Environment | CFSAN024089 | IVb-v1 | AscP0/ApaP0 | SRR1571515 |
| CFSAN024077 | IVb-v1 | AscP0/ApaP0 | SRR1571519 | |
| CFSAN024082 | IVb-v1 | AscP0/ApaP0 | SRR1571539 | |
| CFSAN024090 | 1/2b | AscP2/ApaP4 | SRR1571543 | |
| CFSAN024081 | 1/2b | AscP2/ApaP2 | SRR1571521 | |
| CFSAN024083 | 1/2b | AscP3/ApaP3 | SRR1571546 | |
| CFSAN024087 | 1/2b | AscP2/ApaP2 | SRR1571523 | |
| CFSAN024084 | 1/2b | AscP1/ApaP1 | SRR1571540 | |
| CFSAN024092 | 1/2b | AscP2/ApaP2 | SRR1571544 | |
| CFSAN024091 | 1/2b | AscP3/ApaP3 | SRR1571525 | |
| CFSAN024088 | 1/2b | AscP4/ApaP3 | SRR1571524 | |
| CFSAN024086 | 1/2b | AscP2/ApaP2 | SRR1571522 | |
| CFSAN024079 | 1/2b | AscP2/ApaP2 | SRR1571520 | |
| CFSAN024078 | 1/2b | AscP2/ApaP4 | SRR1571538 | |
| CFSAN024080 | 1/2b | AscP3/ApaP5 | SRR1571514 | |
| CFSAN024085 | 1/2b | AscP2/ApaP2 | SRR1571542 | |
| CFSAN024093 | 1/2b | AscP5/ApaP7 | SRR1571545 |
NA, PFGE information not available (i.e., not performed).
Variety information not available.
RESULTS
Enumeration.
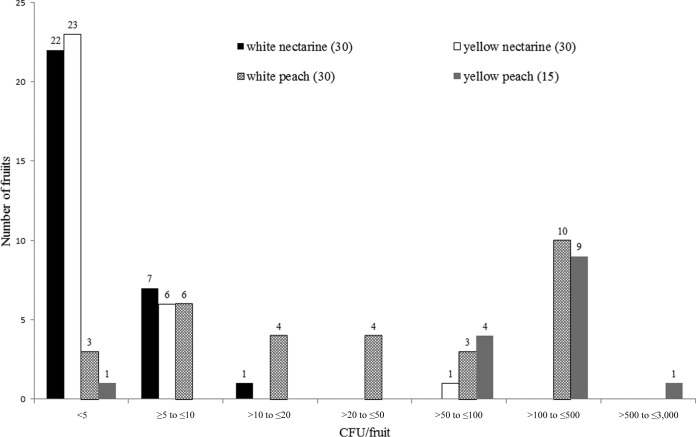
Our enumeration scheme (LOD, 5 CFU/fruit) yielded L. monocytogenes on 53.3% (56/105) of all tested fruits (Table 1). Specifically, 25% of the nectarines (15/60) and 91.1% of the peaches (41/45) yielded L. monocytogenes. The geometric means were 2.95, 3.97, 3.16, and 3.79 CFU/fruit for the 4 lots of nectarines, and 14.45, 73.37, and 156.6 CFU/fruit for the 3 lots of peaches. Overall, peaches had higher levels of L. monocytogenes than nectarines (Table 1). Out of the 60 nectarines, 58 and 59 fruits contained L. monocytogenes at ≤10 and ≤20 CFU/fruit, respectively, with the remaining fruit containing L. monocytogenes at 85 CFU/fruit (Fig. 1). Out of the 45 peaches, 44 fruits contained L. monocytogenes at ≤500 CFU/fruit, and the remaining fruit contained L. monocytogenes at 2,850 CFU/fruit (Fig. 1).
TABLE 1.
Statistical analysis of the enumeration results from seven lots of stone fruits
| Stone fruit | CFU/fruita |
No. of fruits |
||||||
|---|---|---|---|---|---|---|---|---|
| Minimum | Median | Arithmetic mean | Geometric mean | Maximum | Total | Yielding IVb-v1 | Yielding 1/2b | |
| White nectarine (cv. 1) | 2.5b | 2.5 A | 3.5 | 2.95 A | 15 | 15 | 2 | 0 |
| White nectarine (cv. 2) | 2.5 | 2.5 A | 4.8 | 3.97 A | 10 | 15 | 3c | 4c |
| Yellow nectarine | 2.5 | 2.5 A | 8.0 | 3.16 A | 85 | 15 | 1c | 1c |
| Yellow nectarine, organic | 2.5 | 2.5 A | 4.5 | 3.79 A | 10 | 15 | 4c | 3c |
| White peach | 2.5 | 10.0 B | 42.3 | 14.45 B | 325 | 15 | 13c | 1c |
| White peach, organic | 2.5 | 110 C | 135.8 | 73.37 C | 495 | 15 | 10 | 4 |
| Yellow peach, organic | 2.5 | 190 C | 362.5 | 156.6 C | 2,850 | 15 | 13c | 2c |
| Total | 2.5 | 5.0 | 80.2 | 11.28 | 2,850 | 105 | 46 | 15 |
Each fruit weighed 127 to 145 g. Medians were compared by Mann-Whitney test with Holm-Bonferroni correction. Log-transformed values of geometric means were compared with one-way ANOVA. Values with different letters are significantly different (P < 0.05).
Half of the LOD (2.5 CFU/fruit) was assumed as the pathogen level for fruits that did not yield L. monocytogenes.
One of the fruits had one serotype IVb-v1 isolate and one 1/2b isolate.
FIG 1.
Number of fruits containing L. monocytogenes at various levels. Each fruit weighed 127 to 145 g. The numbers are listed on top of each bar. Different cultivars of the same variety are combined for this analysis. The total number of fruits of each variety is listed in parentheses following the variety name.
Serotyping and PFGE.
Out of the 94 isolates from fruits (one isolate from each of the 56 fruits that yielded L. monocytogenes, and one additional isolate from 38 out of these 56 fruits) subject to serotyping, 76 isolates were serotype IVb-v1 (12 from nectarines and 64 from peaches), and 18 isolates were serotype 1/2b (8 from nectarines and 10 from peaches). Serotype IVb-v1, a 4b variant, is recognized as serotype 4b by standard serotyping using antiserum but differs from other serotype 4b isolates by PCR-based serotyping due to acquisition of a 6.3-kb DNA fragment (13, 21). Out of the 38 fruits that had two serotyped isolates per fruit, 5 fruits contained a mixture of both serotype IVb-v1 and 1/2b isolates. The 4 clinical isolates, identified by PulseNet with isolation dates indicating possible exposure to the contaminated fruits (16), were also serotype IVb-v1. Out of the 17 environmental isolates, 3 isolates were serotype IVb-v1, and 14 isolates were serotype 1/2b.
PFGE analysis of serotype IVb-v1 isolates, including the 4 clinical isolates and a subset of food and environmental isolates, revealed an identical profile (Table 2; see also Fig. S1 in the supplemental material). PFGE analysis of a subset of serotype 1/2b isolates from food and environmental samples identified 8 different PFGE profiles which did not match any clinical isolates in PulseNet, with isolation dates indicating possible exposure to the contaminated fruits (16). One environmental isolate (CFSAN024093) had a PFGE profile different from all other isolates (Table 2) and belonged to serotype 1/2b.
SNP-based analyses.
The complete genome of CFSAN023463 consists of a single contig of 2,939,733 bp (G+C content, 38%) representing the complete chromosome with no plasmids. The overall coverage was 139×. The outbreak cluster (serotype IVb-v1) consisted of two clades, designated clade IVb-v1_1, containing the Minnesota clinical isolate, and clade IVb-v1_2, containing the Massachusetts clinical isolate (Fig. 2). Each clade contained isolates from fruits of different varieties/lots and their packing environment. The clinical isolates from Illinois and South Carolina, although indistinguishable by PFGE from other serotype IVb-v1 isolates, were placed outside the outbreak cluster (Fig. 2); this was consistent with the epidemiological findings and the wgMLST phylogeny showing that these patients could not be linked to the contaminated stone fruits (16).
FIG 2.
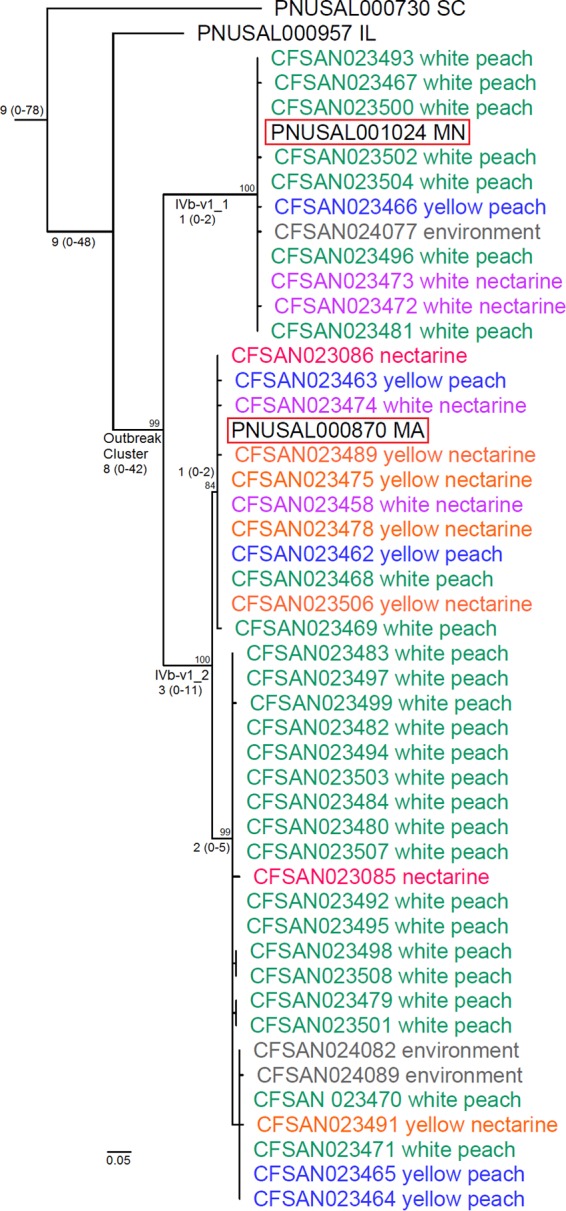
Phylogenetic tree of serotype IVb-v1 isolates constructed from SNPs identified by the CFSAN SNP Pipeline, using CFSAN023463 as the reference. All isolates in the tree were indistinguishable by PFGE. Clinical isolate identification (ID) is followed by the abbreviation of the state where it was isolated. For other isolates, the isolate ID is followed by the food and environmental source of the isolate. Isolates were from patients (black), white peaches (green), yellow peaches (blue), white nectarines (purple), yellow nectarines (orange), nectarines of unidentified variety (red), and environment (gray). The outbreak-associated clinical isolates are highlighted in red boxes. The median, minimum, and maximum pairwise SNP differences among isolates of major clades are shown near the root of each clade, with the minimum and maximum in parentheses. Bootstrap values are shown near major nodes.
The pairwise SNP differences ranged from 0 to 2 (median, 1 difference) among clade IVb-v1_1 isolates, and these ranged from 0 to 11 (median, 3 differences) among clade IVb-v1_2 isolates (Fig. 2). The pairwise SNP differences ranged from 35 to 42 (median, 38 differences) between clade IVb-v1_1 isolates and clade IVb-v1_2 isolates. The pairwise SNP differences ranged from 47 to 48 (median, 47.5 differences) between the Illinois clinical isolate and clade IVb-v1_1 isolates, and these ranged from 40 to 46 differences (median, 43 differences) between the Illinois clinical isolate and clade IVb-v1_2 isolates. The pairwise SNP differences ranged from 70 to 78 between the South Carolina clinical isolate and the stone fruit isolates, indicating that this isolate was more distant from the outbreak cluster (Fig. 2).
In general, it did not appear that the WGS clades or subclades were strongly associated with fruit varieties because isolates from different fruit varieties were distributed across both clades IVb-v1_1 and IVb-v1_2 (Fig. 2). The facility locations of environmental samples were not made available for us to establish any association between WGS clades and different facility locations or to determine whether the contamination was persistent or transient. Twenty-six SNPs (12 nonsynonymous, 13 synonymous, and 1 noncoding) and 56 SNPs (29 nonsynonymous, 24 synonymous, and 3 noncoding) distinguished the outbreak cluster from the Illinois clinical isolate and the South Carolina clinical isolate, respectively (see Table S1 in the supplemental material). Some SNPs were in proteins, including internalins, flagellar hook proteins E and L, and a few surface proteins with yet-unidentified functions (see Table S1). Among all 47 outbreak-associated isolates, we identified 58 polymorphic loci (containing 37 nonsynonymous, 14 synonymous, and 7 noncoding SNPs), some of which were in proteins involved in bacteriocin protection, stress response, and surface anchoring (see Tables S2 and S3 in the supplemental material).
The complete genome of CFSAN023459 consists of three replicons: a circular chromosome of 3,039,887 bp (G+C content, 38.1%), a 12,949-bp plasmid (G+C content, 36.4%), and a 52,687-bp plasmid (G+C content, 35.1%). The overall coverage was 119×. The MLST-matched outgroup (CFSAN010068, as described below) and serotype 1/2b isolates, excluding CFSAN024093, differed by at least 250 SNPs in the chromosome, indicating a relatively distant relationship (see Fig. S2 in the supplemental material). We therefore removed CFSAN010068 and analyzed only the stone fruit 1/2b isolates, except CFSAN024093, for the precise determination of SNPs (Fig. 3). The pairwise SNP differences in the chromosome among the serotype 1/2b isolates except CFSAN024093 ranged from 0 to 17 differences (median, 7 differences), indicating a very close relationship among all isolates, despite possessing 7 PFGE profiles. There were 40 polymorphic loci in the chromosome among serotype 1/2b isolates (see Table S4 in the supplemental material), which contained 24 nonsynonymous, 10 synonymous, and 6 noncoding SNPs (see Table S5 in the supplemental material). CFSAN024093 was genetically very distant from other serotype 1/2b isolates during preliminary analysis (data not shown) and thus was not included in the analyses illustrated in Fig. 2 and 3.
FIG 3.
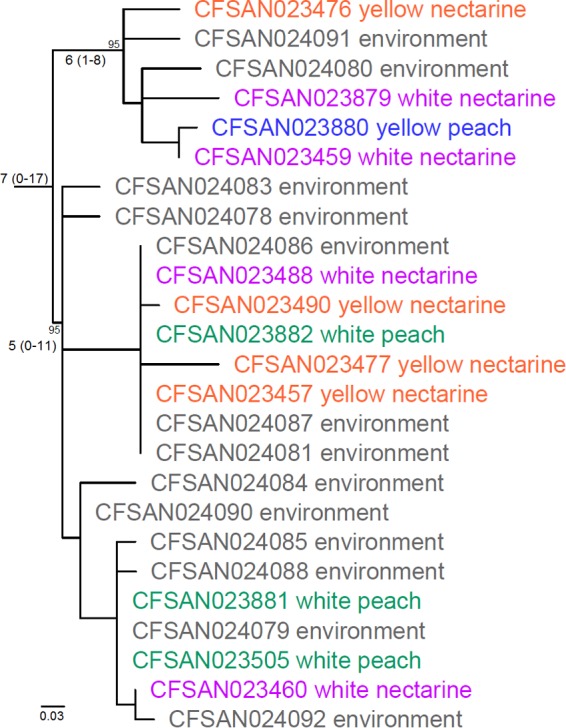
Phylogenetic tree of serotype 1/2b isolates, except CFSAN024093, constructed from SNPs identified by the CFSAN SNP Pipeline, using CFSAN023459 as the reference. The isolate ID is followed by the food and environmental source of the isolate. Isolates were from white peaches (green), yellow peaches (blue), white nectarines (purple), yellow nectarines (orange) and environment (gray). The median, minimum, and maximum pairwise SNP differences among isolates of major clades are shown near the root of each clade, with the minimum and maximum in parentheses. Bootstrap values are shown near major nodes.
Prophage, clonal group, inlA, and methylation analyses.
One incomplete putative prophage of CFSAN023463 was identified by PHAST (36) (Table 3). BLAST analysis of this putative prophage against all serotype IVb-v1 draft genomes, including those from the two epidemiologically unrelated clinical isolates, had >99% query coverage and 100% sequence identity. This was also consistent with the SNP analyses showing that none of the SNPs among serotype IVb-v1 isolates were in the putative prophages (see Tables S1 to S3 in the supplemental material). Four putative prophages of CFSAN023459 were identified by PHAST (36) (Table 3). BLAST analysis of these 4 putative prophages against serotype 1/2b draft genomes, except CFSAN024093, yielded >95% coverage and >99% sequence identity. The variations were mostly due to small insertions/deletions that could be caused by incomplete coverage of prophages by draft sequencing. This is also consistent with the WGS analyses showing that only one of the SNPs among serotype 1/2b isolates except CFSAN024093 was in the putative prophages (see Tables S4 and S5 in the supplemental material). In contrast, BLAST analysis of CFSAN023459 prophages 1, 2, 3, and 4 against CFSAN024093 yielded 100%, 3%, 0%, and 35% coverage, respectively, indicating significant prophage divergence between CFSAN024093 and other stone fruit-associated serotype 1/2b isolates. BLAST analysis of CFSAN023459 prophages 1, 2, 3, and 4 against the MLST-matched outgroup (CFSAN010068, GenBank accession no. NZ_CP014250.1) yielded 100%, 5%, 0%, and 43% coverage, respectively, indicating significant divergence between CFSAN010068 and stone fruit-associated serotype 1/2b isolates.
TABLE 3.
Putative prophages in the complete genomes of CFSAN023463 and CFSAN023459 identified by PHAST (36)
| Strain | Prophage ID | Length (kbp) | Prophage completeness | Positionsa | Most common match, accession no. |
|---|---|---|---|---|---|
| CFSAN023463 | 1 | 22.9 | Incomplete | 134411–157386 | Listeria A118, NC_003216 |
| CFSAN023459 | 1 | 22.9 | Incomplete | 118524–141499 | Listeria A118 |
| 2 | 32.4 | Intact | 689648–722048 | Listeria A118 | |
| 3 | 27.6 | Incomplete | 1799035–1826710 | Lactobacillus iA2, NC_028830 | |
| 4 | 57.7 | Intact | 2439092–2496851 | Listeria vB_LmoS_293, NC_028929 |
Position based on each complete genome.
In silico MLST analysis showed that all serotype IVb-v1 isolates had ST382, and all but one serotype 1/2b isolates had ST5, part of clonal complex 5 (CC5). ST382 is a singleton, not a CC, because no other isolates that differ from ST382 by one allele have been observed in the Pasteur MLST database (http://bigsdb.pasteur.fr/listeria/listeria.html) as of August 2016. ST382 was alternatively designated epidemic clone IX (13), and CC5 was alternatively designated epidemic clone VI (13, 37). One serotype 1/2b isolate, CFSAN024093, belonged to singleton ST392. The inlA sequences in the serotypes IVb-v1 and 1/2b isolates did not have premature stop codons. The methyltransferases in both CFSAN023463 and CFSAN023459 used the N6-methyladenine (m6A) methylation (see Table S7 in the supplemental material).
DISCUSSION
This incident represented the first reported outbreak and recall associated with fresh whole stone fruits due to the contamination of L. monocytogenes.
Fresh whole fruits have now been confirmed to be transmission vehicles for L. monocytogenes, with outbreaks and recalls linked to contaminated cantaloupes (38), apples (3), mangoes (39), sliced apples (40), and melons (41). Prior to this outbreak, the risk of L. monocytogenes contamination in stone fruit had not been extensively studied, probably because stone fruits have characteristics that do not support the growth of L. monocytogenes. Peach and nectarine pulp generally have a pH below 4.0 (42), which is not favorable for Listeria growth. Collignon and Korsten (43) showed that L. monocytogenes artificially inoculated onto peach surfaces at initial levels of 3 and 5 log CFU/fruit rapidly decreased during the first 6 days of refrigerated storage, although they also established that the L. monocytogenes levels did not significantly drop from the 6th day to the 20th day (final day) of storage at refrigerated temperatures.
In this study, the levels of L. monocytogenes in 99% of the fruits (104/105) were <500 CFU/fruit. Given that the average rinsing recovery rate was 75% to 85% (our unpublished data), the actual levels of L. monocytogenes on the fruits were likely <700 CFU/fruit. The remaining fruit contained L. monocytogenes of 2,850 CFU/fruit, and thus the actual level was likely <4,000 CFU/fruit. Due to the relatively long incubation period of listeriosis and relatively short shelf-life of many ready-to-eat foods, it is difficult to obtain samples from the same lots as the food consumed by the case-patients. Thus, we collected fruits from different lots that were produced in the implicated packing facility and determined the prevalence and levels of L. monocytogenes. It is important to note that the fruits analyzed in our study were not fully ripe and would have needed extra days to reach a stage of ripeness for human consumption. These fruits had also not been distributed to commerce, so it is possible that the behavior of L. monocytogenes on these fruits might be different from that on fruits released to commerce. These variables need to be taken into account when using this data set for future risk assessment. Further, the diversity among L. monocytogenes subpopulations suggests that the observation in this study may only apply to subpopulations of L. monocytogenes most similar to those involved in this outbreak, not necessarily to the entire species. In this outbreak, the Massachusetts patient was between 80 and 89 years old, and the Minnesota patient was between 70 and 79 years old (metadata provided in the GenomeTrakr database under the BioSample ID). A recent study on the 2010 to 2015 U.S. multistate ice cream outbreak showed that the levels of L. monocytogenes were ≤20, ≤50, and ≤100 most probable number (MPN)/g in 92.3%, 98.4%, and 99.8% of the ice cream, respectively (44, 55). That study showed that the geometric means of L. monocytogenes in 7 contaminated lots of ice cream ranged from 0.15 to 7.1 MPN/g. These ice cream samples were linked to a cluster of 4 elderly patients hospitalized with underlying conditions prior to exposure to the contaminated ice cream (44, 55). Contaminated meat or cheese linked to a 2012-2013 Austria outbreak contained L. monocytogenes at <30 CFU/g (45). Contaminated cheese linked to a 2012 U.S. multistate outbreak contained L. monocytogenes at ∼9.0 × 103 to ∼3.75 × 106 CFU/g (median, 4.77 × 104 CFU/g) (4). Contaminated foie gras from 3 unopened samples linked to a 2012-2013 Spain outbreak contained L. monocytogenes at 5.2 × 104 CFU/g (56). These data could help us gain better understanding of the risk associated with L. monocytogenes contamination.
The use of ALOA and RAPID'L. mono agar was critical in the enumeration. We investigated esculin-based agars, e.g., polymyxin-acriflavin-lithium chloride-ceftazidime-aesculin-mannitol (PALCAM) and modified Oxford agar, but the background flora on the fruits made the plate counting difficult. In contrast, ALOA and RAPID'L. mono agar allowed easier differentiation between L. monocytogenes and background flora. We observed presumptive Listeria spp. that were not L. monocytogenes. A Listeria innocua isolate was also recovered from the company A environment (data not shown), confirming the value of using L. monocytogenes-specific chromogenic agars.
WGS helped linking clinical isolates to the recalled fruits and separated them from PFGE-matched, but epidemiologically unrelated, isolates.
Following the positive pathogen findings in fruits, initial epidemiological investigation, and PFGE analyses, WGS was performed in real time to help identify clinical cases linked to the contaminated fruits (14, 16). Our study illustrates how using SNP-based WGS analyses enabled us to exclude two epidemiologically unrelated clinical cases that were indistinguishable by PFGE from outbreak-associated isolates. The CFSAN SNP Pipeline is a reference-based SNP identification tool. Technically, a reference genome could be one of the isolates associated with the outbreak or an existing genome in the GenomeTrakr database that is genetically close to the isolates being analyzed; in any case, using a complete genome could maximize the resolution power. In the present study, we chose complete genomes of the stone fruit isolates as the references.
The WGS analyses described in the present study and that in the epidemiological report (16) were based on completely different phylogenetic algorithms. They generated identical phylogeny for the same set of isolates, further increasing the confidence in the WGS analyses. The nationwide screening of illnesses based on onset dates and PFGE matches did not identify any patients possibly linked to serotype 1/2b isolates from the stone fruits (16). This could be partially attributed to the lower incidence of serotype 1/2b (18 isolates) than serotype IVb-v1 (76 isolates) in stone fruits with our sampling scheme, especially in peaches (10 serotype 1/2b isolates and 64 serotype IVb-v1 isolates), which had higher levels of L. monocytogenes than those in nectarines. Nonetheless, we still could not exclude the possibility that serotype 1/2b isolates caused unreported illnesses, especially considering that isolates from the same clonal group have been repeatedly involved in previous listeriosis outbreaks as discussed below. With only two colonies picked from each of the 38 fruits, we identified 5 fruits containing both serotypes IVb-v1 and 1/2b; thus, it is possible that more than 5 fruits were contaminated by both serotypes. Therefore, we could not exclude the possibility that the case-patients were coinfected with both serotypes, but that only serotype IVb-v1 was isolated, partially due to its higher incidence. The environmental isolates collected from company A clustered with the food and clinical isolates, indicating that the packing facility was contaminated; however, available environmental samples were only from the packing facility, and specific sampling locations were not available; thus, we cannot determine whether fruit contamination was due to bacteria persistent in the facility or transient contamination originating from sources outside the facility, e.g., fruit orchards.
In most previous listeriosis outbreaks, clusters of illnesses were recognized as an outbreak, and patients were interviewed before the food or environmental source was confirmed by microbial source tracking. In contrast, the investigation and recognition of this outbreak started from the positive pathogen findings in foods (14, 16). One limitation with such an approach is that the identification of clinical cases can be affected by the sampling bias of the food products. We sampled 105 fruits from 7 lots and also obtained fruit and environmental isolates collected by company A; however, we did not have any evidence to determine whether there were L. monocytogenes strains of other genotypes in other lots of the fruits.
WGS data in the present study also provided valuable information about the genetic diversity among isolates involved in a common-source outbreak or isolates associated with foods manufactured in a single packing facility. Previous studies have identified such diversity to be 0 to 5 SNPs (6, 46), 5 to 10 SNPs (47, 48), 10 to 20 SNPs (7), and 20 to 30 SNPs (9), although we need to keep in mind that different studies might target slightly different regions of the genome and employ different bioinformatics tools or parameters to identify SNPs. The Illinois clinical isolate and the outbreak-associated isolates differed by 40 to 48 SNPs, similar to the number of SNP differences (up to 42) among outbreak-associated isolates. However, the WGS phylogeny clearly placed the Illinois clinical isolate outside the outbreak cluster. Similarly, the number of wgMLST allelic differences (up to 47) between the Illinois clinical isolate and the outbreak cluster was similar to the number of allelic differences (up to 43) among isolates in the outbreak cluster, but the Illinois clinical isolate was clearly placed outside the outbreak cluster by wgMLST (16). This indicates that the SNP threshold or the genetic diversity of isolates should not serve as the purpose of outbreak case definition; rather, it should always be combined with WGS phylogeny and epidemiologic evidence to identify strain relationships.
The SNP differences (≤48 SNPs) between outbreak-associated isolates and PFGE-matched, but epidemiologically unrelated, isolates were smaller than those between the stone fruit-associated serotype 1/2b isolates and MLST-matched outgroup (>250 SNPs) and those between the outbreak-associated isolates and MLST-matched, but epidemiologically unrelated, isolates in several previous studies (6, 7). This is consistent with the observation that PFGE possessed more discriminatory power than MLST (37). Therefore, the inclusion of PFGE-matched and epidemiologically unrelated isolates demonstrated the high resolution of WGS and also allowed the identification of SNPs highly specific to the outbreak isolates; many of the SNPs were in surface proteins (see Tables S1, S3, and S5 in the supplemental material), possibly associated with virulence (49).
Putative prophages were conserved among isolates from the fruits and their packing environment.
Our WGS analyses revealed one putative prophage in the complete genome of CFSAN023463 and four putative prophages in the complete genome of CFSAN023459. The PHAST-identified prophage 4 included a 43.1-kbp comK prophage (between positions 2453301 and 2496449), indicating that the PHAST results need to be manually examined before any future in-depth analysis. These putative prophages showed a heavily mosaic nature, because each region contained proteins that were similar to multiple different Listeria phages and phages from other species (see Table S6 in the supplemental material). We used fully closed reference genomes to maximize the resolution power in identifying prophage variations. The >95% query coverage of BLAST analyses between the reference genomes and all but one draft genome indicated that our draft sequencing provided good coverage for prophage regions. BLAST analyses showed that all isolates except CFSAN024093 contained these putative prophages, and the SNP-based WGS analyses revealed 0 and 1 SNP in the putative prophages among serotype IVb-v1 isolates and 1/2b isolates, respectively (see Tables S1 to S5 in the supplemental material). Our finding is consistent with previous observations suggesting that prophages could serve as markers for epidemiology of L. monocytogenes. Specifically, for MLST-matched isolates, prophages were conserved among those involved in a single outbreak/incident or those resident in a facility for a relatively short period of time, but they were more diverse among those involved in different outbreaks/incidents or persistent in a facility for a relatively long period of time (7, 48, 50, 51). In this study, there were no major variations in the putative prophages between the outbreak-associated isolates and the two PFGE-matched, but epidemiologically unrelated, isolates, consistent with some previous findings suggesting that prophage variations and PFGE possessed similar discriminatory power (50, 51). The prophages of all but one stone fruit-associated serotype 1/2b isolate were conserved, but they differed significantly from the MLST-matched outgroup CFSAN010068 from a different outbreak, confirming previous observations that prophages among MLST-matched isolates from different incidents could have significant divergence (50, 51). More whole-genome sequences, especially complete genomes, are needed in order to better understand the Listeria prophage variations among isolates in the same outbreak or those from the same facility.
WGS data provided information on clonal groups of the isolates from fruits and environment.
The serotype IVb-v1 isolates in this study belonged to singleton ST382, which was not observed in two large-scale surveys studying over 8,000 L. monocytogenes isolates from multiple sources and geographic locations (52, 53). However, ST382 had been associated with a 2014-2015 U.S. multistate caramel apple outbreak and a 2015-2016 U.S. multistate packaged leafy green salad outbreak (13). The singleton ST382 thus represents an emerging clonal group of L. monocytogenes. All but one serotype 1/2b isolate in this study were MLST ST5, part of CC5. The same ST had been involved in a 2011 U.S. multistate cantaloupe outbreak (37, 38), a 2013 U.S. Hispanic-style cheese outbreak (13, 32), a 2010 to 2015 U.S. multistate ice cream outbreak (1, 13), a 2012-2013 Austria outbreak linked to meat and/or cheese (45) (in silico MLST performed in our study using published WGS but not described in Materials and Methods), and a 1996 Canada imitation crabmeat outbreak (37, 54). Thus, CC5 appears to be a widely distributed clone contaminating a variety of food products and processing environments and causing outbreaks.
Conclusions.
The SNP-based WGS analysis provided further discrimination between outbreak-associated isolates and epidemiologically unrelated clinical isolates that were indistinguishable by PFGE. The study highlights the importance to combine WGS with epidemiological evidence to for identifying outbreak-associated isolates. The enumeration data could be useful for future risk-based characterization of L. monocytogenes.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded in part by an appointment to the Research Participation Program at the U.S. Food and Drug Administration (FDA) Center for Food Safety and Applied Nutrition, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the FDA.
We thank Cheryl Tarr and Suzana Kucerova at the U.S. Centers for Disease Control and Prevention for sharing the sequences and PFGE profiles of the four clinical isolates. We also thank the FDA San Francisco District Office for collecting the stone fruits used in this study. We are grateful to Lili Fox Vélez for her editorial assistance on this paper.
The conclusions, findings, and opinions are those of the authors and do not necessarily represent the official position of the U.S. FDA.
Funding Statement
This work, including the efforts of Anna Maounounen-Laasri, was funded in part by DOE| LDRD | Oak Ridge Institute for Science and Education (ORISE).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01486-16.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2015. Multistate outbreak of listeriosis linked to Blue Bell Creameries products (final update). Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/listeria/outbreaks/ice-cream-03-15/. [Google Scholar]
- 2.Chen Y, Zhang W, Knabel SJ. 2005. Multi-virulence-locus sequence typing clarifies epidemiology of recent listeriosis outbreaks in the United States. J Clin Microbiol 43:5291–5294. doi: 10.1128/JCM.43.10.5291-5294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2015. Multistate outbreak of listeriosis linked to commercially produced, prepackaged caramel apples made from Bidart Bros. apples (final update). Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/listeria/outbreaks/caramel-apples-12-14/. [Google Scholar]
- 4.Heiman KE, Garalde VB, Gronostaj M, Jackson KA, Beam S, Joseph L, Saupe A, Ricotta E, Waechter H, Wellman A, Adams-Cameron M, Ray G, Fields A, Chen Y, Datta A, Burall L, Sabol A, Kucerova Z, Trees E, Metz M, Leblanc P, Lance S, Griffin PM, Tauxe RV, Silk BJ. 2015. Multistate outbreak of listeriosis caused by imported cheese and evidence of cross-contamination of other cheeses, USA, 2012. Epidemiol Infect 144:2698–2708. doi: 10.1017/S095026881500117X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, Bulach DM, Stinear TP, Seemann T, Howden BP. 2016. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J Clin Microbiol 54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kvistholm Jensen A, Nielsen EM, Bjorkman JT, Jensen T, Muller L, Persson S, Bjerager G, Perge A, Krause TG, Kiil K, Sorensen G, Andersen JK, Molbak K, Ethelberg S. 2016. Whole-genome sequencing used to investigate a nationwide outbreak of listeriosis caused by ready-to-eat delicatessen meat, Denmark, 2014. Clin Infect Dis 63: 333–33. doi: 10.1093/cid/ciw192. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Holmes N, Martinez E, Howard P, Hill-Cawthorne G, Sintchenko V. 2015. It is not all about single nucleotide polymorphisms: comparison of mobile genetic elements and deletions in Listeria monocytogenes genomes links cases of hospital-acquired listeriosis to the environmental source. J Clin Microbiol 53:3492–3500. doi: 10.1128/JCM.00202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pettengill JB, Luo Y, Davis S, Chen Y, Gonzalez-Escalona N, Ottesen A, Rand H, Allard MW, Strain E. 2014. An evaluation of alternative methods for constructing phylogenies from whole genome sequence data: a case study with Salmonella. PeerJ 2:e620. doi: 10.7717/peerj.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilmour MW, Graham M, Van Domselaar G, Tyler S, Kent H, Trout-Yakel KM, Larios O, Allen V, Lee B, Nadon C. 2010. High-throughput genome sequencing of two Listeria monocytogenes clinical isolates during a large foodborne outbreak. BMC Genomics 11:120. doi: 10.1186/1471-2164-11-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, Harmsen D, Mellmann A. 2015. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol 53:2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis S, Pettengill JB, Luo Y, Payne J, Shpuntoff A, Rand H, Strain A. 2015. CFSAN SNP Pipeline: an automated method for constructing SNP matrices from next-generation sequence data. PeerJ Comput Sci 1:e20. doi: 10.7717/peerj-cs.20. [DOI] [Google Scholar]
- 12.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Gonzalez-Escalona N, Hammack TS, Allard M, Strain EA, Brown EW. 2016. Core genome multilocus sequence typing for identification of globally distributed clonal groups and differentiation of outbreak strains of Listeria monocytogenes. Appl Environ Microbiol 82:6258–6272. doi: 10.1128/AEM.01532-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, Carleton H, Katz LS, Stroika S, Gould LH, Mody RK, Silk BJ, Beal J, Chen Y, Timme R, Doyle M, Fields A, Wise M, Tillman G, Defibaugh-Chavez S, Kucerova Z, Sabol A, Roache K, Trees E, Simmons M, Wasilenko J, Kubota K, Pouseele H, Klimke W, Besser J, Brown E, Allard M, Gerner-Smidt P. 2016. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin Infect Dis 63:380–386. doi: 10.1093/cid/ciw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allard MW, Strain E, Melka D, Bunning K, Musser SM, Brown EW, Timme R. 2016. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol 54:1975–1983. doi: 10.1128/JCM.00081-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson BR, Salter M, Tarr C, Conrad A, Harvey E, Steinbock L, Saupe A, Sorenson A, Katz L, Stroika S, Jackson KA, Carleton H, Kucerova Z, Melka D, Strain E, Parish M, Mody RK. 2015. Notes from the field: listeriosis associated with stone fruit—United States, 2014. Morb Mortal Wkly Rep 64:282–283. [PMC free article] [PubMed] [Google Scholar]
- 17.Hitchins AD, Jinneman K, Chen Y. 2016. Food and Drug Administration bacteriological analytical manual chapter 10, detection and enumeration of Listeria monocytogenes in foods. US Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm071400.htm. [Google Scholar]
- 18.Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand J Statist 6:65–70. [Google Scholar]
- 19.Whitney J. 1997. Notes on methodology—testing for differences with the nonparametric Mann-Whitney U test. J Wound Ostomy Continence Nurs 24:12. doi: 10.1016/S1071-5754(97)90044-9. [DOI] [PubMed] [Google Scholar]
- 20.Bewick V, Cheek L, Ball J. 2004. Statistics review 9: one-way analysis of variance. Crit Care 8:130–136. doi: 10.1186/cc2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burall LS, Simpson AC, Datta AR. 2011. Evaluation of a serotyping scheme using a combination of an antibody-based serogrouping method and a multiplex PCR assay for identifying the major serotypes of Listeria monocytogenes. J Food Prot 74:403–409. doi: 10.4315/0362-028X.JFP-10-355. [DOI] [PubMed] [Google Scholar]
- 22.Graves LM, Swaminathan B. 2001. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int J Food Microbiol 65:55–62. doi: 10.1016/S0168-1605(00)00501-8. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Chen Y, Ayers S, Melka D, Laasri A, Payne JS, Zheng J, Son I, Timme R, Kastanis G, Hammack TS, Strain E, Allard MW, Evans PS, Brown EW. 2015. Draft genome sequence of Salmonella enterica subsp. enterica serovar Give, isolated from an imported chili powder product. Genome Announc 3(4):e00726-15. doi: 10.1128/genomeA.00726-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allard MW, Muruvanda T, Strain E, Timme R, Luo Y, Wang C, Keys CE, Payne J, Cooper T, Luong K, Song Y, Chin CS, Korlach J, Roberts RJ, Evans P, Musser SM, Brown EW. 2013. Fully assembled genome sequence for Salmonella enterica subsp. enterica serovar Javiana CFSAN001992. Genome Announc 1(2):e00081-13. doi: 10.1128/genomeA.00081-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 26.Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Feng Z, Losic B, Mahajan MC, Jabado OJ, Deikus G, Clark TA, Luong K, Murray IA, Davis BM, Keren-Paz A, Chess A, Roberts RJ, Korlach J, Turner SW, Kumar V, Waldor MK, Schadt EE. 2012. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol 30:1232–1239. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts RJ, Vincze T, Posfai J, Macelis D. 2010. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. 2009. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Ph.D. dissertation. The University of Texas at Austin, Austin, TX. [Google Scholar]
- 32.Centers for Disease Control and Prevention. 2014. Multistate outbreak of listeriosis linked to Roos Foods dairy products (final update). Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/listeria/outbreaks/cheese-02-14/. [Google Scholar]
- 33.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lomonaco S, Verghese B, Gerner-Smidt P, Tarr C, Gladney L, Joseph L, Katz L, Turnsek M, Frace M, Chen Y, Brown E, Meinersmann R, Berrang M, Knabel S. 2013. Novel epidemic clones of Listeria monocytogenes, United States, 2011. Emerg Infect Dis 19:147–150. doi: 10.3201/eid1901.121167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCollum JT, Cronquist AB, Silk BJ, Jackson KA, O'Connor KA, Cosgrove S, Gossack JP, Parachini SS, Jain NS, Ettestad P, Ibraheem M, Cantu V, Joshi M, DuVernoy T, Fogg NW Jr, Gorny JR, Mogen KM, Spires C, Teitell P, Joseph LA, Tarr CL, Imanishi M, Neil KP, Tauxe RV, Mahon BE. 2013. Multistate outbreak of listeriosis associated with cantaloupe. N Engl J Med 369:944–953. doi: 10.1056/NEJMoa1215837. [DOI] [PubMed] [Google Scholar]
- 39.Food Safety News. 2014. Organic mangoes recalled for possible Listeria contamination. Food Safety News, Seattle, WA: http://www.foodsafetynews.com/2014/05/organic-mangos-recalled-for-possible-listeria-contamination/#.Ve-UkZc-Yg4. [Google Scholar]
- 40.Food Safety News. 2013. Apple slices recalled for possible Listeria contamination. Food Safety News, Seattle, WA: http://www.foodsafetynews.com/2013/11/apple-slices-recalled-for-potential-listeria-contamination/#.Ve-Xc5c-Yg4. [Google Scholar]
- 41.Food Safety News. 2012. Listeria cantaloupe recall expanded to 188,000 melons. Food Safety News, Seattle, WA: http://www.foodsafetynews.com/2012/08/listeria-cantaloupe-recall-expanded-to-188000-melons/#.Ve-Ym5c-Yg4. [Google Scholar]
- 42.US Food and Drug Administration. 2015. BBB—pH values of various foods. US Food and Drug Administration, Silver Spring, MD: http://www.fda.gov/Food/FoodborneIllnessContaminants/CausesOfIllnessBadBugBook/ucm122561.htm. [Google Scholar]
- 43.Collignon S, Korsten L. 2010. Attachment and colonization by Escherichia coli O157:H7, Listeria monocytogenes, Salmonella enterica subsp. enterica serovar Typhimurium, and Staphylococcus aureus on stone fruit surfaces and survival through a simulated commercial export chain. J Food Prot 73:1247–1256. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y, Allard E, Wooten A, Hur M, Sheth I, Laasri A, Hammack TS, Macarisin D. 2016. Recovery and growth potential of Listeria monocytogenes in temperature abused milkshakes prepared from naturally contaminated ice cream linked to a listeriosis outbreak. Front Microbiol 7:764. doi: 10.3389/fmicb.2016.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmid D, Allerberger F, Huhulescu S, Pietzka A, Amar C, Kleta S, Prager R, Preussel K, Aichinger E, Mellmann A. 2014. Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011–2013. Clin Microbiol Infect 20:431–436. doi: 10.1111/1469-0691.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holch A, Webb K, Lukjancenko O, Ussery D, Rosenthal BM, Gram L. 2013. Genome sequencing identifies two nearly unchanged strains of persistent Listeria monocytogenes isolated at two different fish processing plants sampled 6 years apart. Appl Environ Microbiol 79:2944–2951. doi: 10.1128/AEM.03715-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orsi RH, Borowsky ML, Lauer P, Young SK, Nusbaum C, Galagan JE, Birren BW, Ivy RA, Sun Q, Graves LM, Swaminathan B, Wiedmann M. 2008. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9:539. doi: 10.1186/1471-2164-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, Li C, Strain EA, Knabel SJ, Allard MW, Brown EW. 2012. Whole genome analysis of Listeria monocytogenes epidemic clone II isolates from food processing plants. Abstr 112th Gen Meet Am Soc Microbiol, San Francisco, CA, 16 to 19 June 2012. [Google Scholar]
- 49.Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol 10:238–245. doi: 10.1016/S0966-842X(02)02342-9. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Knabel SJ. 2008. Prophages in Listeria monocytogenes contain single-nucleotide polymorphisms that differentiate outbreak clones within epidemic clones. J Clin Microbiol 46:1478–1484. doi: 10.1128/JCM.01873-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verghese B, Lok M, Wen J, Alessandria V, Chen Y, Kathariou S, Knabel S. 2011. comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Appl Environ Microbiol 77:3279–3292. doi: 10.1128/AEM.00546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haase JK, Didelot X, Lecuit M, Korkeala H, L. monocytogenes MLST Study Group, Achtman M. 2014. The ubiquitous nature of Listeria monocytogenes clones: a large-scale multilocus sequence typing study. Environ Microbiol 16:405–416. doi: 10.1111/1462-2920.12342. [DOI] [PubMed] [Google Scholar]
- 53.Maury MM, Tsai YH, Charlier C, Touchon M, Chenal-Francisque V, Leclercq A, Criscuolo A, Gaultier C, Roussel S, Brisabois A, Disson O, Rocha EP, Brisse S, Lecuit M. 2016. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet 48:308–313. doi: 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farber JM, Daley EM, MacKie MT, Limerick B. 2000. A small outbreak of listeriosis potentially linked to the consumption of imitation crab meat. Lett Appl Microbiol 31:100–104. doi: 10.1046/j.1365-2672.2000.00775.x. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, Burall L, Macarisin D, Pouillot R, Strain E, De Jesus A, Laasri AM, Wang H, Ali A, Tatavarthy A, Zhang G, Hu L, Day J, Kang J, Sahu S, Srinivasan D, Klontz K, Parish M, Evans P, Brown EW, Hammack TS, Zink D, Datta A. 2016. Prevalence and level of Listeria monocytogenes in ice cream linked to a listeriosis outbreak in the United States. J Food Prot 79:1828–1832. [DOI] [PubMed] [Google Scholar]
- 56.Perez-Trallero E, Zigorraga C, Artieda J, Alkorta M, Marimon JM. 2014. Two outbreaks of Listeria monocytogenes infection, Northern Spain. Emerg Infect Dis 20:2155–2157. doi: 10.3201/eid2012.140993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.