Abstract
Macrolide antibiotic binding to the ribosome inhibits catalysis of peptide bond formation between specific donor and acceptor substrates. Why particular reactions are problematic for the macrolide-bound ribosome remains unclear. Using comprehensive mutational analysis and biochemical experiments with synthetic substrate analogs, we find that the positive charge of these specific residues and the length of their side chains underlie inefficient peptide bond formation in the macrolide-bound ribosome. Even in the absence of antibiotic, peptide bond formation between these particular donors and acceptors is rather inefficient, suggesting that macrolides magnify a problem present for intrinsically difficult substrates. Our findings emphasize the existence of functional interactions between the nascent protein and the catalytic site of the ribosomal peptidyl transferase center.
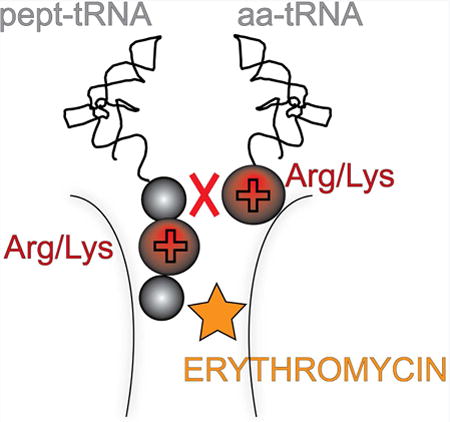
Graphical abstract
Macrolide antibiotics stall ribosomes at the Arg/Lys-X-Arg/Lys motif. Sothiselvam et al. find that the charge and size of key amino acid side chains in this motif make peptide bond formation inefficient. Antibiotics greatly magnify the problem of these intrinsically difficult donor-acceptor pairs.
Introduction
The ribosome is an incredibly complex and sophisticated protein synthesis machine which represents one of the major antibiotic targets in bacterial cells (Wilson, 2009). While many ribosome-targeting inhibitors completely abolish protein synthesis, some of them interfere with translation in a context-specific manner. Macrolides, such as erythromycin (ERY) and its derivatives, represent the best-studied examples of inhibitors whose action critically depends on the amino acid sequence of the synthesized protein (Davis et al., 2014; Hardesty et al., 1990; Kannan et al., 2014; Kannan et al., 2012; Starosta et al., 2010). Depending on the nature of the nascent chain and the structure of the drug, synthesis of the polypeptide can be arrested during very early rounds (Otaka and Kaji, 1975; Tenson et al., 2003), at the later stages of translation elongation (Davis et al., 2014; Kannan et al., 2014), or not at all (Kannan et al., 2012; Starosta et al., 2010). While optimizing clinical outcomes of antibiotic therapy hinges on achieving an understanding of the mode of drug action, a mechanistic explanation for the context specificity of macrolides is lacking.
Macrolide antibiotics target the large ribosomal subunit. The drugs bind in the nascent peptide exit tunnel (NPET) at a short distance from the peptidyl transferase center (PTC). Binding of the antibiotic obstructs the NPET and restricts placement of the nascent chain in the tunnel and its progression from the PTC to the tunnel exit (Bulkley et al., 2010; Dunkle et al., 2010; Schlunzen et al., 2001; Tu et al., 2005). When the newly-synthesized peptide chain grows to around 4 amino acids, it reaches the site of antibiotic binding (Arenz et al., 2014a; Arenz et al., 2014b). Synthesis of some proteins by the drug-bound ribosome is interrupted at this stage, especially if the macrolide, like ERY, contains a C3-bound cladinose sugar that protrudes into the lumen of the tunnel (Bulkley et al., 2010; Dunkle et al., 2010; Schlunzen et al., 2001). However, some nascent peptides are able to bypass the antibiotic, allowing their continued synthesis in spite of the presence of a bulky drug molecule in the NPET (Hardesty et al., 1990; Kannan et al., 2012; Starosta et al., 2010). Ketolide drugs, which lack a C3 cladinose, rarely inhibit translation at the early stages, and synthesis of many proteins continues past the initial rounds of protein elongation (Kannan et al., 2014; Kannan et al., 2012).
Although macrolides do not inhibit synthesis of many proteins at the early rounds, translation of the majority of polypeptides by the drug-bound ribosome is eventually interrupted at the later stages of elongation (Davis et al., 2014; Kannan et al., 2014). Strikingly, abrogation of translation does not occur randomly, but rather, drug-bound ribosomes become arrested at specific, well-defined mRNA sites. Ribosome profiling analysis carried out in Gram positive and Gram negative bacteria treated with macrolides helped identify the major sites of late translation arrest and allowed for initial classification of problematic sequences (Davis et al., 2014; Kannan et al., 2014). Several amino acid motifs conducive to macrolide-induced arrest emerged from these studies. One of the most prevalent motifs conforms to the consensus R/K-X-R/K, where R and K represent arginine and lysine, respectively, and ‘X’ represents any amino acid. In vitro biochemical testing supported the conclusion drawn from the profiling analysis that the ribosome stalls when the codon specifying the middle amino acid (X) of the motif enters the P site. Accordingly, the first residue of the consensus (R or K) represents the penultimate amino acid of the nascent peptide chain while the last consensus sequence residue (also R or K) corresponds to the A site-bound aminoacyl-tRNA (Davis et al., 2014; Kannan et al., 2014; Sothiselvam et al., 2014).
Importantly, the context specificity of antibiotic action is exploited for regulation of expression of resistance genes (reviewed in (Weisblum, 1995)). The inducible macrolide-resistance genes remain silent in the absence of the antibiotic, but are activated in its presence. Activation relies on programmed ribosome stalling at a precise, evolutionarily defined site of the regulatory upstream ORF (uORF) that precedes the resistance gene (Ramu et al., 2009; Subramanian et al., 2011; Weisblum, 1995). The ribosomes arrested at the uORF alter the folding of the mRNA, activating the expression of the downstream resistance cistron. The structure of the antibiotic and the sequence of the leader peptide are the two main factors that determine the site of programmed translation arrest (Arenz et al., 2014a; Arenz et al., 2014b; Gryczan et al., 1980; Horinouchi and Weisblum, 1980; Ramu et al., 2011; Vazquez-Laslop, 2010; Vazquez-Laslop et al., 2008). Strikingly, the R/K-X-R/K sequence, identified as one of the major motifs of macrolide-induced translation arrest (Davis et al., 2014; Kannan et al., 2014), is present in many of the regulatory uORFs of macrolide resistance genes, including the well-studied ermDL ORF that controls expression of the ermD gene (Kwak et al., 1991; Kwon et al., 2006; Sothiselvam et al., 2014). In all the tested regulatory uORFs with the R/K-X-R/K motif, the ribosome stalls when the second codon of the consensus enters the ribosomal P site (Almutairi et al., 2015; Sothiselvam et al., 2014).
Strikingly, in the stalled ribosome, the amino acid residues of the R/K-X-R/K motif are located at the PTC active site rather than being juxtaposed next to the antibiotic molecule bound in the NPET (Arenz et al., 2014a; Arenz et al., 2014b). Similarly, other stalling motifs identified in ribosome profiling experiments were also confined to the C-terminal residues of the nascent chain and the incoming amino acid (Davis et al., 2014; Kannan et al., 2014). These observations led to the conclusion that, at least at the late elongation stage, macrolides do not hinder translation by simply barricading the NPET, but rather act as context-specific PTC inhibitors (Kannan et al., 2014). Indeed, RNA chemical probing showed that binding of macrolides to the NPET of the vacant ribosome was sufficient to allosterically induce structural changes in the PTC (Sothiselvam et al., 2014). In agreement with this view, translation of the 5′-terminally truncated ermDL ORF, which encodes the peptide starting with the MRLR sequence, was efficiently arrested by macrolides at the Leu3 codon, when the nascent chain was only three amino acids long and had only limited contacts with the drug. These results suggested that inhibition of translation elongation by macrolides does not necessarily require blocking the progression of the nascent chain in the drug-obstructed NPET, but rather, that binding of the antibiotic allosterically renders the PTC more restrictive towards its substrates, thus predisposing the ribosome for translation arrest when certain combinations of donors and acceptors meet at the catalytic center (Kannan et al., 2014; Sothiselvam et al., 2014).
Despite the importance of understanding the precise molecular mechanisms of antibiotic action for the development of better drugs and for the exploration of ribosome functions, the basis for macrolide specificity remains obscure. It is poorly understood why certain combinations of donor and acceptor substrates, defined by the identified consensus sequences, are troublesome for the drug-bound ribosome. In particular, for the translation arrest directed by the R/K-X-R/K sequence, one could envision that either the positive charge or the extended length of the side chains of the arginine and lysine residues of the motif could be the dominant factor contributing to stalling. It is also unclear whether the binding of macrolide antibiotics to the ribosome causes some normally ‘easy’ substrates to become ‘difficult’, or if the drugs simply exacerbate the challenging nature of intrinsically problematic substrates.
In order to gain further mechanistic insight into the mode of action of macrolide antibiotics, in this work we investigated the effect of the drug on the ability of the ribosome to catalyze the transfer of the short nascent MRL peptide to acceptor substrates that differ in their physicochemical properties. The lack of an extended nascent chain in this stalled complex allowed us to minimize the contribution of the context preceding the stalling motif on translation arrest (Davis et al., 2014; Elgamal et al., 2014; Kannan et al., 2014; Starosta et al., 2014; Woolstenhulme et al., 2013). We found that two factors, the length of the side chains of the amino acid residues in the donor and acceptor substrates and even more importantly, the positive charge of the critical residues contribute to macrolide-induced arrest. Interestingly, our results indicate that the model substrates that react extremely slowly in the presence of antibiotic appear to be problematic donor-acceptor pairs even for the drug-free ribosome.
Results
Antibiotic-mediated translation arrest at the MRLR ORF is independent of the mRNA sequence and tRNA structure
Our previous experiments showed that macrolide antibiotics arrest translation of the truncated ermDL gene (MRLR…), when the Leu3 codon is placed in the ribosomal P site and the Arg4 codon enters the decoding site (Sothiselvam et al., 2014). In order to test whether macrolide-induced ribosome stalling is influenced by the structures of mRNA or tRNA, we changed codons 2-4 of the truncated ermDL template to several different combinations of synonymous triplets that not only alter the mRNA sequence, but also direct binding of different aminoacyl-tRNA isoacceptors (Figure S1A). Toeprinting analysis showed that these alterations in mRNA sequence had minimal effects on the efficiency of ERY-directed arrest (Figure S1A). Therefore, neither the mRNA sequence nor the structure of the tRNA body affect macrolide-dependent ribosome stalling at the truncated ermDL ORF, leaving the nature of the nascent peptide sequence as the likely primary determinant of translation arrest.
Only the templates encoding Arg or Lys in the second and fourth codons are conducive to ERY-induced translation arrest
In the ribosome stalled at the truncated ermDL template, the MRL tripeptide esterifies the P site tRNALeu and the A site codon specifies for Arg-tRNA. We wanted to understand how the chemical nature of the peptidyl donor and aminoacyl acceptor affects catalysis of peptide bond formation by the drug-bound ribosome. First, we investigated the contributions of the second and third amino acids of the peptidyl moiety of the donor substrate, MRL-tRNA, to macrolide-dependent translation arrest. Codons 2 and 3 of the MRLR ORF, which encode the penultimate and the C-terminal residues of the nascent peptide, were mutated to specify all other 19 amino acids, and ERY-induced ribosome stalling in these templates was analyzed by toeprinting. Most of the alterations of the penultimate amino acid (Arg2) abolished stalling. Only the Arg2-to-Lys replacement maintained efficient translation arrest (Figure 1A). In contrast, the nature of the C-terminal residue was of a much lesser importance: only changes of Leu3 to Trp or Pro showed a somewhat increased bypass of the arrest site, whereas the majority of the mutants directed ribosome stalling as readily as the wild type sequence (Figure 1B).
Figure 1. Amino acid residues critical for ERY dependent translation arrest in the MRLR ORF.
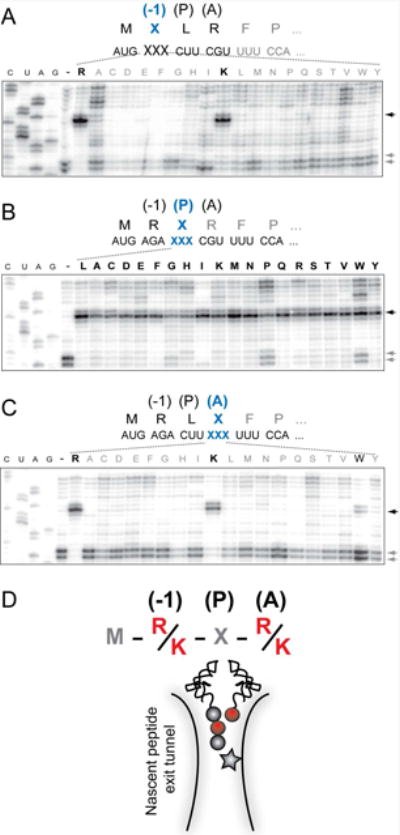
Toeprinting analysis of ERY-dependent ribosome stalling in the MRLR ORFs. The codons specifying the residues of the RLR motif located in the penultimate (-1) position of the MRL tripeptide (A), the peptide's C-terminal residue located in the PTC P site (B) or the amino acid to be placed in the A site (C) were mutated to code for each of the other 19 canonical amino acids. Black arrows indicate toeprint bands representing ERY-dependent translation arrest at the third codon. Lanes marked ‘-‘ represent the reactions with wt template but lacking ERY. Gray arrows point to the toeprint bands corresponding to the ribosomes, which bypassed the third codon and were captured at the downstream Pro6 codon due to the presence of mupirocin, an IleRS inhibitor. In the case of Ile mutations in the MRLR peptide (lanes ‘I’), the Ile7 codon was replaced with the Trp codon and mupirocin was substituted with indlomycin, the TrpRS inhibitor. Symbols representing amino acids conducive to ERY-dependent arrest are italicized and shown in bold. Sequencing lanes are marked as C, U, A, G. Shown gels are representative of two independent experiments. (D) Representation of the ERY-stalled ribosome with the tripeptidyl MRL-tRNALeu in the P site and aminoacyl-tRNA in the A site. Amino acid residues critical for drug dependent ribosome stalling are depicted by red spheres and the antibiotic molecule is indicated by a star. See also Figures S1 and S2.
We then examined the role of the acceptor substrate. By mutating the fourth codon of the MRLR ORF, we directed binding of aminoacyl-tRNAs linked to each of the 20 different amino acids to the ribosomal A site. Similar to the effect observed for the penultimate position of the nascent peptide, only the Arg4-to-Lys (and to a much lesser extent the Arg4-to-Trp) substitution was conducive to ERY-induced stalling (Figure 1C). Thus, ERY promotes formation of a stable stalled ribosome complex preferentially on those short templates where the second and fourth codons specify either Arg or Lys.
Because individual substitutions of Arg2 or Arg4 with Lys were compatible with ERY-induced stalling, we also tested whether simultaneous replacement of both Arg codons with Lys would still support translation arrest. Indeed, we found that the majority of the ribosomes that reached the third codon of the MKLK template became arrested (Figure S1B, black arrowheads, lane ‘L’). In addition, similar to the results obtained with the MRLR template, mutations of the Leu3 codon, sandwiched between the two Lys codons in the MKLK template, had little effect on ERY-induced arrest (Figure S1B). An exception was the MKPK sequence. In this template, drug-bound ribosomes could not reach the 3rd codon because they stalled at the Lys2 codon (Figure S1B, lane ‘P’), likely due to the reported propensity of macrolides to inhibit peptide bond formation between a donor peptide with a C-terminal lysine and prolyl-tRNA (Davis et al., 2014; Kannan et al., 2014).
Altogether, our mutational studies showed that the antibiotic bound in the ribosomal exit tunnel efficiently and specifically prevents the transfer of nascent tripeptides that have an Arg or Lys as their penultimate residues to an Arg or Lys acceptor (Figure 1D).
The presence of Arg or Lys in the donor and acceptor substrates is critical for translation arrest prompted by diverse macrolide antibiotics
Stalling peptides that regulate expression of erm genes often exhibit strong selectivity towards macrolide antibiotics containing C3 cladinose (e.g. ERY), but do not respond to cladinose-lacking ketolides (Vazquez-Laslop et al., 2011; Vazquez-Laslop et al., 2008). In contrast, both cladinose-containing macrolides as well as ketolides arrest translation of the truncated ErmDL (Sothiselvam et al., 2014). We asked whether the identity of the second and fourth codons of the MRLR template, which is critical for ERY-induced arrest (Figure 1), is also important for ribosome stalling directed by other macrolide antibiotics. Therefore, we tested the response of ErmDL mutants to the fluoro-ketolide solithromycin (SOL) and the cladinose-containing 15-member ring, azithromycin (AZI) (Figure S2A).
Similar to the effects observed with ERY, the mutations of Arg2 or Arg4 to Ala completely abolished arrest induced by SOL or AZI. Furthermore, similar to the effect on ERY-mediated arrest, altering the third codon (Leu) to Ala allowed for efficient arrest directed by AZI or SOL (although this mutation caused some read-through of the SOL-bound ribosome) (Figure S2B). We concluded that the nature of the penultimate residue of the short nascent peptide and the incoming amino acid are the most critical for arrest induced by a broad range of macrolide antibiotics.
Antibiotic exacerbates the difficulty of peptide bond formation between intrinsically problematic ribosomal substrates
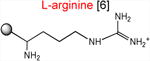
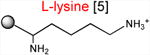
The results presented above strongly suggest that the ribosome with a macrolide antibiotic bound in the exit tunnel stalls when it needs to catalyze the transfer of tripeptides MRX or MKX to an Arg or Lys acceptor. In order to substantiate this conclusion, we followed the kinetics of peptide bond formation between the donor MRL-tRNA and different model acceptor substrates. For these experiments, we synthesized chemically stable substrates mimicking the aminoacyl-tRNA acceptor end (Graber et al., 2010; Moroder et al., 2009) (Table 1). In these aminoacyl-tRNA analogs with the general structure ACCA-X, various amino acid residues (X) were linked to the 3′ carbon atom of the universal tRNA sequence ACCA via a stable amide bond (Supplementary Information). Binding of these substrates to the ribosomal A site does not require the elongation factor EF-Tu or interaction with the mRNA codon in the decoding center and their acceptor ability depends exclusively on the nature of the amino acid residue. In the initial series of experiments, we used analogs that contained canonical amino acid residues that are either conducive to stalling (X = Arg or Lys) or abolish the arrest (X = Ala) (Table 1).
Table 1. Reactivity of acceptor substrate analogs with the MRL-tRNA and MAL-tRNA donors in the ERY-bound and drug-free ribosome.
| Acceptor Substrate (a) | Donor Substrate | |||
|---|---|---|---|---|
| MRL | MAL | |||
| No drug kapp (min-1) |
ERY kapp (min-1) |
No drug kapp (min-1) |
ERY kapp (min-1) |
|
|
0.09 ± 0.15 | 0.01 ± 0.02 | 0.96 ± 0.23 | 0.87 ± 0.21 |
|
0.15 ± 0.03 | 0.01 ± 0.01 | 0.74 ± 0.14 | 1.17 ± 0.23 |
|
0.08 ± 0.02 | < 0.01 b) | 1.03 ± 0.29 | 0.81 ± 0.22 |
|
> 1.8 * c) | 0.61± 0.16 | ND d) | ND |
|
> 1.8 * | 0.32 ± 0.08 | > 1.8 * | > 1.8 * |
|
> 1.8 * | 0.31 ± 0.07 | > 1.8 * | > 1.8 * |
|
> 1.8 * | 0.34 ± 0.10 | > 1.8 * | > 1.8 * |
|
> 1.8 * | > 1.8 * | > 1.8 * | > 1.8 * |
|
> 1.8 * | > 1.8 * | > 1.8 * | > 1.8 * |
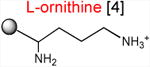
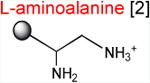
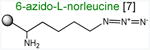
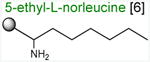
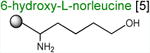
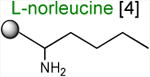
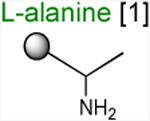
Acceptor substrates conform to the general formulae pACCA-N-X where X corresponds to different canonical or non-canonical amino acids. The pACCA-N moiety of the substrates is represented by a sphere and the chemical structure of the aminoacyl residue is shown. The names of the substrates whose side chain carries a positive charge are shown in red and the substrates with neutral net charge of the side chain are in green. The number of non-hydrogen atoms in the side chain skeleton is indicated in brackets next to the substrate name.
The reaction was too slow to accurately determine its kapp.
Reaction rates with kapp exceeding 1.8 min-1 could not be measured in the experimental setup used for the kinetics experiments. The kapp values for the corresponding reactions are indicated as ‘> 1.8’ and are marked by an asterisk.
not determined
The ribosome/mRNA/MRL-tRNA complex was prepared by translating the MRL peptide from a synthetic tri-codon mRNA in the PURE cell-free translation system (Shimizu et al., 2001). Because the mRNA lacks a stop codon, the ribosome stalls at the third codon carrying the tripeptidyl MRL-tRNALeu bound in the P site and a vacant A site regardless of the presence of ERY (Figure 2A). The stalled ribosome complexes were then mixed with high concentrations (0.7 mM) of the acceptor substrates and the progression of the peptidyl transfer reaction was monitored by quantifying the amount of unreacted MRL-tRNALeu resolved in Bis-Tris polyacrylamide gels (Figures 2B and S3A). Upon addition of the ACCA-N-Ala acceptor substrate, irrespective of the presence of ERY, the reaction was completed within 30 s (the shortest incubation time we could reliably measure with our experimental setup) (apparent rate constant kapp > 1.8 min-1) (Figure 2B-C and Table 1). This indicates that both the drug-free and ERY-bound ribosomes efficiently catalyze transfer of MRL to the non-stalling Ala acceptor. In striking contrast, in the presence of ERY, the majority of MRL-tRNALeu remained unreacted with the ACCA-N-Arg or ACCA-N-Lys acceptors after 30 min of incubation (Figure 2 B-C). Interestingly, while the absence of the antibiotic caused the reaction with these analogs to accelerate nearly 10-fold (kapp = 0.9-1.5 min-1), it remained notably slower in comparison with the ACCA-N-Ala substrate (Figure S3A-B and Table 1). The marked reduction in the reaction rate between MRL-tRNA and ACCA-N-Arg or ACCA-N-Lys acceptors in the absence of antibiotics was not manifested in the conventional toeprinting experiments (‘no antibiotic’ lanes in Figure 1A-C) likely because the putative ribosome pausing was too transient to be captured by this assay. Nevertheless, the results of biochemical testing clearly show that the transfer of the MRL donor to the Arg- or Lys- acceptors is intrinsically difficult for the ribosome and that the presence of the antibiotic in the exit tunnel dramatically intensifies the problem, essentially halting peptide bond formation.
Figure 2. Arg and Lys are poor acceptors of the MRL-peptide in the stalled ribosome.
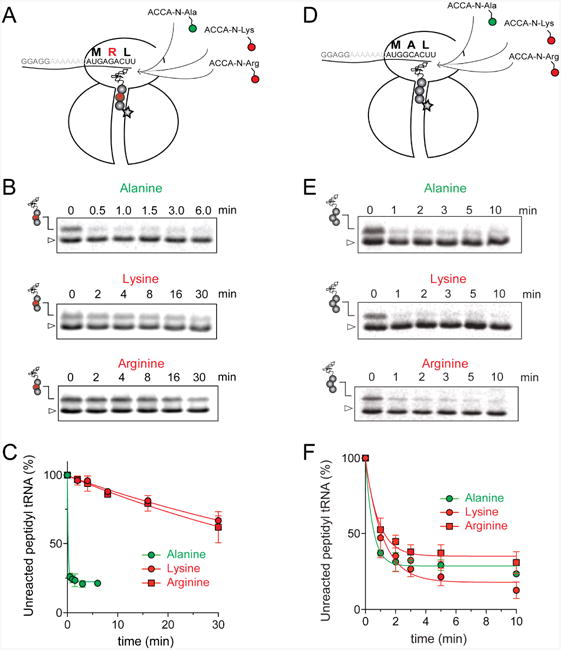
(A) and (D) Cartoon depicting the experimental design: ERY-bound ribosomes carrying MRL-tRNALeu (A) or MAL-tRNALeu (D) in the P-site and an empty A-site are reacted with model A-site substrates ACCA-N-Arg and ACCA-N-Lys (red circles) or ACCA-N-Ala (green circle). Antibiotic bound in the NPET is represented by a star. (B) and (E) Gel electrophoresis analysis of the 35S-labeled MRL-tRNALeu (B) or MAL-tRNALeu (E) (marked by peptidyl-tRNA drawings) remaining unreacted upon addition of ACCA-N-Ala, ACCA-N-Lys or ACCA-N-Arg. The band corresponding to fMet-tRNAfMet present in the reaction is indicated by a triangle. Gels are representative of at least two independent experiments. (C) and (F) Results of the quantification of the amount of unreacted MRL-tRNALeu (C) or MAL-tRNALeu (F) in the gels shown in (B) and (E), respectively. The amount of unreacted peptidyl-tRNA at 0 time point was set as 100%. Error bars show deviation from the mean in two independent experiments for each donor substrate. See also Figure S3.
The positive charge of the A site substrates is the predominant factor that makes them poor acceptors of the MRL peptide
Arg and Lys are unique among the 20 canonical amino acids because they contain the longest side chains, which are positively charged at physiological pH. We considered that the size, the charge, or the combination of these two features could make it difficult for the ribosome to use Arg-tRNA or Lys-tRNA as acceptors of the MRL peptide, hence leading to translation arrest. To dissect the contribution of length and charge of the acceptor's side chain to antibiotic-mediated ribosome stalling, we prepared a new series of synthetic substrate analogs that varied in the length of the side chain and in the presence or absence of the positive charge (Table 1). We then tested the acceptor activity of these substrates in the same experimental setup that we used before with the Ala-, Lys-, or Arg-acceptor analogs. In the presence of ERY, the transfer of the MRL peptide to ACCA-N-6-azido-L-norleucine, ACCA-N-6-hydroxy-L-norleucine, or ACCA-N-6-ethyl-L-norleucine, whose side chain skeletons range in length from 5 to 7 non-hydrogen atoms but lack the net positive charge, occurs with the apparent rate constant kapp of ca. 0.3 min-1 (Figures 3 and Table1). Remarkably, shortening the uncharged side chain to 4 carbon atoms (in ACCA-N-norleucine) significantly accelerated the reaction, making it too fast to accurately determine the rate constant in our experiments (kapp > 1.8 min-1) (Figures 3 and Table1). Likewise, in the absence of the antibiotic, all uncharged acceptor substrates reacted with kinetics too rapid to be accurately measured (Figure S3B and Table 1). These results showed that the length of the acceptor side chain contributes to the arrest mechanism because macrolides notably slow down the transfer of the MRL peptide with acceptors having extended side chains. Nevertheless, the reaction of the MRL peptide with electroneutral acceptors still proceeded at a considerable rate indicating that the extended length of the acceptor side chain is insufficient to support formation of a stable stalled complex.
Figure 3. Positive charge and extended size of the acceptor amino acid side chain prevent efficient peptide bond formation with the MRL peptide in ERY-bound ribosomes.
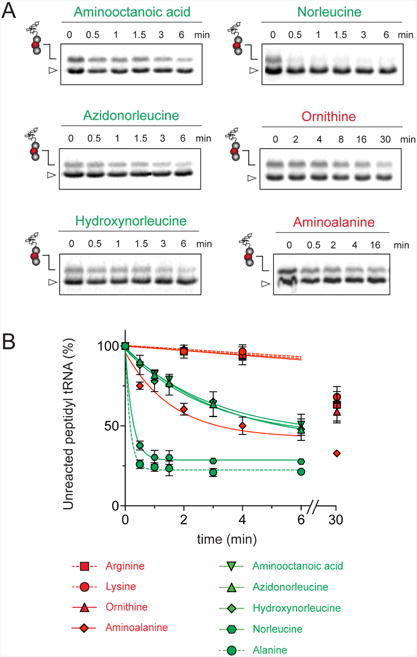
Kinetics of reaction of the MRL-tRNALeu in the P site of ERY-bound ribosomes (as illustrated in the cartoon in Figure 2A) with acceptor substrate analogs carrying non-canonical amino acids. Quantification of the amount of unreacted MRL-tRNALeu is from the gels shown in Figure S5. The amount of unreacted MRL-tRNALeu at 0 time point was set as 100%. Error bars show deviation from the mean calculated from two independent experiments. The 30 min time point for the ACCA-N-aminoalanine substrate was assayed in a single experiment. The reactivity plots of the substrate analogs with canonical amino acids (Arg, Lys and Ala) are those from Figure 2C and are shown here by dashed lines for comparison.
Introduction of the positive charge to the side chain on the acceptor substrate exhibited a more dramatic effect on the rate of reaction with the MRL donor. Replacement of the terminal methyl group of norleucine with a positively charged amino group in ACCA-N-ornithine resulted in a precipitous drop of the MRL transfer rate, essentially abolishing the reaction in the presence of ERY (kapp < 0.01 min-1) (Figures 3 and Table 1). In addition, consistent with what was observed for Arg and Lys acceptors, even in the absence of ERY, the transfer of the MRL peptide to ACCA-N-ornithine was rather inefficient (kapp 0.08 ± 0.02 min-1) (Figure S3B and Table 1). These results clearly indicate that the major obstacle for catalysis of the transfer of the MRL peptide to the acceptor in the PTC of the macrolide-bound ribosome is imposed by the charge of the A site amino acid. However, in agreement with the notion of the importance of the size of the side chain, ACCA-N-aminoalanine, with a postively-charged side chain as short as 2 non-hydrogen atoms, reacted faster than any of the other tested positively charged acceptors (kapp 0.XX ± 0.XX min-1) (Figure 3 and Table 1), yet much slower than any of the uncharged substrates with a side chain shorter than 5 atoms.
The sluggish transfer of the MRL peptide to positively charged substrates ACCA-N-Lys, ACCA-N-Arg, and ACCA-N-ornithine critically depends on the presence of Arg (or Lys) in the penultimate position of the nascent chain. The non-stalling donor peptide MAL swiftly reacted with the acceptor substrates containing Ala, Lys or Arg residues, irrespective of the presence of antibiotic (kapp ≥ 0.74 min-1) (Figures 2D-F, S3C-D, and Table 1). Consequently, removal of the long positively-charged side chain of the penultimate residue of the donor substrate allows for efficient peptidyl transfer, even to positively-charged A site substrates. Thus, it appears that the simultaneous presence of the charged long side chains in the penultimate position of the donor substrate and in the aminoacyl acceptor present the obstacle for catalysis of peptide bond formation by the macrolide-bound ribosome.
One possible model that follows from our findings is that a simple electrostatic repulsion between the penultimate residue of the peptidyl donor and the aminoacyl moiety of the acceptor may be sufficient to hamper the catalysis of the peptidyl transfer reaction in the macrolide-bound ribosome. If this were the case, then simultaneous replacement of the positively charged residues encoded in codons 2 and 4 of the MRLR ORF with negatively charged amino acids would be comparably detrimental for translation. We tested this hypothesis in a straightforward toeprinting experiment exploiting two new templates, in which the second and the fourth codons of the MRLR ORF were mutated to specify peptides MDLD and MELE, which carry negatively charged amino acids in the critical positions (Figure 4). However, neither of these peptides were able to direct efficient arrest of the macrolide-bound ribosome. Thus, it is not the simple repulsion of two equivalent charges, but the explicit presence of positive charges at the extended side chains of the penultimate residue of the P site peptidyl donor and of the A site aminoacyl acceptor that obstructs peptide bond formation in the ribosome with a macrolide antibiotic bound in the NPET.
Figure 4. Simultaneous replacement of the first and last amino acids of the R/K-x-R/K motif to the negatively charged amino acids prevents ERY-dependent translation arrest.
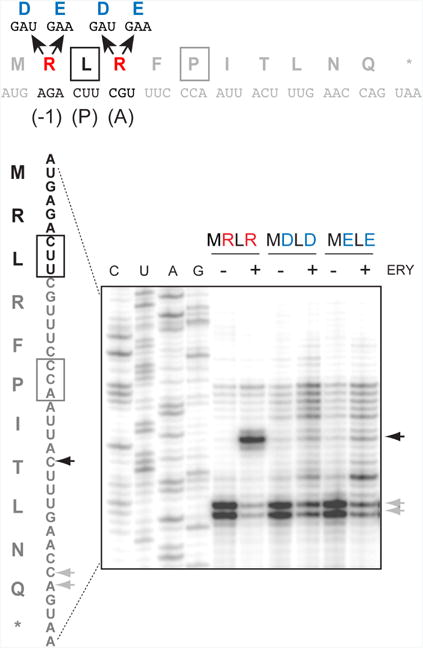
Toeprinting analysis of ERY-dependent ribosome stalling at the templates encoding MDLD and MELE peptides. The full sequences of the wt and mutant ermDL ORF and the encoded peptides are shown above the gel. The penultimate (-1) amino acid residue of the nascent chain in the drug-stalled ribosome, the P site, and the A site residues are marked above the peptide sequence. Black arrows indicate ERY dependent translation arrest at the Leu codon. Gray arrows indicate the toeprint bands representing ribosomes captured at the downstream Pro5 codon due to the presence of mupirocin in the reaction mixture, which inhibits the Ile-tRNA formation. Sequencing lanes are marked. The shown gel is representative of two independent experiments.
Discussion
In our experiments, we interrogated one of the most common problematic motifs for the macrolide-bound ribosome, R/K-X-R/K, and found that specific physicochemical properties of substrates participating in peptide bond formation are the major contributing factors to drug-induced translation arrest.
During translation elongation, macrolides act as inhibitors of peptide bond formation between specific combinations of PTC donor and acceptor substrates (Kannan et al., 2014). This mode of action is possibly mediated by allosteric changes in the PTC induced by the drug (Sothiselvam et al., 2014). In addition, direct interactions between the nascent chain and the antibiotic in the NPET may also modulate antibiotic action (Arenz et al., 2014a; Arenz et al., 2014b; Vazquez-Laslop, 2010) and may at times allow the drug-bound ribosome to bypass a problematic motif (Sothiselvam et al., in preparation). Such influence of the context likely explains why in the macrolide-treated cells no significant increase in ribosome density is observed at some of the R/K-X-R/K sequences (Davis et al., 2014; Kannan et al., 2014). Our use of the shortest known peptide capable of directing macrolide-dependent ribosome stalling, MRL (Sothiselvam et al., 2014), made it possible to significantly simplify the system by minimizing the influence of the N-terminal context on the drug-induced arrest. Furthermore, the small size of the stalling peptide made it possible to probe the contribution of each of its residues (with the exception of the N-terminal formyl-methionine) to translation arrest.
Strikingly, in the context of the short donor substrate, the physicochemical characteristics of only one of its residues, the penultimate amino acid, are critical. The properties of the C-terminal amino acid, which directly participates in peptide bond formation, are of lesser importance. Our comprehensive mutational analysis showed that the simultaneous presence of Lys or Arg residues in the penultimate position of the peptide and in the acceptor substrate is sufficient for preventing fast peptide bond formation in the macrolide-bound ribosome. Subsequent biochemical studies, which utilized different peptidyl donors (MRL and MAL) and a series of synthetic acceptors, showed that the positive charge of the critical amino acid residues and the size of their side chains are the key factors that render peptide bond formation inefficient in the drug-bound ribosome. The positively charged, long-side-chain acceptors of the MRL peptide (ACCA-N-Arg, -Lys, or -ornithine) reacted extremely slowly, whereas shortening of the positively charged side chain (ACCA-N-aminoalanine) or removal of the charge increased the reaction rate, with the most dramatic acceleration achieved with short uncharged acceptor substrates (Figure 3). Importantly, substituting the electroneutral Ala for Arg in the penultimate position of the donor (MAL) had a similar effect.
Although the contribution of the size of the side chain in natural substrate analogs could be generally concealed due to the dominance of the charge effect, it might nevertheless play a role in the effects of the drugs on cellular translation. Indeed, although in our in vitro experiments both ACCA-N-Lys and ACCA-N-Arg reacted too slowly to accurately distinguish the difference in their reactivity, ribosome profiling analysis showed a more pronounced enrichment of Arg containing sequences of the R/K-X-R/K motif in the sites of macrolide-induced translation stalling (Davis et al., 2014; Kannan et al., 2014). Because the guanidinium group of Arg and ε-amino group of Lys are both completely protonated at physiological pH, it is generally possible that the longer side chain of Arg (6 atoms) compared to Lys (5 atoms) accounts for a more severe translation arrest at the Arg-containing motifs.
Our findings provide an explanation for a long-known fact that translation of poly(A) RNA, encoding poly-lysine, is efficiently inhibited by erythromycin and results in accumulation of short (di- to penta-lysine) peptides (Mao and Robishaw, 1971; Otaka and Kaji, 1975; Vazquez, 1966). Because the poly(Lys) sequence conforms to the R/K-X-R/K motif, it is not surprising that its synthesis would be interrupted by macrolides at the very early rounds, because the drugs should block the transfer of even very short nascent chains carrying Lys in the penultimate position to the incoming Lys-tRNA.
Our results show that the interplay of the penultimate residue of the nascent peptide and the incoming amino acid has a dramatic effect on macrolide-induced arrest at the R/K-X-R/K sites. The nature of the C-terminal residue of the donor peptide that resides in the PTC P site appears to be far less significant, at least in the case of ERY-induced arrest with the minimal peptide (Figure 1B). Nevertheless, from the somewhat varying relative intensity of the arrest bands in the toeprint gels (Figure 1B), it could be inferred that the C-terminal residue does modulate, to some extent, the efficiency of peptide bond formation in the macrolide-bound ribosome. The influence of the C-terminal peptide residue could account for the varying extent of enrichment of different R/K-X-R/K sequences at the sites of ERY-induced arrest observed in ribosome profiling experiments (Kannan et al., 2014). Furthermore, the nature of the tunnel-bound antibiotic affects the importance of the C-terminal residue of the nascent chain for translation arrest. While Leu3-to-Ala replacement had only a negligible effect on the transfer of the MRL peptide to Arg-tRNA in the ERY- or AZI-bound ribosome, the same mutation notably diminished translation arrest when the fluoroketolide SOL was bound (Figure S2). This result is reminiscent of our recent finding that the nature of the C-terminal residue of another stalling regulatory peptide, ErmBL, which conforms to a different stalling motif, XDK (Kannan et al., 2014), determines whether macrolides or ketolides are recognized as the stalling cofactor (Gupta et al., 2016).
Several scenarios could account for the slow transfer of a peptidyl donor with a positively charged penultimate residue to a positively charged acceptor in the ERY-bound ribosome. One possibility is that electrostatic repulsion between the incoming amino acid and the penultimate nascent peptide residue, possibly exacerbated by direct steric hindrance and a close proximity of the charges located at the ends of the extended side chains, prevents proper accommodation of the aminoacyl-tRNA acceptor in the PTC A site (Arenz et al., 2014a; Arenz et al., 2014b; Gupta et al., 2016; Ramu et al., 2011). This could lead to either an aberrant placement of the stably bound aminoacyl-tRNA or to rapid dissociation of Arg- or Lys-tRNA from the A site. It is also possible that binding of the positively charged substrate to the A site displaces the peptidyl-tRNA in the P site, if its penultimate position is occupied by a positively charged residue. Finally, both the donor and acceptor substrates could be mutually misaligned in the PTC active site, which would be detrimental for efficient peptide bond formation.
While the charge of the amino acids in the donor and acceptor substrates plays the key role in macrolide-induced arrest, only positively charged residues were detrimental for peptide bond formation. Simultaneous replacement of both of the critical residues with negatively charged amino acids was not conducive to translation arrest (Figure 4). It is thus obvious that the polarity of the charge of the donor and acceptor is central to the mechanism of stalling. In agreement with this conclusion, while R/K-X-R/K is one of the most predominant macrolide stalling motifs in Gram-positive and Gram-negative bacteria (Davis et al., 2014; Kannan et al., 2014), no particular enrichment of the D/E-X-D/E sequences at the sites of drug-induced translation arrest has been noted. The strict polarity requirement suggests that electrostatic interactions of the substrates with additional charged group(s) in the ribosome or its ligands could be involved in the mechanism of stalling. It is conceivable that the positive charge at the end of the extended side chain of the (-1) residue of the peptidyl donor or aminoacyl acceptor could interact with the electronegative phosphate group of one of the neighboring 23S rRNA nucleotides. The generally electronegative potential of the NPET could be also a contributing factor (Lu et al., 2007). Alternatively, the protonated 3′ dimethyl-amino group of the macrolide molecule could influence the placement of the positively charged substrates in spite of the ca. 8Å distance that separates it from the PTC active site.
The rate of the catalytic step of peptide bond formation in the drug-free ribosome depends on the nature of the reacting substrates (Bourd et al., 1982; Johansson et al., 2011; Monro et al., 1968; Muto and Ito, 2008; Wohlgemuth et al., 2008). However, this difference in reactivity is normally masked by the slow rate of aminoacyl-tRNA binding and accommodation, which is the rate-limiting step of translation elongation (Wintermeyer et al., 2004). It is conceivable, however, that if the general catalytic capacity of the PTC is decreased, then formation of peptide bonds between certain generally ‘sluggish’ substrates could slow down sufficiently to become rate limiting. From this standpoint, it is remarkable that even in the absence of antibiotic, the reaction of the MRL peptide with the acceptor analogs carrying positively charged amino acids with extended side chains was notably slower that the reaction with electroneutral acceptors (Table 1). This observation hints that peptide bond formation between substrates conforming to the R/K-X-R/K motif is intrinsically slow. Therefore, macrolide antibiotics do not convert well-behaved substrates into slow reactants. Rather, the drugs amplify the problem of the substrates which are inherently ‘difficult’, making peptide bond formation rate limiting. We are fully aware, however, that the use of the artificial acceptor substrates may significantly aggravate the difference in their reactivity (Brunelle et al., 2006; Katunin et al., 2002). Furthermore, the kinetic parameters obtained in the cell-free setting with the use of model substrates reflect only the general trend in relative substrate reactivity rather than the actual kinetics of transpeptidation, which should be much faster when the full length tRNA delivers the acceptor amino acid to the PTC A site (Wohlgemuth et al., 2008). Indeed, the lack of pronounced ribosome stalling at the R/K-X-R/K motif in the absence of antibiotic (Davis et al., 2014; Kannan et al., 2014; Li et al., 2012; Mohammad et al., 2016) suggests that even for these ‘slow’ substrates the rate of transpeptidation in the living cell is not rate limiting. Therefore, it is possible that inhibition of translation by macrolide antibiotics reveals a general but concealed phenomenon of the dependence of the rate of peptidyl transfer on the interplay of the penultimate residue of the nascent chain and the incoming amino acid.
Other sequence motifs conducive to macrolide action, which do not conform to the R/K-X-R/K consensus, are also confined to amino acid residues residing at or near the PTC. The features of those substrates that make them difficult for the drug-bound ribosome are currently unknown. Interestingly, however, one of the identified ‘macrolide motifs’ (XPX) (Davis et al., 2014; Kannan et al., 2014) includes proline, a particularly slow participant in peptide bond formation (Johansson et al., 2011) especially in specific contexts (Doerfel et al., 2013; Ude et al., 2013). Therefore, our conclusion that macrolides exacerbate the problematic nature of the inherently difficult sequences could expand beyond the specific example of the R/K-X-R/K motif explored in this paper. Further exploration of the context specificity of macrolide action might reveal new hidden features of the general mechanisms of protein synthesis.
Materials and Methods
Reagents
Erythromycin, azithromycin and mupirocin were from Sigma-Aldrich, solithromycin was provided by Dr. Fernandes (CEMPRA Pharmaceuticals), indolmycin was from Santa Cruz Biotechnology. Reverse transcriptase used in toeprinting analysis was purchased from Sigma-Aldrich. All the DNA primers (listed in a table in Supplementary Experimental Procedures) were synthesized by IDT.
Synthesis of ACCA-N-X substrates is described in Supplementary Experimental Procedures.
Toeprinting assay
Linear DNA templates (0.5–1 pmol) encoding the ORF of interest preceded by the T7 promoter sequence were generated by PCR using primers indicated in Table S1. In order to capture the ribosomes that escaped ERY-dependent arrest at the RLR sequence, an Ile codon (Ile6) was introduced after the Pro5 codon and all the reactions were carried out in the presence of 50 mM mupirocin, an IleRS inhibitor. In the templates the codons of the motif were mutated to specify Ile, the Ile6 codon was replaced with a Trp codon and, in these cases, reactions were carried out in the presence of indolmycin, an inhibitor of TrpRS.
Templates were used to direct coupled transcription–translation in the PURExpress cell-free system (New England Biolabs). The reactions were performed in a total volume of 5 µL and, where indicated, were supplemented with antibiotics (50 mM final concentration). Following 10 min incubation at 37 °C, the primer extension reaction was initiated by addition of the primer NV1 (Table S1) and 3 U of reverse transcriptase. The resulting cDNA products along with sequencing reactions were separated in a 6% sequencing gel and visualized with a Typhoon imager (GE).
In vitro peptidyl transfer reaction
Synthetic mRNAs (synthesized by Thermo Fisher), encoding MRL peptide (ATAAGGAGGAAAAAATATGAGACUU, coding sequence is underlined) or MAL peptide (ATAAGGAGGAAAAAATATGGCACUU), were used for in vitro translation in the PURE system. The 5 µL reactions containing 75 pmol of the mRNA template were supplemented with 1µCi [35S]-methionine (specific activity 1175 Ci/mmol) and, where indicated, 50 µM ERY. Following 7 min incubation at 37 °C, thiostrepton was added to the final concentration of 50 mM in order to prevent further rounds of translation. The synthetic acceptor substrates, ACCA-N-X, dissolved in water, were then added to a final concentration of 0.7 mM. Aliquots (5 ml) were withdrawn at different times and immediately mixed with an equal volume of SDS-containing tricine sample buffer (Bio-Rad Laboratories). Reaction products were resolved in 16% Bis-Tris polyacrylamide gel as previously described (Vazquez-Laslop et al., 2008). Gels were fixed, dried, exposed to a phosphorimaging screen and visualized with a Typhoon imager (GE).
Quantification of the peptidyl-tRNA bands was performed using ImageJ (Schneider et al., 2012). Because the reactions contain excess of [35S]-methionine and fMet-tRNAMet is continuously regenerated in the reaction, the fMet-tRNAMet band served as the loading control and normalization factor. The normalized peptidyl-tRNA band at t=0, that represents the total amount of unreacted peptidyl-tRNA before addition of the A-site substrates, was set at 100%. Progress of the reaction was monitored by quantifying the amount of unreacted peptidyl-tRNA at experimental time points. The data were fitted to one phase exponential decay model in Graphpad PRISM using the equation Y=(Y0 - PL)*exp(-k*X) + PL, where Y is the amount of unreacted peptidyl-tRNA (%) at any given time point, Y0 is the initial amount of unreacted peptidyl-tRNA at t=0, PL (plateau) is the Y value at infinite times (calculated in PRISM), k is the rate constant, and X is time in minutes. The error range of the apparent rate constants shown in Table 1 represents the standard error of mean.
Supplementary Material
Highlights.
Macrolide antibiotics induce ribosome stalling at Arg/Lys-X-Arg/Lys motifs
Size and charge of key residue side chains hinder peptide bond formation
Properties of the nascent protein impact the catalytic capacity of the ribosome
Acknowledgments
We thank Emanuel Ehmki, for contribution to chemical synthesis, Axel Innis and Daniel Wilson for stimulating discussions, Prabha Fernandes for providing solithromycin and Lisa Smith for editing the manuscript. This work was supported by grant GM104370 from the National Institute of General Medicine, NIH (to A.S.M. and N.V.-L.) and the grants P27947 and I1040 from Austrian Science Fund FWF (to R.M.).
Footnotes
Author Contributions: S.S., N.V.-L., and A.S.M. conceived the project, designed the experiments, analyzed the data and wrote the manuscript. S.S. performed the experiments. D.K. performed some experiments. R.M. conceived and designed the synthesis of the acceptor substrates. S.N. and L.R. carried out the synthesis and chemical analysis of the acceptor substrates.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almutairi MM, Sung Ryeol Park SR, Rose S, Hansenb DA, Vázquez-Laslop N, Douthwaite S, Sherman DH, Mankin AS. Resistance to ketolide antibiotics by coordinated expression of rRNA methyltransferases in a bacterial producer of natural ketolides. Proc Natl Acad Sci USA. 2015;112:12956–12961. doi: 10.1073/pnas.1512090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenz S, Meydan S, Starosta A, Berninghausen O, Beckmann R, Vázquez-Laslop N, Wilson DN. Drug sensing by the ribosome induces translational arrest via active site perturbation. Mol Cell. 2014a;56:446–452. doi: 10.1016/j.molcel.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenz S, Ramu H, Gupta P, Berninghausen O, Beckmann R, Vazquez-Laslop N, Mankin AS, Wilson DN. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nature Commun. 2014b;5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourd S, Victorova L, Kukhanova M. On substrate specificity of the donor site of the Escherichia coli ribosomal peptidyl transferase center. Synthesis of dipeptides from 3′-terminal fragments of aminoacyl-tRNA. FEBS Lett. 1982;142:96–100. doi: 10.1016/0014-5793(82)80227-5. [DOI] [PubMed] [Google Scholar]
- Brunelle JL, Youngman EM, Sharma D, Green R. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA. 2006;12:33–39. doi: 10.1261/rna.2256706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulkley D, Innis CA, Blaha G, Steitz TA. Revisiting the structures of several antibiotics bound to the bacterial ribosome. Proc Natl Acad Sci USA. 2010;107:17158–17163. doi: 10.1073/pnas.1008685107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AR, Gohara DW, Yap MN. Sequence selectivity of macrolide-induced translational attenuation. Proc Natl Acad Sci USA. 2014;111:15379–15384. doi: 10.1073/pnas.1410356111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci USA. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgamal S, Katz A, Hersch SJ, Newsom D, White P, Navarre WW, Ibba M. EF-P dependent pauses integrate proximal and distal signals during translation. PLoS Genet. 2014;10:e1004553. doi: 10.1371/journal.pgen.1004553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber D, Moroder H, Steger J, Trappl K, Polacek N, Micura R. Reliable semi-synthesis of hydrolysis-resistant 3′-peptidyl-tRNA conjugates containing genuine tRNA modifications. Nucl Acids Res. 2010;38:6796–6802. doi: 10.1093/nar/gkq508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan TJ, Grandi G, Hahn J, Grandi R, Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucl Acids Res. 1980;8:6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Liu B, Klepacki D, Gupta V, Schulten K, Mankin AS, Vázquez-Laslop N. Nascent peptide assists the ribosome in recognizing chemically distinct small molecules. Nat Chem Biol. 2016;12:153–158. doi: 10.1038/nchembio.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardesty B, Picking WD, Odom OW. The extension of polyphenylalanine and polylysine peptides on Escherichia coli ribosomes. Biochim Biophys Acta. 1990;1050:197–202. doi: 10.1016/0167-4781(90)90166-y. [DOI] [PubMed] [Google Scholar]
- Horinouchi S, Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci USA. 1980;77:7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Ieong KW, Trobro S, Strazewski P, Aqvist J, Pavlov MY, Ehrenberg M. pH-sensitivity of the ribosomal peptidyl transfer reaction dependent on the identity of the A-site aminoacyl-tRNA. Proc Natl Acad Sci USA. 2011;108:79–84. doi: 10.1073/pnas.1012612107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Kanabar P, Schryer D, Florin T, Oh E, Bahroos N, Tenson T, Weissman JS, Mankin AS. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci USA. 2014;111:15958–15963. doi: 10.1073/pnas.1417334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan K, Vázquez-Laslop N, Mankin AS. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell. 2012;151:508–520. doi: 10.1016/j.cell.2012.09.018. [DOI] [PubMed] [Google Scholar]
- Katunin VI, Muth GW, Strobel SA, Wintermeyer W, Rodnina MV. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol Cell. 2002;10:339–346. doi: 10.1016/s1097-2765(02)00566-x. [DOI] [PubMed] [Google Scholar]
- Kwak JH, Choi EC, Weisblum B. Transcriptional attenuation control of ermK, a macrolide-lincosamide-streptogramin B resistance determinant from Bacillus licheniformis. J Bacteriol. 1991;173:4725–4735. doi: 10.1128/jb.173.15.4725-4735.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon AR, Min YH, Yoon EJ, Kim JA, Shim MJ, Choi EC. ErmK leader peptide : amino acid sequence critical for induction by erythromycin. Arch Pharm Res. 2006;29:1154–1157. doi: 10.1007/BF02969307. [DOI] [PubMed] [Google Scholar]
- Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Kobertz WR, Deutsch C. Mapping the Electrostatic Potential within the Ribosomal Exit Tunnel. J Mol Biol. 2007;371:1378–1391. doi: 10.1016/j.jmb.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Mao JCH, Robishaw EE. Effects of macrolides on peptide-bond formation and translocation. Biochemistry. 1971;10:2054–2061. doi: 10.1021/bi00787a014. [DOI] [PubMed] [Google Scholar]
- Mohammad F, Woolstenhulme CJ, Green R, Buskirk AR. Clarifying the Translational Pausing Landscape in Bacteria by Ribosome Profiling. Cell Rep. 2016;14:686–694. doi: 10.1016/j.celrep.2015.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monro RE, Cerna J, Marcker KA. Ribosome-catalyzed peptidyl transfer: substrate specificity at the P-site. Proc Natl Acad Sci USA. 1968;61:1042–1049. doi: 10.1073/pnas.61.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroder H, Steger J, Graber D, Fauster K, Trappl K, Marquez V, Polacek N, Wilson DN, Micura R. Non-hydrolyzable RNA-peptide conjugates: a powerful advance in the synthesis of mimics for 3′-peptidyl tRNA termini. Angew Chem Int Ed Engl. 2009;48:4056–4060. doi: 10.1002/anie.200900939. [DOI] [PubMed] [Google Scholar]
- Muto H, Ito K. Peptidyl-prolyl-tRNA at the ribosomal P-site reacts poorly with puromycin. Biochem Biophys Res Commun. 2008;366:1043–1047. doi: 10.1016/j.bbrc.2007.12.072. [DOI] [PubMed] [Google Scholar]
- Otaka T, Kaji A. Release of (oligo) peptidyl-tRNA from ribosomes by erythromycin A. Proc Natl Acad Sci USA. 1975;72:2649–2652. doi: 10.1073/pnas.72.7.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol Microbiol. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- Ramu H, Vázquez-Laslop N, Klepacki D, Dai Q, Piccirilli J, Micura R, Mankin AS. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell. 2011;41:321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- Sothiselvam S, Liu B, Han W, Ramu H, Klepacki D, Atkinson GC, Brauer A, Remm M, Tenson T, Schulten K, et al. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc Natl Acad Sci USA. 2014;111:9804–9809. doi: 10.1073/pnas.1403586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starosta AL, Karpenko VV, Shishkina AV, Mikolajka A, Sumbatyan NV, Schluenzen F, Korshunova GA, Bogdanov AA, Wilson DN. Interplay between the ribosomal tunnel, nascent chain, and macrolides influences drug inhibition. Chem Biol. 2010;17:504–514. doi: 10.1016/j.chembiol.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Starosta AL, Lassak J, Peil L, Atkinson GC, Virumae K, Tenson T, Remme J, Jung K, Wilson DN. Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Res. 2014;42:10711–10719. doi: 10.1093/nar/gku768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian SL, Ramu H, Mankin AS. Inducible resistance to macrolide antibiotics. In: Dougherty TJ, Pucci MJ, editors. Antibiotic Drug Discovery and Development. New Yoork, NY: Springer Publishing Company; 2011. [Google Scholar]
- Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol. 2003;330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- Vazquez D. Antibiotics affecting chloramphenicol uptake by bacteria. Their effect on amino acid incorporation in a cell-free system. Biochim Biophys Acta. 1966;114:289–295. doi: 10.1016/0005-2787(66)90310-8. [DOI] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Klepacki D, Mulhearn DC, Ramu H, Krasnykh O, Franzblau S, Mankin AS. Role of antibiotic ligand in nascent peptide-dependent ribosome stalling. Proc Natl Acad Sci USA. 2011;108:10496–10501. doi: 10.1073/pnas.1103474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. The key role of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29:3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Weisblum B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother. 1995;39:797–805. doi: 10.1128/aac.39.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Peske F, Beringer M, Gromadski KB, Savelsbergh A, Rodnina MV. Mechanisms of elongation on the ribosome: dynamics of a macromolecular machine. Biochem Soc Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem. 2008;283:32229–32235. doi: 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA. 2013;110:E878–887. doi: 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


