Abstract
Bloodstream infections with Staphylococcus aureus are clinically significant and are often treated with empirical methicillin resistance (MRSA, methicillin-resistant S. aureus) coverage. However, vancomycin has associated harms. We hypothesized that MRSA screening correlated with resistance in S. aureus bacteremia and could help determine the requirement for empirical vancomycin therapy. We reviewed consecutive S. aureus bacteremias over a 5-year period at two tertiary care hospitals. MRSA colonization was evaluated in three ways: as tested within 30 days of bacteremia (30-day criterion), as tested within 30 days but accounting for any prior positive results (ever-positive criterion), or as tested in known-positive patients, with patients with unknown MRSA status being labeled negative (known-positive criterion). There were 409 S. aureus bacteremias: 302 (73.8%) methicillin-susceptible S. aureus (MSSA) and 107 (26.2%) MRSA bacteremias. In the 167 patients with MSSA bacteremias, 7.2% had a positive MRSA test within 30 days. Of 107 patients with MRSA bacteremia, 68 were tested within 30 days (54 positive; 79.8%), and another 21 (19.6%) were previously positive. The 30-day criterion provided negative predictive values (NPV) exceeding 90% and 95% if the prevalence of MRSA in S. aureus bacteremia was less than 33.4% and 19.2%, respectively. The same NPVs were predicted at MRSA proportions below 39.7% and 23.8%, respectively, for the ever-positive criterion and 34.4% and 19.9%, respectively, for the known-positive criterion. In MRSA-colonized patients, positive predictive values exceeded 50% at low prevalence. MRSA screening could help avoid empirical vancomycin therapy and its complications in stable patients and settings with low-to-moderate proportions of MRSA bacteremia.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is a clinically significant pathogen that often requires therapy with antimicrobial agents, such as vancomycin, which have the potential for harm. Depending on the geographic region, MRSA colonization rates vary, with the overall colonization prevalence estimated at 4 to 7% in the United States (1) and 4.2% in hospitalized patients in Canada (2). Due to the significant clinical impact of MRSA (3), empirical vancomycin use is often initiated if a bloodstream infection with a Gram-positive pathogen is suspected. Vancomycin use is associated with increased drug and monitoring costs, as well as drug-induced acute kidney injury in up to 15% of patients, especially when the drug is used to achieve the higher trough levels recommended for serious MRSA infections (4). Furthermore, the empirical use of vancomycin may lead to the inadvertent omission of beta-lactam therapy, which is superior for the treatment of methicillin-sensitive S. aureus (MSSA) bacteremia (5). More judicious use of empirical MRSA coverage could therefore reduce costs, adverse events, and the delay to administration of beta-lactam therapy while potentially also decreasing selection pressure for organisms such as vancomycin-resistant enterococci (6).
Since MRSA colonization is a risk factor for the development of subsequent MRSA infection, we hypothesized that a patient's known MRSA status could identify patients who might and might not benefit from empirical MRSA coverage in the context of S. aureus bacteremia. The ability of MRSA screening swabs to predict methicillin-resistance in S. aureus bacteremia would fundamentally depend on the proportion of S. aureus bacteremia that is methicillin resistant (here referred to as the MRSA proportion).
MATERIALS AND METHODS
We conducted a retrospective review of all consecutive S. aureus bloodstream infections from 1 April 2010 to 1 April 2015 at the McGill University Health Centre (832 beds; 2 hospitals; catchment area population, 850,000). Only the first positive culture per patient was included. S. aureus susceptibilities were determined using a Vitek-2 automated system (bioMérieux, France) and interpreted in accordance with guidelines from the Clinical and Laboratory Standards Institute. Methicillin resistance was confirmed using a 30-μg cefoxitin disk.
We employ universal MRSA screening on admission to medical wards and critical care units, as well as targeted screening in other units. Patients usually have MRSA screening swabs collected from the nares, but clinical samples are also accepted (e.g., perianal region, open wounds, and catheter sites). A flowchart of this process is included in File S1 in the supplemental material. Briefly, swabs are inoculated on MRSA chromogenic plates (Bio-Rad) and in a Staphylococcus broth that contains 2.5% NaCl and 8 mg/liter of aztreonam (Oxoid). Typical colonies on the chromogenic plates are confirmed to be S. aureus via wet mount and latex agglutination testing (Bio-Rad). Atypical colonies undergo confirmatory testing via real-time PCR in a LightCycler 480 instrument (Roche) in order to ascertain that methicillin resistance is conferred by the mecA gene. Broth from specimens with negative plates at 24 h subsequently undergoes an internally validated PCR test to detect the presence of S. aureus genes (see File S2) (7). A negative PCR result substantiates the absence of Staphylococcus aureus, whereas broth with a positive PCR is subcultured on a blood agar plate (Oxoid) and chromogenic agar to confirm the presence or absence of MRSA as described above.
For the purpose of this analysis, MRSA screening swab status was categorized in three ways (Table 1). First, a patient was considered negative for MRSA screening if we obtained a documented negative MRSA swab result within 30 days prior to the positive blood culture being collected (30-day criterion). Second, we relabeled any patient with any prior positive MRSA specimen as positive (ever-positive criterion). Finally, we categorized all patients who were not previously known to be colonized or infected as MRSA negative, including those never screened (known-positive criterion). The last method simulates a “worst-case” sensitivity analysis whereby all patients with unknown MRSA carriage status are assumed to be negative. Missing data were not inferred for the first two tests. MRSA carrier status was determined from the hospital electronic medical record.
TABLE 1.
Patient criterion assignment
| Criterion | Patient criterion assignment |
||
|---|---|---|---|
| Positive | Negative | Excluded from analysis | |
| 30-Day criterion | Positive MRSA swab within 30 days of blood culture | Negative MRSA swab within 30 days of blood culture | No MRSA swab within 30 days of blood culture |
| Ever-positive criterion | Positive MRSA swab at any time before the blood culture | Negative MRSA swab within 30 days of blood culture | No MRSA swab result available at any time |
| Known-positive criterion | Positive MRSA swab at any time before the blood culture | Negative MRSA swab within 30 days of blood culture or no MRSA swab result available | Every patient is included |
Sensitivity, specificity, likelihood ratios, and negative/positive predictive values were calculated using standard formulas. Confidence intervals for the sensitivities and specificities were computed using Clopper-Pearson confidence intervals. Likelihood ratio and negative/positive predictive value confidence intervals were obtained by simulating the appropriate binomial random variables 1,000 times using the parametric bootstrap method (8) (see File S3 in the supplemental material). Analyses were performed in R (version 3.2.0) and ggplot2 (version 1.0.1).
RESULTS
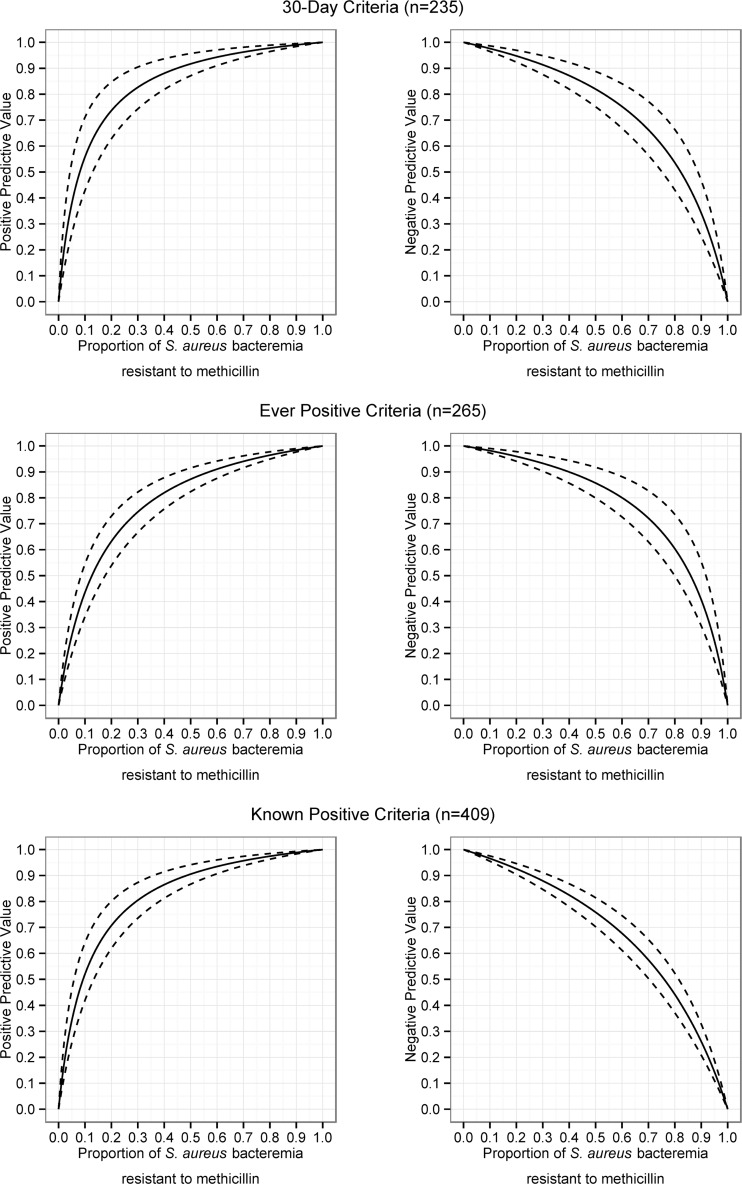
In total, there were 409 patients with Staphylococcus aureus bacteremias. Of those, 302 (73.8%) patients had a methicillin-susceptible S. aureus bacteremia, while the rest had an MRSA bloodstream infection. Of the 302 MSSA infections, 155 (51.3%) had a negative MRSA screening swab within 30 days, 12 (3.97%) had a positive MRSA screening swab, and an additional 10 (3.31%) were previously known to be MRSA positive (Table 2). In the 167 tested MSSA patients, the overall positivity of MRSA screening swabs within 30 days was 7.2%. There were 107 episodes of MRSA bacteremia (MRSA proportion, 26.2%), of whom 68 (63.6%) were screened within 30 days (54 positive, or 79.4%, and 14 negative, or 20.6%), and an additional 21 (19.6%) were previously positive. A total of 144 of the 409 (35.2%) S. aureus bacteremia patients were never screened prior to their bacteremia; these patients are included in the known-positive criterion. Table 3 gives the sensitivity, specificity, likelihood ratios, and positive/negative predictive values for the three methods. Plots of positive and negative predictive values as a function of MRSA proportion are included in Fig. 1. Negative predictive values exceeding 90% were obtained at MRSA proportions of 33.4%, 39.7%, and 34.4% for the 30-day, ever-positive, and known-positive criteria, respectively. Similarly, at MRSA proportions of 19.2%, 23.8%, and 19.9%, respectively, the negative predictive values exceeded 95%. The positive predictive value exceeded 50% even at low MRSA proportions.
TABLE 2.
Cross-comparison of test results
| Criterion and test result | No. of MRSA-positive results | No. of MRSA-negative results |
|---|---|---|
| 30-Day criterion | ||
| MRSA in blood culture | 54 | 14 |
| MSSA in blood culture | 12 | 155 |
| Ever-positive criterion | ||
| MRSA in blood culture | 75 | 13 |
| MSSA in blood culture | 22 | 155 |
| Known-positive criterion | ||
| MRSA in blood culture | 75 | 32 |
| MSSA in blood culture | 22 | 280 |
TABLE 3.
Diagnostic properties for the three criteria for interpretation of MRSA screening results
| Test parametera | 30-Day criterion (n = 235)b | Ever-positive criterion (n = 265) | Known-positive criterion (n = 409) |
|---|---|---|---|
| Sensitivity (% [95% CI]) | 79.4 (67.9–88.3) | 85.2 (76.0–91.9) | 70.1 (60.4–78.6) |
| Specificity (% [95% CI]) | 92.8 (87.8–96.2) | 87.6 (81.8–92.0) | 92.7 (89.2–95.4) |
| Positive LR (% [95% CI]) | 12.4 (6.82–23.6) | 7.09 (4.76–11.1) | 10.1 (6.65–15.5) |
| Negative LR (% [95% CI]) | 0.22 (0.12–0.33) | 0.17 (0.08–0.26) | 0.32 (0.23–0.42) |
| MRSA proportion (%) below which NPV is >90% | 33.4 | 39.7 | 34.4 |
| MRSA proportion (%) below which NPV is >95% | 19.2 | 23.8 | 19.9 |
LR, likelihood ratio; NPV, negative predictive value.
n, number of patients with available results.
FIG 1.
Positive and negative predictive values (solid line, median; 95% confidence interval, dashed lines) for 30-day, ever-positive, and known-positive criteria based on the proportion of Staphylococcus aureus bacteremias having methicillin resistance.
DISCUSSION
We demonstrate the usefulness of MRSA screening results for predicting MRSA bacteremia in centers with low-to-moderate MRSA proportions of 20 to 40%. In general, patients with positive MRSA screening swabs (at any time) are at high risk of MRSA infection in the context of a presumed S. aureus bacteremia and should receive empirical vancomycin therapy. Conversely, in clinically stable patients, if one accepts a risk of initially undertreating 5 to 10% of MRSA infections, the presence of a negative screening test within 30 days supports forgoing empirical vancomycin use, provided MRSA makes up less than 20 to 40% of local S. aureus bloodstream infections. This approach would avoid vancomycin exposure in the 90 to 95% of patients without MRSA bacteremia, while still ensuring empirical vancomycin therapy in severe cases or as definitive therapy. It would also follow that the number of patients experiencing unnecessary renal injury would diminish. As expected, the 30-day criterion was both less sensitive and more specific than the ever-positive criterion. However, all criteria performed similarly, and thus older results can remain helpful, depending on the local MRSA proportion. To help physicians compare results to their local epidemiology, we have provided negative predictive values for each criterion at various MRSA proportions (Fig. 1).
Rapid MRSA detection tests are fast alternatives to bacterial cultures and have proved useful in MRSA control (9). However, the use of MRSA screening tests to guide antibiotic therapy remains poorly studied. Prior studies have evaluated their use in multiple situations, including intra-abdominal (10), postoperative (11), respiratory (12, 13), and overall documented clinical infections (14). There is little published on the use of MRSA screening in bloodstream infection. Bai et al. (15) also studied the predictive ability of MRSA screening tests in S. aureus bloodstream infection. They obtained overall sensitivity and specificity values that differed from those of our study (56% and 98%, respectively), but despite the poorer test performance at some of their centers, the overall negative predictive values of the test also exceeded 90%. The different sensitivities could be explained by key methodological differences between the two studies. First, we incorporated a previously known positive status into our prediction, which increased sensitivity and therefore the negative predictive value. Second, we considered only MRSA screening results which were already available at the time the blood culture was taken, whereas Bai et al. included screening isolates taken at the same time as the blood culture. Third, our study involved only tertiary care health centers, and we used a different laboratory protocol. In particular, whereas the previous study relied solely on selective medium to exclude MRSA, in cases with negative chromogenic agar, our laboratory also performed PCR on staphylococcal broth with subsequent subculture so as to improve the diagnostic sensitivity of screening.
With regard to MRSA screening, we believe our results reinforce the need for MRSA control programs as we have demonstrated a strong association between MRSA colonization status and methicillin resistance in subsequent S. aureus bacteremia. As observed from our MRSA proportion plots, efforts at curtailing MRSA spread could result in a clinically significant decrease in MRSA bacteremia and thus in the need for empirical vancomycin therapy.
One of the major limitations of our study is the absence of specific patient-level data; more information (e.g., comorbidities, physical exam, and other laboratory testing) could help more accurately predict MRSA bacteremia. Generalizability is also an issue as our study is limited to two hospitals at one academic medical center; however, the key message regarding the negative/positive predictive values of screening is compatible with all of the other cited studies.
In conclusion, while the retrospective nature of studies such as ours does not provide proof of clinical benefit, we nonetheless suggest that MRSA screening tests could help guide the appropriate use of empirical antibiotic therapy in suspected Gram-positive bloodstream infections. Combining available MRSA screening results with local epidemiology could help clinicians make more educated decisions regarding empirical vancomycin use so as to reduce adverse events while ensuring adequate antimicrobial coverage.
Supplementary Material
ACKNOWLEDGMENTS
We have no conflicts of interest to declare.
All authors had access to the data and a role in writing the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01751-16.
REFERENCES
- 1.Jarvis WR, Jarvis AA, Chinn RY. 2012. National prevalence of methicillin-resistant Staphylococcus aureus in inpatients at United States health care facilities, 2010. Am J Infect Control 40:194–200. doi: 10.1016/j.ajic.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Public Health Agency of Canada. 2015. Canadian antimicrobial resistance surveillance system—report 2015. Public Health Agency of Canada, Ottawa, Canada. [Google Scholar]
- 3.Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. 2003. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis 36:53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 4.Pritchard L, Baker C, Leggett J, Sehdev P, Brown A, Bayley KB. 2010. Increasing vancomycin serum trough concentrations and incidence of nephrotoxicity. Am J Med 123:1143–1149. doi: 10.1016/j.amjmed.2010.07.025. [DOI] [PubMed] [Google Scholar]
- 5.McDanel JS, Perencevich EN, Diekema DJ, Herwaldt LA, Smith TC, Chrischilles EA, Dawson JD, Jiang L, Goto M, Schweizer ML. 2015. Comparative effectiveness of beta-lactams versus vancomycin for treatment of methicillin-susceptible Staphylococcus aureus bloodstream infections among 122 hospitals. Clin Infect Dis 61:361–367. doi: 10.1093/cid/civ308. [DOI] [PubMed] [Google Scholar]
- 6.Siegel JD, Rinehart E, Jackson M, Chiarello L, Healthcare Infection Control Practices Advisory Committee. 2006. Management of multidrug-resistant organisms in healthcare settings, 2006. http://www.cdc.gov/hicpac/pdf/guidelines/mdroguideline2006.pdf. [DOI] [PubMed]
- 7.Lebel P, Fenn S, Loo VG. 2001. Rapid PCR detection of Staphylococcus aureus from selective broths inoculated with surveillance swabs, p 153. Abstr 101st Gen Meet Am Soc Microbiol. American Society for Microbiology, Washington, DC. [Google Scholar]
- 8.Efron B, Tibshirani RJ. 1993. An introduction to the bootstrap. CRC Press, Boca Raton, FL. [Google Scholar]
- 9.Calfee DP, Salgado CD, Milstone AM, Harris AD, Kuhar DT, Moody J, Aureden K, Huang SS, Maragakis LL, Yokoe DS. 2014. Strategies to prevent methicillin-resistant Staphylococcus aureus transmission and infection in acute care hospitals: 2014 update. Infect Control Hosp Epidemiol 35:772–796. doi: 10.1086/676534. [DOI] [PubMed] [Google Scholar]
- 10.Hennessy SA, Shah PM, Guidry CA, Davies SW, Hranjec T, Sawyer RG. 2015. Can nasal methicillin-resistant Staphylococcus aureus screening be used to avoid empiric vancomycin use in intra-abdominal infection? Surg Infect (Larchmt) 16:396–400. doi: 10.1089/sur.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsubara Y, Uchiyama H, Higashi T, Edagawa A, Ishii H, Nagata S, Hashimoto K, Eguchi D, Kawanaka H, Okuyama T, Tateishi M, Korenaga D, Takenaka K. 2014. Nasal MRSA screening for surgical patients: predictive value for postoperative infections caused by MRSA. Surg Today 44:1018–1025. doi: 10.1007/s00595-013-0648-8. [DOI] [PubMed] [Google Scholar]
- 12.Langsjoen J, Brady C, Obenauf E, Kellie S. 2014. Nasal screening is useful in excluding methicillin-resistant Staphylococcus aureus in ventilator-associated pneumonia. Am J Infect Control 42:1014–1015. doi: 10.1016/j.ajic.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 13.Boyce JM, Pop OF, Abreu-Lanfranco O, Hung WY, Fisher A, Karjoo A, Thompson B, Protopapas Z. 2013. A trial of discontinuation of empiric vancomycin therapy in patients with suspected methicillin-resistant Staphylococcus aureus health care-associated pneumonia. Antimicrob Agents Chemother 57:1163–1168. doi: 10.1128/AAC.01965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.MacFadden DR, Elligsen M, Robicsek A, Ricciuto DR, Daneman N. 2013. Utility of prior screening for methicillin-resistant Staphylococcus aureus in predicting resistance of S. aureus infections. CMAJ 185:E725–E730. doi: 10.1503/cmaj.130364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai AD, Burry L, Showler A, Steinberg M, Ricciuto D, Fernandes T, Chiu A, Raybardhan S, Tomlinson GA, Bell CM, Morris AM. 2015. Usefulness of previous methicillin-resistant Staphylococcus aureus screening results in guiding empirical therapy for S. aureus bacteremia. Can J Infect Dis Med Microbiol 26:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.