Abstract
Candida albicans is an important fungal pathogen with a diploid genome that can adapt to caspofungin, a major drug from the echinocandin class, by a reversible loss of one copy of chromosome 5 (Ch5). Here, we explore a hypothesis that more than one gene for negative regulation of echinocandin tolerance is carried on Ch5. We constructed C. albicans strains that each lacked one of the following Ch5 genes: CHT2 for chitinase, PGA4 for glucanosyltransferase, and CSU51, a putative transcription factor. We demonstrate that independent deletion of each of these genes increased tolerance for caspofungin and anidulafungin, another echinocandin. Our data indicate that Ch5 carries multiple genes for negative control of echinocandin tolerance, although the final number has yet to be established.
INTRODUCTION
Candida albicans is a unicellular budding fungus that lives as part of normal human gut or genital microflora. It is also a major opportunistic pathogen in immunocompromised individuals. Naturally occurring strains of C. albicans are usually diploids with eight pairs of chromosomes. However, aneuploidy is well tolerated and is a means to introduce phenotypic diversity in a cell population (1). Moreover, the copy number of a particular chromosome can control adaptation to a specific adverse environment (1), including the development of resistance to fluconazole, a major antifungal from the azole class (1–3). The best-studied regulation due to chromosome copy number is the reversible loss of chromosome 5 (Ch5) controlling resistance to l-sorbose, a toxic sugar that kills C. albicans or other fungi in a manner similar to that of echinocandins (reviewed in reference 4). This regulation is complex, including multiple CSU (control of sorbose utilization) genes scattered along Ch5 that are organized in two functionally redundant pathways (5). The expression of at least two such genes, CSU51 (orf19.1105.2) and CSU53 (orf19.3931), is finely tuned by antisense regulation (6). Recently, we used laboratory mutants to demonstrate that the reversible loss of Ch5 also controls tolerance to the major echinocandin caspofungin, such that strains with one copy acquire caspofungin tolerance whereas strains that spontaneously duplicate monosomic Ch5 revert to susceptibility (4). Based on the model system of sorbose resistance, Ch5 can carry multiple genes encoding negative regulators of echinocandin susceptibility.
Unlike research into the negative control of sorbose resistance, the study of negative control of echinocandin tolerance due to loss of one Ch5 is in its beginning. It was previously reported that disruption of both copies of the Ch5 gene PGA4 (orf19.4035) confers increased caspofungin tolerance (7). PGA4 encodes a glycosylphosphatidylinositol (GPI)-anchored cell surface protein called 1,3-β-d-glucanosyltransferase, which resembles the GEL family of oligosaccharide transferases in Aspergillus fumigatus. However, this result needs reevaluation as the gene was disrupted in the genetic background of the BWP17 strain, which is unstable and responds to genetic manipulations in a nonconventional fashion (8). Most importantly, one Ch5 in BWP17 lacks an ∼36.8-kb portion adjacent to the right telomere (8) that encompasses PGA4. Another Ch5 gene that is a strong candidate for negative control is CHT2 (orf19.3895), which encodes a GPI-anchored chitinase involved in hydrolysis of cell wall chitin. CHT2 is repressed in the core caspofungin response (9, 10). Of a total of four C. albicans genes for chitinases, only CHT2 and CHT3 (orf19.7586) were reported to be downregulated after treatment of C. albicans biofilm with micafungin, another echinocandin, which allowed the authors to suggest that CHT2 and CHT3 are involved in the cell wall's tolerance to stress caused by micafungin and the induction of chitin synthesis (11). Earlier, mutations of CHT2 and CHT3 were found in a laboratory mutant that became highly tolerant to caspofungin and exhibited high chitin content but had no FKS1 mutations causing clinical caspofungin resistance (12). The authors suggested that mutations of CHT2 and CHT3 could result in increased chitin and could affect susceptibility to caspofungin.
In this work, we prepared and characterized deletion strains lacking an entire open reading frame (ORF) of either PGA4 or CHT2 and deletion strains lacking another putative GPI anchor, CSU51. The latter encodes a predicted transcription factor of the helix-loop-helix class, which, as described above, was previously found to be a negative regulator of sorbose resistance (5, 6). We demonstrated that independent deletion of PGA4, CHT2, or CSU51 conferred increased tolerance to the echinocandins caspofungin and anidulafungin. Our data indicate that C. albicans Ch5 carries multiple genes for the negative control of susceptibility to caspofungin and anidulafungin drugs; however, the final number of these genes still needs to be determined.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The C. albicans Ura− strain CAF4-2, a derivative of the reference strain SC5314 (13), was used as a recipient strain to delete genes. This and other strains generated in this study are listed in Table 1. Cells were routinely maintained at 37°C. To prevent induction of chromosome alterations, all strains used were preserved in 15% (vol/vol) glycerol solution at −70°C (2, 8, 14, 15).
TABLE 1.
C. albicans strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| CAF4-2 | ura3Δ::imm434/ura3Δ::imm434 | 13 |
| ER503 (fragmentation site 12) | 394.22-kb truncation of right arm of Ch5 | 5 |
| ER506 (fragmentation site 12) | Same as above, but independent truncation | 5 |
| NCS8 (csu51+/−) | csu51Δ::URA3-FLPa/CSU51 | This study |
| NCS6 (csu51+/−) | csu51Δ::URA3-FLP/CSU51 | This study |
| NCS5 (csu51+/−) | csu51Δ::URA3-FLP/CSU51 | This study |
| NACS1 (csu51−/−) | csu51Δ::URA3-FLP/csu51Δ::NAT1-FLPb | This study |
| NACS8 (csu51−/−) | csu51Δ::URA3-FLP/csu51Δ::NAT1-FLP | This study |
| NACS19 (csu51−/−) | csu51Δ::URA3-FLP/csu51Δ::NAT1-FLP | This study |
| NC136 (cht2+/−) | cht2Δ::URA3-FLP/CHT2 | This study |
| NC72 (cht2+/−) | cht2Δ::URA3-FLP/CHT2 | This study |
| NC133 (cht2+/−) | cht2Δ::URA3-FLP/CHT2 | This study |
| NAC4 (cht2−/−) | cht2Δ::URA3-FLP/cht2Δ::NAT1-FLP | This study |
| NAC12 (cht2−/−) | cht2Δ::URA3-FLP/cht2Δ::NAT1-FLP | This study |
| NAC7 (cht2−/−) | cht2Δ::URA3-FLP/cht2Δ::NAT1-FLP | This study |
| NP6 (pga4+/−) | pga4Δ::URA3-FLP/PGA4 | This study |
| NP3 (pga4+/−) | pga4Δ::URA3-FLP/PGA4 | This study |
| NP5 (pga4+/−) | pga4Δ::URA3-FLP/PGA4 | This study |
| NAP88 (pga4−/−) | pga4Δ::URA3-FLP/pga4Δ::NAT1-FLP | This study |
| NAP86 (pga4−/−) | pga4Δ::URA3-FLP/pga4Δ::NAT1-FLP | This study |
| NAP76 (pga4−/−) | pga4Δ::URA3-FLP/pga4Δ::NAT1-FLP | This study |
| JRCT1 | Clinical isolate | 4 |
| JMC200-3-3 | Same as above, but a single Ch5, MTLα | F. Yang and E. Rustchenko, unpublished data |
| JMC200-3-3-R | Same as above, but Ch5 duplicated | F. Yang and E. Rustchenko, unpublished data |
| SC5314 | Clinical isolate | 4 |
| SMC60-2-5 | Same as above, but a single Ch5, MTLa | F. Yang and E. Rustchenko, unpublished data |
| SMC60-2-5-R | Same as above, but Ch5 duplicated | F. Yang and E. Rustchenko, unpublished data |
| BWP17 | ura3Δ::imm434/ura3Δ::imm434 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/argΔ::hisG | 27 |
| DAY286 | ura3Δ::imm434/ura3Δ::imm434 his1::hisG/his1::hisG pARG4::URA3::arg4::hisG/arg4::hisG | 27 |
| FJS5 | ura3Δ::imm434/ura3Δ::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG cht2::Tn7-UAU1c/cht2::Tn7-URA3 | 27 |
| CAF2-1 | ura3Δ::imm434/URA3 | 28 |
| DSY1768 | cht2Δ::hisG-URA3-hisG/cht2Δ::hisG | 28 |
| pga4−/− strain | ura3Δ::imm434/ura3Δ::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG pga4::Tn7-UAU1/pga4::Tn7-URA3 | 7 |
URA3-FLP denotes the FRT-SAP2P–FLP-URA3-FRT cassette (URA3 flipper).
NAT1-FLP denotes the FRT-SAP2P–FLP-NAT1-FRT cassette (NAT1 flipper).
UAU1 denotes the ura3-ARG4-ura3 cassette (33).
Cells of Escherichia coli DH5α were used for plasmid amplifications. Synthetic dextrose (SD), yeast extract-peptone-dextrose (YPD), and l-sorbose media were prepared as previously described (16, 17). Media were solidified with 2% (wt/vol) agar. Nourseothricin, 150 μg/ml (Werner Bioagents, Jena, Germany); uridine, 50 μg/ml (Sigma, St. Louis, MO, USA); and caspofungin, 120 ng/ml or 200 ng/ml (Merck Sharp & Dohme Corp., Kenilworth, NJ, USA) or anidulafungin, 15 ng/ml (Pfizer Inc., New York, NY, USA), were added when needed.
All primers and plasmids used in this study are presented in Tables 2 and 3, respectively.
TABLE 2.
Primers used in this study
| Primer purpose | Gene | Primer name and sequence |
|---|---|---|
| Amplification of | ||
| ORF of indicated gene | URA3 | NN17 F, CTCATGCCTCACCAGTAGCA |
| NN18 R, AACCCTCTTGGCTCTTGGTT | ||
| NAT1 | NN19 F, ACCATCGGAAGCAGTACCAT | |
| NN20 R, TGTTCCAGGTGATGCTGAAG | ||
| CSU51 | NN1 F, TTGCTTCCATCAACGCTAAA | |
| NN2 R, CAGCACCAACAGCAGCTAAA | ||
| CHT2 | NN9 F, CTTCTGGGGCTTGGTTGAAC | |
| NN10 R, TGGACATCAGTGACGGTTGA | ||
| PGA4 | NN13 F, TGTCAATACCCCGCACTCTT | |
| NN14 R, TTAAGTATCCACCGTCGCCA | ||
| Flanking regions of indicated gene | CSU51 | NN21 F, AAAAAAGGTACCCCCACCTTGTGTACAGGAAT |
| NN22 R, AAAAAAGGGCCCTATGTGTAATTGATGGAAT | ||
| NN23 F, AAAAAACCGCGGGCACAACAATACATTATAAGA | ||
| NN24 R, AAAAAAGAGCTCTCTTCCCATAGGAATAATATGAATAAA | ||
| CHT2 | NN25 F, AAAAAAGGTACCTTGTTTCATTTTGGTGGAAGC | |
| NN26 R, AAAAAAGGGCCCTTTGGCTTGTTTTGTTAAGGGTA | ||
| NN27 F, AAAAAACCGCGGAAGGCTTTCCGCCAATATG | ||
| NN28 R, AAAAAAGAGCTCATCCCATTGACCACGAGAAT | ||
| PGA | NN29 F, AAAAAAGGTACCTGGTTGTCCTCTTTCCCACT | |
| NN30 R, AAAAAAGGGCCCGGAGAAATGAACGAATGAATTG | ||
| NN31 F, AAAAAACCGCGGGCCTATAGCGTCAACCTCTTC | ||
| NN32 R, AAAAAAGAGCTCTTGCAAAAGAGAATTATTGAGCA | ||
| Junctions between chromosome and deletion cassette | CSU51 | NN3 F, TCTTTTTGGGTGTTGGAAAAAA |
| NN4 R, AATGGTGATGTCTAGTGGGTT | ||
| NN5 F, CAGTTGAAGAAAGAAATAGAA | ||
| NN6 R, AAAAACCCCTTTATTGTTGGAAA | ||
| NN7 F, GCACGTCAAGACTGTCAAGG | ||
| NN6 R, AAAAACCCCTTTATTGTTGGAAA | ||
| NN3 F, TCTTTTTGGGTGTTGGAAAAAA | ||
| NN8 R, AAAGTCAAAGTTCCAAGGGG | ||
| CHT2 | NN11 F, TGAATATTAGCCCGCTTTGC | |
| NN4 R, AATGGTGATGTCTAGTGGGTT | ||
| NN5 F, CAGTTGAAGAAAGAAATAGAA | ||
| NN12 R, TCATCATGACCCCAACTCA | ||
| NN7 F, GCACGTCAAGACTGTCAAGG | ||
| NN12 R, TCATCATGACCCCAACTCA | ||
| NN11 F, TGAATATTAGCCCGCTTTGC | ||
| NN8 R, AAAGTCAAAGTTCCAAGGGG | ||
| PGA4 | NN15 F, TCAATTCGAGTTGTTGTTGGA | |
| NN4 R, AATGGTGATGTCTAGTGGGTT | ||
| NN5 F, CAGTTGAAGAAAGAAATAGAA | ||
| NN16 R, GGAATCGGCAGAGTACAAGG | ||
| NN7 F, GCACGTCAAGACTGTCAAGG | ||
| NN16 R, GGAATCGGCAGAGTACAAGG | ||
| NN15 F, TCAATTCGAGTTGTTGTTGGA | ||
| NN8 R, AAAGTCAAAGTTCCAAGGGG | ||
| Semiquantitative RT-PCR | REX2 control gene | HR1 F, GGTTGATTGTGAGATGACAGGATTAGATG |
| HR2 R, TCTTTCGTCTCTTCTTCCAGCT | ||
| CSU51 | HR3 F, TGCAATTCACCAAAGTTATCGC | |
| HR4 R, AGCACCAACAGCAGCTAAAG | ||
| CHT2 | HR5 F, GGTGCTGGTGGTCAAGAAAG | |
| HR6 R, AGGGTAGGAAGTGGTTTGGC | ||
| PGA4 | HR7 F, GCCAAAGCCGGTATTTACGTG | |
| HR8 R, AAGCATCGAACCCATGACCAG |
TABLE 3.
Plasmids used in this study
| Plasmid | Descriptiona | Source |
|---|---|---|
| pSFU1 | URA3-FLP cassette | 20 |
| pJK863 | CaNAT1-FLP cassette carrying nourseothricin resistance gene | 21 |
| pNN1 | Same as pSFU1, but URA3-FLP is flanked by 5′ and 3′ CSU51NCR for CSU51 first allele knockout | This study |
| pNN3 | Same as pSFU1, but URA3-FLP is flanked by 5′ and 3′ CHT2NCR for CHT2 first allele knockout | This study |
| pNN5 | Same as pSFU1, but URA3-FLP is flanked by 5′ and 3′ PGA4NCR for PGA4 first allele knockout | This study |
| pNN2 | Same as pJK863, but CaNAT1-FLP is flanked by 5′ and 3′ CSU51NCR for CSU51 second allele knockout | This study |
| pNN4 | Same as pJK863, but CaNAT1-FLP is flanked by 5′ and 3′ CHT2NCR for CHT2 second allele knockout | This study |
| pNN6 | Same as pJK863, but CaNAT1-FLP is flanked by 5′ and 3′ PGA4NCR for PGA4 second allele knockout | This study |
NCR, noncoding region; Ca, C. albicans.
Gene deletions.
We sequentially deleted two copies of the entire ORF of each gene in the genetic background of the recipient strain (CAF4-2) using a cloning-based method (18, 19). The first copy was deleted with the cassette carrying URA3, whereas the second copy was deleted with the cassette carrying NAT1. We independently generated strains lacking CHT2, CSU51, or PGA4. Also, deletion strains lacking each gene were generated from at least three independent experiments to ensure that the phenotype of interest was related to the deleted gene and not a mutation.
Deletion cassettes were prepared either in plasmid pSFU1 carrying the URA3 flipper (20) or in pJK863 carrying the NAT1 flipper (21) by subcloning various sequences of approximately 300 bp that flanked ORFs of target genes. Flanking sequences were amplified by PCR from the total genomic DNA of CAF4-2. The 5′ untranslated region (UTR) of the target gene was amplified with primers which introduced KpnI and ApaI restriction sites (Table 2) and subsequently subcloned in either the pSFU1 or pJK863 plasmid linearized with KpnI/ApaI restriction enzymes. The 3′ UTR sequence was amplified with primers which introduced SacII and SacI restriction sites (Table 2) and subsequently subcloned in either the pSFU1 or pJK863 plasmid linearized with SacII/SacI restriction enzymes. The deletion cassette was released by restriction digest with KpnI and SacI enzymes and used to transform CAF4-2 cells. SD medium was used to select transformants carrying the URA3 flipper, whereas YPD medium supplemented with 150 μg/ml of nourseothricin was used to select transformants for putative null mutants carrying both the URA3 and NAT1 flippers. Transformants were randomly picked up and purified, and the proper integration of the deletion cassettes was confirmed with PCR and sequencing.
Spot assay.
Cells from −70°C freezer stocks were streaked on YPD plates and incubated at 37°C until young colonies (approximately 2 × 105 cells per colony) appeared. Cells were collected, appropriate dilutions were prepared, and 5 μl of each dilution was spotted on control YPD solid medium supplemented with 50 μg/ml of uridine and either 120 or 200 ng/ml of caspofungin. The plates were incubated at 37°C and photographed with a Molecular Imager Gel Doc XR+ system (Bio-Rad, Hercules, CA).
Broth microdilution assay for determination of MICs.
To determine MICs, we performed a broth microdilution test in accordance with the CLSI reference M27-A3 broth microdilution method for yeasts (22). An inoculum of 1 × 104 cells/ml of each strain was prepared in RPMI 1640 medium buffered to pH 7.0 with 0.165 M morpholinepropanesulfonic acid (MOPS). A series of 2-fold dilutions of the drug were prepared directly in 96-well flat-bottom polystyrene microtiter plates. Subsequently, 100 μl of the cell suspension was added to each well to give a final concentration of 1 × 103 cells/ml in a total volume of 200 μl. The microtiter plates were incubated for 24 h at 35°C. Both negative (inoculum-free) and positive (drug-free) controls were included. Each strain was tested in duplicate on a microtiter plate. The turbidities were determined using a microplate reader (Spectra Max M5; Molecular Devices Corp.) at 600 nm.
Determination of MICs.
The data generated by the microplate reader (see above) were used to calculate MICs at 50% of the inhibition of growth compared with growth in the drug-free control well. For this purpose, we applied nonlinear least-squares regression analysis according to a dose-response curve model using an equation,
| (1) |
where A is range of optical densities at 600 nm (OD600), x is drug concentration, M is MIC, h is Hill slope value, and b is percent inhibition (23). Then, the best-fit curve was generated using Microsoft Excel software.
In addition, projected MIC values were calculated at different degrees of inhibition: 70%, 80%, and 90%.
Determination of 1,3-β-glucan content in the cell wall.
Cells were cultured in YPD broth up to log phase and then harvested and washed with TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) twice. The same amount of cells from each strain was prepared. The 1,3-β-glucan levels were determined by the aniline blue assay, as previously described (4, 24). Briefly, 500 μl of cell suspensions from each strain was resuspended with 100 μl of 6 M NaOH, followed by incubation at 80°C for 30 min. After solubilization of a glucan, 2.1 ml of aniline blue mix (0.03% aniline blue, 0.18 M HCl, and 0.49 M glycine-NaOH, pH 9.5) was added to each sample, which was incubated at 50°C for 30 min and then cooled to room temperature for 30 min. After incubation, fluorescence was measured in a black 96-well microplate using a fluorescence plate reader (Spectra Max M5; Molecular Devices Corp.) at an excitation wavelength of 400 nm, an emission wavelength of 460 nm, and a wavelength cutoff of 455 nm.
Determination of chitin content in the cell wall.
The chitin content was determined by measuring the absorbance of glucosamine released by acid hydrolysis of the purified cell wall as described previously (4, 25). Briefly, approximately 3 × 103 CFU was plated on YPD plates and incubated at 37°C. Young colonies of each strain (approximately 2 × 105 cells per colony) were harvested from the surface of plates with autoclaved distilled water and collected by centrifugation. After being washed with water, cells were disrupted with 0.5-mm glass beads (11079105; BioSpec Products, Inc., Bartlesville, OK) using a mini-Beadbeater (BioSpec Products, Inc., Bartlesville, OK). Then, the pellet was washed five times with 1 M NaCl, extracted in SDS-MerOH extraction buffer (50 mM Tris, 2% sodium dodecyl sulfate, 0.3 M mercaptoethanol, 1 mM EDTA, pH 8.0) at 100°C for 10 min, and then washed three times in distilled water (dH2O). Cells were dried with a SpeedVac concentrator (Phoenix Equipment Inc., Rochester, NY) and weighed. Dry samples were suspended in 1 ml of 6 M HCl and boiled for 17 h. The acid was evaporated at 65°C. The hydrolyzed samples were resuspended in 1 ml of sterile dH2O. A 100-μl portion of the sample was mixed with 100 μl of 1.5 M Na2CO3 in 4% acetyl acetone. The mixture was boiled for 20 min, and then 700 μl of 96% ethanol and 100 μl of p-dimethylaminobenzaldehyde solution in a 1:1 mixture of ethyl alcohol and concentrated HCl were added, followed by 1 h of incubation at room temperature. Optical densities were read at 520 nm with a plate reader (Spectra Max M5; Molecular Devices Corp., Sunnyvale, CA). Glucosamine (Sigma-Aldrich, St. Louis, MO) was used as a standard for measurement of chitin content. The final chitin level in each sample was calculated as a percentage of the cell wall dry weight.
PCR.
For cloning purposes, PCR was conducted using Ex Taq DNA polymerase (TaKaRa Biomedicals, Otsu, Shiga, Japan) according to the manufacturer's instruction. Other PCRs were conducted using Dream Taq DNA polymerase (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions. PCR products were electrophoresed on 1% agarose at 95 V for 45 min and stained with 1 μg/ml ethidium bromide for 10 min. Gel images were obtained using the Molecular Imager Gel Doc XR+ system (Bio-Rad, Hercules, CA).
Semiquantitative reverse transcription-PCR (RT-PCR).
To determine expression of genes of interest, we standardized the growth of C. albicans cells. Briefly, petri dishes with synthetic medium in which glucose was substituted for sorbitol were seeded with ∼3,000 CFU per plate and incubated at 37°C for 20 to 40 h until colonies contained ∼105 cells/colony. We prepared three batches of total RNA from three independent cultures of each strain. RNA extraction and reverse transcription were conducted using an RNeasy minikit (Qiagen, Valencia, CA) and a high-capacity cDNA reverse transcription kit (Applied Biosystems; Thermo Fisher Scientific, Waltham, MA), respectively, according to the manufacturer's instructions. PCRs were performed using Dream Taq DNA polymerase (Thermo Scientific, Rockford, IL) according to manufacturer recommendations. The gene REX2 (orf19.1466) was used as an internal control. Pilot PCRs were conducted and showed that 28 to 31 amplification cycles produce amplicons with a linear increase of DNA. In final PCRs, approximately 20 to 80 ng each of cDNA for a gene of interest and a control gene were used as the templates to amplify the genes for the above number of cycles.
Semiquantitative RT-PCR was previously described by us (6). Briefly, PCR products from several consecutive cycles in the exponential phase were electrophoresed on 1% agarose at 95 V for 45 min and stained with 1 μg/ml ethidium bromide for 10 min. The stained DNA bands were photographed using the Molecular Imager Gel Doc XR+ system (Bio-Rad, Hercules, CA), and band intensities were detected using Image Lab software (Bio-Rad, Hercules, CA). The gene of interest was normalized against the control gene REX2 by calculating the mutant/parent ratio of densitometry values.
Miscellaneous.
Plasmid DNA purification from the gel was done with the QIAquick gel extraction kit (Qiagen, Valencia, CA). For transformation of C. albicans cells, the lithium acetate method was used as described previously (26). Sequencing by the Sanger method was done in the Genomic Research Center of the University of Rochester. Plates were photographed with the Molecular Imager Gel Doc XR+ system (Bio-Rad, Hercules, CA).
RESULTS
Truncation of Ch5 right arm confers an increased caspofungin tolerance.
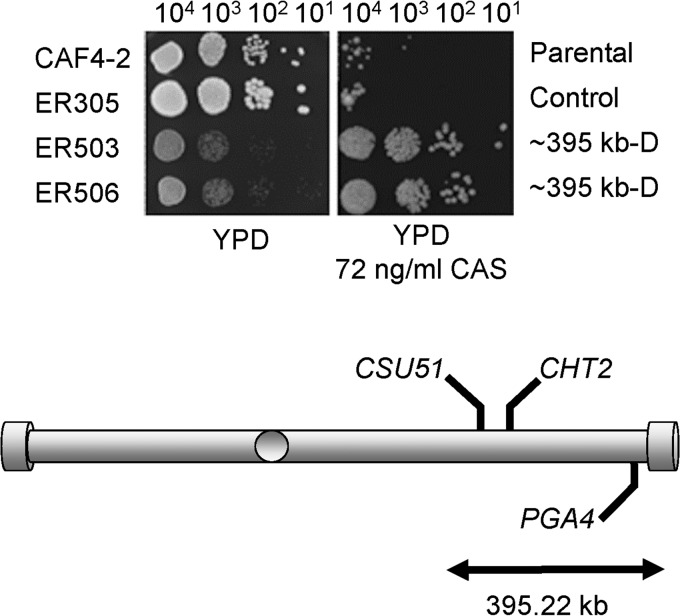
As we reported earlier, truncating one Ch5 by deleting a 395.22-kb portion adjacent to the right telomere caused resistance to sorbose similar to that with the loss of an entire Ch5 (5). This is because the truncation cuts the dose of the critical genes encoding negative regulators of sorbose resistance in half. We tested two independently derived strains, ER503 and ER506, which both lacked the 395.22-kb portion on one Ch5, for growth on caspofungin-supplemented medium and found that the truncated Ch5 also causes caspofungin tolerance, compared to parental and control strains (Fig. 1). In contrast, small deletions on the Ch5 right arm had no effect on caspofungin susceptibility (data not shown). This result indicated that the portion of 395.22 kb on Ch5 encompasses a genetic element(s) that acts to suppress caspofungin tolerance. We examined genes carried within this portion and found PGA4, which has been previously reported to negatively control caspofungin susceptibility (7), as well as CHT2 and CSU51, which could be candidates for negative regulation (see the introduction for more). Because sorbose and caspofungin kill C. albicans in similar manners, the presence of multiple Ch5 genes for negative regulation of caspofungin tolerance is in agreement with our previous finding of multiple Ch5 genes for negative regulation of sorbose resistance.
FIG 1.
Spot assay for caspofungin (CAS) susceptibility of two separately derived mutants, ER503 and ER505, each carrying a 395.22-kb deletion adjacent to the right telomere on one Ch5 (Table 1). Shown is the comparative growth of ER503 and ER505 versus growth of the parental strain CAF4-2 and the control strain ER305 with integrated empty vector. Media and caspofungin concentration are indicated. From left to right, 104, 103, 102, and 101 cells were spotted on a YPD control plate or a YPD plate supplemented with caspofungin and incubated for 3 days at 37°C. Strains and their genotypes are indicated on the left and right, respectively. Note that strains ER503 and ER505 with a truncated Ch5 grow slower than control strain CAF4-2 or ER305 on a control YPD plate. However, in the presence of caspofungin, ER503 and ER505 grow well whereas control strains fail to grow. Also shown is a cartoon of Ch5 indicating positions of PGA4, CHT2, and CSU51.
The cht2/cht2, csu51/csu51, and pga4/pga4 mutants acquired caspofungin and anidulafungin tolerance.
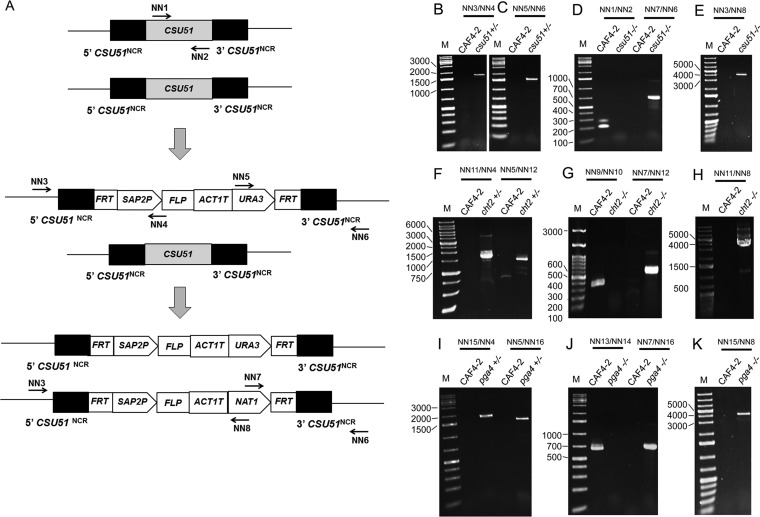
We prepared independent deletion strains lacking one or two copies of CHT2 (cht2+/− or cht2−/−), CSU51 (csu51+/− or csu51−/−), or PGA4 (pga4+/− or pga4−/−) (Table 1) as described in Materials and Methods and illustrated in Fig. 2. We took care to prepare three separately derived mutants of each kind, cht2+/− or cht2−/−, csu51+/− or csu51−/−, and pga4+/− or pga4−/− (Table 1). All deletion strains were analyzed with an agar-based spot assay for susceptibility to caspofungin, and one representative double-deletion mutant of each kind was also analyzed for susceptibility to anidulafungin. Each measurement was repeated three times.
FIG 2.
Sequential deletion of a gene with the URA3 flipper deletion cassette followed by the NAT1 flipper deletion cassette and verification of gene deletion by a PCR screening method. (A) Diagram showing sequential use of deletion cassettes and position of PCR primers using CSU51 as an example. (B and C) Deletion of the first CSU51 copy. PCR amplicons represent the 5′ and 3′ junctions, respectively, of the URA3 flipper cassette. (D and E) Deletion of the second CSU51 copy. The PCR amplicon in CAF4-2 corresponds to CSU51. PCR amplicons in csu51−/− represent the 3′ and 5′ junctions, respectively, of the NAT1 flipper cassette. (F) Deletion of the first CHT2 copy. The PCR amplicon on the left or on the right represents, respectively, the 5′ or 3′ junction of the URA3 flipper cassette. (G and H) Deletion of the second CHT2 copy. The PCR amplicon in CAF4-2 represents CHT2. PCR amplicons in cht2−/− represent the 3′ and 5′ junctions, respectively, of the NAT1 flipper cassette. (I) Deletion of the first PGA4 copy. The PCR amplicon on the left or the right represents the 5′ or 3′ junctions, respectively, of the URA3 flipper cassette. (J and K) Deletion of the second PGA4 copy. The PCR amplicon in CAF4-2 represents PGA4. PCR amplicons in pga4−/− represent the 3′ and 5′ junctions, respectively, of the NAT1 flipper cassette. Primers (Table 2) or representative deletion strains (Table 1) are indicated on top. Lanes M contain a 1-kb Plus DNA ladder (Goldbio, St. Louis, MO, USA) with some bands indicated on the left in base pairs. NCR, FRT, SAP2P, FLP, and ACT1T stand for, respectively, noncoding region, FLP recombinase target, SAP2 promoter, flippase, and transcription termination sequence of the ACT1 gene. Note that PCR verification of deletions was done for all single- or double-deletion mutants (Table 1); data for representative mutants are shown here. Numbers at left of panels represent size in base pairs.
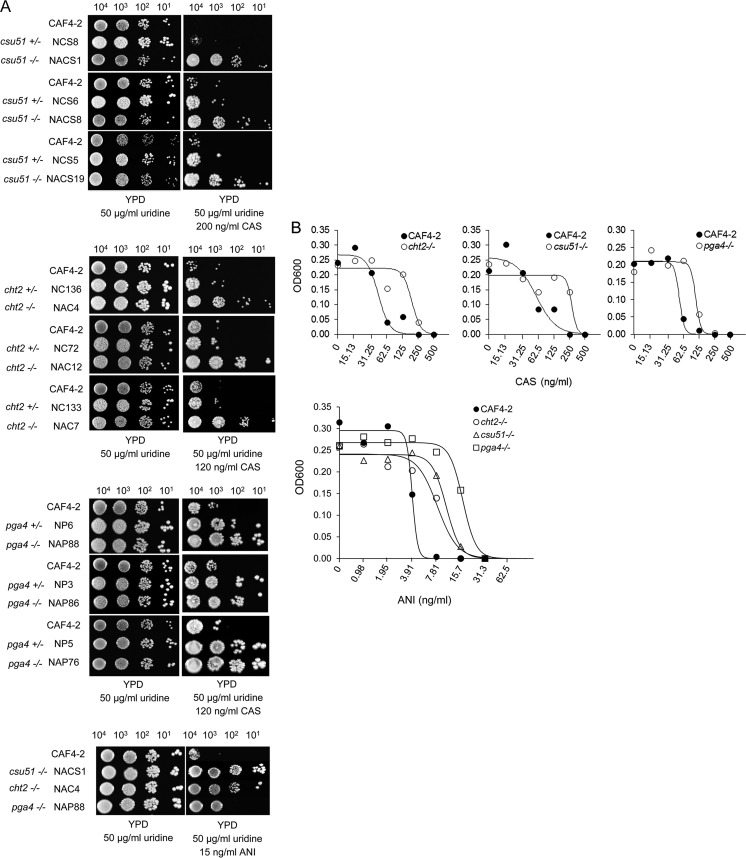
By conducting pilot experiments, we found optimal drug concentrations that revealed growth differences based on the number of growing spots between the parental strain CAF4-2 and mutants: 120 ng/ml or 200 ng/ml for caspofungin and 15 ng/ml for anidulafungin. For example, no growth of CAF4-2 versus the double-deletion mutant lacking CSU51 in any of the four spots on the plate with caspofungin or greatly reduced growth of CAF4-2 versus either a single- or double-deletion mutant lacking PGA4 in the first one or two spots from left to right and no growth in the two remaining spots (Fig. 3A) was considered suppressed growth of the parental strain compared to that of the mutants. Data from one representative experiment are shown in Fig. 3A; the two other repeats are shown in Fig. S1A and B in the supplemental material.
FIG 3.
Analysis of caspofungin or anidulafungin susceptibility phenotype. (A) Spot assay shows comparative growth of all single- or double-deletion mutants that are listed in Table 1 versus their parental strain CAF4-2 on control YPD medium, as well as on YPD medium supplemented with caspofungin (CAS), as indicated. Also shown is the comparative growth of representative double-deletion mutants on control YPD medium, as well as on YPD medium supplemented with anidulafungin (ANI), as indicated. Strains are indicated on the left. For more details, see the legend to Fig. 1. Note that each mutant was tested in three independent spot assays, of which one assay is shown here. See Fig. S1 in the supplemental material for the other two assays. Numbers above panels are numbers of cells. (B) Broth microdilution assay shows comparative growth of representative double-deletion mutants NAC4 (cht2−/−), NACS1 (csu51−/−), and NAP88 (pga4−/−), after 24 h of incubation in media containing different concentrations of either caspofungin or anidulafungin, as indicated. The best-fit curves in each experiment were generated as explained in Materials and Methods. Note that each mutant was tested in three independent assays, of which a representative assay is shown here. See Fig. S2 and S3 in the supplemental material for the other two assays. Also, note that assays with anidulafungin were performed on the same microtiter plate.
We found that all mutants lacking both copies of CHT2 or CSU51 showed growth in all four spots in the presence of caspofungin, unlike the parental strain CAF4-2, which had no growth. Mutants lacking a single copy showed, predominantly, growth only in the first spot on the left. All mutants lacking either one or both copies of PGA4 grew up better than CAF4-2, and their growth levels were similar, thus indicating haploinsufficiency. A representative mutant lacking PGA4 also consistently formed larger colonies than the parental strain (data not shown). In addition, all representative mutants lacking both copies of CHT2, CSU51, or PGA4 showed better growth in the presence of anidulafungin than CAF4-2.
We then used one representative double-deletion mutant of each kind, cht2−/−, csu51−/−, and pga4−/−, to perform a quantitative broth microdilution assay for determination of caspofungin or anidulafungin MICs (Materials and Methods). Each experiment was independently repeated three times. MICs were determined and the best-fit curves were generated in each experiment as explained in Materials and Methods. In representative experiments with caspofungin (Fig. 3B), CAF4-2 displayed a MIC90 of 62.5 ng/ml on each graph, whereas deletion mutants displayed higher MICs (see Fig. S2A and B in the supplemental material for two more repeats). In addition, we used one representative experiment to calculate projected MIC values at different degrees of inhibition: 70%, 80%, and 90% (Materials and Methods). As expected, projected MICs of mutants were higher than those of CAF4-2 (Table 4). The differences were evaluated with Student's t test and were all <0.005. In a representative experiment with anidulafungin (Fig. 3B), CAF4-2 displayed a MIC50 of 3.91 ng/ml at approximately 50% inhibition, whereas deletion mutants displayed higher MICs (see Fig. S3A and B in the supplemental material for two more repeats). Regarding calculated projected MIC values at different degrees of inhibition, values for 70%, 80%, and 90% inhibition of mutants were higher than those of CAF4-2 (Table 5). The differences were evaluated with Student's t test and were all <0.001.
TABLE 4.
Calculated values of caspofungin MICs for double-deletion mutants and CAF4-2a
| Strain | MIC (ng/ml) |
|||
|---|---|---|---|---|
| MIC50 | MIC70 | MIC80 | MIC90 | |
| CAF4-2 | 43 ± 7 | 53 ± 8 | 60 ± 10 | 73 ± 12 |
| NAC4 (cht2−/−) | 188 ± 37 | 221 ± 43 | 246 ± 48 | 288 ± 56 |
| CAF4-2 | 57 ± 17 | 84 ± 24 | 101 ± 29 | 151 ± 44 |
| NACS1 (csu51−/−) | 277 ± 32 | 306 ± 36 | 325 ± 38 | 356 ± 42 |
| CAF4-2 | 60 ± 1 | 61 ± 1 | 63 ± 1 | 67 ± 2 |
| NAP88 (pga4−/−) | 124 ± 1 | 124 ± 5 | 125 ± 1 | 139 ± 8 |
MIC50, MIC70, MIC80, or MIC90 refers to the concentration of caspofungin at which 50%, 70%, 80%, or 90% of growth is inhibited, respectively. See Materials and Methods for the calculation of MIC values. The differences between mutants and CAF4-2 were evaluated with Student's t test, and all P values were <0.005. Note that parental CAF4-2 was assayed together with each deletion mutant.
TABLE 5.
Calculated values of anidulafungin MICs for double-deletion mutants and CAF4-2a
| Strain | MIC (ng/ml) |
|||
|---|---|---|---|---|
| MIC50 | MIC70 | MIC80 | MIC90 | |
| CAF4-2 | 3.9 ± 0.1 | 4.1 ± 0.1 | 4.2 ± 0.1 | 4.3 ± 0.1 |
| NAC4 (cht2−/−) | 8.0 ± 0.9 | 10.2 ± 1.1 | 11.9 ± 1.3 | 15.0 ± 1.6 |
| NACS1 (csu51−/−) | 10.3 ± 0.6 | 12.2 ± 0.7 | 13.6 ± 0.8 | 16.0 ± 0.9 |
| NAP88 (pga4−/−) | 16.6 ± 1.0 | 19.5 ± 0.7 | 21.6 ± 0.8 | 25.1 ± 0.9 |
MIC50, MIC70, MIC80, or MIC90 refers to the concentration of caspofungin at which 50%, 70%, 80%, or 90% of growth is inhibited, respectively. See Materials and Methods for the calculation of MIC values. The differences between mutants and CAF4-2 were evaluated with Student's t test, and all P values were <0.001.
We also tested a pga4−/− disruption mutant in which the Tn7-UAU1 disruption cassette was inserted in PGA4 (7) (Table 1). We also tested two CHT2 mutants: the FJS5 mutant, in which the Tn7-UAU1 disruption cassette was inserted in CHT2 (27), and the DSY1768 mutant, in which 490 nucleotides (nt) was deleted between nt +587 (with respect to the first ATG) and nt +1076 in the CHT2 ORF (28) (Table 1). We found that the pga4−/− mutant grows better than the control strain in the presence of caspofungin (see Fig. S4 in the supplemental material), i.e., indicating greater caspofungin tolerance, similarly to our deletion mutants. However, neither DSY1768 or FJS5 mutants acquired higher tolerance (see Fig. S4). Importantly, in the DSY1768 mutant, of a total CHT2 ORF (1,752 nt) only 490 nt, or approximately one-third, of the middle portion of the ORF was deleted, leaving intact 587 nt at the beginning and 676 nt at the end of the ORF. Apparently, this deletion was not sufficient to abolish CHT2 function. We believe that incomplete removal of the CHT2 ORF in the DSY1768 mutant and disruption of CHT2 instead of deletion in the FJS5 mutant allowed retention of CHT2 function and determined the phenotypic difference between these mutants and our mutants lacking a full ORF of CHT2. Overall, in this study, we confirmed PGA4's involvement in the negative control of caspofungin tolerance in different genetic backgrounds (see reference 7). We also provided evidence of CHT2 and CSU51 involvement in the negative control of caspofungin tolerance, and we provided evidence of CHT2, CSU51, and PGA4 involvement in the negative control of anidulafungin tolerance.
CHT2, CSU51, and PGA4 are downregulated on monosomic Ch5.
We examined whether Ch5 copy number controls expression of CHT2, CSU51, and PGA4 in caspofungin-tolerant mutants, as could be expected for the genes encoding negative regulators of caspofungin tolerance, i.e., decreased expression on the monosomic Ch5 but not on the reduplicated Ch5. For this purpose, we prepared a sequential series of genetically related strains including parental strain JRCT1 followed by Ch5 monosomic mutant JMC200-3-3 followed by Ch5 reduplicated derivative JMC200-3-3R (F. Yang and E. Rustchenko, unpublished data). The construction of a series of matched strains with a copy number of Ch5 alternating between two and one is a well-established procedure in our laboratory, as described in detail previously (4, 29). Briefly, a Ch5 monosomic mutant was obtained by plating cells of JRCT1 on YPD medium supplemented with a lethal amount of caspofungin, whereas the Ch5 reduplicated derivative was obtained due to spontaneous duplication of the monosomic Ch5 when growing cells of JMC200-3-3 on YPD solid medium in the absence of selection. Expression changes were determined with semiquantitative RT-PCR (Materials and Methods). Gels with representative amplicons that were quantitated are shown in Fig. S5 in the supplemental material. We found that CHT2, CSU51, and PGA4 are downregulated on the monosomic Ch5 but not on the duplicated Ch5 (Table 6). This experiment establishes the relationship between negative control due to the loss of one Ch5 and Ch5-residing genes CHT2, CSU51, and PGA4.
TABLE 6.
Expression change of CHT2, CSU51, or PGA4 carried on the monosomic or duplicated Ch5 versus normal disomic Ch5 of the parental strain, as determined with semiquantitative RT-PCR from three independent RNA preparationsa
| Gene | Ch5 monosomic |
Ch5 duplicated |
||||||
|---|---|---|---|---|---|---|---|---|
| JMC200-3-3/JRCT1 |
SMC60-2-5/SC5314 |
JMC200-3-3R/JRCT1 |
SMC60-2-5R/SC5314 |
|||||
| Mutant/parent ratio | Mean ± SD | Mutant/parent ratio | Mean ± SD | Mutant/parent ratio | Mean ± SD | Mutant/parent ratio | Mean ± SD | |
| CHT2 | 0.61, 0.70, 0.54 | 0.62 ± 0.08b | 0.10, 0.04, 0.06 | 0.07 ± 0.03b | 1.40, 1.32, 1.61 | 1.45 ± 0.15 | 1.14, 0.84, 0.96 | 0.98 ± 0.15 |
| CSU51 | 0.57, 0.67, 0.51 | 0.58 ± 0.08b | 0.54, 0.61, 0.73 | 0.68 ± 0.06b | 0.88, 0.80, 1.12 | 0.93 ± 0.17 | 1.13, 1.39, 1.48 | 1.33 ± 0.18 |
| PGA4 | 0.82, 0.82, 0.70 | 0.78 ± 0.07b | 0.65, 0.81, 0.85 | 0.77 ± 0.10b | 0.91, 0.92, 0.94 | 0.92 ± 0.02 | 0.87, 1.13, 0.90 | 0.97 ± 0.14 |
Expression change was determined as mutant/parent ratio for each gene using two series of sequential derivatives: JRCT1, parent → JMC200-3-3, Ch5 monosomic → JMC200-3-3R, Ch5 duplicated, or SC5314, parent → SMC60-2-5, Ch5 monosomic → SMC60-2-5R, Ch5 duplicated.
The differences in expression between derivatives with monosomic or duplicated Ch5 and parent strain, JRCT1 or SC5314, were evaluated with Student's t test. P values were <0.05.
CHT2, CSU51, or PGA4 controls the decrease of 1,3-β-glucan and increase of chitin in the cell wall.
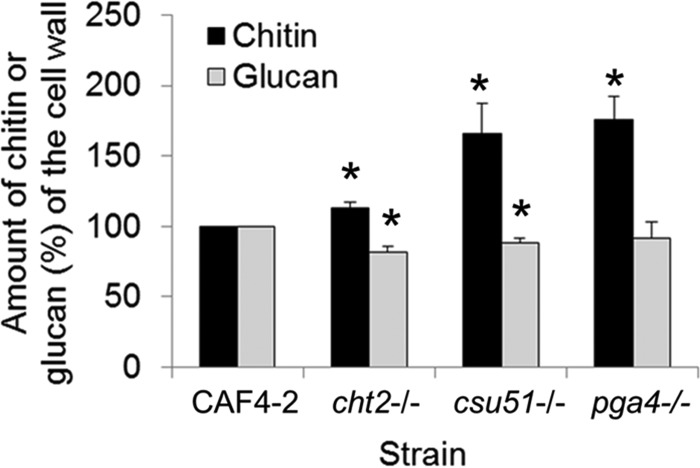
Caspofungin tolerance is associated with cell wall remodeling, including the decrease of 1,3-β-glucan and increase of chitin (4, 12, 30). Thus, we asked whether the same is true for the CHT2, CSU51, or PGA4 deletion strains (see Materials and Methods for the assays). As shown in Fig. 4, the cht2−/−, csu51−/−, and pga4−/− representative deletion strains possessed significantly higher cell wall chitin contents than the parental strain. Glucan, however, was substantially lower in the cht2−/− and csu51−/− strains and marginally decreased in the pga4−/− strain.
FIG 4.
Levels of cell wall chitin or glucan in representative double-deletion mutants NAC4 (cht2−/−), NACS1 (csu51−/−), and NAP88 (pga4−/−), compared with the parental strain CAF4-2. Results were averaged from three independent experiments. The amount of chitin or glucan in the parental strain is set as 100%. The asterisks indicate a P value of <0.05, determined using Student's t test.
Only CSU51 controls susceptibility to both sorbose and caspofungin.
The CSU51 gene has been previously reported as encoding the negative regulator of growth on sorbose medium (5, 6). The csu51−/− mutant, thus, is expected to grow on sorbose medium, as we confirmed here (Fig. 5). However, the cht2−/− or pga4−/− mutant showed no growth on sorbose plates (Fig. 5). This indicates that at least some CSU genes can negatively control both sorbose resistance and echinocandin tolerance. However, genes that negatively control echinocandin tolerance do not necessarily control sorbose resistance.
FIG 5.
Growth of representative double-deletion mutants NAC4 (cht2−/−), NACS1 (csu51−/−), and NAP88 (pga4−/−), on medium containing l-sorbose as a sole source of carbon. (A) Growth of streaks after 5 days of incubation at 37°C on control SD and sorbose media, as indicated. (B) Spot assay for growth with the strains shown in panel A after incubation for 6 days at 37°C on control SD and sorbose media, as indicated. Strains indicated on the left in panels A and B are the parental strain CAF4-2 for a negative control, Ch5 monosomic mutant Sor1210(60) (4) for a positive control, and the abovementioned mutants. The Fig. 3A legend contains more details. Note that only the mutant lacking CSU51 grows on sorbose medium. Numbers above the panels are numbers of cells.
DISCUSSION
To investigate the relation of Ch5 gene CHT2, CSU51, or PGA4 to the caspofungin or anidulafungin susceptibility phenotype, we developed a rigorous approach to independently delete the coding region of two copies of these genes. Our approach included the use of a stable strain to perform deletions (8); the consecutive use of two different deletion cassettes to avoid the excision of the first integrated cassette, a procedure that can be mutagenic; the design of flanking sequences for a deletion cassette that are approximately 300 bp long to avoid disturbing the flanking genes (18); and the generation of three separately derived double-deletion constructs for each gene to eliminate any potential phenotype confusion caused by an undesirable mutation that mimics the phenotype of the deleted gene.
We found that mutants lacking either CHT2, CSU51, or PGA4 acquired greater relative tolerance to caspofungin or anidulafungin. The deletion mutants also acquired remodeled cell walls with decreased content of 1,3-β-glucan but increased chitin content, which is typical for echinocandin-tolerant strains (4, 12, 30). We also determined that, as could be expected, the abovementioned genes are downregulated on the monosomic Ch5 in the representative tolerant mutants that were generated by exposure to caspofungin, but these genes were not downregulated when Ch5 was reduplicated. This is significant, as genes on the monosomic Ch5 can be also upregulated to the disomic or above the disomic level (31).
Because of the criteria used to select three genes for our study (see the introduction), we believe that these genes are not the only Ch5 genes encoding negative regulators of caspofungin or anidulafungin tolerance. Also, multiple genes for the same phenotype are expected to reside on the same chromosome, because C. albicans can control adaptation to adverse environments by varying the copy number of chromosomes, i.e., making an entire chromosome behave as a single regulatory unit. Such regulation presumes a biased distribution of genes over chromosomes, so that many of the genes relevant for the same phenotype can be controlled by a single event of the change of chromosome copy number. Only 140 genes, or approximately 25%, out of a total of 523 genes residing on Ch5 are characterized to date. The remaining 367 genes, and also possibly some of the characterized genes, comprise a sufficiently large pool to search for more genes for negative regulation of caspofungin tolerance. Primary candidates could be Ch5 GPI-anchored genes similar to GPI-anchored CSU51, CHT2, and PGA4.
We emphasize that independent deletion of a single copy of CHT2 or CSU51 conferred practically no increase in tolerance, although lack of a single copy of PGA4 was sufficient to increase tolerance. It seems that a combined loss of one copy of many genes on Ch5 with or without haploinsufficiency can lead to a cell acquiring a relatively robust level of tolerance that is sufficient for adaptation at least in vitro. This matter needs further clarification.
The Ch5 genes analyzed here encode GPI-anchored chitinase Cht2p and GPI-anchored glucanosyltransferase Pga4p for cell wall biosynthesis, as well as putative GPI-anchored transcription factor Csu51p, thus indicating that various metabolic pathways are involved in the negative control of echinocandin susceptibility. The function of CHT2, PGA4, and CSU51 needs further clarification in order to fully understand each gene's role in echinocandin tolerance. For example, CSU51 controls susceptibility to sorbose and echinocandins, but CHT2 and PGA4 control susceptibility only to echinocandins. Furthermore, CHT2 is downregulated, sometime dramatically, on the monosomic Ch5 in either sorbose- or caspofungin-generated mutants (4; this work). However, despite this downregulation, sorbose-generated mutants often do not acquire tolerance to caspofungin (4). Another laboratory recently reported that the level of Cht2p in C. albicans decreased upon heat stress (32). We conclude that CHT2 possibly has a broader function in a more generalized response to stress. The spectrum of stressors to which each such gene responds will be the subject of future studies.
In summary, our data indicate that Ch5 of C. albicans carries at least three genes encoding negative regulators of caspofungin and anidulafungin tolerance. There is a possibility that Ch5 carries more such genes. The final number of these genes is still to be determined. This is similar to previously reported multiple Ch5 CSU genes for negative control of resistance to a toxic sugar sorbose that kills C. albicans in a manner similar to that of echinocandins (5). Loss of one copy of Ch5, leading to echinocandin tolerance, is prominent in laboratory mutants. Thus, it is possible that similar Ch5 rearrangement can be found in clinical isolates. Such loss of one copy of Ch5 can contribute to the evolution of resistance, as based on point mutations in the FKS1 gene. Clinically, it may eventually prove to be possible to improve the efficacy of echinocandins by enhancing products of Ch5 genes that repress echinocandin tolerance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Feng Yang for providing us with two sequential series of genetically related strains, as well as testing two mutants with truncated Ch5 for caspofungin tolerance. We thank Julia R. Köhler for the gift of plasmid pJK863 and protocols. We thank Joachim Morschhäuser for the gift of plasmid pSFU1. We also thank Louise Walker and Carol A. Munro for sharing pga4−/− and DSY1768 mutants and Aaron P. Mitchell for sharing the FJS5 mutant with us. We thank Mark Dumond for critically reading the manuscript. We are grateful to Bogdan Polevoda for consulting. We also thank Louis DiDone for technical assistance and Damian J. Krysan for sharing a plate reader with us. We thank Merck Sharp & Dohme Corp. for the generous donation of caspofungin and Pfizer Inc. for the generous donation of anidulafungin via a compound transfer program.
These studies were supported by grant AI 110764 to E. Rustchenko.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01888-16.
REFERENCES
- 1.Rustchenko E. 2007. Chromosome instability in Candida albicans. Yeast 7:2–11. doi: 10.1111/j.1567-1364.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- 2.Perepnikhatka V, Fischer FJ, Niimi M, Baker RA, Cannon RD, Wang YK, Sherman F, Rustchenko E. 1999. Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J Bacteriol 181:4041–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selmecki A, Forche A, Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang F, Kravets A, Bethlendy G, Welle S, Rustchenko E. 2013. Chromosome 5 monosomy of Candida albicans controls susceptibility to various toxic agents, including major antifungals. Antimicrob Agents Chemother 57:5026–5036. doi: 10.1128/AAC.00516-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kabir MA, Ahmad A, Greenberg JR, Wang YK, Rustchenko E. 2005. Loss and gain of chromosome 5 controls growth of Candida albicans on sorbose due to dispersed redundant negative regulators. Proc Natl Acad Sci U S A 102:12147–12152. doi: 10.1073/pnas.0505625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmad A, Kravets A, Rustchenko E. 2012. Transcriptional regulatory circuitries in the human pathogen Candida albicans involving sense-antisense interactions. Genetics 190:537–547. doi: 10.1534/genetics.111.136267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plaine A, Walker L, Da Costa G, Mora-Montes HM, McKinnon A, Gow NA, Gaillardin C, Munro CA, Richard ML. 2008. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet Biol 10:1404–1414. doi: 10.1016/j.fgb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahmad A, Kabir MA, Kravets A, Andaluz E, Larriba G, Rustchenko E. 2008. Chromosome instability and unusual features of some widely used strains of Candida albicans. Yeast 25:433–448. doi: 10.1002/yea.1597. [DOI] [PubMed] [Google Scholar]
- 9.Liu TT, Lee REB, Barker KS, Lee RE, Wei L, Homayouni R, Rogers PD. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob Agents Chemother 49:2226–2236. doi: 10.1128/AAC.49.6.2226-2236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruno VM, Kalachikov S, Subaran R, Nobile CJ, Kyratsous C, Mitchell AP. 2006. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog 2:e21. doi: 10.1371/journal.ppat.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaneko Y, Ohno H, Kohno S, Miyazaki Y. 2010. Micafungin alters the expression of genes related to cell wall integrity in Candida albicans biofilms. Jpn J Infect Dis 63:355–357. [PubMed] [Google Scholar]
- 12.Drakulovski P, Dunyach C, Bertout S, Reynes J, Mallié MA. 2011. Candida albicans strain with high MIC for caspofungin and no FKS1 mutations exhibits a high chitin content and mutations in two chitinase genes. Med Mycol 49:467–474. doi: 10.3109/13693786.2010.538732. [DOI] [PubMed] [Google Scholar]
- 13.Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rustchenko-Bulgac EP. 1991. Variations of Candida albicans electrophoretic karyotypes. J Bacteriol 173:6586–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YK, Das B, Huber DH, Wellington M, Kabir MA, Sherman F, Rustchenko E. 2004. Role of the 14-3-3 protein in carbon metabolism of the pathogenic yeast Candida albicans. Yeast 21:685–702. doi: 10.1002/yea.1079. [DOI] [PubMed] [Google Scholar]
- 16.Rustchenko EP, Howard DH, Sherman F. 1994. Chromosomal alterations of Candida albicans are associated with the gain and loss of assimilating functions. J Bacteriol 176:3231–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherman F. 2002. Getting started with yeast. Methods Enzymol 350:3–41. doi: 10.1016/S0076-6879(02)50954-X. [DOI] [PubMed] [Google Scholar]
- 18.Wolyniak MJ, Sundström P. 2007. Role of actin cytoskeletal dynamics in activation of the cyclic AMP pathway and HWP1 gene expression in Candida albicans. Eukaryot Cell 6:1824–1840. doi: 10.1128/EC.00188-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YL, Montedonico AE, Kauffman S, Dunlap JR, Menn FM, Reynolds TB. 2010. Phosphatidylserine synthase and phosphatidylserine decarboxylase are essential for cell wall integrity and virulence in Candida albicans. Mol Microbiol 75:1112–1132. doi: 10.1111/j.1365-2958.2009.07018.x. [DOI] [PubMed] [Google Scholar]
- 20.Morschhäuser J, Michel S, Staib P. 1999. Sequential gene disruption in Candida albicans by FLP-mediated site-specific recombination. Mol Microbiol 32:547–556. doi: 10.1046/j.1365-2958.1999.01393.x. [DOI] [PubMed] [Google Scholar]
- 21.Shen J, Guo W, Köhler JR. 2005. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect Immun 73:1239–1242. doi: 10.1128/IAI.73.2.1239-1242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. 2008. M27-A3 reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard, 3rd ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 23.Motulsky HJ, Christopoulos A. 2003. Fitting models to biological data using linear and nonlinear regression. A practical guide to curve fitting, p 256–289. GraphPad Software, Inc, San Diego, CA. [Google Scholar]
- 24.Shedletzky E, Unger C, Delmer DP. 1997. A microtiter-based fluorescence assay for (1,3)-beta-glucan synthases. Anal Biochem 249:88–93. doi: 10.1006/abio.1997.2162. [DOI] [PubMed] [Google Scholar]
- 25.Munro CA, Selvaggini S, de Bruijn I, Walker L, Lenardon MD, Gerssen B, Milne S, Brown AJ, Gow NA. 2007. The PKC, HOG and Ca2+ signalling pathways coordinately regulate chitin synthesis in Candida albicans. Mol Microbiol 63:1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Köhler JR, Fink GR. 1996. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci U S A 93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, Edwards JE, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog 2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selvaggini S, Munro CA, Paschoud S, Sanglard D, Gow NA. 2004. Independent regulation of chitin synthase and chitinase activity in Candida albicans and Saccharomyces cerevisiae. Microbiology 150:921–928. doi: 10.1099/mic.0.26661-0. [DOI] [PubMed] [Google Scholar]
- 29.Janbon G, Sherman F, Rustchenko E. 1998. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc Natl Acad Sci U S A 95:5150–5155. doi: 10.1073/pnas.95.9.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker LA, Gow NA, Munro CA. 2010. Fungal echinocandin resistance. Fungal Genet Biol 47:117–126. doi: 10.1016/j.fgb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kravets A, Qin H, Ahmad A, Bethlendy G, Gao Q, Rustchenko E. 2010. Widespread occurrence of dosage compensation in Candida albicans. PLoS One 5:e10856. doi: 10.1371/journal.pone.0010856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heilmann CJ, Sorgo AG, Mohammadi S, Sosinska GJ, de Koster CG, Brul S, de Koning LJ, Klis FM. 2013. Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot Cell 12:254–264. doi: 10.1128/EC.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enloe B, Diamond A, Mitchell AP. 2000. A single-transformation gene function test in diploid Candida albicans. J Bacteriol 182:5730–5736. doi: 10.1128/JB.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.