SUMMARY
Cells assemble and maintain functionally distinct actin cytoskeleton networks with various actin filament organizations and dynamics through the coordinated action of different sets of actin binding proteins. The biochemical and functional properties of diverse actin binding proteins, both alone and in combination, have been increasingly well studied. Conversely, how different sets of actin binding proteins properly sort to distinct actin filament networks in the first place is not nearly as well understood. Actin binding protein sorting is critical for the self-organization of diverse dynamic actin cytoskeleton networks within a common cytoplasm. Using in vitro reconstitution techniques including biomimetic assays and single molecule multi-color TIRF microscopy, we discovered that sorting of the prominent actin bundling proteins fascin and α-actinin to distinct networks is an intrinsic behavior, free of complicated cellular signaling cascades. When mixed, fascin and α-actinin mutually exclude each other by promoting their own recruitment and inhibiting recruitment of the other, resulting in the formation of distinct fascin- or α-actinin-bundled domains. Subdiffraction-resolution light microscopy and negative staining electron microscopy revealed that fascin domains are densely packed, while α-actinin domains consist of widely spaced parallel actin filaments. Importantly, other actin binding proteins such as fimbrin and espin show high specificity between these two bundle types within the same reaction. Here we directly observe that fascin and α-actinin intrinsically segregate to discrete bundled domains that are specifically recognized by other actin binding proteins.
Graphical abstract
INTRODUCTION
Cells simultaneously assemble and disassemble multiple actin filament (F-actin) networks from a common pool of proteins [1]. The filament organization and dynamics of these networks are tailored to facilitate different fundamental cellular processes such as motility, cytokinesis and endocytosis. Functionally diverse F-actin networks are optimized to perform specific tasks through the coordinated actions of numerous actin binding proteins (ABPs) with complementary biochemical properties [1]. F-actin networks obtain their particular properties by recruiting unique sets of ABPs that specifically shape the networks by directly regulating the F-actin geometric organization, turnover dynamics and protein composition [2].
Although the behavior of individual ABPs has been well studied, we know far less about how they segregate to the proper F-actin network in the first place. At least three non-mutually exclusive mechanisms have been suggested [2]. First, signaling cascades, such as through Rho family GTPases, control the formation of different types of networks by regulating ABPs. For example, the Arp2/3 complex nucleation promoting factors WASP/WAVE and Diaphanous-related formins induce branched or straight filament organizations in response to different signals [3, 4]. Second, the F-actin binding affinities of ABPs may be altered locally, such as through the local chemical environment (e.g. pH or ion concentration). Third, actin filaments within different networks may selectively bind different ABPs through recognition of different F-actin features (e.g. branched vs. straight architecture) or specific F-actin conformations [2, 5–7].
Metazoan cells contain two general types of bundled F-actin networks that display different mechanical properties as a consequence of their association with unique sets of ABPs. The first type of network contains tightly packed, parallel-aligned filaments that drive membrane protrusions such as filopodia, microvilli and stereocilia. These networks are associated with one or more compact bundlers such as fimbrin/plastin, espin/forked, and fascin [8]. The second type of network contains widely spaced filaments of mixed polarity. These networks make up contractile elements such as stress fibers, muscle sarcomeres and cytokinetic rings. They are often associated with the bundling protein α-actinin, an elongated but rigid homodimer [9].
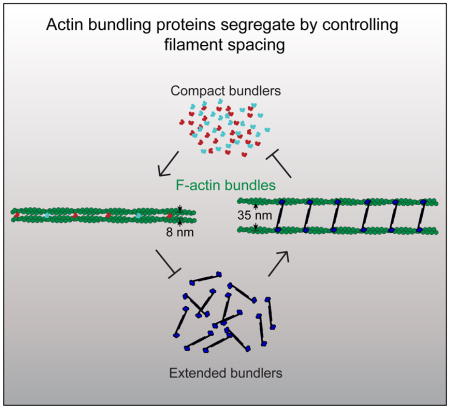
Since these bundling proteins facilitate the formation of F-actin networks with fundamentally different mechanical properties and vastly different architectures, we posit that their segregation could occur without the need for any complex signaling mechanisms. We demonstrate through in vitro reconstitution and multi-color Total Internal Reflection Fluorescence Microscopy (TIRFM) imaging that self-segregation is an intrinsic property of fascin and α-actinin. Fascin and α-actinin compete for F-actin not through simple mass action competition, but rather they each drive the formation of F-actin bundles that enhance their own recruitment while excluding the other. Therefore, there is a segregation mechanism intrinsically built into these classes of bundlers driving them to different types of F-actin networks. Additionally, we show that other ABPs discriminate between these types of bundles, likely through recognition of a structural or architectural feature of the F-actin bundle. We provide evidence that suggests this feature is the spacing between the actin filaments.
RESULTS
Fascin and α-actinin bundle F-actin cooperatively and with similar efficiency
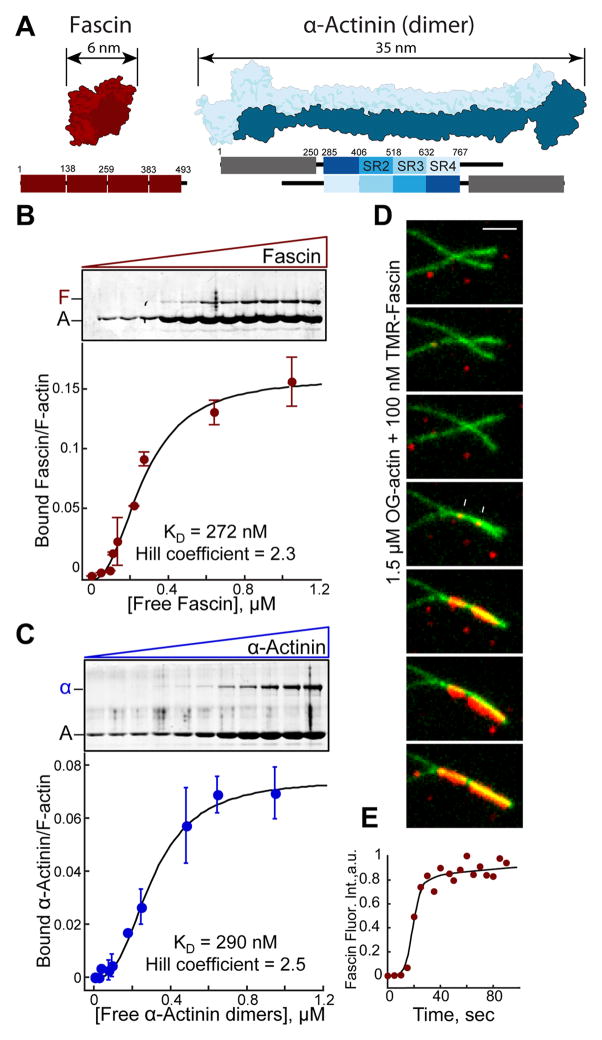
Fascin1 and α-actinin-4 are widely expressed F-actin bundling proteins with very different structural organization. Fascin is a globular protein (diameter ~5–6 nm) that uses four tandem β-trefoil domains to bind F-actin (Figure 1A) [10–13], whereas α-actinin is a dimer consisting of two calponin homology F-actin binding domains separated by a rod formed of spectrin repeats producing an elongated molecule on the order of 35 nm (Figure 1A) [14–16]. Given that fascin and α-actinin are found at very different types of bundled F-actin networks in cells, we considered whether their segregation is an intrinsic property or requires other cellular factors. Therefore, we analyzed their behavior when mixed in F-actin bundling assays in vitro. We first investigated the bundlers alone. Fascin and α-actinin bundle F-actin with moderate cooperativity (Hill coefficient: 2.3 and 2.5) and with similar bundling affinity (KD = 272 and 290 nM) in low speed sedimentation assays (Figures 1B and 1C). Cooperative F-actin bundling was directly visualized with two-color TIRFM assays. Fascin-mediated bundling initiates slowly from discrete spots (Figure 1D), but then progresses rapidly, with fascin intensity saturating within seconds (Figure 1E). This suggests two limiting steps in bundling, whereby filaments must come into close proximity by diffusion and bundler concentration must be above a critical threshold. This bundling threshold is ~25 to 50 nM for fascin and α-actinin, since lower concentrations form transient bundles that fall apart rapidly (not shown). Although structurally very different, we conclude that Fascin and α-actinin bundle F-actin cooperatively and with similar efficiency.
Figure 1. Fascin and α-actinin each bundle F-actin cooperatively.
(A) Structural fold (top) and domain organizations (bottom) showing the four β-trefoil domains of fascin (PDB #3P53; [11]) (left) and an α-actinin dimer (PDB #1SJJ; [15]) (right). ABD, Actin-binding domain; SR, spectrin repeat.
(B and C) Low speed (10,000 × g) sedimentation assays. F-actin was assembled from 3.0 μM Mg-ATP actin monomers with increasing concentrations of (B) fascin or (C) α-actinin. Coomassie-stained SDS-PAGE gels (top), and graphs (bottom) of the amount of actin in pellets. Error bars indicate SEM; n=2.
(D and E) Two-color TIRFM of 1.5 μM actin (15% Oregon green-actin) and 100 nM TMR-fascin.
(D) Merged time-lapse micrographs (Scale bar=2 μm). Arrows indicate two initial TMR-fascin loading events.
(E) Accumulation of TMR-fascin on the actin bundle over time.
Fascin and α-actinin segregate to different F-actin networks on biomimetic beads
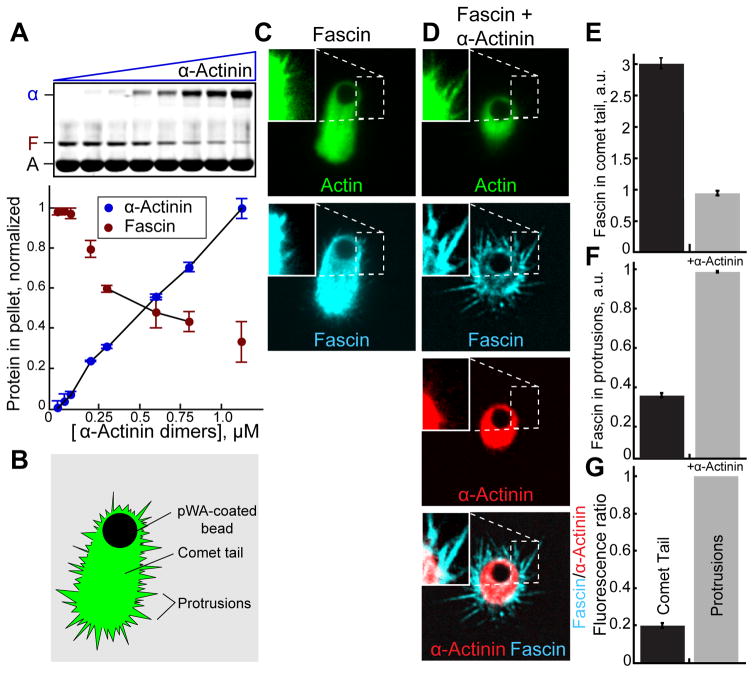
Since fascin and α-actinin make bundles with unique properties and associate with different F-actin networks in cells, we tested whether they compete for F-actin in simplified systems using purified components. When mixed in sedimentation assays, increasing concentrations of α-actinin reduce the amount of fascin in F-actin bundles, revealing a competitive relationship (Figure 2A). We then asked whether fascin and α-actinin intrinsically segregate to different F-actin networks assembled in vitro, free of cellular signaling networks (Figures 2B–2G) [17]. We generated a mixture of short-branched and protruding filopodia-like F-actin networks on biomimetic beads (Figure 2B). Actin (1% Oregon green-actin) was assembled from beads coated with the Arp2/3 complex activator GST-pWA in reactions containing capping protein, profilin and fascin in either the absence or presence of α-actinin. In these conditions beads form long comet tails (Figures 2B and 2C) and become motile (not shown). The periphery contains protrusive F-actin networks that are detectable with enhanced image brightness (Figure 2C, inset). In the absence of α-actinin, Cy5-fascin associates with both networks (Figure 2C). When TMR-α-actinin is included in the reaction, comet tails are shorter and TMR-α-actinin localizes more intensely to comet tails than protrusions (Figure 2D). Importantly, in the presence of α-actinin, fascin is depleted in comet tails and enriched in protrusions (Figures 2D–2G). Therefore α-actinin and fascin appear to have intrinsic properties that are sufficient to drive their segregation in the absence of the complex cellular environment. Because we are observing large-scale network effects at low resolution, and filament architecture is complex, the mechanism(s) driving segregation is difficult to elucidate using biomimetic bead assays.
Figure 2. Fascin and α-actinin compete and segregate to different F-actin networks reconstituted on motile beads.
A) Low speed (10,000 × g) sedimentation of F-actin assembled from 3.0 μM Mg-ATP actin monomers with 600 nM fascin and a range of α-actinin concentrations. Coomassie-stained SDS-PAGE gel (top), and graph (bottom) of the amount of actin (A), fascin (F) or α-actinin (α) in pellets. Error bars indicate SEM; n≥2.
(B–G) Addition of GST-pWA-coated beads to polymerization reactions containing 4 μM Mg-ATP actin (1% Oregon green-actin), 5 μM profilin, 100 nM Arp2/3 complex, 20 nM capping protein (CP), 150 nM Cy5-fascin (blue), with and without 50 nM TMR-α-actinin (red).
(B) Cartoon of comet tail and protrusive networks formed on a motile bead.
(C and D) Fluorescent confocal micrographs of beads in the presence of (C) Cy5-fascin or (D) Cy5-fascin and TMR-α-actinin. Shown are representative single planes taken from the center of a confocal z-stack with brightness enhanced in the inset for visualization of protrusions. Scale bar=2 μm.
(E and F) Comparison of the ratio of Cy5-fascin to Oregon green-actin fluorescence in (E) comet tails or (F) protrusions in the absence and presence of α-actinin. Error bars indicate SE; n=2 experiments with >20 beads each.
(G) Ratio of Cy5-Fascin to TMR-α-actinin fluorescence in protrusions and comet tails.
Fascin and α-actinin separate into mutually exclusive domains on F-actin bundles
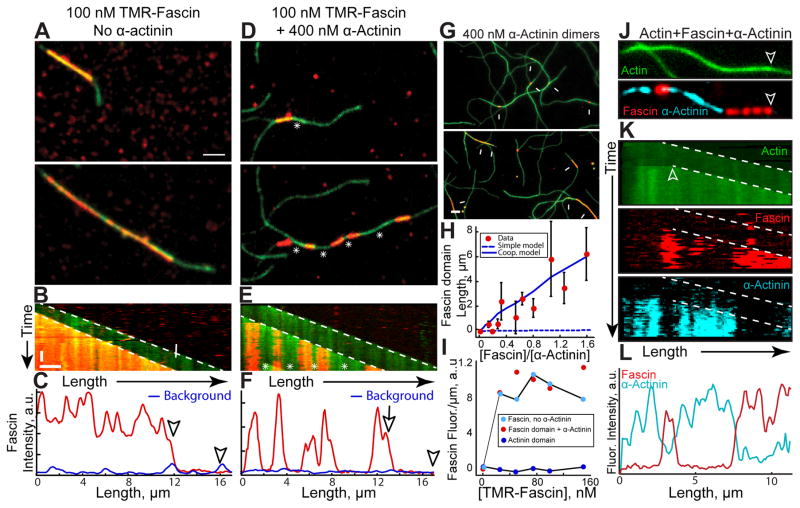
To better understand the mechanisms by which α-actinin and fascin segregate, we employed multi-color TIRFM to directly visualize bundle formation of elongating actin filaments (15% Oregon green-actin). To simplify analysis, we looked exclusively at two-filament bundle formation. Two-filament bundles contain unbundled leading barbed ends (marked by arrowheads in all figures) and trailing barbed ends (marked by arrows) of the bundle (see Figure 3A). With TMR-fascin alone, the trailing barbed end is rapidly bundled, as there is no resolvable lag between TMR-fascin signal and the elongating trailing barbed end (Figures 3A and 3B; Movie S1). TMR-fascin associates with the entire length of the two-filament bundle (Figures 3A–3C). Conversely, when unlabeled α-actinin was included in the reaction, TMR-fascin separates into discrete domains of the two-filament bundle (Figures 3D–3F; Movie S1). In the presence of α-actinin, fascin-decorated two-filament bundles are punctuated by long stretches of bundle devoid of fascin (Figures 3D and 3E, asterisks). Line scans revealed that TMR-fascin signal within these undecorated regions is not above levels associated with either single filaments or background binding (Figure 3F). Elongation rates of the trailing filament barbed ends are reduced ~10% in Fascin domains, but not α-actinin domains (Figure S1), perhaps because of a structural difference in fascin bundles [18, 19], or the dense packing of filaments in fascin bundles blocks access of actin monomers. Intrinsic segregation is a conserved property because TMR-fascin also forms domains in the presence of C. elegans α-actinin (Figure S2A–S2B, and Movie S2).
Figure 3. Fascin and α-actinin segregate into mutually exclusive domains on two-filament bundles.
(A–I) Two-color TIRFM of 1.5 μM actin (15% Oregon green-actin) with TMR-fascin (red).
(A–F) 100 nM TMR-fascin (red) in the absence (A–C) or presence (D–F) of 400 nM α-actinin dimers. Arrowheads and arrows mark leading and trailing barbed ends of parallel two-filament bundles. See also Movie S1 and S2
(A, D) Merged time-lapse micrographs. Scale bar: 2 μm.
(B, E) Merged kymographs of filament length (x-axis scale bar=2 μm) over time (y-axis scale bar=50 sec).
(C, F) Line scans of TMR-fascin intensity across the two-filament bundles at 300 sec. Blue line scans were generated from fascin intensity on single filaments.
(G–I) 400 nM α-actinin dimers with a range of TMR-fascin (red) concentrations.
(G) Merged micrographs in the presence of 50 nM (top) or 500 nM (bottom) TMR-fascin. Scale bar: 2 μm. Arrows indicate TMR-fascin domains on two-filament bundles.
(H) Dependence of TMR-fascin domain length on TMR-fascin concentration. Error bars indicate SEM; n=2 experiments each containing >10 domains. Dashed blue line indicates domain sizes predicted from a simple kinetic model of competition for binding sites, whereas the solid blue line includes a kinetic barrier to exchange of 4.8 kBT (see supplemental experimental procedures).
(I) Dependence of TMR-fascin intensity on TMR-fascin concentration within pure fascin bundles (cyan circles), fascin domains (red circles) or α-actinin domains (blue circles).
(J–L) Three-color TIRFM of 1.5 μM actin (15% Oregon green-actin), 200 nM TMR-fascin (red) and 400 nM Cy5-α-actinin dimers (cyan). See also Movie S3
(J) Micrographs of actin (top) with fascin and α-actinin (bottom). Scale bar=2 μm. Arrows indicate leading edge of a two-filament bundle.
(K) Kymographs of filament length (x-axis scale bar=2 μm) over time (y-axis scale bar=50 sec) for Oregon green-actin (top), TMR-fascin (middle) and Cy5-α-actinin (bottom). Arrowheads and arrow indicate leading and trailing barbed ends.
(L) Line scan of TMR-fascin and Cy5-α-actinin intensity across the two-filament bundle from J. See also Figure S1, S2, S3, S4 and S5.
While Fascin and α-actinin may directly compete for binding sites, domain formation cannot be fully explained by this type of simple mass action competition. Control reactions with both TMR- and Cy5-fascin illustrate that binding site competition does not produce resolvable domains (Figures S2C and S2D). We next mixed α-actinin with a range of TMR-fascin concentrations to determine the dependence of fascin domain length on TMR-fascin concentration. Fascin domain length is exponentially distributed (Figure S2E and S2F), and higher TMR-fascin concentrations form longer domains as expected (Figures 3G and 3H, Figure S2E and S2F). Fascin domains are significantly longer than those predicted from a Monte Carlo simulation based on simple mass action kinetics (Figure 3H, dashed blue line), whereas including cooperativity by adding an energetic barrier of 4.8 kBT for switching from one type of crosslinker to the other gives a reasonable fit to the experimental data (Figure 3H, solid blue line, and Figure S3) (see methods). Interestingly, although the fascin domains become longer, the density of fascin within the domains stays constant as TMR-fascin concentration increases (Figure 3I). Since increasing the ratio of fascin to α-actinin increases domain length while having no effect on the density of fascin binding within domains, we conclude that competition occurs at the leading edge of the trailing barbed end as the two-filament bundle assembles rather than within a domain once it is formed.
To better understand the intrinsic sorting of α-actinin and fascin we used three-color TIRFM to simultaneously visualize both bundlers on two-filament actin bundles (Figures 3J–3L; Movie S3). Indeed, fascin and α-actinin segregate to mutually exclusive sections of two-filament F-actin bundles. Whereas fascin only binds parallel bundles, α-actinin also binds antiparallel bundles and weakly to single filaments (Figure S4). We examined single molecule dynamics of fascin and α-actinin on bundles to determine the basis for enhanced association with particular domains (Figure S5). Both fascin and α-actinin are highly dynamic, with similar residence times of ~5 sec on two-filament bundles (Figures S5D and S5E). Although we observed α-actinin molecules binding to single filaments and fascin-bundled domains, we failed to calculate a reliable koff for these binding modes due to the presence of high background in those experiments. On the other hand, we never observed association of fascin with single filaments or α-actinin-bundled domains, even at very fast imaging rates (10 frames/sec) (Figures S5A–S5C). When mixed with α-actinin, single molecules of Cy5-fascin were only observed binding to TMR-fascin domains (Figures S5A–S5C). We conclude that fascin and α-actinin are dynamic bundling proteins and fascin is highly selective for fascin- over α-actinin-bundled domains.
Fascin and α-actinin form bundles with different actin filament spacing
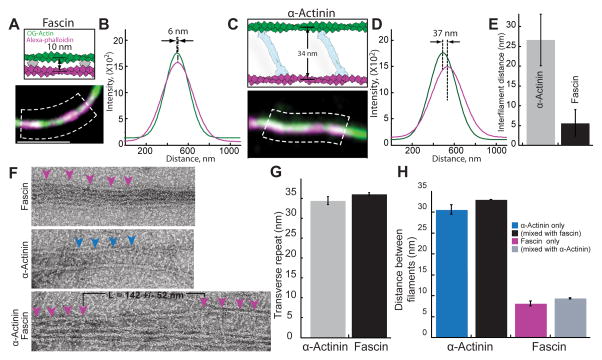
How do fascin and α-actinin mutually exclude each other on F-actin bundles? Ultrastructure studies revealed that fascin and α-actinin form bundles with filaments spaced significantly differently (~8 and ~35 nm respectively) [11, 14]. We therefore visualized two-filament bundles to determine filament spacing in our assays containing mixtures of α-actinin and fascin (Figure 4). First, we employed subdiffraction-resolution light microscopy on two-filament bundles generated by assembling new actin monomers (33% Oregon green-actin) onto preassembled filaments labeled with alexa-647-phalloidin (magenta) (Figures 4A and 4C). Filament distances were measured by fitting a Gaussian function to each filament peak from line scans perpendicular to the bundle (Figures 4A–4D), similar to a previously described method [20]. The average filament distances are ~5 and ~25 nm for fascin and α-actinin domains (Figure 4E), which are probably underestimated because bundles may not always lie flat on the surface. We also visualized bundles by electron microscopy of negatively stained actin filaments (Figures 4F–4H). On their own, bundles are formed with the expected spacing of ~8 nm for fascin and ~32 nm for α-actinin [11, 14]. The distance between bound fascin or α-actinin molecules is about 35 nm, corresponding to a turn of F-actin (Figures 4F and 4G). When fascin and α-actinin were mixed together we observed transitions from filaments spaced 8 nm apart by fascin to 32 nm apart by α-actinin (Figure 4F, lower panel). As expected, we saw the same characteristic interfilament distances when both bundlers were present in the same reaction (Figure 4H).
Figure 4. Actin filaments are spaced differently in bundles formed by fascin and α-actinin.
(A–E) Three-color TIRFM of bundles formed with 100 nM TMR-fascin and 400 nM α-actinin between a preassembled filament stabilized with alexa-647-phalloidin (magenta) and 1.5 μM spontaneously assembled Mg-ATP actin (33% Oregon green-actin).
(A and C) Schematics of predicted outcomes (top) [8, 14], and merged micrographs of two-filament bundles (bottom), for (A) fascin and (C) α-actinin. Scale bar=2 μm.
(B and D) Plots of average fluorescence intensities across a normal line (white lines in dashed boxes (A) and (C)) for two-filament bundles formed by (B) fascin and (D) α-actinin. Distances between the centroids of the bundled filaments were revealed by curve fits to a Gaussian.
(E) Average distance between filaments within α-actinin or fascin domains. Error bars indicate SEM; n≥6 bundles.
(F–H) Electron microscopy of F-actin bundles negatively stained with uranyl acetate, which were formed from 1.5 uM actin.
(F) Micrographs of bundles with 1 μM fascin (top), 800 nM α-actinin (middle) or both (1 μM α-actinin 0.25 μM Fascin (bottom). Magenta and blue arrowheads indicate fascin and α-actinin molecules. L = the length of the transition zone. Scale bar=30 nm.
(G) Distance of transverse repeat in fascin and α-actinin bundles. Error bars indicate SEM; n≥10 bundles.
(H) Distance between filaments in a fascin and α-actinin bundles. Error bars indicate SEM; n≥8 bundles.
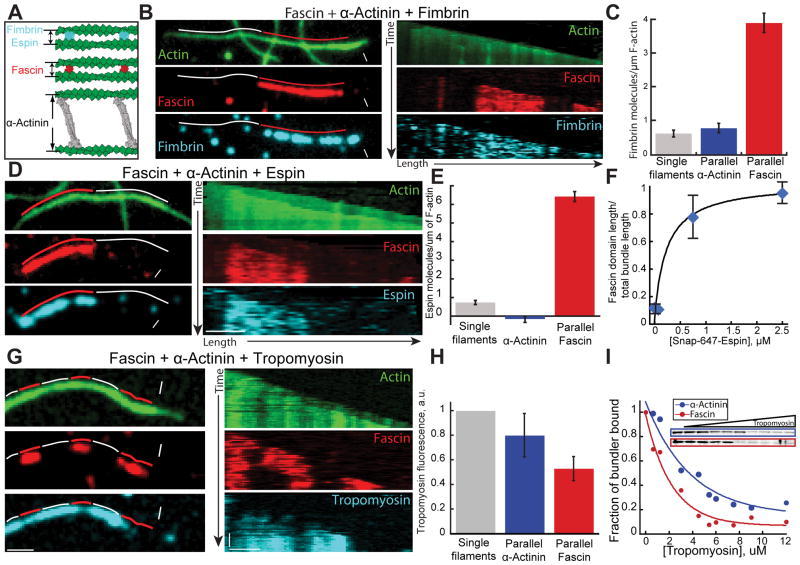
Fimbrin and Espin bind to fascin but not α-actinin bundles
To test the idea that filament spacing is important for segregation of fascin and α-actinin, we investigated other F-actin bundling proteins that often co-segregate with fascin in cells. Fimbrin is a globular protein like fascin with an actin bundling core ~6–8 nm in diameter (Figure 5A) [21], and is also often present with fascin in densely packed protrusive actin structures like microvilli, filopodia and hair cell stereocilia [22, 23]. However, fimbrin is actually in the same bundling protein family as α-actinin because they share a conserved tandem CH actin binding domain (ABD) [24], and evidence suggests they stabilize a similar actin filament structural conformation [25–27]. An important difference between fimbrin and α-actinin, is that α-actinin’s ABDs are separated by a long (~25 nm) spectrin repeat rod (Figure 5A) [28]. We first looked at fimbrin and fascin in the absence of α-actinin using three-color TIRF microscopy. Fascin and fimbrin do not mutually exclude each other, but instead colocalize on parallel two-filament bundles (Figure S6A). Fascin binding to parallel bundles decreases as a function of TMR-fimbrin concentration, but we never observed segregation of fascin and fimbrin into domains (Figure S6A and S6B). We hypothesized that fimbrin and fascin colocalize on two-filament bundles due to their similar size (5–8 nm diameter), and that fimbrin, like fascin, does not bind well to bundles facilitated by α-actinin. Therefore, we used three-color TIRFM to follow the assembly of actin (15% Oregon green-actin) with TMR-fascin, unlabeled α-actinin and Cy5-fimbrin (Figures 5B and 5C; Movie S4). Six-fold more fimbrin associates with fascin bundles than with parallel α-actinin bundles (Figures 5B and 5C). A low amount of fimbrin does associate with α-actinin bundles, but this is likely due to association with single filaments given that fimbrin binds similarly to α-actinin parallel bundles and single filaments (Figure 5C). To determine if this segregation feature extends to other proteins that generate compact bundles, we also looked at espin using the same three-color TIRFM assay. Espin associates with protrusive F-actin structures, and in hair-cell stereocilia it colocalizes with both fimbrin and fascin to generate tight parallel bundles [29], (Figure 5A). In the absence of α-actinin, espin isoform 2B colocalizes (and competes at high concentrations) with fascin in parallel bundles (data not shown). When α-actinin is included in the reaction, Cy5-espin 2B selectively associates with fascin domains over α-actinin domains (Figures 5D–5F; Movie S5). Additionally, espin 2B enhances fascin’s ability to compete with α-actinin. As increasingly more espin 2B is included in the reaction, the fascin domain length expands (Figure 5F). We conclude that fimbrin, espin 2B and fascin recognize a similar bundle type that is different from those formed by α-actinin, which facilitates their intrinsic segregation into distinct bundled domains. Importantly, fimbrin, Espin and α-actinin all bind to single filaments with a much lower affinity than bundled filaments.
Figure 5. Fimbrin and espin segregate to filaments bundled by fascin.
Three-color TIRFM of two-filament bundles formed by fascin and α-actinin with (A–C) fimbrin, (D–F) espin, or (G and H) muscle tropomyosin. Red and white lines indicate fascin and α-actinin domains. White arrowheads and arrows indicate leading and trailing barbed ends.
(A) Schematic of bundles formed by fimbrin, espin, fascin and α-actinin [8, 14].
(B–C) 1.5 μM actin (15% Oregon green-actin), 400 nM α-actinin dimers, 75 nM Cy5-fascin (red) and 15 nM TMR-fimbrin (Cyan). See also Movie S4
(B) Micrographs (left) and corresponding kymographs (right) of a parallel two-filament bundle, with Oregon green-actin (top), TMR-fascin (middle) and Cy5-fimbrin (bottom). Scale bars=2 μm.
(C) Cy5-fimbrin molecules on single filaments (n=6), two-filament parallel α-actinin bundles (n=23) and two-filament fascin bundles (n=16). Error bars indicate SEM.
(D–F) 1.5 μM actin (15% OG-labeled), 100 nM Cy5-fascin (red), 400 nM α-actinin dimers and a range of concentrations of SNAP-647-espin-2B (Cyan). See also Movie S5
(D) Micrographs (left) and corresponding kymographs (right) of a parallel two-filament bundle, with Oregon green-actin (top), TMR-fascin (middle) and SNAP-647-espin-2B (bottom). Scale bars=2 μm.
(E) Number of SNAP-647-espin-2B molecules per μm of single filament (n=15), parallel α-actinin bundles (n=15) or fascin bundles (n=17). Error bars indicate SEM.
(F) Ratio of the total domain length of TMR-fascin two-filament parallel bundles over the total length of two-filament parallel bundles as a function of SNAP-647-espin-2B concentration. Error bars indicate SEM; n=2 movies.
(G–H) 1.5 μM actin (10% Alexa-488-labeled), 100 nM Cy5-fascin (red), 150 nM α-actinin and 100 nM skeletal muscle α/β TMR-tropomyosin (Cyan).
(G) Micrographs (left) and corresponding kymographs (right) of a parallel two-filament bundle, with Alexa-488-actin (top), Cy5-fascin (middle) and TMR-tropomyosin (bottom). Scale bars = 2 μm.
(H) Normalized Tropomyosin fluorescence on control actin filament, α-actinin and Fascin parallel bundles. Error bars indicate SEM.
(I) High speed co-sedimentation assays with 5 μM Mg-ATP actin, 600 nM fascin (blue) or α-actinin (red), and an increasing concentration of skeletal muscle α/β tropomyosin. Gels (inset) show fascin or α-actinin depletion from the pellet with increasing tropomyosin. Graph is the fraction of total fascin or α-actinin in the pellet as a function of tropomyosin concentration. Normalized curves were fit to a single exponential decay function.
See also Figure S6
Fascin, α-actinin, espin and fimbrin all have a significantly higher affinity for bundled filaments than single filaments (for example, see Figure 1D). A sorting model based on filament spacing distance predicts that ABPs that bind only one filament would not be sensitive to spacing. We tested this possibility by looking at labeled skeletal muscle α/β tropomyosin. While tropomyosin competes with and inhibits both fascin and α-actinin binding at high concentrations (Figure 5I), at lower concentrations tropomyosin binds to both fascin and α-actinin domains as predicted (Figure 5G and 5H).
DISCUSSION
Fascin and α-actinin contain intrinsic properties that drive their segregation
Our work directly shows that α-actinin and fascin contain intrinsic properties that drive their segregation in the absence of complex cellular signaling pathways. It has been appreciated that actin filaments within different networks are not identical because combinations of different ABPs promote particular filament and network properties. Here we show that bundlers do not generically crosslink filaments together, but rather, generate specific bundled features that result in both cooperative and competitive binding effects that influence the association of other ABPs.
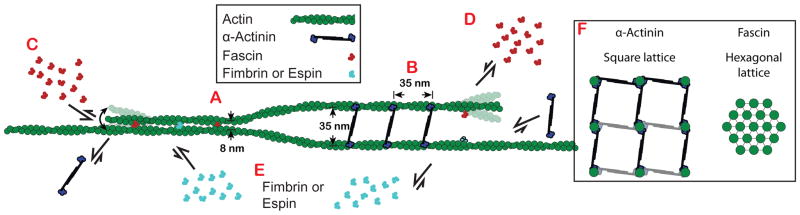
Mechanism of segregation
Bundlers generally bind to bundled F-actin with a much higher affinity than single filaments, presumably because within the particular bundles, both actin-binding sites are easily engaged to adjacent filaments. We propose that filament spacing within bundles may facilitate the engagement of both binding sites for specific bundlers, and therefore recruit or inhibit the binding of bundler classes. α-Actinin-4 and fascin 1 both bind in exclusive runs that are ~50x longer than predicted by simulations based on simple mass action competition for binding sites (Figure 3H). Our results could be explained by a mechanism whereby α-actinin and fascin specifically promote their own binding by stabilizing different filament structural conformations [2]. However, fimbrin may actually stabilize an F-actin conformation similar to α-actinin [23–25], yet they segregate from each other on two-filament bundles, suggesting that individual filament conformation is not the primary mechanism of sorting. Therefore, we hypothesize that filament spacing, rather than filament conformation, is the dominant feature for several reasons (Figure 6). First, both fimbrin and espin make very condensed bundles with similar filament spacing to fascin bundles (~10 nm), and both are highly selective for fascin bundles over α-actinin bundles. Second, fimbrin and espin display affinities for α-actinin two-filament bundles that are similar to their affinities for single, non-bundled filaments (Figures 5C and 5E), which suggests that these bundlers associate with only one of the filaments within the two-filament α-actinin bundle. Third, filaments would need to be highly bent to accommodate heterogeneous binding of long and short bundlers (Figures 6C and 6D). The interfilament spacing of an α-actinin bundle (Figure 6A) is about the same distance (~35 nm) that separates binding sites along a filament (Figure 6B). Fascin bundles the trailing filament rapidly (Figure 1E; Figures 3A–F), and as new binding sites appear on growing filaments (~35 nm from last binding site) they are rapidly bound by fascin (Figure 6C). This rapid binding can be observed in the cooperative binding of these bundlers (Figures 1B–1E), which is likely due to the newly assembled binding site being well-oriented for fascin to make contacts with both filaments. Conversely, due to their small size, fascin (and other small bundlers) can only associate with one of the two filaments in an α-actinin bundle (Figure 6D). Small bundlers like fimbrin and espin can exchange readily within fascin bundles where they can make connections to both filaments (Figure 6E).
Figure 6. Model for segregation of fascin and α-actinin on F-actin bundles.
(A) Fascin (red) and α-actinin (blue) form bundles with interfilament spacing of ~8 and ~35 nm, respectively [11, 14].
(B) The transverse repeat (distance between proteins along the length within a bundle) of both fascin and α-actinin is ~35 nm, about the same distance as a complete turn of the actin filament.
(C) Fascin molecules add cooperatively to a polymerizing fascin bundle, whereas α-actinin’s affinity for fascin bundles is much lower.
(D) α-actinin molecules add cooperatively to a polymerizing α-actinin bundle, whereas fascin molecules do not.
(E) Short filament spacing bundlers such as fimbrin and espin (cyan) associate well within fascin bundles, but not α-actinin bundles.
(F) Fascin and α-actinin drive the formation of square- and hexagonally-packed multifilament bundles
Scaling of segregation to multifilament bundles
Much of our work is based on two-filament bundles. Although we do not know how these principles scale with bundles containing greater than two filaments, we found that these proteins segregate efficiently in our reconstituted biomimetic system where bundles presumably contain many filaments (Figures 2B–2G). However, it is difficult in the biomimetic system to know the mechanism of segregation. Looking at two filament bundles shows that these bundlers segregate intrinsically, and we therefore reasonably speculate that similar mechanisms are involved in the biomimetic assay. However, filaments in the different parts of the network may display different levels of alignment. Filaments in the comet tail may be more highly branched than those in protrusions, and α-actinin may crosslink unaligned filaments more efficiently than fascin [30]. Bundlers may also differently recognize branched filament architectures. For example, fimbrin does not efficiently bundle short-branched 2D networks [31], but we imagine that filament architectures are far more complicated in 3D networks.
There are several ways we imagine segregation could scale to multifilament bundles. First, a particular actin conformation could be recognized by ABPs that are incorporating new filaments into the bundle. Second, multifilament bundles pack into very distinct lattice structures. Fascin bundles at steady state have ~20–30 filaments in vitro and assemble into highly-ordered hexagonally-packed bundles [32] whereas α-actinin generates a square lattice with filaments spaced 30 nm apart [33] (Figure 6F). We imagine that either type of bundled structure is stable, whereas a mixed structure is not. α-Actinin crosslinks may impose unfavorable and energetically costly defects in hexagonally packed structures, and vice versa. Classic experiments showed that multiple compact bundlers coordinate to efficiently assemble large protrusive F-actin structures such as bristles and microvilli [8, 34]. For example, in the assembly of Drosophila bristles, the espin homolog forked is thought to generate F-actin bundles early on that are poised in an orientation to be maximally crosslinked by fascin [34].
We propose that filament spacing significantly contributes to the segregation of ABPs to different F-actin networks, which is a previously unappreciated mechanism. We imagine that other signals are necessary to initiate either fascin or α-actinin networks. Phosphorylation of both fascin and α-actinin regulates their bundling properties [9, 35], and α-actinin bundling is sensitive to Ca2+ [9]. Furthermore, actin assembly factors may generate filament conformations, and/or filament spacing, which promote binding of particular bundlers. For example, densely branched networks of short filaments generated by the Arp2/3 complex in lamellipodia may not be favorable to fascin binding, while formin and/or Ena/VASP activity may allow filaments to grow long enough to become bundled by fascin. For example, we recently discovered that Ena/VASP and fascin have a synergistic relationship [36]. Conversely, α-actinin can bind to Arp2/3 complex-generated networks in the lamellipodia [32], which could position it to become incorporated into transverse arcs [37]. However, our work shows that once fascin or α-actinin bundled networks are initiated, they promote their own binding along with specific sets of ABPs.
EXPERIMENTAL PROCEDURES
Protein purification and labeling
Fission yeast fimbrin SpFim1 was purified as described [38]. Human α-actinin-4, C. elegans α-actinin, SNAP-espin-2B was expressed in bacteria E. coli BL21-Codon Plus(DE3)-RP (Stratagene) cells and purified with affinity chromatography. Actin was purified from rabbit muscle acetone powder (Pel Freez Biologicals) and labeled on Cys374 with Oregon green (Life Technologies, Grand Island, NY) as described [39, 40]. Mouse capping protein, human fascin 1 and human profilin HPRO1 were expressed in bacteria and purified as described [41–43]. Arp2/3 complex was purified from calf thymus by WASp(VCA) affinity chromatography [44]. WASP fragment construct GST-human WASp pWA was purified by Glutathione-Sepharose affinity chromatography [45]. Human fascin and both Human and C. elegans α-actinin were labeled with either Cy5-Monomaleimide (GE Healthcare) or TMR-6-Maleimide (Life Technologies, Grand Island, NY). Skeletal muscle tropomyosin was purified from rabbit muscle acetone powder (Pel Freez Biologicals) and labeled as described [46]. Proteins containing SNAP fusions were labeled with SNAP-surface-549 or SNAP-surface-647 (New England BioLabs). Proteins were incubated with the dye overnight at 4°C according to manufacturer’s protocols.
Actin filament sedimentation assay
Actin filaments were preassembled for 45 min at 25°C from 15 μM Mg-ATP actin monomers. Assembled actin was then incubated with accessory protein(s) for 20 minutes at 25°C, and spun at 10,000g (low-speed) for 20 minutes at 25°C. In α-actinin-fascin competition assays, α-actinin was introduced first and incubated for 20 min and then fascin was added and incubated for an additional 20 min. Equal volumes of supernatant and pellet were separated by 12.5% SDS-polyacrylamide gel electrophoresis, stained with Coomassie Blue for 20 minutes, and destained for 16 hours. Gels were analyzed by densitometry on an Odyssey Infrared Imager (LI-COR Biosciences, Lincoln, NE).
TIRFM assay
TIRF microscopy and flow chamber construction was performed as described previously [36].
Bead motility assay
Polystyrene microspheres (Polysciences, Eppelheim, Germany) were coated with GST-pWA [47]. Motile beads were imaged after 15 minutes of polymerization on an inverted microscope (Ti-E; Nikon, Melville, NY) with a confocal scan head (CSU-X; Yokogawa Electric, Musashino, Tokyo, Japan), 491, 561, and 642 laser lines (Spectral Applied Research, Richmond Hill, Ontario, Canada) and an HQ2 CCD camera (Roper Scientific, Trenton, NJ). Z-stacks were acquired, reconstructed and analyzed using ImageJ. Fluorescence ratios were determined using the central, single plane of motile beads. Background-subtracted fluorescence values were measured for each fluorescent protein in the comet tail region and in the protrusion region. The fluorescence ratios (Figure 2E and 2F) were determined by dividing fascin fluorescence by actin fluorescence. The ratio for fascin/α-actinin fluorescence was determined similarly.
Interfilament distance measurement by TIRFM
To generate two-filament bundles where each filament was a different color, we first let 5 μM of Mg-ATP-actin assemble for 45 min at room temperature. These filaments were then incubated with an equimolar amount of Alexa-647-phalloidin, diluted in TIRF buffer and flowed into the chamber. We then initiated assembly of 1.5 μM actin, 33% OG-labeled, plus TMR-fascin and α-actinin and gently flowed this mixture into the chamber. A fraction of the bundles that formed contained one Alexa-647-filament and one OG-actin filament. If the two-filament bundle had TMR-signal we scored it as a fascin bundle, if no TMR-signal then we scored it as an α-actinin bundle. The distance between actin filaments within both fascin and α-actinin bundles is expected to be less than the pixel size (< 100 nm) of our images. To calculate this sub-pixel value we analyzed the data similar to a previously described method [20].
Negative staining electron microscopy
1.5 μM actin was polymerized with either 250–500 nM α-actinin, 1–2 μM fascin, or both bundling proteins for 30 minutes. This solution was then applied to Formvar and carbon-coated 400 mesh copper grids for 1 min, washed and negatively stained with 1% (w/v) uranyl acetate for 1 min, blotted, and dried. Visualization of the bundles using transmission electron microscopy was performed on a FEI Tecnai G2 Spirit microscope at 120 kV. Images were captured on a Gatan 2k × 2k CCD camera. Bundle parameters were measured using ImageJ.
Cooperative kinetic model
We choose to model the extension of the bundle as a simple kinetic process on a lattice of binding sites where only the next open site can be modified. For the data in Fig 3H, we created a lattice of 400 virtual binding sites, corresponding to two actin filaments coming together of length 400*37nm ~ 15 μm. In the simulations, there are two types of crosslinkers that can occupy a site, A and B. We associate with each a rate of addition to a bundle of like-type, and . In the case where a crosslinker of type A occupies a site, the rate of addition for crosslinker B is changed to , where kB is Boltzmann’s constant and T is temperature. Similarly, occupancy of site B means the addition rate of A will be changed to .
Supplementary Material
Acknowledgments
This work was supported by NIH R01 GM079265 and ACS RSG-11-126-01-CSM (to D.R.K.), NIH MCB Training Grant T32 GM0071832 (to A.J.H. and J.R.C.), NSF Graduate Student Fellowship DGE-1144082 (to A.J.H. and J.R.C.), NIH Ruth L. Kirschstein NRSA F32 GM113415-01 (to G.M.H.), and NIH R01 DC004314 (to J.R.B.). Additional support was provided to D.R.K. and G.A.V. by the Chicago MRSEC, which is funded by NSF through grant DMR-1420709. We thank Ben Glick and GSL Biotech for the use of SnapGene for plasmid construction, Jared Winkelman for helpful discussions, and Joe Austin and Yimei Chen of The University of Chicago Advanced Electron Microscopy Facility.
Footnotes
AUTHOR CONTRIBUTIONS
J.D.W., C.S. and D.R.K. designed research; J.D.W., C.S., G.M.H., A.J.H., A.N.M., J.R.C. and J.R.B. performed and analyzed research; J.D.W., C.S., G.M.H., G.A.V., J.R.B. and D.R.K. interpreted data; and J.D.W. and D.R.K. wrote the manuscript with input from C.S., G.M.H., G.A.V and J.R.B.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin Dynamics, Architecture, and Mechanics in Cell Motility. Physiological Reviews. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 2.Michelot A, Drubin DG. Building Distinct Actin Filament Networks in a Common Cytoplasm. Current Biology. 2011;21:R560–R569. doi: 10.1016/j.cub.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends in Cell Biology. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Michelot A, Costanzo M, Sarkeshik A, Boone C, Yates JR, Drubin DG. Reconstitution and protein composition analysis of endocytic actin patches. Curr Biol. 2010;20:1890–1899. doi: 10.1016/j.cub.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brawley CM, Rock RS. Unconventional myosin traffic in cells reveals a selective actin cytoskeleton. PNAS. 2009;106:9685–9690. doi: 10.1073/pnas.0810451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagy S, Rock RS. Structured Post-IQ Domain Governs Selectivity of Myosin X for Fascin-Actin Bundles. J Biol Chem. 2010;285:26608–26617. doi: 10.1074/jbc.M110.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartles JR. Parallel actin bundles and their multiple actin-bundling proteins. Current Opinion in Cell Biology. 2000;12:72–78. doi: 10.1016/s0955-0674(99)00059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foley KS, Young PW. The non-muscle functions of actinins: an update. Biochemical Journal. 2014;459:1–13. doi: 10.1042/BJ20131511. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Yang S, Jakoncic J, Zhang JJ, Huang XY. Migrastatin analogues target fascin to block tumour metastasis. Nature. 2010;464:1062–1066. doi: 10.1038/nature08978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansen S, Collins A, Yang C, Rebowski G, Svitkina T, Dominguez R. Mechanism of Actin Filament Bundling by Fascin. J Biol Chem. 2011;286:30087–30096. doi: 10.1074/jbc.M111.251439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sedeh RS, Fedorov AA, Fedorov EV, Ono S, Matsumura F, Almo SC, Bathe M. Structure, Evolutionary Conservation, and Conformational Dynamics of Homo sapiens Fascin-1, an F-actin Crosslinking Protein. Journal of Molecular Biology. 2010;400:589–604. doi: 10.1016/j.jmb.2010.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S, Huang FK, Huang J, Chen S, Jakoncic J, Leo-Macias A, Diaz-Avalos R, Chen L, Zhang JJ, Huang XY. Molecular Mechanism of Fascin Function in Filopodial Formation. J Biol Chem. 2013;288:274–284. doi: 10.1074/jbc.M112.427971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hampton CM, Taylor DW, Taylor KA. Novel Structures for α-Actinin:F-Actin Interactions and their Implications for Actin–Membrane Attachment and Tension Sensing in the Cytoskeleton. Journal of Molecular Biology. 2007;368:92–104. doi: 10.1016/j.jmb.2007.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Taylor DW, Taylor KA. A 3-D Reconstruction of Smooth Muscle α-Actinin by CryoEm Reveals Two Different Conformations at the Actin-binding Region. Journal of Molecular Biology. 2004;338:115–125. doi: 10.1016/j.jmb.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 16.de Ribeiro EA, Pinotsis N, Ghisleni A, Salmazo A, Konarev PV, Kostan J, Sjöblom B, Schreiner C, Polyansky AA, Gkougkoulia EA, et al. The Structure and Regulation of Human Muscle α-Actinin. Cell. 2014;159:1447–1460. doi: 10.1016/j.cell.2014.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Achard V, Martiel JL, Michelot A, Guérin C, Reymann AC, Blanchoin L, Boujemaa-Paterski R. A “Primer”-Based Mechanism Underlies Branched Actin Filament Network Formation and Motility. Current Biology. 2010;20:423–428. doi: 10.1016/j.cub.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 18.Claessens MMaE, Semmrich C, Ramos L, Bausch AR. Helical twist controls the thickness of F-actin bundles. PNAS. 2008;105:8819–8822. doi: 10.1073/pnas.0711149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin H, Drew KRP, Bartles JR, Wong GCL, Grason GM. Cooperativity and Frustration in Protein-Mediated Parallel Actin Bundles. Phys Rev Lett. 2009;103:1–4. doi: 10.1103/PhysRevLett.103.238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487:64–69. doi: 10.1038/nature11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein MG, Shi W, Ramagopal U, Tseng Y, Wirtz D, Kovar DR, Staiger CJ, Almo SC. Structure of the Actin Crosslinking Core of Fimbrin. Structure. 2004;12:999–1013. doi: 10.1016/j.str.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Bretscher A, Weber K. Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J Cell Biol. 1980;86:335–340. doi: 10.1083/jcb.86.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glenney JR, Kaulfus P, Matsudaira P, Weber K. F-actin binding and bundling properties of fimbrin, a major cytoskeletal protein of microvillus core filaments. J Biol Chem. 1981;256:9283–9288. [PubMed] [Google Scholar]
- 24.Matsudalra P. Modular organization of actin crosslinking proteins. Trends in Biochemical Sciences. 1991;16:87–92. doi: 10.1016/0968-0004(91)90039-x. [DOI] [PubMed] [Google Scholar]
- 25.Hanein D, Matsudaira P, DeRosier DJ. Evidence for a Conformational Change in Actin Induced by Fimbrin (N375) Binding. J Cell Biol. 1997;139:387–396. doi: 10.1083/jcb.139.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodgkinson JL, EL-Mezgueldi M, Craig R, Vibert P, Marston SB, Lehman W. 3-D image reconstruction of reconstituted smooth muscle thin filaments containing calponin: Visualization of interactions between F-actin and Calponin1. Journal of Molecular Biology. 1997;273:150–159. doi: 10.1006/jmbi.1997.1307. [DOI] [PubMed] [Google Scholar]
- 27.McGough A, Way M, DeRosier D. Determination of the alpha-actinin-binding site on actin filaments by cryoelectron microscopy and image analysis. J Cell Biol. 1994;126:433–443. doi: 10.1083/jcb.126.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ylänne J, Scheffzek K, Young P, Saraste M. Crystal Structure of the α-Actinin Rod Reveals an Extensive Torsional Twist. Structure. 2001;9:597–604. doi: 10.1016/s0969-2126(01)00619-0. [DOI] [PubMed] [Google Scholar]
- 29.Shin JB, Krey JF, Hassan A, Metlagel Z, Tauscher AN, Pagana JM, Sherman NE, Jeffery ED, Spinelli KJ, Zhao H, et al. Molecular Architecture of the Chick Vestibular Hair Bundle. Nat Neurosci. 2013;16:365–374. doi: 10.1038/nn.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courson DS, Rock RS. Actin Cross-link Assembly and Disassembly Mechanics for α-Actinin and Fascin. J Biol Chem. 2010;285:26350–26357. doi: 10.1074/jbc.M110.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skau CT, Courson DS, Bestul AJ, Winkelman JD, Rock RS, Sirotkin V, Kovar DR. Actin Filament Bundling by Fimbrin Is Important for Endocytosis, Cytokinesis, and Polarization in Fission Yeast. J Biol Chem. 2011;286:26964–26977. doi: 10.1074/jbc.M111.239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. The Journal of cell biology. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelletier O, Pokidysheva E, Hirst LS, Bouxsein N, Li Y, Safinya CR. Structure of actin cross-linked with α-actinin: a network of bundles. Physical review letters. 2003;91:1–4. doi: 10.1103/PhysRevLett.91.148102. [DOI] [PubMed] [Google Scholar]
- 34.Tilney LG, Connelly PS, Vranich KA, Shaw MK, Guild GM. Why Are Two Different Cross-linkers Necessary for Actin Bundle Formation In Vivo and What Does Each Cross-link Contribute? J Cell Biol. 1998;143:121–133. doi: 10.1083/jcb.143.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams JC. Roles of fascin in cell adhesion and motility. Current Opinion in Cell Biology. 2004;16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Winkelman JD, Bilancia CG, Peifer M, Kovar DR. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with Fascin. PNAS. 2014:4121–4126. doi: 10.1073/pnas.1322093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skau CT, Kovar DR. Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr Biol. 2010;20:1415–1422. doi: 10.1016/j.cub.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spudich JA, Watt S. The Regulation of Rabbit Skeletal Muscle Contraction I. BIOCHEMICAL STUDIES OF THE INTERACTION OF THE TROPOMYOSIN-TROPONIN COMPLEX WITH ACTIN AND THE PROTEOLYTIC FRAGMENTS OF MYOSIN. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 40.Kuhn JR, Pollard TD. Real-Time Measurements of Actin Filament Polymerization by Total Internal Reflection Fluorescence Microscopy. Biophysical Journal. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmgren S, Ojala PJ, Wear MA, Cooper JA, Lappalainen P. Interactions with PIP2, ADP-actin monomers, and capping protein regulate the activity and localization of yeast twinfilin. J Cell Biol. 2001;155:251–260. doi: 10.1083/jcb.200106157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vignjevic D, Yarar D, Welch MD, Peloquin J, Svitkina T, Borisy GG. Formation of filopodia-like bundles in vitro from a dendritic network. J Cell Biol. 2003;160:951–962. doi: 10.1083/jcb.200208059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu J, Pollard TD. Profilin Binding to Poly-l-Proline and Actin Monomers along with Ability to Catalyze Actin Nucleotide Exchange Is Required for Viability of Fission Yeast. Mol Biol Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier MF. Activation of the Cdc42 Effector N-Wasp by the Shigella flexneri Icsa Protein Promotes Actin Nucleation by Arp2/3 Complex and Bacterial Actin-Based Motility. J Cell Biol. 1999;146:1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-Related Protein, Activates Nucleation of Actin Filaments by the Arp2/3 Complex. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsiao JY, Goins LM, Petek NA, Mullins RD. Arp2/3 Complex and Cofilin Modulate Binding of Tropomyosin to Branched Actin Networks. Curr Biol. 2015;25:1573–1582. doi: 10.1016/j.cub.2015.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reymann AC, Suarez C, Guérin C, Martiel JL, Staiger CJ, Blanchoin L, Boujemaa-Paterski R. Turnover of Branched Actin Filament Networks by Stochastic Fragmentation with ADF/Cofilin. Mol Biol Cell. 2011;22:2541–2550. doi: 10.1091/mbc.E11-01-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.