ABSTRACT
For over 20 years, bacterial multidrug resistance (MDR) efflux pumps have been studied because of their impact on resistance to antimicrobials. However, critical questions remain, including why produce efflux pumps under non-antimicrobial treatment conditions, and why have multiple pumps if their only purpose is antimicrobial efflux? Salmonella spp. possess five efflux pump families, including the resistance-nodulation-division (RND) efflux pumps. Notably, the RND efflux pump AcrD has a unique substrate profile, distinct from other Salmonella efflux pumps. Here we show that inactivation of acrD results in a profoundly altered transcriptome and modulation of pathways integral to Salmonella biology. The most significant transcriptome changes were central metabolism related, with additional changes observed in pathogenicity, environmental sensing, and stress response pathway expression. The extent of tricarboxylic acid cycle and fumarate metabolism expression changes led us to hypothesize that acrD inactivation may result in motility defects due to perturbation of metabolite concentrations, such as fumarate, for which a role in motility has been established. Despite minimal detectable changes in flagellar gene expression, we found that an acrD mutant Salmonella enterica serovar Typhimurium isolate was significantly impaired for swarming motility, which was restored by addition of fumarate. The acrD mutant outcompeted the wild type in fitness experiments. The results of these diverse experiments provide strong evidence that the AcrD efflux pump is not simply a redundant system providing response resilience, but also has distinct physiological functions. Together, these data indicate that the AcrD efflux pump has a significant and previously underappreciated impact on bacterial biology, despite only minor perturbations of antibiotic resistance profiles.
IMPORTANCE
Efflux pumps in Gram-negative bacteria are studied because of their important contributions to antimicrobial resistance. However, the role of these pumps in bacterial biology has remained surprisingly elusive. Here, we provide evidence that loss of the AcrD efflux pump significantly impacts the physiology of Salmonella enterica serovar Typhimurium. Inactivation of acrD led to changes in the expression of 403 genes involved in fundamental processes, including basic metabolism, virulence, and stress responses. Pathways such as these allow Salmonella to grow, survive in the environment, and cause disease. Indeed, our data show that the acrD mutant is more fit than wild-type Salmonella under standard lab conditions. We hypothesized that inactivation of acrD would alter levels of bacterial metabolites, impacting traits such as swarming motility. We demonstrated this by exogenous addition of the metabolite fumarate, which partially restored the acrD mutant’s swarming defect. This work extends our understanding of the role of bacterial efflux pumps.
INTRODUCTION
Resistance-nodulation-division (RND) efflux pumps in Gram-negative bacteria confer intrinsic multidrug resistance (MDR) by exporting a broad range of antimicrobial compounds out of the bacterial cell. Traditionally, Enterobacteriaceae such as Escherichia coli and Salmonella have five families of efflux pumps, the ABC and MFS superfamilies and the SMR, MATE, and RND families (1). Recently, the PACE family of efflux pumps was discovered in Acinetobacter species and is present in a range of Proteobacteria (2, 3). Salmonella enterica serovar Typhimurium (here, Salmonella Typhimurium) is used extensively as a model pathogen for many reasons, including easy genetic manipulation, good infection models, and relevance to other pathogens (4, 5). Salmonella Typhimurium has five RND MDR efflux systems: AcrAB, AcrAD, AcrEF, MdtABC, and MdsABC (5, 6). AcrB, and its homologues in other Gram-negative pathogens (e.g., MexB in Pseudomonas aeruginosa and CmeB in Campylobacter jejuni) is considered the most important RND system to human health because it transports a wide range of structurally varied antimicrobials and is more abundant within the cell than other efflux pumps (for a review, see reference 7). Furthermore, inactivation of acrB in Salmonella (and its homologues in other bacteria) confers multidrug hypersusceptibility, while single deletions of the other RND efflux pump genes have little or no effect on susceptibility to most antimicrobial agents (5, 8, 9). Interestingly, AcrD has 70% nucleotide and 79% amino acid similarity with AcrB (9), yet it possesses a distinct substrate profile that includes aminoglycoside antibiotics (10, 11). AcrB and AcrD also differ in the structures of their proximal binding pockets, which may underpin the differences in their substrate profiles (12). We have previously shown that the AcrD efflux pump impacts biofilm formation, as evidenced by an acrD mutant that showed significantly reduced biofilm formation and reduced expression of key biofilm proteins encoded by csgBD (13).
Little is known about the natural and potentially homeostatic functions of efflux pumps in pathogen biology. Efflux pumps are evolutionarily ancient proteins which long predate the use of antibiotics (14). In Salmonella, the AcrAB-TolC efflux pump is involved in protection from and efflux of bile and toxic bile salts (15–21). In E. coli, a physiological substrate is the siderophore enterobactin, which is effluxed by AcrAB-TolC, AcrAD-TolC, and MdtABC pumps (22–24). To further understand the natural function of AcrB in Salmonella, we previously determined the transcriptome of a Salmonella acrB mutant; the effect was profound and correlated with an altered basic biology of the organism (25). Furthermore, we previously showed that inactivation of individual or multiple acr efflux pump genes results in increased expression of the remaining efflux pump genes, suggesting a sensing mechanism to detect and regulate relative expression levels of the different pumps (9). When acrB was deleted, acrD expression increased (9). While the physiological role of AcrB is becoming clearer, much less is known about the functions of AcrD. Therefore, we sought to understand how loss of AcrD impacted the biology of the organism and whether loss of this pump led to changes to Salmonella physiology.
In the current study, we determined the transcriptome of a Salmonella mutant, constructed and tested previously (8, 9), in which acrD had been genetically inactivated. Here, we demonstrate that multiple genes, including those involved in metabolism, stress responses, and virulence, were altered by acrD inactivation. Using these transcriptomic data as a guide, we examined phenotypic differences between the parental strain and the acrD mutant. Specifically, we explored the ability of an acrD Salmonella mutant to swim and swarm, grow on various carbon sources, grow under anaerobic conditions, and invade polarized epithelial cells, and we tested the fitness of the acrD mutant relative to the parental strain. Our data suggest that AcrD has a different role in the biology of the organism than previously assumed.
RESULTS
Inactivation of acrD leads to distinct transcriptional changes.
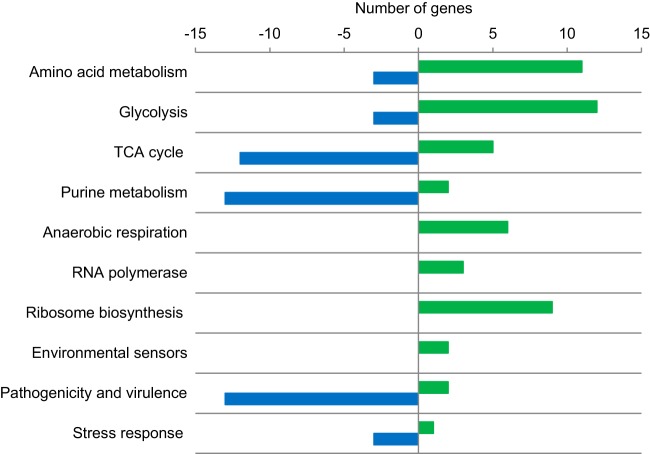
The transcriptome of the acrD mutant revealed significantly altered expression of 403 genes compared to the parental SL1344 strain (for a list of genes with significantly altered expression, see Table S1 in the supplemental material; full microarray data are available at http://www.ebi.ac.uk/arrayexpress/experiments/E-MEXP-2975/samples/). Specifically, in the acrD mutant expression of genes encoding proteins involved in anaerobic growth, ATP synthesis, amino acid metabolism, sugar transport, glycolysis/gluconeogenesis, ribosomal subunit biosynthesis, RNA polymerase, and oxidative phosphorylation were generally increased (Fig. 1). Expression of genes associated with pathogenicity, sigma factors, stress response, the tricarboxylic acid (TCA) cycle, and purine metabolism were generally decreased (Fig. 1). Surprisingly, very few of the transcriptomic changes could be directly linked to drug export.
FIG 1 .
The gene expression changes of SL1344 acrD::aph compared to wild-type SL1344. Genes with altered expression in the acrD mutant were categorized according to function. The horizontal axis indicates the number of genes in each category, with negative values indicating genes that were downregulated and positive values indicating genes that were upregulated.
It seemed unlikely that the changes observed at the transcriptomic level were simply due to the lack of a large membrane protein. If this were the case, we would expect the deletion of acrD and acrB to result in similar changes and also changes in membrane stress response genes, e.g., cpxAR and baeSR (26–28). Both CpxR and BaeR are known to induce expression of efflux pumps, including AcrD (26, 29, 30). In E. coli, inactivation of tolC activates the Cpx and Bae pathways (28). However, there was no significant change in expression of either the cpx or bae gene in the acrD mutant.
Carbon metabolism is largely unaltered by loss of acrD.
Since many of the observed transcriptional changes involved genes that encode products involved in carbon utilization, the Biolog phenotypic microarray system (Biolog, Inc., USA) was used to measure the respiration of the wild type and acrD mutant on various carbon sources (PM1 and PM2A). The respiration levels on different carbon sources for the acrD mutant and the wild-type strain were very similar, with only one major difference observed. The acrD mutant had significantly higher respiration levels on saccharate relative to that of wild-type Salmonella (P = 0.0037).
In order to validate the results from the phenotypic microarrays and to determine if the change in respiration was also associated with a change in growth, selected carbon sources were tested. Growth in M9 minimal medium supplemented with glucose, fructose, succinate, aspartate, pyruvate, or saccharate was examined (see Fig. S1 in the supplemental material). As a control, growth of the acrD mutant and wild-type Salmonella in LB broth was measured, and no differences were found in generation time or the final optical density (OD) at 19 h (Fig. S2). The generation time of the acrD mutant was not significantly different when grown with glucose, fructose, succinate, aspartate, pyruvate, or saccharate as a carbon source (Fig. S3a). Likewise, measurement of the final OD of the culture indicated that the acrD mutant and wild-type strain grew to similar final densities on the carbon sources tested (Fig. S3b).
Inactivation of acrD leads to increased expression of nitrate reductase and nitrite reductase genes.
In the acrD mutant, the expression of six genes (napACF and nirBCD) involved in anaerobic growth was increased between 3.06- and 22.83-fold (Table S1). However, under anaerobic conditions, there was no significant difference in the generation time (Fig. S4a) or the final OD (Fig. S4b) of the acrD mutant culture compared to the wild-type parental strain. Additionally, there was no difference in growth in 1 mM sodium nitrate, nor was there a difference in survival in 10 mM acidified sodium nitrite (data not shown).
Inactivation of acrD results in reduced virulence gene expression.
Inactivation of acrD led to decreased expression of 13 genes known to be involved in Salmonella virulence. The expression levels of genes from Salmonella pathogenicity islands 1 (SPI-1), -2, -3, -10, and -18 were significantly decreased (0.05- to 0.5-fold, which corresponds to a 20- to 2-fold reduction) (see Table S1). This included reduced expression of genes encoding secreted proteins (cigR and sseBCDE) and virulence protein chaperones (sicP and sscAB), while expression of a gene encoding a needle apparatus sorting platform component (orgA) was increased.
Genes from SPI-1 have been shown to be expressed by only a portion of the total population (31–33), so techniques such as microarrays and reverse transcription-PCR (RT-PCR), which measure gene expression across a whole population, only provide partial information about gene expression of individual bacteria in a population. The expression of the SPI-1 gene prgH, which encodes the essential basal component of the needle complex of the SPI-1 type III secretion system, has been used previously to measure expression of SPI-1 in single cells (31, 34). Therefore, strains containing the promoter of prgH fused to the green fluorescent protein gene gfp were used to compare the percentage of cells in the population that expressed SPI-1 in the acrD mutant versus in the wild type, as described previously (31, 34). This showed that when acrD was inactivated, fewer cells expressed SPI-1 than did wild-type Salmonella cells (Fig. 2). These findings correlate with the decreased detection of SPI-1 transcripts in the acrD mutant microarray analysis (Table S1).
FIG 2 .
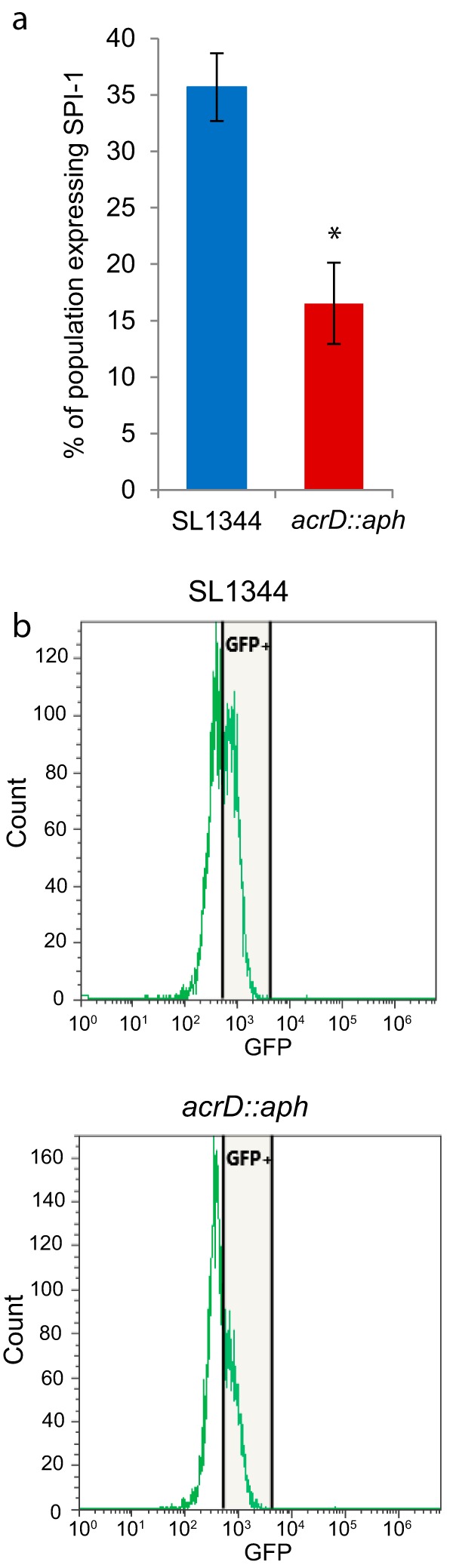
SPI-1 expression of the acrD::aph strain. Strains containing the promoter of prgH (SPI-1) fused to gfp were analyzed by flow cytometry and the number of fluorescent and nonfluorescent cells were enumerated. (a) Mean results ± standard deviations. *, P < 0.05. (b) A representative example of the flow cytometry results for SL1344 and SL1344 acrD::aph cells.
Intriguingly, this reduction in the proportion of bacteria expressing SPI-1 did not correlate with decreased invasion of the acrD mutant into Caco-2 polarized epithelial cells. There was no difference in the virulence of the acrD mutant compared to the parental SL1344 strain; as there was no difference between wild-type SL1344 and the acrD::aph strain in association (2 h), invasion (4 h), and persistence (8 and 24 h) in Caco-2 polarized epithelial cells (P = 0.745, P = 0.846, P = 0.942, and P = 0.685, respectively) (Fig. S5).
Inactivation of acrD reduces motility.
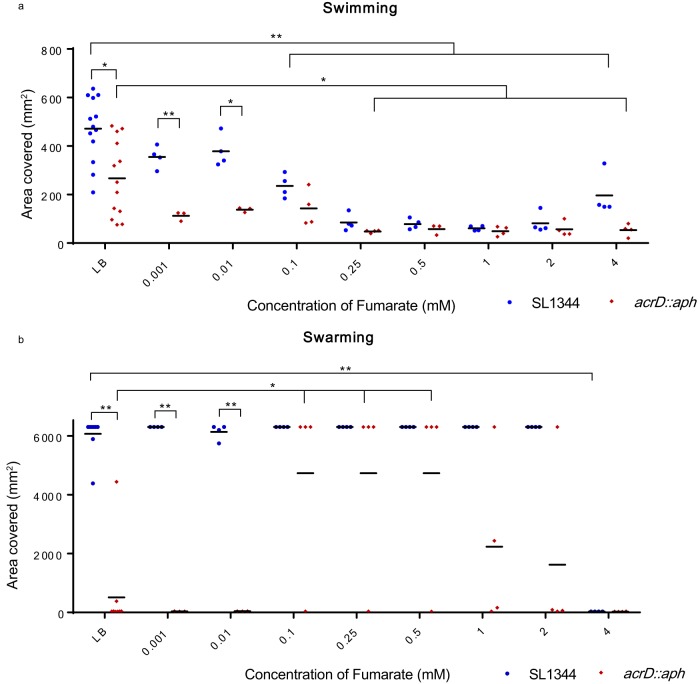
Compared to SL1344, the acrD mutant was impaired for swimming. SL1344 swam to cover an average area of 471.5 mm2, while the acrD mutant covered 266.7 mm2 (P = 0.0018) (Fig. 3a). Likewise, the acrD mutant was impaired for swarming compared with SL1344, which covered an average area of 6,090 mm2 while the acrD mutant covered 510 mm2 (P < 0.001) (Fig. 3b). The swimming assay produces results that are inherently variable, and so we minimized this impact by performing 13 replicates.
FIG 3 .
Impact of fumarate on motility of acrD::aph mutant cells compared to SL1344 cells. Areas covered by SL1344 and SL1344 acrD::aph mutant cells for swimming (a) and swarming (b) motility in LB alone and in the presence of increasing concentrations of fumarate are summarized. Data presented are from 13 replicates for LB alone and 4 replicates for fumarate, with mean values indicated. *, P < 0.05; **, P < 0.001.
Since there were no changes in expression of common motility genes found in the transcriptome (e.g., flagellar genes), yet we observed a drastic impact on motility, we explored the effect of metabolites on motility. This was because expression levels of many metabolism-associated genes were altered in the transcriptome. Fumarate has been shown to impact the switching of the direction of flagellum rotation (35–37). Furthermore, the transcriptomic data showed increased expression of frdABCD and aspA, as well as decreased expression of sdhCDA and fumC (see Table S1), the gene products of which impact fumarate levels. To determine if fumarate could restore the motility defect seen in the acrD mutant, exogenous fumarate was added to the culture medium. At concentrations of 0.1, 0.25, 0.5, 1, 2, and 4 mM, fumarate inhibited swimming motility of wild-type SL1344 (P = 0.004, P < 0.001, P < 0.001, P < 0.001, P < 0.001, P = 0.0016, respectively), while concentrations of 0.01 and lower did not impact swimming (Fig. 3a). For the acrD mutant, concentrations of 0.25, 0.5, 1, 2, and 4 mM fumarate inhibited swimming motility (P = 0.015, P = 0.041, P = 0.016, P = 0.020, P = 0.018, respectively) (Fig. 3a). Fumarate concentrations between 0.1 and 0.5 mM increased the swarming ability of the acrD mutant (P < 0.05 for 0.1, 0.25, and 0.5 mM fumarate) (Fig. 3b) compared with LB alone. Our data indicate that the ability to swarm is either on or off. In LB alone, 9 replicates showed that the wild type swarmed very well and only one replicate showed moderate swarming; for the acrD mutant, 8 replicates did not swarm at all and 2 replicates swarmed moderately (Fig. 3b). At concentrations of 0.1, 0.25, and 0.5 mM fumarate, the proportion of acrD mutants which did swarm increased (P < 0.05) (Fig. 3b). At 4 mM fumarate, swarming of the wild type was inhibited (P < 0.001) (Fig. 3b). One limitation of this experiment was that the swarming wild-type bacteria reached the edge of the plate, and we were unable to determine if fumarate increased swarming of SL1344 by the same magnitude as in the mutant.
Inactivation of acrD impacts bacterial fitness.
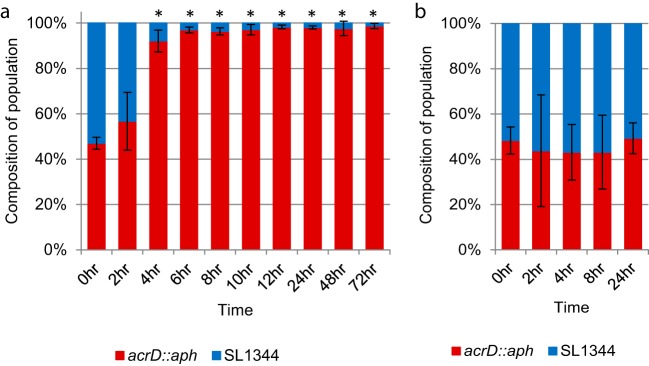
Since no differences were seen in growth when evaluated under a variety of conditions, yet expression levels of 403 genes were altered, we hypothesized that these numerous gene expression changes would alter the fitness of the acrD mutant in a competitive environment. Bacterial fitness can be determined in a number of different ways; initially we tested fitness under standard laboratory conditions in vitro (LB broth, 37°C with aeration, inoculation with a 50:50 ratio of wild-type Salmonella to acrD mutant). Under these conditions, the acrD mutant outcompeted the wild type as early as 4 h postinoculation (Fig. 4a). After 2 h of growth, the acrD mutant composed 57% of the population, and by 4 h 92% of the bacteria were acrD mutants (P < 0.001). This high proportion rose to 97% at 6 h (P < 0.001) and then varied between 96% and 99% (P < 0.001) for the remainder of the 72 h, which included passaging into fresh medium at 12, 24, and 48 h.
FIG 4 .
Impact of acrD inactivation on bacterial fitness. (a) Fitness under standard laboratory conditions in vitro. LB broth was inoculated with a 50:50 mixture of wild-type and acrD mutant cells and grown at 37°C with aeration. Samples were taken at the indicated time points, and fresh cultures were inoculated every 24 h. Data presented are the means of three independent experiments with three biological replicates each, ± standard deviations. (b) Competitive infection of polarized Caco-2 epithelial cells. After infection with a 50:50 mixture of wild-type and acrD mutant cells, association was determined at 2 h postinfection prior, to gentamicin treatment. A gentamicin protection assay was used to calculate invasion and persistence at later time points. Replica plating was used to determine the proportion of wild-type versus acrD::aph bacteria. Data presented are the means of three independent experiments performed, with 4 replicates each, ± standard deviations. *, P < 0.05; **, P < 0.001.
To measure fitness under in vivo conditions, competitive infection experiments were carried out. Polarized Caco-2 epithelial cells were infected with a 50:50 ratio of wild-type Salmonella to the acrD mutant. At 2, 4, 8, and 24 h postinfection, the proportion of the acrD mutant to the wild-type SL1344 was not significantly altered (Fig. 4b). This indicated there was no fitness differential associated with the loss of acrD during the infection of polarized Caco-2 epithelial cells (Fig. 4b).
DISCUSSION
Our data suggest that the AcrD efflux pump has a unique biological role. This was demonstrated by the significant changes observed in the transcriptome. Comparison of the transcriptomes of the acrD mutant (presented in this work; further information is available at http://www.ebi.ac.uk/arrayexpress/experiments/E-MEXP-2975/samples/) with the previously published acrB mutant transcriptome (25) showed that the effect was very distinct, which supports the hypothesis that AcrD is not a “backup” efflux pump but has its own physiological purpose in the cell. This comparison identified 232 significant gene expression changes specific to the inactivation of acrD and that were not affected by the disruption of acrB. Compared to the acrB mutant transcriptome, 169 genes were differentially expressed by both acrB and acrD mutants. Of these, expression of 91 genes was altered in the same way (e.g., increased in both mutants), and 78 genes were expressed in an opposite manner (e.g., increased in one but decreased in the other). There is experimental evidence that AcrB and AcrD efflux pumps have distinctive substrate profiles with respect to aminoglycoside antibiotics (10–12). Our data showed that deletion of either acrB or acrD has different impacts on the bacterial transcriptome, which supports the suggestion that these pumps have distinct roles. We speculate that accumulation of certain metabolic intermediates in the bacterial cell may trigger feedback mechanisms which alter gene expression and metabolism in different ways (23, 38).
Some of the genes identified in the microarray were drastically altered in the acrD mutant; for example, adh expression increased 70-fold and fruB increased 45-fold. On the other hand, some genes had <10-fold changes but these changes were still statistically significant. These data are included in Table S1 in the supplemental material for several reasons. First, depending on gene function, small changes in expression can translate into significant changes in the biology of the organism (39, 40). Second, multiple small changes which occur in the same or related pathways can have strong impacts on phenotypes (41, 42). In the data presented here, the expression changes in genes involved in fumarate metabolism (frdABCD and sdhCDA) were below 10-fold, yet fumarate had a direct and significant impact on the acrD mutant’s motility (Fig. 3). Third, these data, including the large and small changes, will likely prove useful for the scientific community.
Nearly 100 genes associated with metabolism were altered by the inactivation of acrD. Table S1 lists genes with significantly altered expression; selected genes and operons are discussed below. Expression of the pyruvate formate lyase I gene, pflB, increased. PflB is involved in glucose metabolism, converting pyruvate to formate and acetate, and is preferentially used under anaerobic conditions (43, 44). Abernathy et al. (44) found that deletion of pflB led to increased intracellular replication in intestinal epithelial cells as a result of increased SPI-1 expression. In our study, the acrD mutant had increased pflB expression and reduced SPI-1 expression, indicating another connection between efflux pump gene expression, metabolism, and virulence. The fructose operon (fruBKA), under the control of FruR (45), was upregulated in the acrD mutant. However, there was no difference between growth of the acrD mutant and wild type when fructose was used as a carbon source. Another metabolism gene, ilvC, had higher expression in the acrD mutant. IlvC is a component of the pathway Salmonella uses to synthesize isoleucine and valine, and it is encoded by ilvGEDAYC (46). ilvGEDA is an operon, but transcription of ilvC is separate and dependent on IlvY (46). We were surprised that the Biolog phenotypic microarray data did not indicate significantly altered respiration of the acrD strain on carbon sources other than saccharate. We hypothesize that the effect of the transcriptional changes may be compensatory changes within the mutant to ameliorate the effect of loss of AcrD.
Six genes, napACF and nirBCD, associated with nitrate and nitrite reduction were upregulated when acrD was inactivated. This contrasts with the reduction in expression of these same genes in the acrB mutant (25). Salmonella has two nitrate reductases, one located in the cytoplasm and the other in the periplasm (NapACF) (47). NirBD is an NADH-dependent nitrite reductase located in the cytoplasm, while NirC is a nitrite/proton antiporter in the bacterial membrane, that allows transport of nitrite into the cytoplasm for detoxification (48, 49), and contributes to Salmonella virulence in both mice and macrophages (50). However, there was no difference in the ability of the acrD mutant to grow anaerobically compared to wild-type Salmonella, nor was there a significant difference in the ability of the mutant to grow in the presence of sodium nitrate or survive in acidified nitrite. It seems plausible that deletion of these genes, as in studies performed by other groups, has a greater effect on phenotype than the level of the increased expression (3.5- to 23-fold) seen in our acrD mutant.
Among the differences in the transcriptomes, the expression levels of genes involved in virulence were reduced in the acrD mutant. Salmonella has several pathogenicity islands, including SPI-1 and SPI-2, which contribute to invasion of nonphagocytic cells as well as survival and replication within host cells (4, 51). The SPI-1-associated genes, which were downregulated in the acrD mutant, were different from the SPI-1 genes downregulated in the acrB mutant (25). Our transcriptomic data indicate that inactivation of acrD led to reduced expression of SPI-1- and SPI-2-associated chaperones, translocon components, and effectors. Despite the reduced expression of some SPI-1-associated genes and the reduced proportion of bacteria expressing the SPI-1 apparatus, neither association, invasion, nor persistence in Caco-2 treated cells was impacted by the loss of acrD. A previous study also showed no impact of acrD inactivation on adhesion or invasion of INT-407 epithelial cells or on colonization and persistence of 1-day-old and 2-week-old chicks between the acrD mutant and wild-type strain SL1344 (52). In line with these findings, Nishino et al. (2006) showed inactivation of acrD had minimal impact on BALB/c mouse survival (5). In contradiction to the findings of Buckley et al. (52) and Nishino et al. in 2006 (5), another study found that the acrD strain was attenuated in an INT-407 model of infection (9). However the polarized Caco-2 epithelial cells used in our study are more physiologically relevant. Furthermore, our competitive infection data confirm that the acrD mutant is as fit as wild-type Salmonella during infection. Salmonella invasion of polarized epithelial cells occurs in a highly cooperative manner with multiple bacteria engulfed by Salmonella induced membrane ruffles (53). We hypothesize that this cooperative entry accounts for this apparent contradiction, as cells that express SPI-1 induce ruffles, which engulf both SPI-1 expressing and non- SPI-1-expressing bacteria.
The acrD mutant was able to outcompete the wild-type Salmonella in vitro in the fitness experiment in LB culture medium. We postulate that the metabolic shifts highlighted by the transcriptomic data provide an advantage to the acrD mutant in the LB fitness experiment. The cumulative effect of changes to metabolic gene expression does not impair the ability of the mutant to outcompete the parental strain.
Salmonella species are highly motile, and are capable of swimming and swarming in a flagella dependent manner (54, 55). Swimming is associated with counterclockwise rotation of the flagella, and is often interspersed with tumbles, brought about by switching direction of flagellar rotation (56). This ability to switch rotation direction is also important for swarming motility, and is controlled by the chemotaxis response regulator CheY (56). Swarming Salmonella move within a wet “slime” layer composed of polysaccharides, surfactants, and peptides (57, 58). In a gene expression study conducted by Wang et al. 2004 (57), the flagella operons were not upregulated during swarming. Swarming Salmonella also have distinct changes in basic metabolism, with glucose a key energy source for the process (59). We made similar observations in this study; expression of multiple metabolic but not flagella specific genes was altered in the acrD mutant, yet significant changes to swarming were detected.
Recent studies have focused on finding novel genes and/or pathways which mediate swarming (60-62). A screen of a mutant library containing 1023 mutants in S. typhimurium 14028s found 21 mutants with impaired swimming and swarming, forty nine with impaired swimming, but normal swarming, and also 49 with impaired swarming but normal swimming, and 39 hyper-motile mutants (61). Further exploration of these novel motility genes identified by Bogomolnaya et al. 2014 is needed. Future work uncovering more pathways and proteins directly involved in swarming may shed further light on our data.
Despite the drastic reduction in swarming motility of the acrD mutant, we did not see significant changes in the expression of genes known to impact motility and swarming, including cheY. We observed significant changes in expression of genes associated with the production of fumarate. The key enzymes in the interconversion of succinate and fumarate are FrdABCD and SdhCDAB (63). Under aerobic conditions, sdh is upregulated and these proteins catalyze the oxidation of succinate at higher frequency (64). In the acrD mutant, the sdhCDA genes were downregulated. Under anaerobic conditions, frd genes are upregulated and fumarate can act as a terminal electron acceptor; however it is a low energy yielding acceptor (63-66). In the acrD mutant the frdABCD operon was upregulated, yet no difference in anaerobic growth was seen. Together with the upregulation of aspA and downregulation of fumC we hypothesized that the acrD mutant has altered intracellular levels of fumarate.
Fumarate is associated with flagella rotation switching in a model which uses cytoplasm free envelopes of both Salmonella and E. coli where flagella are present, but do not normally change directions (35). However, the addition of 1 mM fumarate restores the switching of flagella, even in the absence of CheY (35, 37). With the addition of fumarate the flagella rotated in the clock-wise direction more often; fumarate was shown to target the switch motor complex (37). In line with these findings, when we added fumarate (0.001 to 4 mM) to the swarming assay medium, there was a clear increase in swarming of the acrD mutant at concentrations of 0.1 to 0.5 mM. The intracellular concentration of fumarate in S. typhimurium has previously been shown to be 0.23 µM (± 0.12) (67), thus the exogenous levels of fumarate needed to impact swarming (0.1 to 1 mM) are higher than those found within the cell. Furthermore, there was an “all-or-nothing” swarming phenotype, and the addition of fumarate increased the proportion of the mutant bacteria that swarmed. Taken together, we hypothesize that inactivation of the AcrD efflux pump leads to altered levels of fumarate, which directly impacts swarming motility.
A recent elegant study examined the localization of AcrB, AcrD and TolC within the membrane (68). In this study they found that AcrB and AcrD formed stabilized foci when bound to TolC and that as levels of AcrD increased, AcrB was displaced from TolC, a phenomenon the authors call transporter exchange (68). This exchange did not occur when AcrB substrates were present (68). Extrapolating this hypothesis provides a potential explanation of our data; when acrD is deleted there are fewer competitors for AcrB to bind with TolC and therefore, the AcrAB-TolC complex is stabilized, even in the absence of AcrB substrates. We postulate this could affect the metabolite concentrations within the cell.
In summary, our study demonstrates important and significant differences in bacterial gene expression occur as a result of inactivation of acrD, and this response is different to the response elicited by the inactivation of acrB. This work highlights a previously underappreciated role of AcrD in the fundamental biology of Salmonella.
MATERIALS AND METHODS
Bacterial strains and culture media.
All strains were derived from Salmonella enterica serovar Typhimurium SL1344 (69), and the efflux pump-inactivated mutant (SL1344 acrD::aph) has been previously described (8, 9). LB broth (Sigma-Aldrich, United Kingdom), M9 minimal medium (components from Sigma-Aldrich, United Kingdom), and morpholinopropanesulfonic acid (MOPS) minimal medium supplemented with 0.2% glucose (Teknova, USA) were used throughout this study.
Microarrays.
Microarray experiments were carried out and results were analyzed exactly as described previously for other strains; they were carried out in parallel to those with an SL1344 acrB::aph mutant (25, 70). Briefly, overnight cultures of S. Typhimurium SL1344 and the mutant strain were grown in MOPS minimal medium (glucose) at 37°C until early logarithmic phase (OD600, ~0.7). For each strain, three biological and two technical replicate RNA preparations were made. Data were analyzed with the Bioconductor (71) and Pathway Tools (72) programs. Data with a B (log odds value) value of ≥0, which corresponds to an adjusted P value of <0.004, were considered significant. The microarray data set is available at http://www.ebi.ac.uk/arrayexpress/experiments/E-MEXP-2975/samples/ (submitted 10 November 2010, released 1 June 2011, last updated 2 May 2014; minimal medium used [http://www.ebi.ac.uk/arrayexpress/protocols/87504/?ref=E-MEXP-2975]).
Phenotypic microarrays.
Salmonella Typhimurium strains were cultured from frozen (−80°C) stocks on LB agar for 24 h at 37°C, aerobically. Bacteria were harvested from agar plates and resuspended into IF-0a inoculating fluid (Biolog, Inc., USA) and adjusted to 42% transmittance (turbidimeter; Biolog). A working cell suspension of 10 ml IF-0a, 2 ml of bacterial suspension, and 120 µl of tetrazolium dye (90 µl dye D, 30 µl dye F; Biolog) was made for the PM1 and PM2A carbon utilization plates. One hundred microliters of suspension was added to each of the 96 wells on the appropriate PM plates. The inoculated PM plates were placed in an Omnilog automatic plate reader (Biolog) and incubated at 37°C, aerobically, for 48 h. Metabolism of the various carbon sources was recorded every 15 min based on the reduction of the tetrazolium violet redox dye, which produces a purple color indicative of active bacterial respiration. Abiotic negative-control plates indicated false-positive results due to autoreduction of the dye observed in PM1 with l-arabinose, d-xylose, d-ribose, or l-lyxose. False-positive results observed in PM2A included d-arabinose, 2-deoxy-d-ribose, d-glucosamine, 5-keto-d-gluconic acid, and dihyroxyacetone.
Each PM plate preparation was repeated in triplicate for each strain. The values of the experimental replicates for each strain for each carbon and nitrogen substrate were compared by a one-way analysis of variance and Tukey post hoc multiple-comparison test, using a 95% family-wise confidence level. Statistical significance was assigned at the P ≤ 0.05 level.
Bacterial growth in different carbon sources.
Bacteria were grown aerobically at 37°C for approximately 16 h in LB broth and then diluted to ~104 CFU/ml in fresh M9 minimal medium. M9 minimal medium was used to coincide with that used in previous similar experiments (73). Growth kinetics were determined in M9 minimal medium supplemented with 6 mM l-histidine (Sigma-Aldrich, United Kingdom) (parental strain SL1344 is a histidine auxotroph) and added carbon source. Carbon sources tested were glucose (0.4%), fructose (0.4%), succinate (0.2%), aspartate (0.4%), pyruvate (0.2%), and saccharate (0.05%) (Sigma-Aldrich, United Kingdom). These concentrations were chosen based on preliminary experiments which determined the concentrations at which bacteria were able to grow. For glucose, fructose, and aspartate, absorbance was measured at 600 nm in a FLUOstar Optima apparatus (BMG Labtech, United Kingdom) at 37°C with agitation before each reading, which was taken every 5 min for 19 h. Data were collected from three biological and three technical replicates of each strain and are presented as the average optical density over time. For succinate, pyruvate, and saccharate, growth in the FLUOstar was poor, and therefore growth kinetics were determined by inoculating 10 ml M9 minimal medium with histidine and the specified carbon source. Cultures were incubated at 37°C with agitation, and absorbance at 600 nm (OD600) was measured initially and at 2, 4, 6, 8, 10, 12, and 24 h. Final density (24 h) and generation time during exponential phase were calculated and analyzed using a two-tailed Student’s t test. P values of ≤0.05 were considered significant.
Anaerobic growth.
Cultures were set up in an anaerobic chamber (anaerobic gas growth mixture of 10% CO2, 10% H2, 80% N2) in MOPS minimal medium supplemented with glucose and 6 mM l-histidine and incubated at 37°C for 24 h. Growth curves in MOPS minimal medium with histidine were started with 4% inoculum from overnight cultures grown anaerobically, and the initial OD600 was measured. The OD600 was measured every 2 h for the first 10 h and again at 24 h. Three independent experiments with three biological replicates were completed. Data are presented as the mean generation time ± the standard deviation. P values of ≤0.05 from a two-tailed Student’s t test were considered significant.
Motility assays.
Bacterial strains were grown overnight at 37°C in LB broth with or without antibiotic, as appropriate. LB broth was used because nutrient-rich broth is required for Salmonella swarming. To prepare inocula, cells were adjusted to an OD600 of 0.5 in phosphate-buffered saline (PBS). Motility plates were prepared (the night before inoculation to allow time to set) by combining LB broth and Difco Bacto agar (BD); for swimming assays, an agar concentration of 0.3% was used, and for swarming assays an agar concentration of 0.6% was used with the addition of 5 g/liter glucose. To inoculate swimming plates, a straight loop was immersed in the inoculum and then stabbed into the center of the swimming plate. These plates were then incubated for 7 h at 37°C. For swarming assays, 5-µl aliquots of inoculum were spotted on the top of swarming plates, which were incubated for 16 h at 37°C. Images were taken using a Syngene Transilluminator box, and ImageJ software was used to quantify the area covered. Data were analyzed using a two-tailed Student’s t test, and P values of ≤0.05 were considered significant.
Infection of tissue culture cells.
Caco-2 epithelial cells were grown in Dulbecco’s minimal essential medium (DMEM; Sigma-Aldrich, United Kingdom) containing 1% (vol/vol) nonessential amino acids (Sigma-Aldrich, United Kingdom), 1% (vol/vol) l-glutamine (Sigma-Aldrich, United Kingdom), and 10% fetal bovine serum (FBS; Sigma-Aldrich, United Kingdom). Cells were polarized by seeding at 1.8 × 105 cells per well and grown for 2 weeks, with fresh DMEM added every 2 to 3 days. Infection assays were carried out as described previously (52). Briefly, overnight cultures of SL1344 wild-type and the acrD::aph mutant were grown at 37°C in LB broth, washed in PBS, and then 1 × 107 bacteria/ml was added to inoculation medium (DMEM without FBS). Caco-2 cells were washed 3 times with warm Hanks’ balanced salt solution (HBSS; Sigma-Aldrich, United Kingdom), and then 1 ml of inoculation medium was added to each well. At 2 h postinfection, all cells were washed with HBSS and for the 2-h time point cells, were lysed using 1% Triton X-100 (Sigma-Aldrich, United Kingdom), serially diluted, and plated on LB agar for bacterial enumeration. For later time points, after washing the inoculation medium containing 100 µg/ml of gentamicin was added to each well and incubated for another 2 h. For the 4-h time point, cells were lysed, serially diluted, and enumerated as described before. For the 8- and 24-h time points, medium was replaced with inoculation medium containing 10 µg/ml gentamicin and cultures were incubated for the remaining time. Cells were lysed, diluted, and enumerated as described above.
Competitive infections were carried out alongside isolated infections (described above), with the following changes. Inoculum was prepared by combining 5 × 106 bacteria/ml of wild-type SL1344 and 5 × 106 bacteria/ml of the mutant acrD::aph strain. Inocula and samples from each time point were serially diluted and plated on LB agar and incubated for 14 to 16 h at 30°C to ensure small colonies. Total colonies were enumerated. Using velvet squares, colonies were replica plated onto LB agar containing 50 µg/ml kanamycin and then incubated for 16 h, aerobically. Kanamycin-resistant colonies were enumerated.
Data are presented as the mean CFU of three independent experiments with four replicates each, ± standard deviations and were analyzed using a two-tailed Student’s t test. P values of ≤0.05 were considered significant.
Competition assays.
Bacterial strains were grown statically at 37°C overnight in LB broth with or without antibiotic, as appropriate. Cells were washed in PBS, and the cultures were adjusted until the OD600 was 0.3. Wild-type SL1344 (100 µl) and mutant acrD::aph cells were added to 10 ml of LB broth (no antibiotics) and incubated at 37°C with aeration. Fresh broth was inoculated every 24 h. CFU were determined by serial dilution and plating on LB agar plates at 0 h (inoculum) and 2, 4, 6, 8, 10, 12, 24, 48, and 72 h. Replica plating with velvet squares onto LB agar plates containing 50 µg/ml of kanamycin was performed to determine the ratio of wild-type to mutant cells, and competition indices were determined. Three independent experiments were completed, with three biological replicates each. Data are presented as mean CFU ± standard deviations and analyzed using a two-tailed Student’s t test. P values of ≤0.05 were considered significant.
SPI-1 reporter assays.
The chromosomal gfp reporter fused to the promoter of prgH has been previously described (31), and we have previously used gfp with the prgH promoter to measure activity of SPI-1 in individual cells (34). In brief, strains SL1344 prgH′-gfp and SL1344 acrB::aph prgH′-gfp were grown to mid-logarithmic phase in MOPS minimal medium. The cells were harvested by centrifugation at 2,200 × g at room temperature, washed, and resuspended in PBS. Bacteria were analyzed by flow cytometry using a FACSAria2 system (BD Biosciences). Cells were illuminated with a 488-nm laser, and scatter and GFP fluorescence data were collected through a 502 LP mirror and 530/30 bandpass filter. For each sample, 10,000 events were collected.
SUPPLEMENTAL MATERIAL
Growth of SL1344 (blue line) and acrD::aph (red line) in M9 minimal medium supplemented with either glucose (0.4%), fructose (0.4%), succinate (0.2%), aspartate (0.4%), pyruvate (0.2%), or saccharate (0.05%). Download
Generation time and final optical density of acrD::aph and SL1344 cultures grown in LB broth. Data presented are the means of three experiments performed in triplicate ± standard deviations. No statistically significant differences were found between groups. Download
Growth of SL1344 and acrD::aph cultures grown in medium containing 6 different carbon sources. Growth in M9 minimal medium supplemented with glucose (0.4%), fructose (0.4%), succinate (0.2%), aspartate (0.4%), pyruvate (0.2%), or saccharate (0.05%) is shown. Data are the mean results of three independent experiments ± standard deviations. (a) Generation time of wild-type SL1344 (blue bars) and SL1344 acrD::aph (red bars) cultures grown in glucose, fructose, succinate, aspartate, pyruvate, or saccharate. (b) Final optical density of wild-type SL1344 (blue bars) and SL1344 acrD::aph (red bars) cultures grown in glucose, fructose, succinate, aspartate, pyruvate, or saccharate. No statistically significant differences were found between groups of data. Download
Anaerobic growth of SL1344 and acrD::aph. (a) Generation time of strains grown anaerobically in MOPS minimal medium. (b) Final optical density of strains grown anaerobically in MOPS minimal medium. Data presented are the mean results of three experiments performed in triplicate ± standard deviations. No statistically significant differences were found between data groups. Download
Invasion of polarized Caco-2 epithelial cells. Association was determined at 2 h postinfection, prior to gentamicin treatment (P = 0.745). A gentamicin protection assay was performed to determine invasion and persistence at 4, 8, and 24 h postinfection (P = 0.846, P = 0.942, and P = 0.685, respectively). Each data point is the mean results for one independent experiment comprising 4 replicates. Download
Significant (adjusted P < 0.004) gene expression changes in acrD::aph cultures compared to the wild type (data are the fold change relative to growth of SL1344; boldface indicates increased gene expression and italics indicate decreased gene expression).
ACKNOWLEDGMENTS
We thank Andrew Bailey for carrying out the microarray experiments and Mark Webber, Xuan Wang Kan, Lee Rosner, and William Shafer for reading and providing constructive criticism of earlier versions of this manuscript.
Funding Statement
This work was funded in part by RCUK | Medical Research Council (MRC) (G0501415).
Footnotes
Citation Buckner MMC, Blair JMA, La Ragione RM, Newcombe J, Dwyer DJ, Ivens A, Piddock LJV. 2016. Beyond antimicrobial resistance: evidence for a distinct role of the AcrD efflux pump in Salmonella biology. mBio 7(6):e01916-16. doi:10.1128/mBio.01916-16.
REFERENCES
- 1.Anes J, McCusker MP, Fanning S, Martins M. 2015. The ins and outs of RND efflux pumps in Escherichia coli. Front Microbiol 6:587. doi: 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassan KA, Jackson SM, Penesyan A, Patching SG, Tetu SG, Eijkelkamp BA, Brown MH, Henderson PJ, Paulsen IT. 2013. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc Natl Acad Sci U S A 110:20254–20259. doi: 10.1073/pnas.1317052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassan KA, Liu Q, Henderson PJ, Paulsen IT. 2015. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 6:e01982-14. doi: 10.1128/mBio.01982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckner MM, Croxen MA, Arena ET, Finlay BB. 2011. A comprehensive study of the contribution of Salmonella enterica serovar Typhimurium SPI2 effectors to bacterial colonization, survival, and replication in typhoid fever, macrophage, and epithelial cell infection models. Virulence 2:208–216. doi: 10.4161/viru.2.3.15894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishino K, Latifi T, Groisman EA. 2006. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol 59:126–141. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 6.Blair JMA, Richmond GE, Piddock LJV. 2014. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol 9:1165–1177. doi: 10.2217/fmb.14.66. [DOI] [PubMed] [Google Scholar]
- 7.Piddock LJV. 2006. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev 19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaves DJ, Ricci V, Piddock LJV. 2004. Expression of acrB, acrF, acrD, marA, and soxS in Salmonella enterica serovar Typhimurium: role in multiple antibiotic resistance. Antimicrob Agents Chemother 48:1145–1150. doi: 10.1128/AAC.48.4.1145-1150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blair JMA, Smith HE, Ricci V, Lawler AJ, Thompson LJ, Piddock LJV. 2015. Expression of homologous RND efflux pump genes is dependent upon AcrB expression: implications for efflux and virulence inhibitor design. J Antimicrob Chemother 70:424–431. doi: 10.1093/jac/dku380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenberg EY, Ma D, Nikaido H. 2000. AcrD of Escherichia coli is an aminoglycoside efflux pump. J Bacteriol 182:1754–1756. doi: 10.1128/JB.182.6.1754-1756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aires JR, Nikaido H. 2005. Aminoglycosides are captured from both periplasm and cytoplasm by the AcrD multidrug efflux transporter of Escherichia coli. J Bacteriol 187:1923–1929. doi: 10.1128/JB.187.6.1923-1929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi N, Tamura N, van Veen HW, Yamaguchi A, Murakami S. 2014. β-Lactam selectivity of multidrug transporters AcrB and AcrD resides in the proximal binding pocket. J Biol Chem 289:10680–10690. doi: 10.1074/jbc.M114.547794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baugh S, Ekanayaka AS, Piddock LJ, Webber MA. 2012. Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J Antimicrob Chemother 67:2409–2417. doi: 10.1093/jac/dks228. [DOI] [PubMed] [Google Scholar]
- 14.Blanco P, Hernando-Amado S, Reales-Calderon JA, Corona F, Lira F, Alcalde-Rico M, Bernardini A, Sanchez MB, Martinez JL. 2016. Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4:14. doi: 10.3390/microorganisms4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikaido E, Yamaguchi A, Nishino K. 2008. AcrAB multidrug efflux pump regulation in Salmonella enterica serovar Typhimurium by RamA in response to environmental signals. J Biol Chem 283:24245–24253. doi: 10.1074/jbc.M804544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol 48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 17.Baucheron S, Nishino K, Monchaux I, Canepa S, Maurel MC, Coste F, Roussel A, Cloeckaert A, Giraud E. 2014. Bile-mediated activation of the acrAB and tolC multidrug efflux genes occurs mainly through transcriptional derepression of Rama in Salmonella enterica serovar Typhimurium. J Antimicrob Chemother 69:2400–2406. doi: 10.1093/jac/dku140. [DOI] [PubMed] [Google Scholar]
- 18.Thanassi DG, Cheng LW, Nikaido H. 1997. Active efflux of bile salts by Escherichia coli. J Bacteriol 179:2512–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Usui M, Asai T, Sato S. 2011. Low expression of AcrB in the deoxycholate-sensitive strains of Salmonella enterica subspecies enterica serovar Pullorum. Microbiol Immunol 55:366–368. doi: 10.1111/j.1348-0421.2011.00321.x. [DOI] [PubMed] [Google Scholar]
- 20.Baucheron S, Mouline C, Praud K, Chaslus-Dancla E, Cloeckaert A. 2005. TolC but not AcrB is essential for multidrug-resistant Salmonella enterica serotype Typhimurium colonization of chicks. J Antimicrob Chemother 55:707–712. doi: 10.1093/jac/dki091. [DOI] [PubMed] [Google Scholar]
- 21.Virlogeux-Payant I, Baucheron S, Pelet J, Trotereau J, Bottreau E, Velge P, Cloeckaert A. 2008. TolC, but not AcrB, is involved in the invasiveness of multidrug-resistant Salmonella enterica serovar Typhimurium by increasing type III secretion system-1 expression. Int J Med Microbiol 298:561–569. doi: 10.1016/j.ijmm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 22.Bleuel C, Grosse C, Taudte N, Scherer J, Wesenberg D, Krauss GJ, Nies DH, Grass G. 2005. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J Bacteriol 187:6701–6707. doi: 10.1128/JB.187.19.6701-6707.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz C, Levy SB. 2014. Regulation of acrAB expression by cellular metabolites in Escherichia coli. J Antimicrob Chemother 69:390–399. doi: 10.1093/jac/dkt352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horiyama T, Nishino K. 2014. AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS One 9:e108642. doi: 10.1371/journal.pone.0108642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webber MA, Bailey AM, Blair JMA, Morgan E, Stevens MP, Hinton JCD, Ivens A, Wain J, Piddock LJV. 2009. The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J Bacteriol 191:4276–4285. doi: 10.1128/JB.00363-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirakawa H, Nishino K, Hirata T, Yamaguchi A. 2003. Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J Bacteriol 185:1851–1856. doi: 10.1128/JB.185.6.1851-1856.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Appia-Ayme C, Patrick E, Sullivan MJ, Alston MJ, Field SJ, AbuOun M, Anjum MF, Rowley G. 2011. Novel inducers of the envelope stress response BaeSR in Salmonella typhimurium: BaeR is critically required for tungstate waste disposal. PLoS One 6:e23713. doi: 10.1371/journal.pone.0023713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosner JL, Martin RG. 2013. Reduction of cellular Stress by TolC-dependent efflux pumps in Escherichia coli indicated by BaeSR and CpxARP activation of spy in efflux mutants. J Bacteriol 195:1042–1050. doi: 10.1128/JB.01996-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. 2005. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol 55:1113–1126. doi: 10.1111/j.1365-2958.2004.04449.x. [DOI] [PubMed] [Google Scholar]
- 30.Hu WS, Chen HW, Zhang RY, Huang CY, Shen CF. 2011. The expression levels of outer membrane proteins STM1530 and OmpD, which are influenced by the CpxAR and BaeSR two-component systems, play important roles in the ceftriaxone resistance of Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother 55:3829–3837. doi: 10.1128/AAC.00216-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hautefort I, Proença MJ, Hinton JCD. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl Environ Microbiol 69:7480–7491. doi: 10.1128/AEM.69.12.7480-7491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt W-D. 2011. The cost of virulence: retarded growth of Salmonella typhimurium cells expressing type III secretion system. PLoS Pathog 7:e1002143. doi: 10.1371/journal.ppat.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark L, Perrett CA, Malt L, Harward C, Humphrey S, Jepson KA, Martinez-Argudo I, Carney LJ, La Ragione RM, Humphrey TJ, Jepson MA. 2011. Differences in Salmonella enterica serovar Typhimurium strain invasiveness are associated with heterogeneity in SPI-1 gene expression. Microbiology 157:2072–2083. doi: 10.1099/mic.0.048496-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blair JM, Richmond GE, Bailey AM, Ivens A, Piddock LJ. 2013. Choice of bacterial growth medium alters the transcriptome and phenotype of Salmonella enterica serovar Typhimurium. PLoS One 8:e63912. doi: 10.1371/journal.pone.0063912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barak R, Eisenbach M. 1992. Fumarate or a fumarate metabolite restores switching ability to rotating flagella of bacterial envelopes. J Bacteriol 174:643–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barak R, Giebel I, Eisenbach M. 1996. The specificity of fumarate as a switching factor of the bacterial flagellar motor. Mol Microbiol 19:139–144. doi: 10.1046/j.1365-2958.1996.365889.x. [DOI] [PubMed] [Google Scholar]
- 37.Prasad K, Caplan SR, Eisenbach M. 1998. Fumarate modulates bacterial flagellar rotation by lowering the free energy difference between the clockwise and counterclockwise states of the motor. J Mol Biol 280:821–828. doi: 10.1006/jmbi.1998.1922. [DOI] [PubMed] [Google Scholar]
- 38.Rosner JL, Martin RG. 2009. An excretory function for the Escherichia coli outer membrane pore TolC: upregulation of marA and soxS transcription and Rob activity due to metabolites accumulated in tolC mutants. J Bacteriol 191:5283–5292. doi: 10.1128/JB.00507-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie X, Zhang H, Zheng Y, Li A, Wang M, Zhou H, Zhu X, Schneider Z, Chen L, Kreiswirth BN, Du H. 2016. RpoE is a putative antibiotic resistance regulator of Salmonella enteric serovar Typhi. Curr Microbiol 72:457–464. doi: 10.1007/s00284-015-0983-7. [DOI] [PubMed] [Google Scholar]
- 40.Antunes LC, Wang M, Andersen SK, Ferreira RB, Kappelhoff R, Han J, Borchers CH, Finlay BB. 2012. Repression of Salmonella enterica phoP expression by small molecules from physiological bile. J Bacteriol 194:2286–2296. doi: 10.1128/JB.00104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Condell O, Power KA, Händler K, Finn S, Sheridan A, Sergeant K, Renaut J, Burgess CM, Hinton JC, Nally JE, Fanning S. 2014. Comparative analysis of Salmonella susceptibility and tolerance to the biocide chlorhexidine identifies a complex cellular defense network. Front Microbiol 5:373. doi: 10.3389/fmicb.2014.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn S, Jung J, Jang IA, Madsen EL, Park W. 2016. Role of glyoxylate shunt in oxidative stress response. J Biol Chem 291:11928–11938. doi: 10.1074/jbc.M115.708149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moreau PL. 2007. The lysine decarboxylase CadA protects Escherichia coli starved of phosphate against fermentation acids. J Bacteriol 189:2249–2261. doi: 10.1128/JB.01306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abernathy J, Corkill C, Hinojosa C, Li X, Zhou H. 2013. Deletions in the pyruvate pathway of Salmonella typhimurium alter SPI1-mediated gene expression and infectivity. J Anim Sci Biotechnol 4:5. doi: 10.1186/2049-1891-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feldheim DA, Chin AM, Nierva CT, Feucht BU, Cao YW, Xu YF, Sutrina SL, Saier MH. 1990. Physiological consequences of the complete loss of phosphoryl-transfer proteins HPr and FPr of the phosphoenolpyruvate:sugar phosphotransferase system and analysis of fructose (fru) operon expression in Salmonella typhimurium. J Bacteriol 172:5459–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blazey DL, Burns RO. 1984. Regulation of Salmonella typhimurium ilvYC genes. J Bacteriol 159:951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rowley G, Hensen D, Felgate H, Arkenberg A, Appia-Ayme C, Prior K, Harrington C, Field SJ, Butt JN, Baggs E, Richardson DJ. 2012. Resolving the contributions of the membrane-bound and periplasmic nitrate reductase systems to nitric oxide and nitrous oxide production in Salmonella enterica serovar Typhimurium. Biochem J 441:755–762. doi: 10.1042/BJ20110971. [DOI] [PubMed] [Google Scholar]
- 48.Lü W, Schwarzer NJ, Du J, Gerbig-Smentek E, Andrade SLA, Einsle O. 2012. Structural and functional characterization of the nitrite channel NirC from Salmonella typhimurium. Proc Natl Acad Sci U S A 109:18395–18400. doi: 10.1073/pnas.1210793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rycovska A, Hatahet L, Fendler K, Michel H. 2012. The nitrite transport protein NirC from Salmonella typhimurium is a nitrite/proton antiporter. Biochim Biophys Acta 1818:1342–1350. doi: 10.1016/j.bbamem.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Das P, Lahiri A, Lahiri A, Chakravortty D. 2009. Novel role of the nitrite transporter NirC in Salmonella pathogenesis: SPI2-dependent suppression of inducible nitric oxide synthase in activated macrophages. Microbiology 155:2476–2489. doi: 10.1099/mic.0.029611-0. [DOI] [PubMed] [Google Scholar]
- 51.Ferreira RB, Buckner MM, Finlay BB. 2012. Genome plasticity in Salmonella enterica and its relevance to host-pathogen interactions, p 84–102. In Hacker J, Dobrindt U, Kurth R (ed), Genome plasticity and infectious diseases. American Society for Microbiology, Washington, DC. [Google Scholar]
- 52.Buckley AM, Webber MA, Cooles S, Randall LP, La Ragione RM, Woodward MJ, Piddock LJV. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol 8:847–856. doi: 10.1111/j.1462-5822.2005.00671.x. [DOI] [PubMed] [Google Scholar]
- 53.Lorkowski M, Felipe-López A, Danzer CA, Hansmeier N, Hensel M. 2014. Salmonella enterica invasion of polarized epithelial cells is a highly cooperative effort. Infect Immun 82:2657–2667. doi: 10.1128/IAI.00023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim W, Surette MG. 2005. Prevalence of surface swarming behavior in Salmonella. J Bacteriol 187:6580–6583. doi: 10.1128/JB.187.18.6580-6583.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wozniak CE, Chevance FF, Hughes KT. 2010. Multiple promoters contribute to swarming and the coordination of transcription with flagellar assembly in Salmonella. J Bacteriol 192:4752–4762. doi: 10.1128/JB.00093-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harshey RM, Partridge JD. 2015. Shelter in a swarm. J Mol Biol 427:3683–3694. doi: 10.1016/j.jmb.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Q, Frye JG, McClelland M, Harshey RM. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol Microbiol 52:169–187. doi: 10.1111/j.1365-2958.2003.03977.x. [DOI] [PubMed] [Google Scholar]
- 58.Toguchi A, Siano M, Burkart M, Harshey RM. 2000. Genetics of swarming motility in Salmonella enterica serovar typhimurium: critical role for lipopolysaccharide. J Bacteriol 182:6308–6321. doi: 10.1128/JB.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim W, Surette MG. 2004. Metabolic differentiation in actively swarming Salmonella. Mol Microbiol 54:702–714. doi: 10.1111/j.1365-2958.2004.04295.x. [DOI] [PubMed] [Google Scholar]
- 60.Deditius JA, Felgner S, Spöring I, Kühne C, Frahm M, Rohde M, Weiß S, Erhardt M. 2015. Characterization of Novel factors involved in swimming and swarming motility in Salmonella enterica serovar Typhimurium. PLoS One 10:e0135351. doi: 10.1371/journal.pone.0135351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bogomolnaya LM, Aldrich L, Ragoza Y, Talamantes M, Andrews KD, McClelland M, Andrews-Polymenis HL. 2014. Identification of novel factors involved in modulating motility of Salmonella enterica serotype typhimurium. PLoS One 9:e111513. doi: 10.1371/journal.pone.0111513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Irazoki O, Mayola A, Campoy S, Barbé J. 2016. SOS system induction inhibits the assembly of chemoreceptor signaling clusters in Salmonella enterica. PLoS One 11:e0146685. doi: 10.1371/journal.pone.0146685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Gong CS, Tsao GT. 1998. Bioconversion of fumaric acid to succinic acid by recombinant E. coli. Appl Biochem Biotechnol 70–72:919–928. doi: 10.1007/BF02920202. [DOI] [PubMed] [Google Scholar]
- 64.Van Hellemond JJ, Tielens AG. 1994. Expression and functional properties of fumarate reductase. Biochem J 304:321–331. doi: 10.1042/bj3040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sonck KA, Kint G, Schoofs G, Vander Wauven C, Vanderleyden J, De Keersmaecker SC. 2009. The proteome of Salmonella typhimurium grown under in vivo-mimicking conditions. Proteomics 9:565–579. doi: 10.1002/pmic.200700476. [DOI] [PubMed] [Google Scholar]
- 66.Cecchini G, Schröder I, Gunsalus RP, Maklashina E. 2002. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim Biophys Acta 1553:140–157. doi: 10.1016/S0005-2728(01)00238-9. [DOI] [PubMed] [Google Scholar]
- 67.White AP, Weljie AM, Apel D, Zhang P, Shaykhutdinov R, Vogel HJ, Surette MG. 2010. A global metabolic shift is linked to Salmonella multicellular development. PLoS One 5:e11814. doi: 10.1371/journal.pone.0011814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamamoto K, Tamai R, Yamazaki M, Inaba T, Sowa Y, Kawagishi I. 2016. Substrate-dependent dynamics of the multidrug efflux transporter AcrB of Escherichia coli. Sci Rep 6:21909. doi: 10.1038/srep21909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wray C, Sojka WJ. 1978. Experimental Salmonella typhimurium infection in calves. Res Vet Sci 25:139–143. [PubMed] [Google Scholar]
- 70.Bailey AM, Ivens A, Kingsley R, Cottell JL, Wain J, Piddock LJV. 2010. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J Bacteriol 192:1607–1616. doi: 10.1128/JB.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reimers M, Carey VJ. 2006. Bioconductor: an open source framework for bioinformatics and computational biology. Methods Enzymol 411:119–134. doi: 10.1016/S0076-6879(06)11008-3. [DOI] [PubMed] [Google Scholar]
- 72.Karp PD, Paley S, Romero P. 2002. The Pathway Tools software. Bioinformatics 18:S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.S225. [DOI] [PubMed] [Google Scholar]
- 73.Antunes LC, Andersen SK, Menendez A, Arena ET, Han J, Ferreira RB, Borchers CH, Finlay BB. 2011. Metabolomics reveals phospholipids as important nutrient sources during Salmonella growth in bile in vitro and in vivo. J Bacteriol 193:4719–4725. doi: 10.1128/JB.05132-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth of SL1344 (blue line) and acrD::aph (red line) in M9 minimal medium supplemented with either glucose (0.4%), fructose (0.4%), succinate (0.2%), aspartate (0.4%), pyruvate (0.2%), or saccharate (0.05%). Download
Generation time and final optical density of acrD::aph and SL1344 cultures grown in LB broth. Data presented are the means of three experiments performed in triplicate ± standard deviations. No statistically significant differences were found between groups. Download
Growth of SL1344 and acrD::aph cultures grown in medium containing 6 different carbon sources. Growth in M9 minimal medium supplemented with glucose (0.4%), fructose (0.4%), succinate (0.2%), aspartate (0.4%), pyruvate (0.2%), or saccharate (0.05%) is shown. Data are the mean results of three independent experiments ± standard deviations. (a) Generation time of wild-type SL1344 (blue bars) and SL1344 acrD::aph (red bars) cultures grown in glucose, fructose, succinate, aspartate, pyruvate, or saccharate. (b) Final optical density of wild-type SL1344 (blue bars) and SL1344 acrD::aph (red bars) cultures grown in glucose, fructose, succinate, aspartate, pyruvate, or saccharate. No statistically significant differences were found between groups of data. Download
Anaerobic growth of SL1344 and acrD::aph. (a) Generation time of strains grown anaerobically in MOPS minimal medium. (b) Final optical density of strains grown anaerobically in MOPS minimal medium. Data presented are the mean results of three experiments performed in triplicate ± standard deviations. No statistically significant differences were found between data groups. Download
Invasion of polarized Caco-2 epithelial cells. Association was determined at 2 h postinfection, prior to gentamicin treatment (P = 0.745). A gentamicin protection assay was performed to determine invasion and persistence at 4, 8, and 24 h postinfection (P = 0.846, P = 0.942, and P = 0.685, respectively). Each data point is the mean results for one independent experiment comprising 4 replicates. Download
Significant (adjusted P < 0.004) gene expression changes in acrD::aph cultures compared to the wild type (data are the fold change relative to growth of SL1344; boldface indicates increased gene expression and italics indicate decreased gene expression).