Abstract
In response to cold, brown adipose tissue (BAT) increases its metabolic rate and expands its mass to produce heat required for survival, a process known as BAT recruitment. The mechanistic target of rapamycin complex 1 (mTORC1) controls metabolism, cell growth and proliferation, but its role in regulating BAT recruitment in response to chronic cold stimulation is unknown. Here, we show that cold activates mTORC1 in BAT, an effect that depends on the sympathetic nervous system. Adipocyte-specific mTORC1 loss in mice completely blocks cold-induced BAT expansion and severely impairs mitochondrial biogenesis. Accordingly, mTORC1 loss reduces oxygen consumption and causes a severe defect in BAT oxidative metabolism upon cold exposure. Using in vivo metabolic imaging, metabolomics and transcriptomics, we show that mTORC1 deletion impairs glucose and lipid oxidation, an effect linked to a defect in tricarboxylic acid (TCA) cycle activity. These analyses also reveal a severe defect in nucleotide synthesis in the absence of mTORC1. Overall, these findings demonstrate an essential role for mTORC1 in the regulation of BAT recruitment and metabolism in response to cold.
In mammals, brown adipose tissue (BAT) serves as a key heat-producing organ required to maintain body temperature and homeothermy1. In response to cold, activation of the sympathetic nervous system (SNS) rapidly turns on lipolysis and thermogenesis along with a specific transcriptional program required for heat production in brown adipocytes1,2. BAT thermogenesis depends on the uncoupling protein 1 (UCP1), a mitochondrial transporter protein that deviates protons across the inner mitochondrial membrane, uncoupling oxidative phosphorylation from ATP synthesis1,3. As a result, energy is dissipated in the form of heat, which facilitates the adaptation to a cold environment.
Until recently, BAT was thought to be present and active only in small mammals and human newborns4,5. With the use of positron emission tomography (PET), several groups confirmed over the last years the existence of BAT in adult humans6,7,8,9. BAT can be found in supraclavicular, cervical, paraspinal and mediastinal regions8,10,11 acting as a heat-producing organ in adult humans9. As demonstrated in rodents, cold exposure rapidly activates the uptake of glucose, non-esterified fatty acids (NEFAs) and triglyceride (TG)-derived fatty acids to support oxidative metabolism and heat production in BAT12,13,14. These results suggest that BAT may be exploited to dissipate energy excess, fight obesity and improve glucose and lipid homeostasis in humans13,15,16,17.
In response to chronic adrenergic stimulation, BAT expands its mass through a combination of hyperplasia and hypertrophy and increases mitochondrial number to support thermogenesis, a phenomenon observed in both rodents1,18,19,20,21,22 and humans23,24. BAT expansion has been associated with improved insulin sensitivity in patients with type 2 diabetes15,24. The signaling events that contribute to BAT expansion during chronic cold exposure are not well characterized. The mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) is a central regulator of cell growth and metabolism25. In response to nutrients and growth factors, mTORC1 activates several anabolic processes including protein and lipid synthesis. Here, we show that mTORC1 activity is robustly increased in BAT of mice exposed to cold. Interestingly, we report that BAT denervation is sufficient to abrogate the elevation in mTORC1 activity, indicating that cold activates mTORC1 through the SNS. Furthermore, adipose-specific loss of Raptor, an essential component of mTORC1, completely blocks cold-induced BAT mass expansion, reduces mitochondrial biogenesis and severely impairs BAT oxidative metabolism. Using a combination of in vivo metabolic imaging, metabolomics and transcriptomics, we show that mTORC1 loss impairs glucose and lipid oxidation, an effect linked to a defect in TCA cycle activity. These analyses also reveal a defect in nucleotide synthesis that may contribute to impair brown adipocyte proliferation. Our findings demonstrate an essential role for mTORC1 in the regulation of BAT recruitment and metabolism in response to cold.
Results
Cold exposure promotes mTORC1 activity in BAT
Cold exposure rapidly activates thermogenesis in BAT1,26. The rapid induction of BAT thermogenic capacity is highly dependent on peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), a key transcriptional regulator that promotes the expression of several thermogenic genes, including Ucp127. Confirming previous findings, we observed that both acute (6 hours) and chronic (2 weeks) cold exposure (10 °C) strongly enhance the expression of Pgc1a and Ucp1 in BAT (Fig. 1A). Other genes known to participate in the metabolic adaptation to cold were also induced in these conditions (Figure S1A).
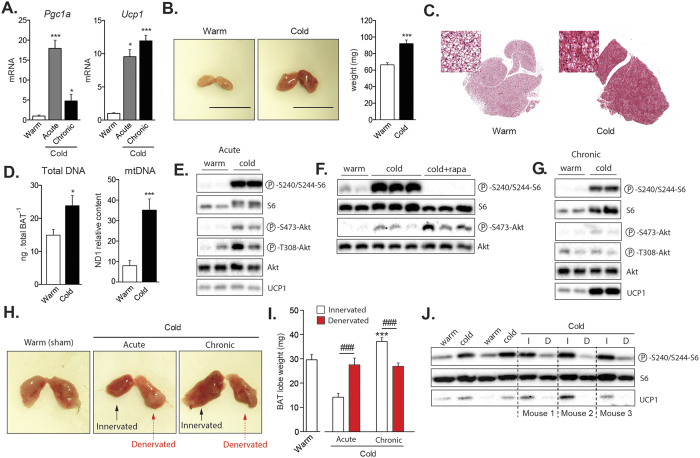
Figure 1. Cold exposure promotes mTORC1 activity in BAT through the sympathetic nervous system.
Mice were either kept at thermoneutrality (warm; 30 °C) or exposed to cold (10 °C) during 2 weeks. Acute cold exposure (6-hour at 10 °C) was performed following 2 weeks at thermoneutrality. (A) Gene expression of Pgc1a and Ucp1 in BAT (n = 8 for warm; n = 5 for acute cold; n = 8 for chronic cold; mean +/− SEM; ANOVA; *P < 0.05; ***P < 0.001). (B) (Left) Representative pictures of BAT collected from mice kept at thermoneutrality or exposed to cold for 2 weeks. (Right) BAT weight measured from the same animals (n = 14–22; mean +/− SEM; t-test; ***P < 0.001). (C) Representative H&E staining of representative BAT collected from mice kept at thermoneutrality or in cold for 2 weeks. (D) (Left) Total DNA content (n = 7–8; mean +/− SEM; t-test; *P < 0.05) and (right) mtDNA content measured in mice kept in warm or cold for 2 weeks. (n = 8; mean +/− SEM; t-test; ***P < 0.05). (E) Western blots performed on BAT collected from mice following warm or acute cold exposure. Mice were fasted during the cold exposure (6-hour period). (F) Western blots performed on BAT collected from mice following warm or acute cold exposure. A group of cold-exposed mice received an acute intraperitoneal injection of rapamycin (2 mg/kg) one-hour prior to the cold exposure (6-hour period). (G) Western blots performed on BAT collected from mice following warm or chronic cold exposure. Mice were fasted during the last 6 hours. (H) Representative pictures of unilateral BAT denervation. (Left) Sham mice kept at thermoneutrality, (middle) unilateral BAT denervation following acute cold or (right) chronic cold exposure. (I) BAT lobe weight following unilateral BAT denervation (n = 8 for Sham-Warm; n = 5 for acute cold; n = 10 for chronic cold; mean +/− SEM; Two-way ANOVA; ***P < 0.001 vs. Sham-Warm lobe; ###P < 0.001 vs. Innervated lobe). (J) Western blots performed on BAT collected from mice with unilateral BAT denervation. Mice were fasted during the last 6 hours. Each mouse was its own control. In this panel, (I) refers to the innervated lobe and and (D) refers to the denervated lobe. All gels have been run under the same experimental conditions.
As previously reported, we observed that chronic exposure to cold induced the expansion of BAT, a process required for the long-term adaptation to cold1,22 (Fig. 1B). Histological and biochemical analyses revealed that chronic cold exposure increased BAT density, total DNA and mitochondrial DNA (mtDNA) content, indicating that the elevation in BAT mass is the result of increased cell number and mitochondrial mass (Fig. 1C,D). Because mTORC1 plays a crucial role in regulating anabolic processes required for cell growth, cell proliferation and mitochondrial biogenesis, we hypothesized that this signaling node may be particularly important for the adaptation of BAT to cold temperature. To specifically test the impact of cold on mTORC1 signaling, mice were acutely or chronically exposed to cold and BAT was collected for analyses. As shown before, we observed that cold exposure rapidly promotes food intake in mice (Figure S1B). Because food intake elevates circulating levels of insulin and nutrients, two well-characterized upstream activators of mTORC1, all the mice were fasted for 6 hours before sacrifice to avoid any contribution of these factors to the modulation of mTOR signaling. As shown in Fig. 1E, we observed that acute cold exposure induced a robust increase in the phosphorylation levels of ribosomal protein S6, a readout of mTORC1 activity. These results confirm the findings of two recent reports showing that adrenergic stimulation and cold both activate mTORC1 in WAT and BAT28,29. Interestingly, pre-treating mice with the mTORC1 inhibitor rapamycin was sufficient to block cold-induced phosphorylation of S6 (Fig. 1F). To test whether the elevation in mTOR signaling was sustained over time, mice were exposed to cold for two weeks. We observed that mTORC1 activity was also highly induced in this condition (Fig. 1G). Altogether, these results indicate that both acute and chronic cold exposure activate mTORC1 in BAT. Interestingly, the activation of mTORC1 by cold was associated with an elevation in the phosphorylation of protein kinase B/Akt (Akt) (Fig. 1E to G), indicating that the phosphoinositide-3 kinase (PI3K)/mTORC2 axis was also activated by cold, as recently suggested30.
Cold-induced mTORC1 activation in BAT depends on the sympathetic nervous system
The elevation in mTORC1 activity observed in response to cold is robust. Such rise is also surprising considering that circulating insulin, a key activator of mTORC1, was not increased in cold-exposed fasted animals. In fact, a reduction in plasma insulin was even observed in mice exposed to cold (Figure S1C). These results strongly support the idea that other factors, not related to the nutritional status, contribute to activate mTORC1 in response to cold. It has long been known that BAT thermogenesis is regulated by the central nervous system through the SNS31. To test the contribution of the SNS to the activation of mTORC1, we first injected mice with CL316,243 (CL), a highly selective β3-adrenergic agonist that activates BAT thermogenesis32,33,34. We observed that CL strongly increased mTORC1 activity in BAT (Figure S1D). However, we also found that CL induced a robust increase in circulating insulin levels following injection (Figure S1E), an effect that has been reported by others35. Because insulin activates mTORC1, a direct role for the SNS in cold-induced mTORC1 activation cannot be demonstrated using this approach. To circumvent this limitation, we performed unilateral BAT denervation and acutely or chronically exposed mice to cold. This approach is extremely powerful as it allows a direct comparison between the innervated and the denervated lobes of BAT within the same mouse, thus ruling out the contribution of humoral factors to the regulation of the pathway. Confirming the validity of the surgical procedure, we observed that blocking the adrenergic drive to BAT reduced glucose uptake, impaired the mobilization of TG and reduced mtDNA content (Figure S1F to S1H). Unilateral BAT denervation completely prevented the expansion of the denervated lobe normally observed in response to chronic cold exposure (Fig. 1H,I). Strikingly, we found that this procedure also prevented the induction of mTORC1 activity observed in response to cold, indicating a key role for the SNS in this effect (Fig. 1J).
Loss of mTORC1 reduces mitochondrial biogenesis and prevents BAT expansion in response to cold
In order to test the importance of mTORC1 in regulating BAT function and expansion in response to cold, we crossed RaptorLox/Lox mice36 with mice expressing the Cre-recombinase under the control of the Adiponectin promoter37. This method was efficient to specifically delete Raptor, an essential component of mTORC1, in adipose tissues (Ad-RaptorKO) (Fig. 2A and Figure S2A). At thermoneutrality and following chronic cold exposure, Ad-RaptorKO mice were slightly heavier than control animals, despite a significant reduction in WAT masses (Figure S2B). Consistent with the reduction in WAT size, Ad-RaptorKO mice showed lower levels of circulating leptin and NEFAs (Figure S2C).
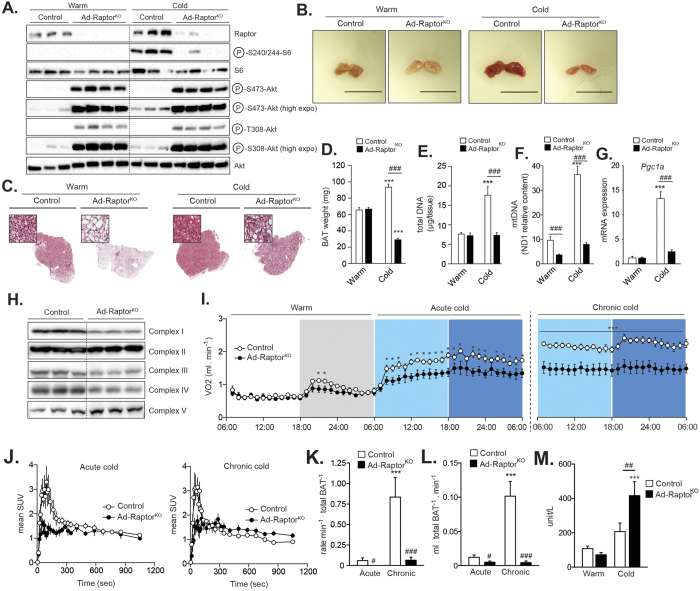
Figure 2. Loss of mTORC1 impairs cold-induced BAT expansion, mitochondrial biogenesis, and oxidative metabolism.
Control and Ad-RaptorKO mice were either kept at thermoneutrality (warm; 30 °C) or exposed to cold (10 °C) for 2 weeks. (A) Western blots performed on BAT following warm or acute cold exposure. All gels have been run under the same experimental conditions. (B) Representative pictures of BAT and (C) H&E staining of representative BAT collected after warm or chronic cold exposure. (D) Total BAT mass (n = 13–17; mean +/− SEM; Two-way ANOVA; ***P < 0.001 vs. Warm; ###P < 0.001 vs. Control). (E) Total DNA content (n = 7–9; mean +/− SEM; Two-way ANOVA; ***P < 0.001 vs. Warm; ###P < 0.001 vs. Control). (F) mtDNA content (n = 7–9; mean +/− SEM; Two-way ANOVA; ***P < 0.001 vs. Warm; ###P < 0.001 vs. Control). (G) Gene expression of Pgc1a (n = 4; mean +/− SEM; Two-way ANOVA; ***P < 0.001 vs. Warm; ###P < 0.001 vs. Control). (H) Western blots performed on BAT collected from control and Ad-RaptorKO mice following chronic cold exposure. All gels were run under the same experimental conditions. (I) Oxygen consumption at thermoneutrality (warm) and during the transition to cold. (n = 6–8; mean +/− SEM; t-test; *P < 0.05; ***P < 0.001). White section: thermoneutrality, when the lights were on; Grey section: thermoneutrality, when the lights were off; Light blue section: cold, when the lights were on; Dark blue section: cold, when the lights were off. (J) Mean SUV time-activity curves of 11C-acetate during (left) acute and (right) chronic cold exposure (n = 4–5; mean +/− SEM). (K) Total BAT oxidative activity index corrected for tissue weight (n = 6–8; mean +/−SEM; Two-way ANOVA; ***P < 0.001 vs. Warm; #P < 0.05 vs. Control; ###P < 0.001 vs. Control). (L) Total BAT blood flow index corrected for tissue weight (n = 4–5; mean +/− SEM; Two-way ANOVA; ***P < 0.001 vs. Warm; #P < 0.05 vs. Control; ###P < 0.001 vs. Control). (M) Plasma CK activity (n = 10–13; mean +/− SEM; Two-way ANOVA; ***P < 0.001 vs. Warm; ###P < 0.001 vs. Control).
As shown in Fig. 2A, the loss of Raptor completely prevented the phosphorylation of S6 induced by acute cold exposure. As previously reported, Raptor deletion caused an important increase in the phosphorylation of Akt (Fig. 2A), an effect linked to the reduction in the negative feedback loops from mTORC1 to PI3K-Akt36,38,39. Morphological, histological and biochemical analyses were next performed in BAT to characterize the physiological consequences linked to chronic loss of mTORC1. We first observed that BAT isolated from Ad-RaptorKO mice housed at thermoneutrality (warm; 30 °C) was whiter than control BAT (Fig. 2B, left panel). Histological analyses revealed the presence of larger lipid droplets accumulated in brown adipocytes of Ad-RaptorKO (Fig. 2C, left panel). Interestingly, the weight of BAT and the total number of cells, as measured by the DNA content, were not different between control and Ad-RaptorKO mice housed in a warm environment (Fig. 2D,E). However, we noticed an important reduction in mtDNA content in BAT of Ad-RaptorKO, indicating that mTORC1 is required to support basal mitochondrial biogenesis (Fig. 2F). The impact of Raptor deletion was also tested in animals exposed to cold for two weeks. Strikingly, we found that loss of mTORC1 completely blocked the expansion of BAT observed in response to chronic cold exposure (Fig. 2B, right panel). We even observed a reduction in BAT weight in Ad-RaptorKO mice exposed to cold, an effect likely linked to the reduction in intracellular TG (Fig. 2C,D). Contrary to control mice, Ad-RaptorKO mice did not show any increase in BAT DNA content following cold exposure (Fig. 2E). These animals also failed to efficiently increase mitochondrial biogenesis, as demonstrated by the severe reduction in mtDNA content (Fig. 2F). This latter effect was associated with a profound defect in the expression of Pgc1a (Fig. 2G) and Ucp1 (Figure S2D). We also measured a reduction in the protein levels of complex I, III and IV of the electron transport in response to Raptor deletion (Fig. 2H). Altogether, these results indicate that mTORC1 controls mitochondrial biogenesis and plays a crucial role in promoting BAT expansion in response to cold.
Loss of mTORC1 impairs BAT oxidative metabolism and non-shivering thermogenesis
To characterize the systemic consequences associated with the loss of mTORC1 in BAT, we performed indirect calorimetry experiments in control and Ad-RaptorKO mice. In a warm environment, a condition in which BAT is not solicited, we did not observe any difference in oxygen consumption between the groups in the light phase (Fig. 2I, white section). We noted that Ad-RaptorKO mice had a tendency for lower oxygen consumption in the dark phase (Fig. 2I, grey section). As reported before40, acute and chronic cold exposure sequentially increased oxygen consumption in control mice, an indicator of increased non-shivering thermogenesis activation and chronic adaptation, respectively (Fig. 2I). In response to acute cold, Ad-RaptorKO mice showed lower oxygen consumption compared to control animals (Fig. 2I). Importantly, the difference between control and Ad-RaptorKO mice were amplified in response to chronic cold exposure (Fig. 2I). The inability of Ad-RaptorKO to activate mitochondrial biogenesis and expand BAT mass upon chronic cold exposure likely explains this result.
To determine whether the reduction in oxygen consumption in Ad-RaptorKO was linked to a specific defect in BAT function, we performed positron emission tomography (PET) experiments using 11C-acetate, a metabolic tracer that allows live measurement of oxidative metabolism by the tricarboxylic acid (TCA) cycle9,14,41. Time-activity curves calculated from 11C-acetate dynamic scans revealed a striking defect in BAT metabolic activity in Ad-RaptorKO mice exposed to cold (Fig. 2J). Mathematical modeling derived from the 11C-acetate time-activity curves confirmed that BAT from Ad-RaptorKO mice displayed a profound defect in oxidative metabolism (Fig. 2K). These differences were even amplified when the data were corrected for total BAT mass, which supports the indirect calorimetry data presented above. Importantly, we did not observe any impact of mTORC1 deletion on oxidative metabolism in any other adipose tissues tested, showing that this effect was specific to BAT (Figure S2E-S2F). Three-compartment kinetic modeling has previously demonstrated that 11C-acetate can be used as a reliable marker of tissue blood flow14,42,43. Using this model, we showed that deleting mTORC1 completely blocked the elevation in blood flow normally observed in BAT chronically exposed to cold (Fig. 2L). Interestingly, we observed that the expression of vascular endothelial growth factor alpha (Vegfa), a key regulator of angiogenesis, was significantly reduced in BAT of Ad-RaptorKO chronically exposed to cold (Figure S2G). Altogether, these results show that mTORC1 in BAT is essential to support oxidative metabolism and the normal physiological adaptation of this tissue to cold temperature.
Despite a severe decrease in BAT oxidative metabolism and systemic oxygen consumption, which supports a reduction in BAT thermogenesis, Ad-RaptorKO mice survived to cold exposure. We observed signs of torpor in Ad-RaptorKO mice, but this effect was marginal, being noticed in only a few mice. These results indicate that shivering may have been activated in skeletal muscle as an alternative to produce heat and preserve body temperature. Supporting this hypothesis, Ad-RaptorKO mice showed higher plasma levels of creatine kinase (CK) activity, which is indicative of enhanced shivering (Fig. 2M).
Live metabolic imaging reveals profound modifications in substrate partitioning in BAT of Ad-RaptorKO mice
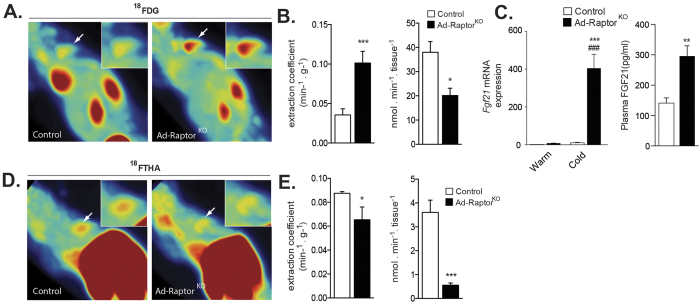
Glucose uptake is rapidly increased in BAT of rodents14,44,45 and humans9,23,46 exposed to cold. Glycolytic intermediates are essential to support lipogenesis and thermogenesis in BAT47,48,49,50. Here, the dynamic uptake of glucose by tissues was assessed using the PET tracer 2-deoxy-2-[18F]-fluoro-D-glucose (18FDG). Upon an acute cold challenge, we observed that 18FDG uptake by BAT was significantly higher in Ad-RaptorKO mice versus controls (Figure S3A and S3B). In mice chronically exposed to cold, we noticed that the extraction coefficient (capacity for one gram of tissue to extract substrate from the circulation) was increased in response to mTORC1 loss (Fig. 3A and B left panel). Such increase in glucose uptake is likely the result of the elevation in Akt activation in BAT of Ad-RaptorKO mice (Fig. 2A), a kinase recognized to play a crucial role in promoting glucose transport in brown adipocytes30,51. Interestingly, we also noted elevated expression of fibroblast growth factor 21 (Fgf21) in BAT of Ad-RaptorKO mice chronically exposed to cold (Fig. 3C), which may have contributed to drive glucose uptake, as reported52,53,54. High circulating FGF21 was also detected in these mice (Fig. 3C). Despite the increase in glucose extraction coefficient, it is important to point out that the total uptake of glucose by the whole BAT was reduced in Ad-RaptorKO (Fig. 3B, right panel). This was likely caused by the severe reduction in BAT mass and by the elevation in glucose uptake in WAT in these mice (Figure S3C).
Figure 3. Loss of mTORC1 affects substrate partitioning in BAT.
Control and Ad-RaptorKO mice were exposed to cold (10 °C) during 2 weeks and fasted for 6 hours before dynamic PET scan procedures. (A) Representative PET images of extraction coefficient uptake of the glucose analog 18FDG between 25–30 min post-injection (last frame). White arrows point to the interscapular BAT that is highlighted in the upper right insert. (B) (Left) Glucose extraction coefficient was determined using the Patlak graphical analysis following a 30-minutes dynamic scan. (Right) Total dynamic glucose uptake corrected for tissue weight was calculated from the extraction coefficient, BAT weight and plasma glucose levels (n = 4–5; mean +/− SEM; t-test; *P < 0.05; ***P < 0.001). (C) Gene expression (left) (n = 4; mean +/− SEM; Two-way ANOVA; ***P < 0.01 vs. Warm; ###P < 0.001 vs. Control) and circulating levels (right) of FGF21 (n = 8; mean +/− SEM; t-test; **P < 0.01). (D) Representative PET images of the uptake of the fatty acid analog 18FTHA between 25–30 min post-injection (last frame). White arrows point to the interscapular BAT that is highlighted in the upper right insert. (E) (Left) NEFA extraction coefficient was determined using the Patlak graphical analysis following a 30-minutes dynamic scan. (Right) Total dynamic NEFA uptake corrected for tissue weight was calculated from the extraction coefficient, BAT weight and plasma NEFA levels (n = 4; mean +/− SEM; t-test; *P < 0.05; ***P < 0.001).
NEFA uptake is strongly enhanced in BAT upon cold exposure9,14. The uptake of NEFA is key to support lipogenesis and the replenishment of intracellular TG pools during acute and chronic cold exposure14. To determine the impact of mTORC1 loss on NEFA uptake by BAT, we performed a set of experiments with the fatty-acid PET tracer analogue, [18F]-fluoro-6-thia-heptadecanoic acid (18FTHA). We first observed that 18FTHA uptake by BAT was not different between control and Ad-RaptorKO mice acutely exposed to cold (Figure S3D,E). However, we measured a slight, but significant reduction in 18FTHA uptake in BAT of Ad-RaptorKO mice exposed to a chronic cold challenge (Fig. 3D,E). We noticed that both the extraction coefficient and the total uptake rate were significantly decreased in BAT of Ad-RaptorKO mice (Fig. 3E). These results are consistent with the fact that BAT of Ad-RaptorKO mice exhibit profound reductions in mitochondrial mass, BAT expansion, and oxidative metabolism in response to chronic cold stimulation.
Loss of mTORC1 severely impairs metabolism in BAT following chronic cold exposure
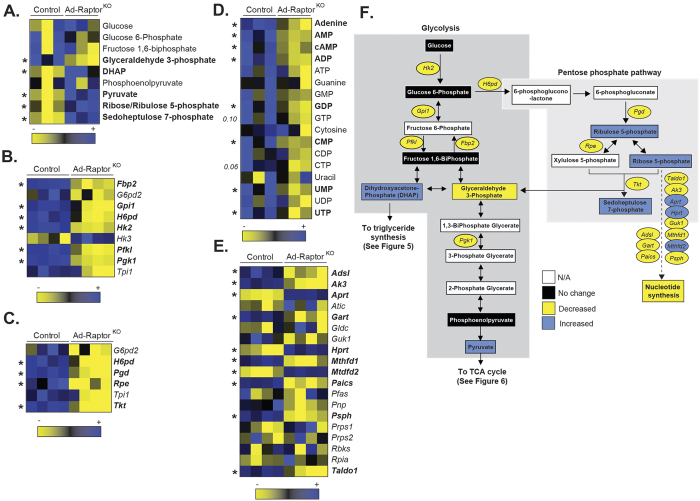
To better define the importance of mTORC1 for BAT metabolic adaptation to cold, we have performed metabolomics and transcriptomics analyses on BAT samples isolated from control and Ad-RaptorKO mice. Supporting the elevation in glucose extraction in response to chronic mTORC1 inhibition (Fig. 3), BAT of Ad-RaptorKO mice markedly accumulated key glycolytic products (Fig. 4A, see also the metabolic map in Fig. 4F). Specifically, we observed a striking elevation in dihydroxyacetone-phosphate (DHAP) and pyruvate levels in Ad-RaptorKO mice (3.6 and 6.4 fold, respectively). Such elevations were observed despite a reduction in the expression of many glycolytic genes, indicating that these changes were not driven by a transcriptional modulation of the glycolytic machinery (Fig. 4B). Supporting the elevation in glucose flux in response to mTORC1 loss, we also measured an increase in the production of ribose/ribulose 5-phosphate and sedoheptulose 7-phosphate, which are key intermediates of the pentose phosphate pathway (PPP) (Fig. 4A). As observed in the case of glycolysis, the elevation in PPP metabolites was associated with a reduction rather than an increase in the expression of genes coding for proteins regulating this pathway (Fig. 4C). The PPP is a key metabolic pathway used for nucleotide biosynthesis and for the generation of NADPH for de novo lipogenesis. Interestingly, we observed a severe decrease in the levels of most nucleotides in BAT of Ad-RaptorKO mice, indicating an inability of the tissue to produce nucleotides despite the accumulation of the several molecules required for their production (Fig. 4D). At the transcriptional level, we observed a mixed profile regarding the expression of genes regulating nucleotide biosynthesis, some genes showing an elevation whereas others showing a reduction (Fig. 4E).
Figure 4. mTORC1 loss affects glucose metabolism in BAT following chronic cold exposure.
Control and Ad-RaptorKO mice were cold exposed during 14 days and fasted for 6 hours before BAT collection. (A) Glycolytic intermediates measured by metabolomics. Metabolites in bold are statistically different between control and Ad-RaptorKO mice (n = 3; mean +/− SEM; t-test; *P < 0.05). (B) Glycolytic gene expression extracted from the microarray dataset. Genes in bold are statistically different between control and Ad-RaptorKO mice (n = 4; mean +/− SEM; SAM t-test; *FDR < 0.05). (C) Genes expression of components of the pentose phosphate pathway extracted from the microarray dataset. Genes in bold are statistically different between control and Ad-RaptorKO mice (n = 4; mean +/− SEM; SAM t-test; *FDR < 0.05). (D) Nucleotides measured by metabolomics. Metabolites in bold are statistically different between control and Ad-RaptorKO mice (n = 3; mean +/− SEM; t-test; *P < 0.05). (E) Expression of genes involved in nucleotide synthesis extracted from the microarray dataset. Genes in bold are statistically different between control and Ad-RaptorKO mice (n = 4; mean +/− SEM; SAM t-test; *FDR < 0.05). (F) Schematic summary of transcriptomics and metabolomics data.
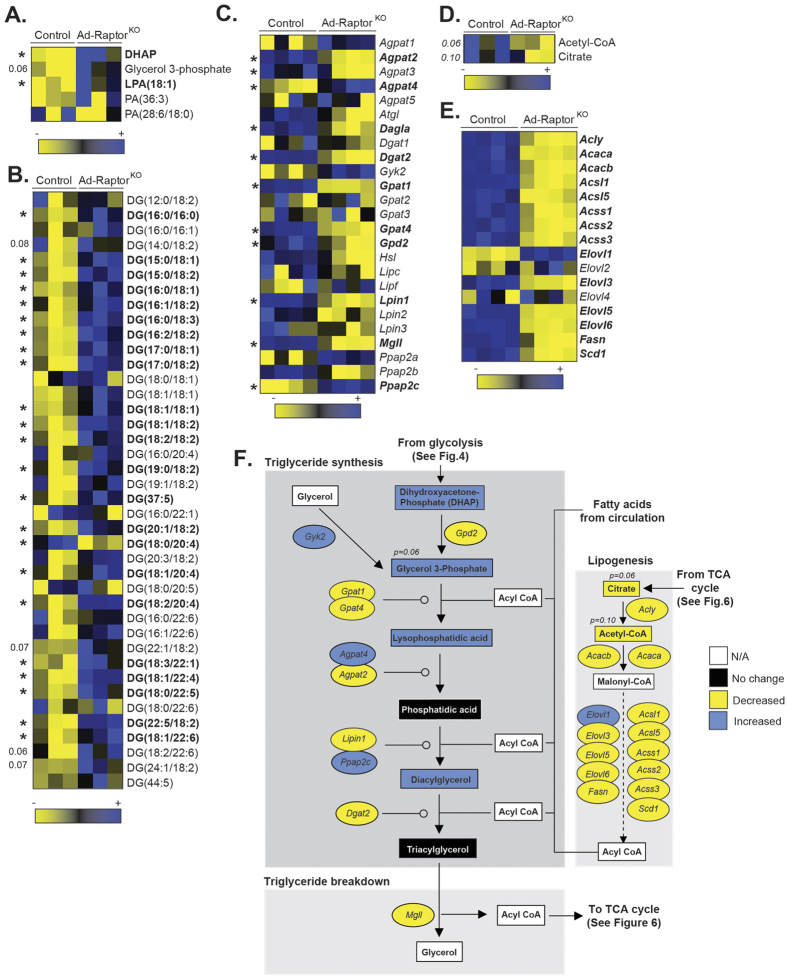
The metabolomics data presented above showed that Ad-RaptorKO mice accumulated high levels of DHAP, a key glycolytic intermediate used for the synthesis of glycerolipids. Downstream of DHAP, we observed an elevation in glycerol 3-phosphate and lysophosphatidic acid (LPA) accumulation in BAT of Ad-RaptorKO mice (Fig. 5A, see also the metabolic map in Fig. 5G). This effect was linked to a dramatic increase in the content of various diacylglycerols (DG) (Fig. 5B). Although several TG species were also higher in BAT of Ad-RaptorKO mice than in control animals, this trend was not as striking as what was observed for DG (Figure S4). Nevertheless, these results indicate that an important fraction of glycolytic intermediates are shuttled onto the glycerol moiety of glycerolipids in Ad-RaptorKO mice. As reported above for glycolysis and PPP, these effects were not caused by an elevation in the expression of genes coding for proteins regulating this process. Indeed, we observed that several genes promoting glycerolipid synthesis were decreased rather than increased in BAT of Ad-RaptorKO mice (Fig. 5C). Triacylglycerol synthesis involves the sequential esterification of acyl-CoA on glycerol 3-phosphate, phosphatidic acid, and DG respectively. The acyl-CoA molecules are provided either by circulating lipids or can be synthesized de novo from the conversion of citrate to malonyl-CoA (see the metabolic map in Fig. 5F). Here, we observed a reduction in citrate and acetyl-CoA levels in BAT of Ad-RaptorKO (Fig. 5D). We also measured a profound defect in the expression of genes coding for proteins required for de novo fatty acid synthesis (Fig. 5E). These results suggest that the fatty acids contributing to the elevation in glycerolipid synthesis in BAT of Ad-RaptorKO mice are most likely derived from circulating lipids rather than de novo lipogenesis.
Figure 5. mTORC1 loss severely impairs lipid metabolism in BAT following chronic cold exposure.
Control and Ad-RaptorKO mice were cold exposed for 14 days and fasted during 6 hours before BAT collection. (A) Metabolites entering in the synthesis of glycerolipids from glucose measured by metabolomics. Metabolites in bold are statistically different between control and Ad-RaptorKO mice (n = 3; mean +/− SEM; t-test; *P < 0.05). (B) Diacylglycerol (DG) species measured by metabolomics. Metabolites in bold are statistically different between control and Ad-RaptorKO mice (n = 3; mean +/− SEM; t-test; *P < 0.05). (C) Expression of genes entering in the synthesis or the breakdown of glycerolipids extracted from the microarray dataset. Genes in bold are statistically different between control and Ad-RaptorKO mice (n = 4; mean +/− SEM; SAM t-test; *FDR < 0.05). (D) Lipogenic metabolites produced from the TCA cycle measured by metabolomics (n = 3; mean +/− SEM; t-test). (E) Lipogenic gene expression extracted from the microarray dataset. Genes in bold are statistically different between control and Ad-RaptorKO mice (n = 4; mean +/− SEM; SAM t-test; *FDR < 0.05). (F) Schematic summary of transcriptomics and metabolomics data.
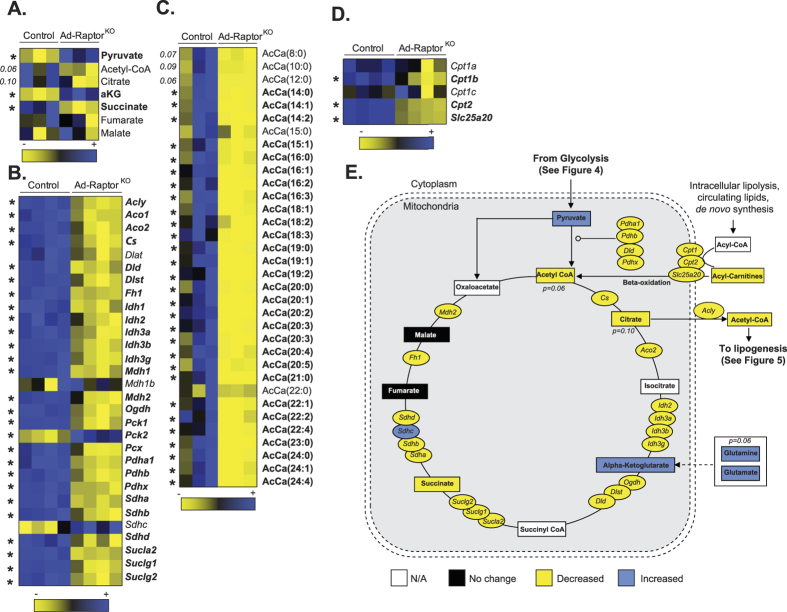
As presented in Fig. 6A (and Fig. 4A), high levels of pyruvate accumulated in BAT of Ad-RaptorKO mice. Pyruvate molecules produced from glycolysis can be used to produce lactate or can be transported into the mitochondria where they are converted to acetyl-CoA. In the mitochondria, acetyl-CoA is used in the TCA cycle to promote cellular respiration and support thermogenesis (see metabolic map in Fig. 6E). Despite high levels of pyruvate in BAT of Ad-RaptorKO mice, we observed no change in lactate levels, but did measure a reduction in the levels of acetyl-CoA and other TCA cycle intermediates such as citrate and succinate, indicating a defect in pyruvate catabolism (Fig. 6A). This effect was associated with a profound reduction in the expression of genes coding for proteins catalyzing the conversion of pyruvate to acetyl-CoA, as well as almost all the genes encoding proteins of the TCA cycle (Fig. 6B). Interestingly, despite the clear impairment in TCA cycle, we observed that alpha-ketoglutarate (aKG) levels were elevated in Ad-RaptorKO mice (Figure S5A and Fig. 6E). This effect may have been caused by the elevation in glutaminolysis, as we observed very high levels of glutamine and glutamate in Ad-RaptorKO mice (Figure S5B). In addition to pyruvate, acetyl-CoA can be produced from the β-oxidation of acyl-CoA in the mitochondria. The import and catabolism of acyl-CoA depends on the production of acyl-carnitines (AcCa) by carnitine acetyltransferases. As shown in Fig. 6C, the production of AcCa was completely impaired in BAT of Ad-RaptorKO mice. We also observed a significant reduction in the expression of the genes encoding proteins required for AcCa formation and mitochondrial transport (Fig. 6D). Altogether, these results indicate that glucose and fatty acid oxidative metabolism are severely reduced in BAT of Ad-RaptorKO mice. An integrative representation of the metabolomics and transcriptomics data is presented in Figure S6.
Figure 6. mTORC1 impairs oxidative metabolism in BAT following chronic cold exposure.
Control and Ad-RaptorKO mice were cold exposed for 14 days and fasted during 6 hours before BAT collection. (A) Metabolites of the TCA cycle measured by metabolomics. Metabolites in bold are statistically different between control and Ad-RaptorKO mice (n = 3; mean +/− SEM; t-test; *P < 0.05). (B) TCA cycle gene expression extracted from the microarray dataset. Genes in bold are statistically different between control and Ad-RaptorKO mice (n = 4; mean +/− SEM; SAM t-test; *FDR < 0.05). (C) Acyl-Carnitine (AcCa) metabolites measured by metabolomics. Metabolites in bold are statistically different between control and Ad-RaptorKO mice (n = 3; mean +/− SEM; t-test; *P < 0.05). (D) Expression of genes controlling Acyl-Carnitine production extracted from the microarray dataset. Genes in bold are statistically different between control and Ad-RaptorKO mice (n = 4; mean +/− SEM; SAM t-test; *FDR < 0.05). (E) Schematic summary of transcriptomics and metabolomics data.
The defects in BAT expansion and metabolism in Ad-RaptorKO are not mediated by a reduction in adrenergic signaling
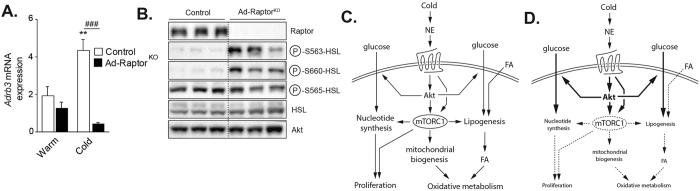
It was recently shown that chronic mTORC1 inhibition with rapamycin reduces the transcription of the β3-adrenergic receptor (Adrb3) in WAT and BAT29. To determine whether a reduction in adrenergic signaling could have contributed to the defect in BAT expansion and metabolism in Ad-RaptorKO mice, we measured mRNA expression of Adrb3. Although mTORC1 loss did not affect Adrb3 expression in BAT of mice kept at thermoneutrality, we observed a severe reduction in the expression of this gene when animals were chronically exposed to cold (Fig. 7A). To evaluate whether this effect was associated with changes in adrenergic signaling, we next performed western blot analyses to measure the phosphorylation status of hormone sensitive lipase (HSL) on S563 and S660, which are key residues phosphorylated downstream of ADRB3. Surprisingly, we observed that, despite a severe reduction in Adrb3 mRNA expression, the phosphorylation of HSL on S563 and S660 was massively increased in Ad-RaptorKO mice. On the other hand, the phosphorylation of HSL on S565, a site phosphorylated by AMP kinase that negatively regulates the activity of HSL, was not modulated in these conditions. These results indicate that defects in BAT observed in response to chronic mTORC1 inhibition are not a consequence of a reduction in adrenergic signaling. In fact, these changes occurred in the face of elevated adrenergic signaling, which further highlights the importance of mTORC1 for BAT recruitment and metabolic adaptation to cold. How mTORC1 inhibition drives adrenergic signaling is unknown, but does not involve any elevation in Adrb1 or Adrb2 expression (Figure S7). A model recapitulating our findings is presented in Fig. 7C,D.
Figure 7. The defects in BAT expansion and metabolism in Ad-RaptorKO are not mediated by a reduction in adrenergic signalling.
Control and Ad-RaptorKO mice were cold exposed for 14 days and fasted during 6 hours before BAT collection. (A) Gene expression of Adrb3 (n = 4; mean +/− SEM; Two-way ANOVA; **P < 0.01 vs. Warm; ###P < 0.001 vs. Control). (B) Western blots performed on BAT collected from control and Ad-RaptorKO mice following chronic cold exposure. All gels have been run under the same experimental conditions. (C) Model summarizing the role of mTORC1 in BAT during cold adaptation. (D) Model summarizing the impact of mTORC1 loss in BAT during cold adaptation. Bold and dashed lines indicate increase and decrease respectively, in comparison to the model presented in (C).
Discussion
In response to chronic cold exposure, BAT mass expands to increase thermogenic capacity1. This process, termed BAT recruitment, plays a crucial role in the long term adaptation to a cold environment. The signaling events that contribute to BAT expansion during chronic cold exposure are not completely characterized. Here we show that mTORC1 activity is highly induced in BAT following cold exposure. This effect was observed in response to acute and chronic cold stimulation, supporting a continuous role for mTORC1 in the response of BAT to cold. Over the years, nutrients and growth factors were identified as the main factors activating mTORC125. The elevation in circulating levels of amino acids, glucose and insulin following food intake rapidly induces mTORC1 in tissues39,55. Here, we provide evidences that mTORC1 activation by cold occurs independently of nutrients and insulin. Hence, cold-driven mTORC1 activity was observed in mice that did not have access to food. In these animals, we did not observe any increase in insulin and glucose levels following cold exposure. Additionally, we observed that cold-induced mTORC1 activity was completely blocked when BAT was denervated. In these experiments, a direct comparison between the innervated and the denervated lobes of BAT was performed within the same mouse, thus ruling out the contribution of any humoral factor to the regulation of the pathway. Together, these results clearly demonstrate that the SNS is primarily responsible for inducing mTORC1 in mice exposed to cold.
Two recent studies reported that β-adrenergic stimulation and cold exposure activate mTORC1, an effect that was shown to be essential to support WAT browning and BAT formation28,29. Importantly, the role of mTORC1 in regulating BAT recruitment and metabolism in vivo in response to chronic cold stimulation, which is the object of the present report, was not addressed in these studies. One of these reports revealed that β-adrenergic stimulation activates mTORC1 by promoting the phosphorylation of Raptor by protein kinase A (PKA), an effect that was reported to be independent of Akt, a key activator of mTORC156,57,58,59. Here, we found that the activation of mTORC1 by cold was associated with an increase in Akt phosphorylation. The reason for these differences is unclear but suggests that the SNS likely activates mTORC1 through Akt-dependent and Akt-independent processes. The phosphorylation of Akt on S473 is an event that is mediated by mTORC260. Consistent with our findings, a recent report shows that the mTORC2/Akt axis is activated in brown adipocytes upon β-adrenergic stimulation and cold exposure30. In this study, adipose-specific loss of mTORC2 was shown to reduce Akt-mediated glucose uptake and to lower resistance to cold30. Another report also showed that adrenergic stimulation promotes mTORC2 activity in brown adipocytes in vitro61. In that case, it was observed that mTORC2 promotes glucose uptake independently of Akt61. Altogether, these evidence indicate that cold sequentially activates mTORC2 and mTORC1, and suggest that each of these complexes plays complementary roles to insure the optimal tissue response to cold. On the one hand, mTORC2 promotes glucose uptake, an event required to support lipogenesis and thermogenesis14,61. On the other hand, mTORC1 promotes BAT expansion, mitochondrial biogenesis and oxidative metabolism, which are all needed to sustain heat production. It is extremely important to take into consideration that cold exposure rapidly drives food intake in rodents to cope with the rise in energy expenditure1. Since the mTOR pathway is very sensitive to activation by insulin and nutrients, it is likely that nutrition synergizes with the adrenergic stimulation to maximize the activation of mTOR signaling in response to cold.
Our findings clearly show that mTORC1 deeply affects mitochondrial mass. We report that cold-induced mitochondrial biogenesis is completely impaired in Ad-RaptorKO mice, an effect associated with an important reduction in the expression of Pgc1a, a transcriptional co-activator that controls mitochondrial biogenesis and function62. Interestingly, mice with heart- or skeletal muscle-specific deletion of Raptor also exhibit a reduction in mitochondrial mass and Pgc1a expression63,64,65. Consistent with the reduction in mitochondrial number, live metabolic imaging and indirect calorimetry experiments revealed a severe defect in BAT oxidative metabolism and systemic oxygen consumption in Ad-RaptorKO mice. Of note, we showed that the rise in oxygen consumption observed in response to acute cold exposure was significantly reduced in Ad-RaptorKO mice. Strikingly, the additional elevation in oxygen consumption in mice chronically exposed to cold, a phenomenon linked to the increase in BAT mass and thermogenic capacity, was completely prevented in mice lacking mTORC1. These results demonstrate that mTORC1 is required to support mitochondrial biogenesis and to increase the thermogenic potential of BAT in response to chronic cold exposure. It is interesting to note that the reduction in mitochondrial biogenesis and the impairment in oxidative metabolism observed in Ad-RaptorKO mice were associated with reductions in Vegfa expression and blood flow in BAT. The importance of mTORC1 in regulating angiogenesis has been demonstrated over the years, especially in the context of tumorigenesis66. Here, our results indicate that mTORC1 exerts similar pro-angiogenic function in BAT in vivo.
Despite a clear defect in BAT oxidative metabolism, all Ad-RaptorKO survived exposure to cold. We noted that a few Ad-RaptorKO mice showed signs of torpor in response to cold exposure but no animal died, indicating that alternative heat-producing processes were taking place to ensure survival. Accordingly, Ad-RaptorKO mice showed high plasma levels of CK activity, a surrogate marker of shivering29,67. Other mouse models with defective BAT thermogenesis have also been shown to survive in the cold by promoting shivering68,69.
Using live metabolic imaging, we showed that glucose uptake was increased in BAT of Ad-RaptorKO mice. The elevation in Akt phosphorylation, an effect linked to the reduction in the negative feedback loops from mTORC1 to PI3K-Akt, likely contributed to stimulate glucose uptake. We also measured a spectacular induction in Fgf21 expression in BAT of Ad-RaptorKO mice exposed to cold. This growth factor, that was identified as a key regulator of glucose uptake and metabolism52,53,54, was previously shown to be produced and secreted by various tissues, including BAT70,71. Interestingly, a very similar induction in Fgf21 expression was observed in BAT of Ucp1KO mice71, suggesting that elevated Fgf21 production may be a common response to impaired BAT thermogenesis. Supporting a rise in glucose flux, we observed that the levels of the glycolytic intermediate DHAP were higher in BAT of Ad-RaptorKO mice. The elevation in DHAP was concomitant to an increase in glycerol 3-phosphate, LPA and DG, indicating that a significant fraction of glycolytic intermediates was channelled to the glycerol moiety of glycerolipids. Importantly, the increase in lipid synthesis was probably not the only factor that has exacerbated lipid deposition in BAT of Ad-RaptorKO mice. The reduction in oxidative metabolism, a direct consequence of impaired mitochondrial biogenesis and TCA cycle activity, has likely contributed to reduce the degradation of lipids in BAT of Ad-RaptorKO mice. In addition to DHAP, we also observed that pyruvate, another glycolytic intermediate, accumulated in BAT of Ad-RaptorKO mice. As observed for DHAP, the rise in pyruvate levels was likely the result of both increased glycolytic flux and reduced pyruvate oxidation. We observed that the conversion of pyruvate to acetyl-CoA, a step required for the oxidation of pyruvate in the mitochondria, was severely blocked in BAT of Ad-RaptorKO mice. Altogether, these findings highlight the severity of the mitochondrial defects induced by chronic mTORC1 deletion.
It was recently shown that cold increases the expression of genes related to PPP in BAT72, a metabolic pathway required to sustain de novo lipogenesis and nucleotide biosynthesis. Here, we confirmed these findings in control mice. Interestingly, Ad-RaptorKO mice showed a significant reduction in the expression of PPP genes. Despite such defects, the amount of PPP intermediates was higher in BAT of these mice. The reason for this apparent discrepancy is unclear, but may be a consequence of elevated glucose flux in BAT of Ad-RaptorKO mice. Interestingly, despite elevated PPP intermediates, we observed a striking reduction in the production of nucleotides in BAT of Ad-RaptorKO mice, suggesting a defect in nucleotide biosynthetic capability in the absence of mTORC1. Interestingly, recent reports indicate that mTORC1 directly regulates nucleotide synthesis73,74. These studies show that mTORC1 stimulates the production of new nucleotides to accommodate an increase in RNA and DNA synthesis needed for ribosome biogenesis and anabolic growth. Here, we provide evidence that mTORC1 plays a crucial role in controlling the biosynthesis of nucleotides in BAT. In addition to its well-established roles in promoting protein and lipid synthesis25, which are both required to support cell growth and proliferation, mTORC1 could regulate BAT expansion also by promoting nucleotide synthesis.
Mice lacking mTORC1 in adipose tissue were previously generated by another group by crossing Raptor floxed mice with Ap2-Cre mice (Ap2-RaptorKO)38. Although the implication of mTORC1 in regulating BAT expansion and metabolism in response to cold was not tested in this report, a few observations were made regarding the role of mTORC1 on systemic metabolism and BAT function. These investigators reported that BAT weight is reduced in Ap2-Cre mice, similar to what is reported here. Nevertheless, no obvious morphological change was reported in BAT of the Ap2-Cre model. Despite a reduction in BAT mass, the Ap2-RaptorKO mice showed enhanced oxygen consumption and protection against high-fat diet induced obesity when housed at room temperature38. Although BAT oxidative metabolism and thermogenic potential were not directly assessed in these mice, these results appear to oppose our findings. The reason for these differences is unclear but could likely be related to the use of the Ap2-Cre model, which was shown to mediate the recombination of floxed alleles in non-adipocyte cell types75,76. Another group recently developed mice lacking mTORC1 in adipose tissue using the adiponectin-cre mouse77, exactly as described here. In this study, Ad-RaptorKO housed at room temperature showed a reduction in WAT mass. As reported by Polak et al., the authors found that Ad-RaptorKO mice are protected against high-fat diet-induced obesity. However, they found that this effect was not associated with change in energy expenditure. This last results is in accordance with our experiments showing that oxygen consumption is minimally affected when Ad-RaptorKO are not exposed to cold. Although Lee et al. did not test the impact of cold on BAT in Ad-RaptorKO mice, they report that mTORC1 loss reduces BAT mass, an effect associated with elevated lipid deposition in brown adipocytes77. Moreover, they observed that deleting Raptor only in brown adipocytes using the UCP1-cre mouse similarly affected BAT mass, indicating that mTORC1 cell-autonomously controls the development of this tissue.
Recent studies reported the impact of tuberous sclerosis complex 1 (Tsc1) loss on WAT and BAT78,79. Because the loss of TSC1 constitutively activates mTORC1, it would be expected that these studies would show results that oppose our findings. Accordingly, Adiponectin-cre mediated recombination of Tsc1 increased BAT mass, an effect associated with a trend towards higher BAT DNA content78. The impact of cold on BAT mass expansion and metabolism was not tested in this report. In another study, Xiang et al. depleted Tsc1 in adipose tissue using the Ap2-cre mice79. In this report, constitutive mTORC1 activation led to a reduction in Ucp1 and Pgc1a expression, a decrease in mitochondrial DNA content and an increase in lipid deposition in BAT. Although these results oppose our findings, it is important to point out that Ap2-Tsc1KO mice die from severe lungs dysfunction 48 hours after birth80. Investigating BAT function and morphology in these conditions was probably not optimal and preclude any comparison with our study.
Altogether, our results indicate that mTORC1 loss impairs BAT recruitment in response to chronic cold exposure, an effect associated with profound defects in mitochondrial biogenesis and metabolism. Although these results clearly establish mTORC1 as an important regulator of BAT function, it is important to point out that these observations were made following the chronic inhibition of mTORC1 in the developing adipocyte, and that we cannot exclude the possibility that some effects reported were secondary to the long-term impairment of mTORC1 signaling. Defining the impact of acute Raptor deletion using an inducible system to deplete mTORC1 only in BAT of adult mice would represent a valuable complement to the present study.
In conclusion, we provide the evidence that mTORC1 plays a critical role in the control of BAT recruitment and metabolism in response to cold. The present results extend the understanding of the molecular mechanisms regulating thermogenesis and provide possible avenues for the development of tools to control energy expenditure, fight obesity and improve glucose and lipid homeostasis in humans.
Experimental Procedures
Animal care
Male C57Bl/6N (10–12 weeks old) mice weighing 20–25 g (Jackson Laboratory, ME, USA) were divided into two groups, which were (i) kept at thermoneutrality (30 °C) or (ii) housed at 10 °C, for 14 days. Raptor floxed mice were developed previously and ordered from Jackson Laboratory (stock number 013188). Adiponectin-cre mice were also ordered from Jackson Laboratory (stock number 010803)39. For all experiments, male adiponectin-Cre; raptorLoxP/LoxP (Ad-RaptorKO) and raptorLoxP/LoxP (control) mice aged between 8 and 14 weeks were used. All animals were kept under a 12 h light/dark cycle (lights on at 0600 hours) with ad libitum access to pelleted chow (Rodent Laboratory Chow 5001, Purina, St. Louis, MO) and tap water. Distinct groups of animals were used for the PET procedures and immunohistochemistry/biochemical measurements described below. All experimental protocols were approved by the Animal Ethics Committee of the Université de Sherbrooke and in accordance with the guidelines of the Canadian Council on Animal Care.
Cold exposition
Following warm (30 °C) or cold exposure (10 °C), mice were individually housed in new cages without access to food but with free access to water. Mice were fasted during 6 h. In some experiments, mice were injected intraperitoneally with rapamycin (2 mg/kg, Sigma #37094) 1 h prior to the 6 h cold exposure.
Unilateral BAT denervation
Unilateral BAT denervation was performed as previously described81,82. Eight weeks old mice were used for this experiment. On the day of surgery, each mouse was weighed and anesthetized with isoflurane. The mouse was shaved and secured on a warm surgical table. Following a standard skin disinfection procedure with ethanol and chlorhexidine (Baxter), a lateral incision was made to expose the interscapular fat pads. A blunt forceps was then used to clamp the caudal edge of the interscapular fat pad and then retract cranially over the head of the mouse. All five branches of intercostal sympathetic nerves connecting to the right BAT fat pad were identified, carefully isolated, and sectioned. After BAT denervation, the interscapular fat pad was returned to its original position. Mice were monitored for 7 days post surgery before being exposed to experimental conditions.
Blood and tissue sampling
Mice were fasted for 6 h (06:00–12:00) and were euthanized through cardiac puncture under isoflurane anaesthesia and samples were transferred into EDTA-coated tubes. Plasma glucose, NEFA and TG levels were measured as described previously14. Insulin, leptin and FGF21 were measured using ELISA kits (#90080 and #90030, Crystal Chem, Downers Grove, IL; #MF2001, R&D Systems, Minneapolis, NE). Creatine kinase activity was measured using the Creatine Kinase Activity Assay Kit (#MAK116, Sigma Aldrich, MO). BAT was carefully excised, dissected on ice and weighed. BAT samples were immediately frozen in liquid nitrogen for protein, RNA and DNA extractions or fixed in 4% (wt/vol) paraformaldehyde for histological analyses.
Western blotting
Tissues were lysed with Triton-X 100 containing lysis buffer (50 mM HEPES, pH 7.4, 2 mM EDTA, 10 mM sodium pyrophosphate, 10 mM sodium glycerophosphate, 40 mM NaCl, 50 mM NaF, 2 mM sodium orthovanadate, 1% Triton-X 100, 0.1% sodium lauryl sulfate, 1% sodium deoxycholate and one tablet of EDTA-free protease inhibitors Roche per 25 ml). Samples were rotated at 4 °C for 10 minutes and then the soluble fractions of cell lysates were isolated by centrifugation at maximum speed for 10 min in a microcentrifuge. Protein levels were then quantified using Bradford reagents and analyzed by Western blotting. Samples were loaded on 4–12% Nupage precast gels or 10% Tris-Glycine gels (Life Technologies). Proteins were transferred to PVDF membranes and incubated with the primary antibody overnight at 4 °C in 5% non-fat milk diluted in PBS-Tween. The following antibodies were used: Akt (Cell Signaling Technology, 4691, dilution 1:1000), phospho-AKT T308 (Cell Signaling Technology, 2965, dilution 1:1000), phospho-AKT S473 (Cell Signaling Technology, 9271, dilution 1:1000), S6 (Cell Signaling Technology, 2217, dilution 1:2500), phospho-S6 S240/244 (Cell Signaling Technology, 5364, dilution 1:1000), Raptor (Cell Signaling Technology, 2280, dilution 1:1000), HSL (Cell Signaling Technology, 4107, dilution 1:1000), phospho-HSL S563 (Cell Signaling Technology, 4139, dilution 1:1000), phospho-HSL S660 (Cell Signaling Technology, 4126, dilution 1:1000), phospho-HSL S565 (Cell Signaling Technology, 4137, dilution 1:1000), UCP1 (Abcam, 10983, dilution 1:1000), and mitochondrial complexes I to V (Abcam, 110413, dilution 1:250). Secondary antibodies were purchased from Cell Signaling Technology and diluted 1:5000. Amersham ECL Western Blotting Detection Reagent (RPN2106) was used to image the blots.
Gene expression analysis
Total RNA was isolated from BAT using PureL and the Aurum™ Total RNA Fatty and Fibrous Tissue Kit (Bio-Rad, 732–6830, Mississauga, ON, Canada). RNA concentration was estimated from absorbance at 260 nm. cDNA synthesis was performed using the iScript Advanced cDNA Synthesis Kit (Bio-Rad, 172–5038, Mississauga, ON, Canada). mRNA extraction and cDNA synthesis were performed following the manufacturer’s instructions. Relative quantification of gene expression was performed on diluted cDNA (1:20) in duplicate samples using a CFX384 touchTM real-time PCR (Bio-Rad, Mississauga, ON, Canada). Chemical detection of the PCR products was achieved with SYBR Green (Bio-Rad, 172–5271, Mississauga, ON, Canada). Specificity of amplification was ensured using melt curve analyses followed by agarose gel electrophoresis of the amplified products. Fold differences in targeted mRNA expression were calculated using 2-delta cycle threshold method and data were normalized on beta-microglobulin (B2m) expression.
Measurement of Mitochondrial DNA Content
Tissues for assessing mitochondrial DNA (mtDNA) content were prepared as described previously83,84. Briefly, total DNA was isolated by proteinase K digestion followed by phenol/chloroform extraction and ethanol precipitation. Ten ng of total DNA was used to amplify mtDNA-encoded NADH dehydrogenase I (ND1) and nuclear DNA-encoded Beta-actin using real-time PCR. MtDNA content was calculated from the ratio of ND1 to genomic Beta-actin.
Histology
BAT samples were fixed overnight in 4% paraformaldehyde in PBS at 4 °C, dehydrated, embedded in paraffin, and cut into 10 μm-thick sections. Sections were stained with hematoxylin and eosin (H&E) to perform general histology. All pictures were taken on an Olympus BX60 microscope (Tokyo, Japan) at a magnification of 40X.
Indirect calorimetry
The Promethion High-Definition Room Calorimetry System was used for the indirect calorimetry studies (GA3, Sable Systems. Las Vegas, NV). Data acquisition and instrument control were coordinated by MetaScreen v. 1.6.2 and the obtained raw data was processed using ExpeData v. 1.4.3 (Sable Systems, Las Vegas, NV) using an analysis script detailing all aspects of data transformation. A standard 12 h light/dark cycle (6:00–12:00) was maintained throughout the calorimetry studies. Prior to data collection, all animals were acclimated to cages for 7 days. Mice were subsequently placed in metabolic cages at thermoneutrality. Mice were prior subjected to a 48 h measurement at 30 °C following a switch at 10 °C for 24 h. At the end of the procedure, mice were returned to a 10 °C environment for 10 days before a second round of measurement at 10 °C.
Small animal PET-CT protocol
All in vivo PET/CT experiments were initiated immediately after the insertion of a cannula in the tail vein for injection of PET tracers. All imaging experiments were performed on the avalanche photodiode-based small animal PET scanner (LabPET/Triumph, Gamma Medica, Northridge, CA) of the Sherbrooke Molecular Imaging Centre, having a 7.5 cm axial field-of-view. The animals were anaesthetized with isoflurane (1.75%, 1.5 L/min) delivered through a nose cone, and were placed in the prone position on the scanner bed with the heart centered within the field-of-view of the scanner to include the interscapular region. Boluses of each radiopharmaceutical compound (10 MBq, in 0.2 ml of 0.9% NaCl) were injected via the caudal vein over 30 seconds after starting PET data acquisition (11C-acetate) and over 60 seconds for 18FDG and 18FTHA. In one set of experiments, a 20-min dynamic data acquisition with 11C-acetate was done to determine tissue blood flow and oxidative metabolism. Just prior to the injection of 11C-acetate (less than 1 minute), an intravenous injection of CL (2 mg/kg, Sigma, #C5976) was performed to measure maximal oxidative activity.
In a second set of experiments, a 30-min dynamic data acquisition with either 18FDG or 18FTHA were used to also determine glucose or NEFA utilization, respectively, as previously described14. List-mode dynamic data acquisition allowed for flexible time framing of the data for kinetic modeling of all tracers. Low dose CT scan imaging was performed using the integrated X-O small-animal CT scanner of the Triumph platform, consisting of a 40 W X-ray tube with a 75 μm focal spot diameter and a 2240 × 2368 CsI flat panel X-ray detector. The detector pixel size was 50 μm, and a 2 × 2 pixel binning scheme was used. Scans were performed at 60 kVp and 230 μA using 512 projections in fly mode to reduce exposure. Blood samples were taken at the end of experiments by heart punctures.
PET imaging data analysis
For 11C-acetate images, dynamic series of 28 frames (1 × 30 s, 12 × 10 s, 8 × 30 s, 6 × 90 s, and 1 × 300 s) were sorted out, whereas 30 frames (1 × 30 s, 12 × 10 s, 8 × 30 s, 6 × 90 s, and 3 × 300 s) were used for 18FDG and 18FTHA imaging. 3D images were reconstructed using 15 iterations of a maximum-likelihood expectation-maximization (MLEM) algorithm incorporating physical description of the detector response functions. Regions of interest (ROIs) were drawn on short-axis images and confirmed with the μCT scan14. Input curves were extracted by means of a ROI drawn on the left ventricular cavity blood pool in summed last-frame images to seek better contrast. The sizes of these almost-circular ROIs were compared with images of eight cylinders of different diameters from which a recovery factor was extracted and applied to the ROIs for partial volume correction85. For 11C-acetate, we used a three-compartment kinetic model that estimates the generation of CO2 from the citric acid cycle using the k2 constant42 and the tissue blood flow through the K1 constant42,43. Glucose and NEFA extraction coefficient were determined using the Patlak graphical analysis86. These values represent the proportion of circulating substrate taken up per mass of tissue. The dynamic glucose and NEFA uptake were determined using the extraction coefficient multiplied by the amount of substrate in circulation.
Metabolomics and lipidomics
Polar and lipid metabolites were extracted using a chloroform-methanol technique. Briefly, 10–30 mg of BAT were crushed to powder in a liquid nitrogen-chilled mortar and pestle. Six hundred μl of LC/MS grade methanol (containing phenylalanine-d8 and valine-d8 as internal standards) was added to BAT powder following by 300 μl LC/MS grade water and 400 μl HPLC grade chloroform (containing internal standards). Top layer containing the polar metabolites and bottom layer containing the organic/lipid metabolites were separated and dried using a speedvac. Once dried, both fractions were stored at −80 °C for further analysis. Polar metabolites were resuspended in 100 μl LC/MS grade water, vortexed for 10 min at 4 °C, centrifuged at 15,000× g in a microcentrifuge at 4 °C, and the supernatant was transferred to an LC/MS vial. 1 μl of each sample was injected for LC/MS analysis as described87. Metabolites were identified by matching exact mass and chromatographic retention time to an in-house library of authentic chemical standards. Data were analyzed using XCalibur QuanBrowser 2.2; raw LC/MS peak areas were normalized to the appropriate internal standard to calculate peak area ratios. A pooled biological sample was generated for quality control purposes and was injected in triplicate at 1 μl, in singlicate at 3 μl, and in singlicate at 1 μl of a 0.1X and a 0.3X dilution in water. The pre-determined quality control cutoffs for each metabolite were CV<0.25 for the peak area ratio across the triplicate injections, and R > 0.975 for the raw peak area across the dilution series (including the average of the triplicate injections). The peak area ratios were further normalized to tissue sample weight.
Organic/lipid extracts were separated on an Ascentis Express C18 2.1 × 150 mm 2.7 μm column (Sigma-Aldrich, St. Louis, MO) connected to a Dionex UltiMate 3000 UPLC system and a QExactive benchtop orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a heated electrospray ionization (HESI) probe. Dried lipid samples were typically dissolved in 50 μl acetonitrile:isopropanol:water (65:30:5) and 5 μl was injected into the LC/MS, with separate injections for positive and negative ionization modes. Mobile phase A in the chromatographic method consisted of water:ACN (60:40) in 10 mM ammonium formate and 0.1% formic acid, and mobile phase B consisted of isopropanol:acetonitrile (90:10), also with 10 mM ammonium formate and 0.1% formic acid. The chromatographic gradient was described previously88. The column oven and autosampler tray were held at 55 °C and 4 °C, respectively. The MS instrument parameters were as described previously89. The spray voltage was set to 4.2 kV, and the heated capillary and the HESI were held at 320 °C and 300 °C, respectively. The S-lens RF level was set to 50, and the sheath and auxiliary gas were set to 35 and 3 units, respectively. These conditions were held constant for both positive and negative ionization mode acquisitions. External mass calibration was performed using the standard calibration mixture every 7 days.
MS spectra of lipids were acquired in full-scan/data-dependent MS2 mode as previously described90.
Lipidomic data analysis
High-throughput identification and relative quantification of lipids was performed separately for positive and negative ionization mode data using LipidSearch software (Thermo Fisher Scientific/Mitsui Knowledge Industries)91,92 using the default parameters for QExactive. Product Search and Alignment. After alignment, raw peak areas for all identified lipids were exported to Excel and filtered according to the following pre-determined quality control criteria: Rej (“Reject” parameter calculated by LipidSearch software) equal to 0; PQ (“Peak Quality” parameter calculated by LipidSearch software) greater than 0.85; CV (standard deviation/average peak area across triplicate injections of a representative [pooled] biological sample) below 0.4; R (linear correlation across a three-point dilution series of the representative [pooled] biological sample) greater than 0.9. Typically ~70% of identified lipids passed all four quality control criteria. Raw peak areas of the filtered lipids were added to generate a “total lipid signal” for each sample, and individual lipid peak areas were normalized to this total signal as a control for extraction efficiency and sample loading.
Microarray analyses
Gene expression was evaluated in BAT of two groups of four mice. Whole-genome gene expression was performed using the Affymetrix GeneChip Mouse Gene 2.0 ST Array. The RNA was labeled and hybridized using a standard Affymetrix protocol performed at the Centre for Applied Genomics, Hospital for Sick Children, Toronto, Canada. The quality of arrays was judged using standard quality control parameters93,94 and all arrays passed the quality control filters. Expression values were extracted using the Robust Multichip Average (RMA) method95 implemented in the oligo package in R96. The Significance Analysis of Microarrays (SAM) method97 was used to identify probesets differentially expressed between groups. The false discovery rate (FDR) and the fold change threshold were set at 5% and 1.5, respectively. All analyses were carried out with the R statistical software version 3.2.3 and Bioconductor packages98.
Statistical analyses
Results are expressed as means ± SEM. Comparisons were done on normally distributed data using Student’s t-test or two-way ANOVA followed by Bonferroni post hoc tests to assess the differences between the various treatments with Graph Pad Prism Software version 6.0 h for Mac (San Diego, CA, USA). Differences of P < 0.05 were considered statistically significant.
Additional Information
How to cite this article: Labbé, S. M. et al. mTORC1 is Required for Brown Adipose Tissue Recruitment and Metabolic Adaptation to Cold. Sci. Rep. 6, 37223; doi: 10.1038/srep37223 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR), the Natural Sciences and Engineering Research Council of Canada (NSERC), Le Fonds de la Recherche Santé Québec (FRQS), Le Réseau de recherche en santé cardiométabolique, diabète et obésité (CMDO), Le Réseau de bio-imagerie du Québec (RBIQ), Diabète Québec, and La Fondation de l’Institut universitaire de cardiologie et de pneumologie de Québec (IUCPQ) to ML. SML is the recipient of a CIHR postdoctoral fellowship. AC held a fellowship from the CIHR Training Program in Obesity/Healthy Body Weight Research and now holds a postdoctoral fellowship from the Canadian Diabetes Association. YB holds a Canada Research Chair in Genomics of Heart and Lung Diseases. WTF hold a grant from FAPESP (2015/19530-5). EF holds a grant from the United States Department of Defense (W81XWH-15-1-0337, Peer-Reviewed Medical Research Program). We are grateful to Jean-François Beaudoin and Maxime Paillé for their helpful PET scan assistance and to Serge Phoenix and Sébastien Tremblay for radiotracers synthesis.
Footnotes
Author Contributions S.M.L. and M.L. designed the project, wrote and edited the manuscript. S.M.L., M.L., M.M., A.C., B.S., G.L., Y.G. performed the experiments. E.F. performed and analysed metabolomics experiments. Y.B. performed transcriptomics analyses. P.C. and W.T.F. provided samples and trained students for the denervation procedures. S.M.L., M.M., A.C., E.F., R.L., W.T.F., R.L., Y.B., D.R. and M.L. interpreted and discussed the results and commented on the manuscript.
References
- Cannon B. & Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 (2004). [DOI] [PubMed] [Google Scholar]
- Morrison S. F., Madden C. J. & Tupone D. Central control of brown adipose tissue thermogenesis. Front Endocrinol (Lausanne) 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls D. G. & Locke R. M. Thermogenic mechanisms in brown fat. Physiol. Rev. 64, 1–64 (1984). [DOI] [PubMed] [Google Scholar]
- Aherne W. & Hull D. Brown adipose tissue and heat production in the newborn infant. J Pathol Bacteriol 91, 223–234 (1966). [DOI] [PubMed] [Google Scholar]
- Enerbäck S. Human brown adipose tissue. Cell Metab. 11, 248–252 (2010). [DOI] [PubMed] [Google Scholar]
- Virtanen K. A. et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360, 1518–1525 (2009). [DOI] [PubMed] [Google Scholar]
- van Marken Lichtenbelt W. D. et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500–1508 (2009). [DOI] [PubMed] [Google Scholar]
- Ouellet V. et al. Outdoor Temperature, Age, Sex, Body Mass Index, and Diabetic Status Determine the Prevalence, Mass, and Glucose-Uptake Activity of 18F-FDG-Detected BAT in Humans. Journal of Clinical Endocrinology & Metabolism 96, 192–199 (2011). [DOI] [PubMed] [Google Scholar]
- Ouellet V. et al. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J. Clin. Invest. 122, 545–552 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard D., Monge-Roffarello B., Chechi K., Labbé S. M. & Turcotte E. E. Control and physiological determinants of sympathetically mediated brown adipose tissue thermogenesis. Front Endocrinol (Lausanne) 3, 36 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondin D. P. et al. A critical appraisal of brown adipose tissue metabolism in humans. Clin. Lipidol. 10, 259–280 (2015). [Google Scholar]
- Bartelt A. et al. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17, 200–205 (2011). [DOI] [PubMed] [Google Scholar]
- Berbée J. F. P. et al. Brown fat activation reduces hypercholesterolaemia and protects from atherosclerosis development. Nat Commun 6, 6356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé S. M. et al. In vivo measurement of energy substrate contribution to cold-induced brown adipose tissue thermogenesis. FASEB J. 29, 2046–2058 (2015). [DOI] [PubMed] [Google Scholar]
- Chondronikola M. et al. Brown adipose tissue improves whole-body glucose homeostasis and insulin sensitivity in humans. Diabetes 63, 4089–4099 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidossis L. & Kajimura S. Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. J. Clin. Invest. 125, 478–486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer A. NRG4: An Endocrine Link between Brown Adipose Tissue and Liver. Cell Metab. 21, 13–14 (2015). [DOI] [PubMed] [Google Scholar]
- Cameron I. L. & Smith R. E. Cytological responses of brown fat tissue in cold-exposed rats. J. Cell Biol. 23, 89–100 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnd T., Skála J. & Lindberg O. Changes in interscapular brown adipose tissue of the rat during perinatal and early postnatal development and after cold acclimation. I. Activities of some respiratory enzymes in the tissue. Comp. Biochem. Physiol. 33, 499–508 (1970). [DOI] [PubMed] [Google Scholar]
- Young J. B., Saville E., Rothwell N. J., Stock M. J. & Landsberg L. Effect of diet and cold exposure on norepinephrine turnover in brown adipose tissue of the rat. J. Clin. Invest. 69, 1061–1071 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster D. O., Depocas F. & Frydman M. L. Noradrenaline-induced calorigenesis in warm- and cold-acclimated rats: relations between concentration of noradrenaline in arterial plasma, blood flow to differently located masses of brown adipose tissue, and calorigenic response. Can. J. Physiol. Pharmacol. 58, 915–924 (1980). [DOI] [PubMed] [Google Scholar]
- Nedergaard J. & Cannon B. UCP1 mRNA does not produce heat. Biochim. Biophys. Acta 1831, 943–949 (2013). [DOI] [PubMed] [Google Scholar]
- Blondin D. P. et al. Increased brown adipose tissue oxidative capacity in cold-acclimated humans. J. Clin. Endocrinol. Metab. 99, E438–E446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen M. J. W. et al. Short-term cold acclimation improves insulin sensitivity in patients with type 2 diabetes mellitus. Nat. Med. (2015). [DOI] [PubMed] [Google Scholar]
- Laplante M. & Sabatini D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J. & CANNON B. The Browning of White Adipose Tissue: Some Burning Issues. Cell Metab. (2014). [DOI] [PubMed] [Google Scholar]
- Klingenspor M. Cold-induced recruitment of brown adipose tissue thermogenesis. Exp. Physiol. 88, 141–148 (2003). [DOI] [PubMed] [Google Scholar]
- Liu D. et al. Activation of mTORC1 is essential for β-adrenergic stimulation of adipose browning. J. Clin. Invest. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran C. M. et al. Rapamycin blocks induction of the thermogenic program in white adipose tissue. Diabetes db150502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert V. et al. mTORC2 sustains thermogenesis via Akt-induced glucose uptake and glycolysis in brown adipose tissue. EMBO Mol Med 8, 232–246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S. F., Madden C. J. & Tupone D. Central neural regulation of brown adipose tissue thermogenesis and energy expenditure. Cell Metab. 19, 741–756 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Umekawa T., Sakane N., Yoshimoto K. & Kondo M. Effect of CL316,243, a highly specific β3-adrenoceptor agonist, on sympathetic nervous system activity in mice. Metabolism 45, 787–791 (1996). [DOI] [PubMed] [Google Scholar]
- Sum F. W. et al. Prodrugs of CL316243: a selective beta3-adrenergic receptor agonist for treating obesity and diabetes. Bioorg. Med. Chem. Lett. 9, 1921–1926 (1999). [DOI] [PubMed] [Google Scholar]
- Klein J. et al. beta(3)-adrenergic stimulation differentially inhibits insulin signaling and decreases insulin-induced glucose uptake in brown adipocytes. J. Biol. Chem. 274, 34795–34802 (1999). [DOI] [PubMed] [Google Scholar]
- Pang Z. et al. GPR40 is partially required for insulin secretion following activation of beta3-adrenergic receptors. Mol. Cell. Endocrinol. 325, 18–25 (2010). [DOI] [PubMed] [Google Scholar]
- Peterson T. R. et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J. et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 13, 249–259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P. et al. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 8, 399–410 (2008). [DOI] [PubMed] [Google Scholar]
- Sengupta S., Peterson T. R., Laplante M., Oh S. & Sabatini D. M. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104 (2010). [DOI] [PubMed] [Google Scholar]
- Cannon B. & Nedergaard J. Nonshivering thermogenesis and its adequate measurement in metabolic studies. Journal of Experimental Biology 214, 242–253 (2010). [DOI] [PubMed] [Google Scholar]
- Bessi V. L. et al. EP 80317, a selective CD36 ligand, shows cardioprotective effects against post-ischaemic myocardial damage in mice. Cardiovasc. Res. 96, 99–108 (2012). [DOI] [PubMed] [Google Scholar]
- van den Hoff J., Burchert W., Wolpers H. G., Meyer G. J. & Hundeshagen H. A kinetic model for cardiac PET with [1-carbon-11]-acetate. Journal of Nuclear Medicine 37, 521–529 (1996). [PubMed] [Google Scholar]
- van den Hoff J. Methodologic aspects of myocardial blood flow quantification with 1-11C-acetate PET. Journal of Nuclear Medicine 42, 1438–1439 (2001). [PubMed] [Google Scholar]
- Mirbolooki M. R., Upadhyay S. K., Constantinescu C. C., Pan M.-L. & Mukherjee J. Adrenergic pathway activation enhances brown adipose tissue metabolism: A [(18)F]FDG PET/CT study in mice. Nuclear Medicine and Biology (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. W. et al. 18F-FDG PET/CT monitoring of β3 agonist-stimulated brown adipocyte recruitment in white adipose tissue. J. Nucl. Med. 56, 153–158 (2015). [DOI] [PubMed] [Google Scholar]
- Blondin D. P. et al. Selective Impairment of Glucose but Not Fatty Acid or Oxidative Metabolism in Brown Adipose Tissue of Subjects With Type 2 Diabetes. Diabetes 64, 2388–2397 (2015). [DOI] [PubMed] [Google Scholar]
- Brito M. N. et al. Brown adipose tissue triacylglycerol synthesis in rats adapted to a high-protein, carbohydrate-free diet. Am. J. Physiol. 276, R1003–R1009 (1999). [DOI] [PubMed] [Google Scholar]
- Moura M. A. et al. Effect of cold acclimation on brown adipose tissue fatty acid synthesis in rats adapted to a high-protein, carbohydrate-free diet. Metab. Clin. Exp. 50, 1493–1498 (2001). [DOI] [PubMed] [Google Scholar]
- Festuccia W. T. et al. The PPARgamma agonist rosiglitazone enhances rat brown adipose tissue lipogenesis from glucose without altering glucose uptake. Am. J. Physiol. Regul. Integr. Comp. Physiol. 296, R1327–R1335 (2009). [DOI] [PubMed] [Google Scholar]
- Festuccia W. T., Blanchard P.-G. & Deshaies Y. Control of Brown Adipose Tissue Glucose and Lipid Metabolism by PPARγ. Front Endocrinol (Lausanne) 2, 84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez R., Teruel T. & Lorenzo M. Akt mediates insulin induction of glucose uptake and up-regulation of GLUT4 gene expression in brown adipocytes. FEBS Lett. 494, 225–231 (2001). [DOI] [PubMed] [Google Scholar]
- Kharitonenkov A. et al. FGF-21 as a novel metabolic regulator. J. Clin. Invest. 115, 1627–1635 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelli B. et al. Interplay between FGF21 and insulin action in the liver regulates metabolism. J. Clin. Invest. 124, 515–527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandanmagsar B. et al. Impaired Mitochondrial Fat Oxidation Induces FGF21 in Muscle. Cell Rep 15, 1686–1699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A. et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Li Y., Zhu T., Wu J. & Guan K.-L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4, 648–657 (2002). [DOI] [PubMed] [Google Scholar]
- Potter C. J., Pedraza L. G. & Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4, 658–665 (2002). [DOI] [PubMed] [Google Scholar]
- Vander Haar E., Lee S.-I., Bandhakavi S., Griffin T. J. & Kim D.-H. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9, 316–323 (2007). [DOI] [PubMed] [Google Scholar]
- Sancak Y. et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25, 903–915 (2007). [DOI] [PubMed] [Google Scholar]
- Sarbassov D. D., Guertin D. A., Ali S. M. & Sabatini D. M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005). [DOI] [PubMed] [Google Scholar]
- Olsen J. M. et al. Glucose uptake in brown fat cells is dependent on mTOR complex 2-promoted GLUT1 translocation. J. Cell Biol. 207, 365–374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P. et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92, 829–839 (1998). [DOI] [PubMed] [Google Scholar]
- Bentzinger C. F. et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 8, 411–424 (2008). [DOI] [PubMed] [Google Scholar]
- Shende P. et al. Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation 123, 1073–1082 (2011). [DOI] [PubMed] [Google Scholar]
- Romanino K. et al. Myopathy caused by mammalian target of rapamycin complex 1 (mTORC1) inactivation is not reversed by restoring mitochondrial function. Proceedings of the National Academy of Sciences 108, 20808–20813 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karar J. & Maity A. PI3K/AKT/mTOR Pathway in Angiogenesis. Front Mol Neurosci 4, 51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abnous K. & Storey K. B. Regulation of skeletal muscle creatine kinase from a hibernating mammal. Arch. Biochem. Biophys. 467, 10–19 (2007). [DOI] [PubMed] [Google Scholar]
- Golozoubova V. et al. Only UCP1 can mediate adaptive nonshivering thermogenesis in the cold. FASEB J. 15, 2048–2050 (2001). [DOI] [PubMed] [Google Scholar]
- Anderson C. M. et al. Dependence of Brown Adipose Tissue Function on CD36-Mediated Coenzyme Q Uptake. Cell Rep (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondares E. et al. Thermogenic activation induces FGF21 expression and release in brown adipose tissue. Journal of Biological Chemistry 286, 12983–12990 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keipert S. et al. Genetic disruption of uncoupling protein 1 in mice renders brown adipose tissue a significant source of FGF21 secretion. Mol Metab 4, 537–542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q. et al. Transcriptome profiling of brown adipose tissue during cold exposure reveals extensive regulation of glucose metabolism. AJP: Endocrinology and Metabolism ajpendo.00277.2014 (2014). [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I., Howell J. J., Asara J. M. & Manning B. D. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339, 1323–1328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I., Hoxhaj G., Ricoult S. J. H., Asara J. M. & Manning B. D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens K., Bottelbergs A. & Baes M. Ectopic recombination in the central and peripheral nervous system by aP2/FABP4-Cre mice: implications for metabolism research. FEBS Lett. 584, 1054–1058 (2010). [DOI] [PubMed] [Google Scholar]
- Lee K. Y. et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes 62, 864–874 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. L., Tang Y., Li H. & Guertin D. A. Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Mol Metab 5, 422–432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdalon J. et al. Constitutive adipocyte mTORC1 activation enhances mitochondrial activity and reduces visceral adiposity in mice. Biochim. Biophys. Acta (2016). [DOI] [PubMed] [Google Scholar]
- Xiang X. et al. TSC1-mTORC1 signaling determines brown-to-white adipocyte phenotypic switch. Diabetes (2014). [DOI] [PubMed] [Google Scholar]
- Xiang X. et al. Deficiency in pulmonary surfactant proteins in mice with fatty acid binding protein 4-Cre-mediated knockout of the tuberous sclerosis complex 1 gene. Exp. Physiol. 98, 830–841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashita N. H. et al. Glycerokinase activity in brown adipose tissue: a sympathetic regulation? Am. J. Physiol. Regul. Integr. Comp. Physiol. 282, R1185–R1190 (2002). [DOI] [PubMed] [Google Scholar]
- Vaughan C. H., Zarebidaki E., Ehlen J. C. & Bartness T. J. Analysis and measurement of the sympathetic and sensory innervation of white and brown adipose tissue. Meth. Enzymol. 537, 199–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia W. T. et al. Depot-specific effects of the PPARgamma agonist rosiglitazone on adipose tissue glucose uptake and metabolism. The Journal of Lipid Research 50, 1185–1194 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro S.-H. et al. Sestrin2 inhibits uncoupling protein 1 expression through suppressing reactive oxygen species. Proceedings of the National Academy of Sciences 111, 7849–7854 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard S. L. et al. Abnormal in vivo myocardial energy substrate uptake in diet-induced type 2 diabetic cardiomyopathy in rats. Am. J. Physiol. Endocrinol. Metab. 298, E1049–E1057 (2010). [DOI] [PubMed] [Google Scholar]
- Patlak C. S. & Blasberg R. G. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. Generalizations. J. Cereb. Blood Flow Metab. 5, 584–590 (1985). [DOI] [PubMed] [Google Scholar]
- Birsoy K. et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C. et al. RPLC-ion-trap-FTMS method for lipid profiling of plasma: method validation and application to p53 mutant mouse model. J. Proteome Res. 7, 4982–4991 (2008). [DOI] [PubMed] [Google Scholar]
- Bird S. S., Marur V. R., Sniatynski M. J., Greenberg H. K. & Kristal B. S. Serum lipidomics profiling using LC-MS and high-energy collisional dissociation fragmentation: focus on triglyceride detection and characterization. Anal. Chem. 83, 6648–6657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tafesse F. G. et al. Disruption of Sphingolipid Biosynthesis Blocks Phagocytosis of Candida albicans. PLoS Pathog. 11, e1005188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T. et al. Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J Chromatogr A 1292, 211–218 (2013). [DOI] [PubMed] [Google Scholar]
- Taguchi R. & Ishikawa M. Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine Lipid Search. J Chromatogr A 1217, 4229–4239 (2010). [DOI] [PubMed] [Google Scholar]
- Heber S. & Sick B. Quality assessment of Affymetrix GeneChip data. OMICS 10, 358–368 (2006). [DOI] [PubMed] [Google Scholar]
- Kauffmann A., Gentleman R. & Huber W. arrayQualityMetrics-a bioconductor package for quality assessment of microarray data. Bioinformatics 25, 415–416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003). [DOI] [PubMed] [Google Scholar]
- Carvalho B. S. & Irizarry R. A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 26, 2363–2367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R. & Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R. C. et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5, R80 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.