Abstract
The consistent and accurate quantification of proteins by mass spectrometry (MS)-based proteomics depends on the performance of instruments, acquisition methods and data analysis software. In collaboration with the software developers, we evaluated OpenSWATH, SWATH2.0, Skyline, Spectronaut and DIA-Umpire, five of the most widely used software methods for processing data from SWATH-MS (sequential window acquisition of all theoretical fragment ion spectra), a method that uses data-independent acquisition (DIA) for label-free protein quantification. We analyzed high-complexity test datasets from hybrid proteome samples of defined quantitative composition acquired on two different MS instruments using different SWATH isolation windows setups. For consistent evaluation we developed LFQbench, an R-package to calculate metrics of precision and accuracy in label-free quantitative MS, and report the identification performance, robustness and specificity of each software tool. Our reference datasets enabled developers to improve their software tools. After optimization, all tools provided highly convergent identification and reliable quantification performance, underscoring their robustness for label-free quantitative proteomics.
Introduction
Mass spectrometry-based quantitative proteomics is an essential tool to elucidate the complex and dynamic nature of proteomes1,2, enabling the in-depth characterization of protein expression changes. Due to their experimental simplicity and capacity to process large cohorts of samples, label-free quantification approaches are most frequently used. While data-dependent acquisition (DDA) selects precursor ions according to their abundances, data-independent acquisition (DIA) approaches implement a parallel fragmentation of all precursor ions, regardless of their intensity or other characteristics, thereby enabling to establish a complete record of the sample3. In recent years, several data-independent acquisition (DIA) mass spectrometric strategies including SWATH-MS4 (sequential window acquisition of all theoretical fragment ion spectra), HDMSE5 (high definition MSE), and AIF6 (all-ion fragmentation) were established that circumvent some of the problems arising from data-dependent acquisition (DDA), such as stochastic and irreproducible precursor ion selection7,8, under-sampling9 and long instrument cycle times8.
In addition to the mass spectrometric method applied, computational methods, e.g. for raw data processing, protein database searching, and statistical analysis of the quantitative data, critically impact the results of quantitative proteomics analyses. As such, evaluating the correctness and relative performance of these methods is essential10. Quantitative proteomics would greatly benefit from an objective comparative benchmarking of the performance and robustness of the various computational approaches and software solutions available or currently in development. Meaningful and unbiased comparisons of software tools and their appropriate use are challenging for a number of reasons11: methods and algorithms may be assessed by scientists lacking relevant expertise, the tested method may suffer from insufficient documentation, or the interpretation of the test results may be subjective12–15. In addition, benchmarking requires high-quality standardized data sets, defined metrics and dedicated software to implement and analyze these metrics, not only to compare existing methods but also to evaluate potential improvements and pitfalls when new methods are developed.
To address these challenges, we developed a computational benchmarking framework for label-free quantitative proteomics, LFQbench, that analyzes and processes data acquired from hybrid proteome samples16 containing several proteomes mixed in defined proportions. To demonstrate the approach, we applied LFQbench to evaluate the performance of the DIA approach SWATH-MS, which provides high-throughput, accurate quantification, and reproducible measurements within a single experimental setup17. We centrally acquired high quality benchmarking data sets using different instrument platforms and acquisition modes, and focused our comparative analysis on five widely-used analysis tools for SWATH-MS data: four ‘peptide-centric’ query17 tools (OpenSWATH18, SWATH2.0, Skyline19, and Spectronaut20) and the ‘data-centric’ approach DIA-Umpire19. While the former use MS/MS libraries to extract groups of signals that reliably represent a specific peptide, followed by statistical methods to distinguish true from false matches14,18,21, the latter assembles “pseudo” tandem MS spectra that can be identified and quantified with conventional database-searching and protein-inference tools without an assay library8,22. All respective software developers participated in our study, ensuring an optimal analysis workflow and parameters for each SWATH software tool.
Results
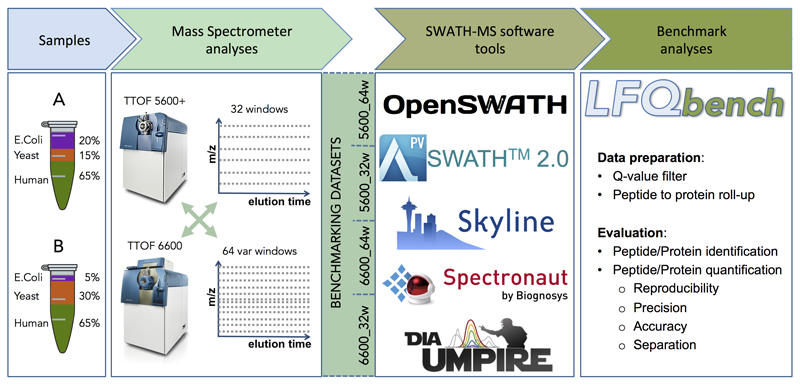
As a benchmarking sample, two hybrid proteome samples consisting of tryptic digests of human, yeast and E.coli proteins were mixed in defined proportions16 (Figure 1) to yield expected peptide and protein ratios of 1:1 for human, 2:1 for yeast, and 1:4 for E.coli proteins if samples A and B are compared. This sample set is referred to as HYE124 (Supplementary Figure 1). While the absolute amounts of individual proteins are not known, these samples provide a defined ground truth for bioinformatics analysis, i.e., defined relative changes between samples, and a sufficiently large number of peptides to enable the in-depth evaluation of both precision and accuracy of relative label-free quantification16. We analyzed HYE124 samples A & B in technical triplicates on two different instrument platforms (TripleTOF 5600 and TripleTOF 6600) using two different SWATH-MS acquisition modes (Supplementary Figure 1), generating a total of four benchmark datasets. To individually address the effects of SWATH window number (32 vs. 64 windows) and window size (fixed vs. variable), we generated a second sample set with higher ratio differences (termed HYE110, see online methods), which was analyzed in four different acquisition modes on the TripleTOF 6600 platform (Supplementary Figure 1). This allowed us to test the performance of the software tools on data generated using a variety of instruments and settings of different sensitivity and co-fragmentation frequency.
Figure 1. Study workflow.
Two proteome-hybrid samples A and B were prepared containing known quantities of peptide digestions of human, yeast, and E.Coli organisms. The samples were analyzed in three technical replicates in SWATH-MS acquisition mode on two different MS instrument platforms (TripleTOF 5600 and TripleTOF 6600) with/using two different swath windows setups (32 fixed size windows and 64 variable size windows). This resulted in four benchmarking datasets. The datasets were analyzed in five software tools: OpenSWATH, SWATH 2.0, Skyline, Spectronaut, and DIA-Umpire. Benchmark analyses of each dataset and software tool were performed based on the output reports generated by the newly developed benchmarking software LFQbench.
Data Evaluation Software: LFQbench
To standardize the complex evaluation process of label-free quantification performance and to make it transparent, we developed the LFQbench software tool. LFQbench is an R-package that implements automated calculation of metrics for precision (coefficients of variation of reported peptide and protein intensities between replicates) and accuracy (deviations from expected abundance ratios) (Supplementary Table 1) of label-free quantification16, as well as the performance in separating proteins ratios for the different species (Table 1 and Supplementary Table 2) in hybrid proteome sample sets. LFQbench directly imports results from label-free quantification software tools, applies filter criteria defined by the software developers of our study and computes protein level quantification information. Next, LFQbench evaluates and graphically represents precision and accuracy of label-free quantification experiments based on hybrid proteome samples. This resource provides current and future software developers with a standardized set of reports on protein and peptide level that enable an in-depth performance evaluation of their software tools. LFQbench is fully compatible with data from hybrid proteome samples acquired on other instrument platforms16,23 enabling the objective assessment of other variables, such as different acquisition schemes, or the comparison of different MS platforms. In addition, LFQbench provides a data simulator, which enables the users to visualize both an “ideal” dataset and the effects of commonly observed problems, e.g., incorrect background subtraction (Supplementary Figure 2). The LFQbench evaluation software is publicly available at https://github.com/IFIproteomics/LFQbench.
Table 1. LFQbench metrics summary of the dataset TripleTOF 6600 - 64w.
Number of protein and peptide identifications, number of valid quantification ratios and calculated hyperbolic arctangents (atanh) transformed AUQC values for separation between (yeast vs. human) and (E.coli vs human) for the dataset TripleTOF 6600 - 64 windows. A valid quantification ratio is defined in LFQbench as the ratio of a peptide or protein present in at least one technical replicate in both samples A and B that falls into a validity range (defined as the range of expectation (a maximum difference of 5 standard deviations from the average) defined by the sample set composition and the spread of the log2-ratios observed for each species. Very few cases (if any) fall out of this range, as it can be checked at the LFQbench logs (metrics file) under the “invalid, out of validity range” section. For better readability of the separation values, a color scale is applied to all separation values, from full red (worst separation value of all shown) to full green (best separation value of all shown). Raw AUQC values for separation are provided in Supplementary Table 2.
| Iteration 1 | Iteration 2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Median CV of Human |
Number of IDs |
Valid quantification ratios |
Overlap Yeast–Human (arctanh) |
Overlap Ecoli–Human (arctanh) |
Median CV of Human |
Number of IDs |
Valid quantification ratios |
Overlap Yeast–Human (arctanh) |
Overlap Ecoli–Human (arctanh) |
||
| peptides | OpenSWATH | 6.5% | 40,726 | 36,098 | 2.12 | 2.24 | 8.2% | 40,728 | 35,944 | 2.10 | 2.26 |
| SWATH2.0 | 6.6% | 35,517 | 35,517 | 2.14 | 2.11 | 6.1% | 35,489 | 26,303 | 2.49 | 2.52 | |
| Skyline | 7.4% | 40,804 | 34,103 | 1.91 | 1.85 | 6.9% | 42,517 | 37,977 | 2.13 | 2.14 | |
| Spectronaut | 5.9% | 42,439 | 37,120 | 1.97 | 1.90 | 6.2% | 42,325 | 36,292 | 2.11 | 2.26 | |
| DIA-Umpire | 13.2% | 36,332 | 28,785 | 1.74 | 1.98 | 12.9% | 36,249 | 25,677 | 1.82 | 2.18 | |
| proteins | OpenSWATH | 5.4% | 4,632 | 4,343 | 2.30 | 2.56 | 6.4% | 4,636 | 4,352 | 2.51 | 2.60 |
| SWATH2.0 | 5.6% | 4,323 | 4,323 | 2.37 | 2.36 | 6.1% | 3,946 | 3,371 | 2.42 | 2.56 | |
| SWATH2.0 (built-in) | 6.0% | 6,178 | 6,178 | 2.23 | 2.03 | ||||||
| Skyline | 5.9% | 4,518 | 4,140 | 2.03 | 2.15 | 5.5% | 4,692 | 4,456 | 2.37 | 2.43 | |
| Spectronaut | 3.3% | 4,692 | 4,346 | 2.13 | 2.18 | 3.3% | 4,675 | 4,300 | 2.31 | 2.50 | |
| DIA-Umpire | 12.0% | 3,795 | 3,379 | 2.12 | 2.30 | 12.3% | 3,673 | 3,111 | 2.13 | 2.85 | |
| DIA-Umpire (built-in) | 13.1% | 4,849 | 4,489 | 1.78 | 1.94 | ||||||
Effects of mass spectrometers and SWATH acquisition modes
First we compared TripleTOF 5600 and TripleTOF6600 systems. The latter provided between 15% and 137% more peptide identifications and between 14% and 102% more protein identifications in the HYE124 sample (Supplementary Table 3), largely due to its higher resolution chromatographic system. Next we analyzed the effect of SWATH window setups on quantification results. For the HYE124 sample, depending on MS instrument and software, the 64w setup provided between 9% and 54% higher numbers of peptide identifications and between 9% and 37% higher numbers of protein identifications (Supplementary Table 3) compared to the 32w setup. Additionally, the 64w setup resulted in approximately 2-fold higher median signal to noise ratio compared to the 32w setup (Supplementary Figure 3). This is most likely due to a decrease in interferences derived from co-fragmentation of other precursors. The HYE110 sample was further acquired with two additional windows schemes (Supplementary Figure 1). Again, we observed the highest median signal to noise ratio for the 64 variable window setup (Supplementary Figure 3). Both the increase in the number of windows and the switch from fixed to variable windows, contributed to these effects to a similar extent. The variable (optimized) windows setups provided between 3% and 29% more peptide identifications and between 4% and 25% more protein identifications compared to fixed windows setups. 64 windows setups provided up to 32% more peptide identifications and up to 21% more protein identifications than 32 windows setups.
The results from the HYE124 dataset showed that the change of swath window size had the largest impact on the results of SWATH2.0 (49%-54% increase in peptides), while the instrument type had the largest effect on results provided by DIA-Umpire (99% - 137% increase). Generally, all tools benefitted more from changing instrument type than from changing window size (Supplementary Table 3). We observed a very high technical reproducibility of reported peptide intensities (R2:0.92-0.99 depending on software tool (Supplementary Table 4), and CVs below 14% in HYE124 sample) within each dataset (Supplementary Figure 4, CVs reported in Table 1 and Supplementary Table 2 and Supplementary Figure 5), and a good correlation (R2:0.78-0.91) between datasets for all library-based tools (Supplementary Figure 6). For all subsequent analyses, we focused on the HYE124 dataset generated by the TripleTOF 6600 using 64 swath windows, which generated the highest number of identifications among the acquired datasets. Results from other settings are provided in Supplementary Figures 7 and 8, and in ProteomeXchange repository.
Label-free quantification performance: First Iteration
We performed two analysis iterations to clearly illustrate the advantages of our benchmarking data set for the future development of software tools for label-free proteomics. In the first iteration, the developers analyzed the datasets with the latest publicly available version of each software tool, using optimized parameters (Supplementary Table 5), and a retention time window width and m/z tolerance agreed on by the developers of library-based tools. We ran LFQbench on the data provided by the developers and sent them the results to identify pitfalls in the software workflows. Next, we initiated an open discussion among developers, who improved their tools by implementing solutions to issues uncovered in the first analysis step, which were then validated in a second iteration.
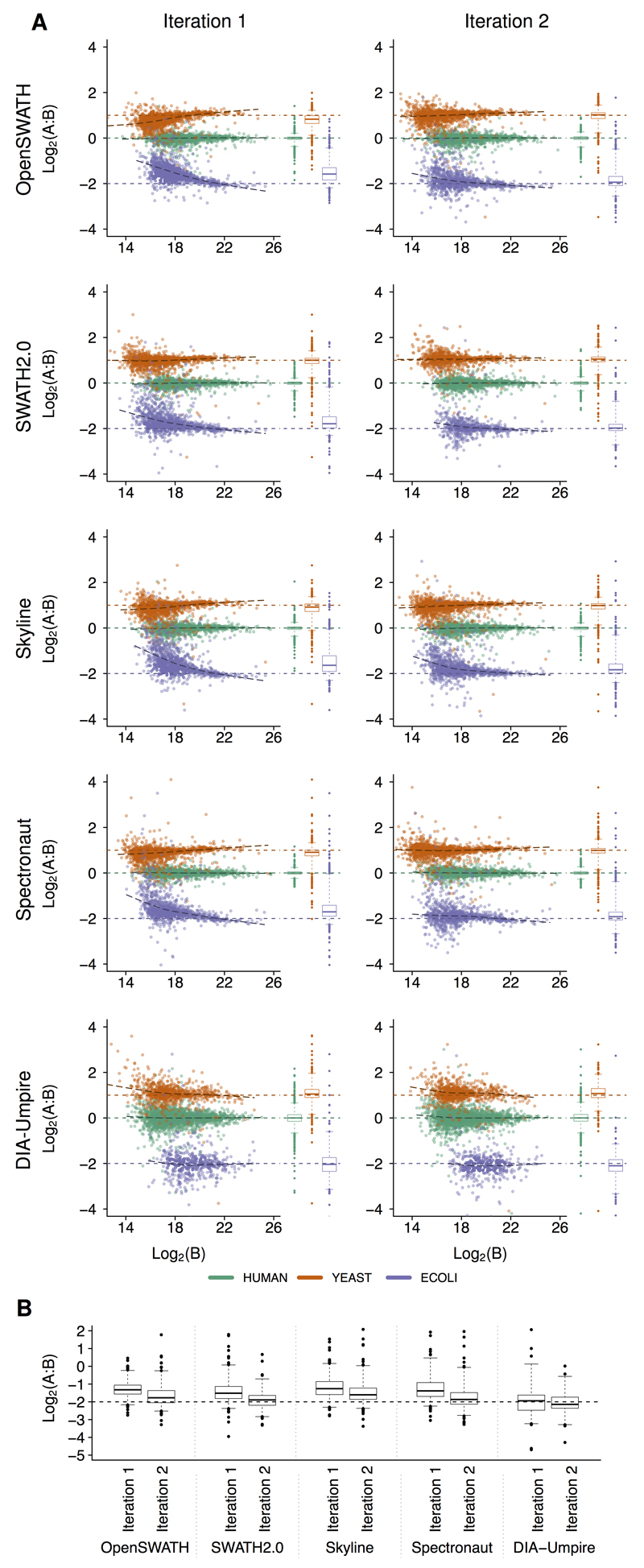
First, we analyzed the relative quantification accuracy of proteins quantified by either single or multiple peptides. Single hit proteins consistently showed a higher quantification variance than proteins defined by multiple peptides (Supplementary Figure 9). Therefore, we required at least two peptides to report a valid quantification value of a protein. Additionally, we required a protein to be quantified in at least two replicates in one sample to reduce the number of false negative proteins (human proteins falsely reported to be exclusive for one of the samples, Supplementary Figure 10). Analysis of results provided by the five software tools in their initial setting revealed both a similar dynamic range in intensities (Supplementary Table 6) and a similar performance in terms of quantification precision and accuracy for high abundance proteins. Across all software tools, protein ratios within the lowest intensity tertile displayed the highest variance (average standard deviation for E. coli proteins distribution = 0.68) and differed most from the expected values (average absolute difference for E. coli distribution = 0.51) (Figure 2 and Supplementary Table 1). Results improved with increasing peptide signals (Supplementary Figures 11-13, and Supplementary Table 1), and the best results were obtained in the highest intensity tertile, as indicated by lowest variances (average standard deviation for E.Coli proteins distribution = 0.38) and deviations from the expected ratios (average absolute difference for E.Coli distribution = 0.06). We observed marked differences in the lower abundance range between the software tools as indicated by systematic deviations from the expected values for low intensity signals in OpenSWATH, Spectronaut, and Skyline (Figure 2, Supplementary Figures 7 and 8). This indicated a potential issue with background subtraction leading to a systematic underestimation of abundance ratios for E. coli and yeast proteins. The observed deviations of the lower abundant proteins impaired the correct separation of the human and yeast low abundance proteins, indicating that it would not be possible to faithfully determine 2-fold expression changes in the lower intensity range. Similar results were obtained on the peptide level (Supplementary Figure 7). Of note, DIA-Umpire and SWATH 2.0 also directly reported quantification results at the protein level (“Built-in”). However, we found that the TOP3 based approach to infer protein level quantification used in this study generally resulted in lower variances and better quantification accuracy for both SWATH 2.0 and DIA-Umpire than the built-in methods (Supplementary Figure 14). When evaluating the absolute quantification results provided by DIA-Umpire, we observed marked differences to values reported by the library-based tools (Supplementary Figure 15). The analysis of the fragments used for quantification revealed that fragment intensity rankings were different between DIA-Umpire and the library based tools (Supplementary Figure 16), resulting in the selection of different fragments for quantification and thus explaining the observed differences.
Figure 2. Protein level LFQbench benchmark results.
After parameter optimization in a first iteration of analyses, intensities reported by each software tool were fitted to PeakView intensity scale using a linear model fixed in the origin (Supplementary Figure 25). Intensities of multiply charged precursors were summed up, and averaged across all technical replicates of each sample. Protein quantities were estimated in each technical replicate by the average of the three most intense peptides reported for each protein. Single hit proteins (a single peptide detected in a protein) were discarded. In the present figure only data derived from TripleTOF 6600 with the 64 swath window setup are displayed. Corresponding data for the other instrument and acquisition setups are shown in Supplementary Figure 8. (a) Log-transformed ratios (log2(A/B)) of proteins (human proteins in green, yeast proteins in orange, and E.Coli proteins in purple) were plotted for each benchmarked software tool over the log-transformed intensity of sample B for the first and second iteration (sample size n between 3,795 and 4,692 proteins). Dashed colored lines represent the expected log2(A/B) values for human, yeast, and E.Coli proteins. Black dashed lines represent the local trend along the x-axis of experimental log-transformed ratios of each population (human, yeast, and E.Coli). For a better understanding of these plots, see plots generated by simulated data (Supplementary Figure 2). (b) (log2(A/B)) of the averages between technical replicates of A and B for E.coli proteins in the lowest intensity tertile. Boxes represent 25% and 75% percentiles, whiskers cover data points between 1% and 99% percentiles. Accuracy could be significantly improved in the second iteration for OpenSWATH, SWATH 2.0, Skyline, and Spectronaut [p < 0.05; One-sided Wilcoxon rank sum tests]. Precision improved significantly in the second iteration for OpenSWATH, Skyline, and Spectronaut in all datasets of HYE124 [p < 0.05 in double-sided F-tests performed for each individual species].
Compared to DIA-Umpire, library-based software tools showed lower numbers of incomplete protein quantification cases (cases with at least one missing value among the six injections (three replicates of A and B samples) (Supplementary Figure 17). In the case of SWATH 2.0, no incomplete cases were observed in the first iteration data, as cross-annotated signals are not required to match the retention time of their corresponding identified seeds, which may lead to false-positive cross-annotations skewing the quantification values.
Label-free quantification performance: Second Iteration
All improved software tools evaluated in the second iteration of the study showed improved precision and/or accuracy of quantification results on both peptide and protein levels compared to their first iteration results (Figure 2 A-B, Table 1, Supplementary Figures 7 and 8, Supplementary Table 1). For a detailed analysis of the improvements, we focused on the reported quantification ratios of E. coli proteins at the lowest intensity tertile as the precision and accuracy of quantification are strongly dependent on the signal intensity24 (Figure 2 E, other species and tertiles shown in Supplementary Figures 11-13). OpenSWATH and Spectronaut improved quantification accuracy of the E. coli peptides and proteins due to an improved background subtraction. DIA-Umpire and SWATH 2.0 improved peptide quantification precision in all tertiles, as wrong quantification values were removed. Skyline generally improved peptide and protein quantification precision. Notably, all four independent datasets of HYE124 produced a very similar pattern of improvements in iteration 2, indicating that no overfitting was generated at the software tools improvement.
To validate the performance of the second iteration tools, we used them for the analysis of the HYE110 sample set, which provides more challenging ratios between the samples, as the signals are closer to the noise threshold in one of the samples. As expected, we observed that precision values of peptides and proteins of yeast and E. coli were better in HYE124 due to the higher ratio differences in HYE110 (Supplementary Figures 7 and 8). The higher ratio samples also produced more incomplete cases, i.e., proteins with fewer than six quantification values. For yeast and E. coli proteins, this rate increased to up to 90% (Supplementary Figure 17). The higher number of incomplete cases also resulted in more human proteins falsely reported to be present exclusively in one of the samples (Supplementary Figures 10 and 18). Spectronaut and OpenSWATH produced the lowest numbers of false reports, followed by Skyline and SWATH2.0 (Supplementary Figure 10), while DIA-Umpire reported the highest numbers. While cross-annotation of signals between runs may reduce this source of potentially false quantification results, stringent control is required, e.g. by retention time alignment, to avoid matching the wrong signals.
Integrated analysis of tools
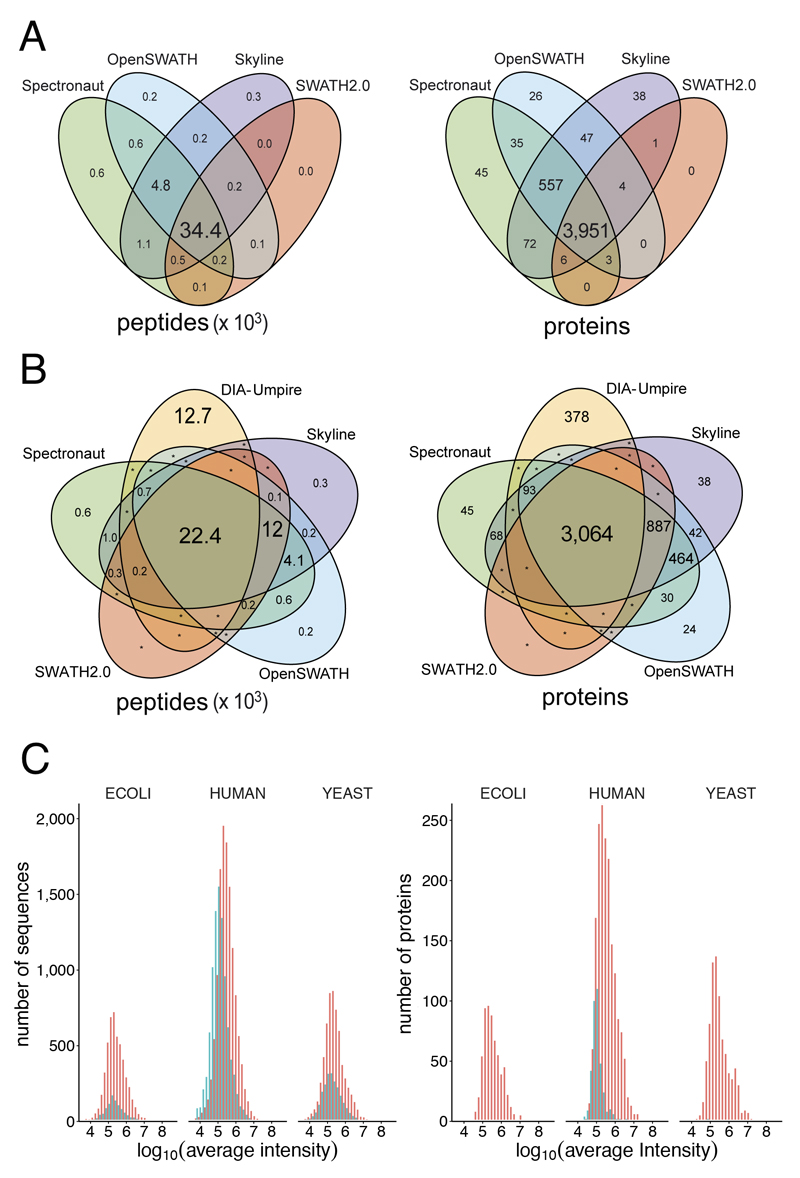
In the HYE124 dataset, the library based tools identified in the iteration 2 between 35,489 and 42,517 peptides mapping to between 3,673 and 4,692 proteins. Notably, we observed an exceptional overlap between these tools, as 93% of all identified peptides and 95% of proteins were identified by at least three out of the four library-based tools (Figure 3 A). The overlap of all five tools was 22,407 peptides and 3,064 proteins (Figure 3 B). On the peptide level, the results provided by the library-based tools covered 65% of the sequences provided by DIA-Umpire, which additionally identified 12,748 sequences not found by the library-based approaches (Figure 3 B), in part due to slightly different search parameters. Only 288 sequences of those identified exclusively by DIA-Umpire were present in the assay library and may potentially be false negative cases for the library-based workflow. Notably, the overlap on protein level was remarkably higher (86%), similarly to the typical overlap between different DDA search engines25, indicating that DIA-Umpire may cover additional peptides not included in the assay library, e.g. singly charged peptide ions (726 peptides), which are usually not triggered for MS/MS in DDA experiments and thus not included in the consensus library. The number of peptides per protein was similar for all library-based tools (Supplementary Figure 19).
Figure 3. Integrated analysis of the five software tools.
(a) Overlap of quantified peptides and proteins for library-based tools. The font size of each element is proportional to the number of peptides or proteins displayed. (b) Overlap of quantified peptides and proteins by all software tools. The font size of each element is proportional to the number of peptides or proteins displayed. An asterisk indicates protein/peptide numbers below ten. (c) Protein abundance distribution of peptides and proteins detected by DIA-Umpire. Red: peptides or proteins shared with other software tools. Turquoise: peptides or proteins detected exclusively by DIA-Umpire.
Notably, most proteins exclusively reported by DIA-Umpire (Figure 3 B) were in the lower intensity range (Figure 3 C) and identified by peptides, which were not included in the transition library used for this study. To exclude possible false positive identifications by DIA-Umpire, we reanalyzed the dataset using a dedicated library covering 6,826 of the 9,813 human peptides identified exclusively by DIA-Umpire (Supplementary file DIAumpire_human_peptides_lib.txt). More than 99% of the 6,826 peptides in this library were detectable by at least two library-based tools (Supplementary Figure 20), which picked the same respective peaks as DIA-Umpire in 98% of the cases (Supplementary Figure 21). This orthogonally validated the peptides identified exclusively by DIA-Umpire and thereby confirmed that our initial library was not complete even as we generated the library from triplicate injections of the three species separately to reduce sample complexity. Library completeness might be further improved e.g. by Offgel-fractionation18,26.
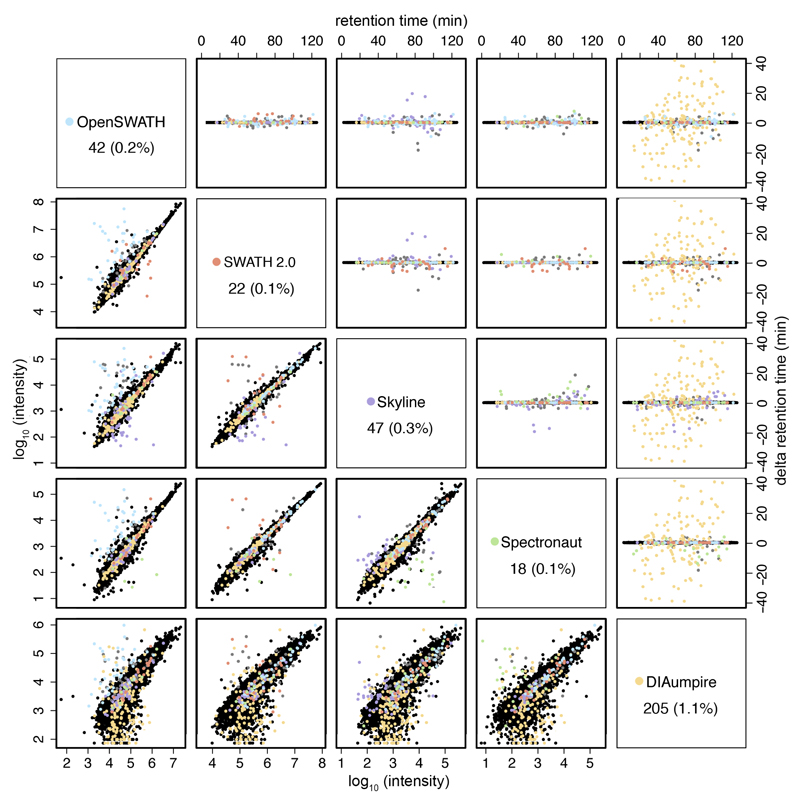
The analysis of common peptides provides a unique opportunity to assess the correctness of the peak picking of each tool. Analyzing one of the injections, we found that all tools pick the same peak (based on retention time) in more than 98% of the cases. All library-based tools each had less than 0.3% of outliers, and DIA-Umpire has approximately 1%. This emphasizes the robustness of SWATH, as even orthogonal identification methods (library-based vs. pseudo-spectra database search) agree in about 99% of the cases (Figure 4). Peptide intensities reported by library-based tools show a very high correlation (R^2: 0.93 – 0.97). The observed differences between DIA-Umpire and any library-based tool (R^2: 0.73 – 0.75) in the first iteration (Supplementary Figure 15) were reduced in the second iteration (R^2: 0.76 – 0.80) (Figure 4). These differences are likely due to the selection of different fragments used for quantification, as about 30% of the top two most intense fragments reported by DIA-Umpire were not included in the DDA library (Supplementary Figure 16). Since DIA-Umpire relies on correct matching of MS1 precursors with their fragments, even high intensity precursors may not be identified by DIA-Umpire due to interferences in the MS1 space (see Supplementary Figure 22). Notably, these differences did not negatively affect the relative quantification accuracy of DIA-Umpire (Figure 2).
Figure 4. Retention time differences and correlation of reported peak intensities between all software tools for the respective matching precursors.
Retention time outliers (upper right panels) are plotted in the color of the outlier software tool (see color legend in the diagonal panels). Diagonal panels show the total number and percentage (to the total number of common detected peptides) of outliers of each respective software tool. Outliers have been defined as producing a standard deviation of the peak retention time greater than 0.2 minutes relative to all other software tools detecting that precursor, after removing ambiguous cases, in which more than one software tool produce a greater standard deviation in the peak retention time. The correlation of reported peak intensities is displayed at the lower left panels. The retention time outliers are also marked in the respective correlation plots.
Discussion
We present a complete methodology to benchmark the robustness of proteomic label-free quantification workflows, based on high-complexity benchmarking samples and the LFQbench software. In contrast to the SGS dataset18, the use of hybrid proteome samples16,23 provides several thousands of proteins present at defined relative ratios, enabling an in-depth statistical evaluation of quantification across a dynamic range of several orders of magnitude16.
Several aspects need to be taken into account when comparing proteomics analysis results. A critical component of a software benchmark is the correct interpretation and optimization of the parameters of each software tool12, e.g. the handling of single hit proteins, which display a higher variance (Supplementary Figure 9). To ensure a consistent evaluation workflow, which can also be directly used by other groups, we developed LFQbench, an R-package that includes all metrics and graphical interpretations agreed upon by all software developers.
The close collaboration with the developers and the two analysis iterations have not only provided objectivity to this study, but also resulted in the improvement of all the software tools, underlining the usefulness of LFQbench and our benchmarking data sets both for developers and end users of the different software tools. In the second iteration, all software tools provided highly accurate relative label-free quantification results (Supplementary Table 1) and achieved a practically perfect separation of the human and yeast proteins (2-fold expression changes), even in the lowest intensity tertile. In the HYE124 dataset, SWATH-MS provided high precision and accuracy of label-free quantification, and allowed to reproducibly quantify close to 5,000 proteins in this highly complex sample set. This level of performance is similar to other label-free quantification approaches16,23, and renders SWATH-MS a valid alternative to isotope-labeling based methods, as similar performance metrics can be achieved24. Future improvements in instrumentation regarding dynamic range, acquisition speed, and analyte separation (by higher resolution chromatography or the inclusion of ion mobility separation) are likely to further boost the performance of SWATH-MS workflows, but will also depend on the availability of deep coverage assay libraries27. Here, discovery workflows such as DIA-Umpire may prove an important orthogonal source for the generation of assay libraries. The observed overlap of peptide and protein identifications provided by the four library-based software tools remarkably exceeded the overlap typically achieved for MS/MS identifications between different DDA search engines25,28,29. The differences between results provided by the library-based tools likely derive from differences in the algorithms used for signal extraction, e.g. using dynamic machine learning to improve identification of the correct peaks, retention time alignment (linear vs. non-linear) and cross-annotation of signals between runs. The strong convergence of the results obtained from the software tools after an optimization cycle indicates that SWATH proteomic results are objectively comparable, even if different software tools are used for their analysis.
In conclusion, our presented methodology provides a rich resource for future improvements of quantitative proteomics software tools, performance control of quantitative proteomics platforms, and also enables benchmarking of algorithms for peak detection interference removal and improved strategies for peptide to protein inference. The LFQbench software and the hybrid proteome samples16 allow to consistently evaluate label-free quantification workflows from any acquisition method or instrument platform.
Online Methods
Sample preparation
Two samples A and B were prepared by following precisely the same steps as in a previous work16: Human cervix carcinoma cell line HeLa was purchased from the German Resource Centre for Biological Material (Braunschweig, Germany) and cultured as described31. Cells were verified to be mycoplasma-free using the VenorGEM mycoplasma detection kit (Sigma, Taufkirchen, Germany). A pure culture of the Saccharomyces cerevisiae bayanus, strain Lalvin EC-1118 was obtained from the Institut Oenologique de Champagne (Epernay, France). Yeast cells were grown in YPD media as described by Fonslow et al.32. Cell lysis and tryptic digestion using a modified filter-aided sample- preparation33 protocol were performed as previously described in detail31. Tryptic digest of Escherichia coli proteins (MassPREP standard) was purchased from Waters Corporation.
To generate the HYE124 hybrid proteome samples, tryptic peptides were combined in the following ratios: Sample A was composed of 65% w/w human, 30% w/w yeast, and 5% w/w E. coli proteins. Sample B was composed of 65% w/w human, 15% w/w yeast, and 20% w/w E. coli proteins (Figure 1). To generate the HYE110 hybrid proteome samples, tryptic peptides were combined in the following ratios: Sample A was composed of 67% w/w human, 30% w/w yeast, and 3% w/w E. coli proteins. Sample B was composed of 67% w/w human, 3% w/w yeast, and 30% w/w E. coli proteins (Supplementary Figure 1). For facilitating retention time alignments among samples, a retention time kit34 (iRT kit from Biognosys, GmbH) was spiked at a concentration of 1:20 v/v in all samples.
Mass spectrometric instrumentation and data acquisition
The LC-MS/MS data acquisition was performed either on (i) a “TTOF5600 system”: a 5600 TripleTOF mass spectrometer (ABSciex, Concord, Ontario, Canada) interfaced with an Eksigent NanoLC Ultra 2D Plus HPLC system (Eksigent, Dublin, CA); or on (ii) a “TTOF6600 system”: a 6600 TripleTOF mass spectrometer (ABSciex, Concord, Ontario, Canada) interfaced with an Eksigent NanoLC Ultra 1D Plus HPLC system (Eksigent, Dublin, CA). For the measurements on the 5600 system, the peptides were separated on a 75 μm-diameter, 20 cm-long fused silica emitter, packed with a Magic C18 AQ 3 µm resin (Michrom BioResources, Auburn, CA, USA). For the measurements on the 6600 system, the peptides were separated on a 75 μm-diameter, 40 cm-long fused silica emitter, packed with a Magic C18 AQ 1.9 µm resin (Michrom BioResources, Auburn, CA, USA). Both systems were operated with the same buffers (buffer A: 2% acetonitrile, 0.1% formic acid; buffer B: 98% acetonitrile, 0.1% formic acid) and the same gradient: linear 2-30% B in 120 minutes, up to 90% B in 1 minute, isocratic at 90% B for 4 minutes, down to 2% B in 1 minute and isocratic at 2% B for 9 minutes.
For shotgun acquisition, 1 µL of the peptide digests for the three organisms (E Coli, Yeast and Human) were injected independently at 1 µg/µL in technical triplicate on the 6600 system operated in shotgun/information dependent acquisition mode. In this mode, the MS1 spectra were collected between 360-1460 m/z for 500 ms. The 20 most intense precursors with charge states 2-5 that exceeded 250 counts per second were selected for fragmentation, and the corresponding fragmentation MS2 spectra were collected between 50-2000 m/z for 150 ms. After the fragmentation event, the precursor ions were dynamically excluded from reselection for 20 s. The precursors were fragmented with the same collision energy equation 0.0625 * m/z -10.5 with a 15 eV collision energy spread for all the precursor charge states to mimic the fragmentation patterns occurring in SWATH MS mode.
For SWATH MS acquisition, 1 µL of the mixed peptide digests (Sample A or Sample B) was injected in technical triplicate on either the 5600 system or the 6600 system. Four window acquisition schemes were used: the original from the work of Gillet et al.4: 32 fixed (“32fixed”) window setup of 25 m/z effective precursor isolation, and a 64 variable (“64var”) window setup optimized on tryptic human cell lysate for equal repartition of the number of precursors that will be co-selected per swath. Those 2 setups were used for the HYE124 acquisition. In addition, acquisition schemes using 32 variable (“32var”) and 64 fixed (“64fixed”) windows setups were performed for the HYE110 sample set to study the effect of fixed versus variable windows. All schemes included an additional 1 m/z window overlap on the lower side of the window. The nominal SWATH windows programmed in both acquisition schemes are provided at the Supplementary Table 7. The SWATH MS2 spectra were collected in high-sensitivity mode from 50 to 2000 m/z, for 100 ms for the 32w setup, and for 50 ms for the 64w setup. Before each SWATH MS cycle an additional MS1 survey scan in high-resolution mode was recorded for 150 ms, resulting in a total duty cycle of ~3.4 s. The collision energy used in SWATH mode was that applied to a doubly charged precursor centered in the middle of the isolation window calculated with the same collision energy equation mentioned above for the shotgun acquisition, and with a spread of 15 eV.
Shotgun data searching and spectral library generation
Profile-mode WIFF files from shotgun data acquisition were converted to mzXML files in centroided format using the qtofpeakpicker algorithm (provided with ProteoWizard/msconvert version 3.0.6141) with the following options: --resolution=20000 --area=1 --threshold=1 --smoothwidth=1.1. The centroided mzXML files were further converted to mgf files using MzXML2Search provided with TPP version 4.7.0. The duplicate shotgun files for each organism were queried each against a customized organism-specific database based on the SwissProt database release from 2014/02/14 and each appended with common contaminants, iRT peptide sequences and the corresponding pseudo-reversed sequence decoys.
The Comet35 (version 2014.02 rev. 0) database search was performed using the following parameters: semi-trypsin digest, up to 2 missed cleavages, static modifications of 57.021464 m/z for cysteines, up to 3 variable modifications of 15.9949 m/z for methionine oxidations (maximal number of variable modifications = 5). The precursor peptide mass tolerance was set to 50 p.p.m. and the fragment bin tolerance set to 0.05 m/z. The Mascot36 (version 2.4.1) database search was performed using the following parameters: semi-tryptic digest, up to 2 missed cleavages, static modifications of carbamidomethyl for cysteines, variable modifications of oxidation for methionine. The precursor peptide mass tolerance was set to +/-25 p.p.m. and the fragment bin tolerance set to +/-0.025 m/z. The identification search results were further processed using PeptipeProphet (with the options: -OAPpdlR -dreverse_) and the results of the search engines per run were combined for each organism using iProphet (TPP version 4.7.0). The search results were finally filtered at 1% protein false discovery rate (FDR) using Mayu37, which resulted in the following iProphet peptide probability cutoffs: 0.319349, 0.92054 and 0.995832 for E.Coli, yeast and human respectively. The MS/MS spectra passing this cutoff for each organism were compiled into three organism-specific redundant spectral libraries with SpectraST38 and the iRT values were computed using the linear iRT regression function embedded in spectrast (option: -c_IRTspectrast_iRT.txt -c_IRR). A consensus library for each organism was finally generated with spectrast. Each organism-specific consensus spectral library was exported to separate assay lists (depending on whether the assay library was used to extract the 32 or 64 fixed or variable SWATH data files, which have different fragment extraction exclusion windows) in TSV format complying to OpenSWATH or SWATH2.0 format using the spectrast2tsv.py script (msproteomicstools version msproteomicstools/master@7527c7b, available from https://github.com/msproteomicstools) using the following options: -l 350,2000 -s y,b -x 1,2 -o 6 -n 6 -p 0.05 -d -e -w 32swaths.txt (respectively 64swaths fixed or variable .txt). The assay libraries for the three organisms were merged at this stage, curated for contaminant, iRT and decoy proteins and saved for downstream targeted SWATH extraction software tools. The consensus library (provided as Supplementary Material: transition library), which contained 44,294 peptides corresponding to 6,903 protein groups. A statistics summary counting number of transitions, peptides, and proteins is provided in Supplementary Table 8.
SWATH MS targeted data extraction
In the sample HYE124, for each tool evaluated, the SWATH files were searched in batches of 6: 3 technical replicate of sample A, and three replicate of sample B, for a given instrument (TTOF5600 system or TTOF6600 system) and for a given SWATH acquisition window scheme (32w or 64w), resulting in 4 result sets per tool and per iteration.
In the sample HYE110, for each tool evaluated, the SWATH files were searched in batches of 6: 3 technical replicate of sample A, and three replicate of sample B, for all four given SWATH acquisition window scheme (32 fixed windows, 32 variable windows, 64 fixed windows, or 64 variable windows), resulting in 4 result sets per tool.
The same retention time extraction window (10 minutes) and fragments mass extraction window (50 p.p.m. and 30 p.p.m. for the TripleTOF 5600 and the TripleTOF 6600 respectively) were used in all software tools. Notably, Spectronaut estimates both parameters dynamically in function of the mass and elution time. Experienced users of Skyline may find at the Supplementary Figures 23 and 24 a benchmark performed with the recommended values for the m/z tolerance (100 p.p.m.).
OpenSWATH targeted data extraction
For OpenSWATH (version OpenMS/develop@4bca6fc) analysis, the different OpenSWATH TSV assay libraries generated above were further converted to TraML39 using the tool ConvertTSVToTraML. Decoy assays were appended to the TraML file using the OpenSwathDecoyGenerator command (option: -method pseudo-reverse -append -exclude_similar). Data analysis using the tool OpenSwathWorkflow was performed on a computer cluster running CentOS release 6.7 through the iPortal40 workflow manager. The SWATH WIFF files were first converted to profile mzXML using msconvert as previously described18. The targeted extraction parameters applied were: 50 ppm (or 30 ppm, see results) for the fragment ion extraction window and 600 seconds for the retention time extraction window. The background subtraction option was either not used (iteration 1) or used with the “original” option with a custom build of OpenMS (iteration 2). After the extraction, pyprophet47 (version 0.13.2) was run on the extraction results to compute the discriminant score using a subset of the scores (main: xx_swath_prelim_score others: library_corr yseries_score xcorr_coelution_weighted massdev_score norm_rt_score library_rmsd bseries_score intensity_score xcorr_coelution log_sn_score isotope_overlap_score massdev_score_weighted xcorr_shape_weighted isotope_correlation_score xcorr_shape) and ten-fold cross-validation for each dataset and to estimate the assay-level q-value (FDR). TRIC (Roest HL et al, in preparation) (version msproteomicstools/master@7527c7b), a cross-run realignment algorithm, was applied to the pyprophet results to correct for potential false peak group ranking in the original peptide identification stage. The default parameters with minor changes (realign_method: lowess, dscore_cutoff: 1, target_fdr: 0.01, max_rt_diff: 30, method: global_best_overall) were used.
SWATH2.0 targeted data extraction
For SWATH2.0 processing, all PeakView TSV assay libraries generated above were appended with iRT peptide assays (protein label [RT-Cal protein]). The iRT peptide assays have been shifted to positive values by adding 62.5 to all values to prevent a known issue of SWATH2.0 with negative iRT values. The SWATH2.0 extraction was performed on a personal computer running Windows 7, PeakView version 2.2 and the SWATH2.0 plug-in “MS/MS(ALL) with SWATH™ Acquisition MicroApp 2.0 Software”. The assay library and the WIFF files were directly loaded into the SWATH2.0, and processed with the parameters specified in the Supplementary Table 5. Peak extraction results were exported to Microsoft Excel files by using the option “Quantitation -> SWATH processing -> Export -> All”.
Skyline targeted data extraction
For Skyline processing, two Skyline document templates corresponding to the four acquisition schemes were generated. Each of these Skyline document templates includes a library (imported from the corresponding SWATH schema library from OpenSWATH) and a retention time predictor that contains the iRT assays of the calibration peptides. The targeted data extractions were performed on a personal computer running Windows 7 and the Skyline-daily version 3.1.1.8669. All parameters were then set as described in the Supplementary Table 5. For the iteration 1, WIFF files were directly imported, and for the iteration 2, WIFF files were converted to centroided mzML files by using the ABsciex MS Data Converter version 1.3 beta. After importing the injection files (either in WIFF or in mzML format), all detected peaks were reintegrated by using the mProphet peak scoring model (each dataset and iteration was trained independently), and the q-value annotation was added to each peak. The resulting weight values of each model are detailed in the Supplementary Table 5. Peak extraction results were exported by using a designed report (SWATHbenchmark report), appended in the Supplementary Material at ProteomeXchange.
Spectronaut targeted data extraction
For Spectronaut processing, all OpenSWATH tsv assay libraries generated above could be used directly. The Spectronaut extraction was performed on a personal computer running Windows 7. Raw WIFF files were converted to HTRMS files with a special converter provided by Biognosys AG able to recognize the older Biognosys iRT retention kit used in our experiments. The HTRMS files are provided as Supplementary Material at ProteomeXchange. For the iteration 1, Spectronaut version 7.0.8065.0.29754 (Nimoy) was used, and Spectronaut 7.0.8065.1.24792 (Nimoy) for the iteration 2. Files in HTRMS format were imported to Spectronaut, and processed with the parameters provided in Supplementary Table 5. In brief, a dynamic window for the XIC extraction window and a non linear iRT calibration strategy were used. The identification was performed by using the normal distribution estimator, including MS1 scoring and the dynamic score refinement. For the quantitation, the interference correction was activated, and a cross run normalization was performed by using the total peak area as normalization base. The profiling strategy was not activated. Peak extraction results were exported by using a designed report (included in the Supplementary Material at ProteomeXchange), which necessarily needs to include the following fields for further processing with LFQbench: EG.Qvalue, FG.NormalizedTotalPeakArea, EG.ProteinId, R.FileName, EG.ModifiedSequence, and FG.Charge. A significance filter of 0.01 was chosen.
DIA-Umpire analysis
The WIFF raw files of the HYE124 sample were first converted into mzML format by the AB MS Data Converter (AB Sciex version 1.3 beta) using the “centroid” option, and then further converted into mzXML format by msconvert.exe from the ProteoWizard package. The mzXML files were processed by the signal extraction (SE) module of DIA-Umpire22 (v1.4) to generate pseudo MS/MS spectra in MGF format. For the HYE110 sample, WIFF raw files were directly converted in to mzXML format by msconvert.exe from the ProteoWizard package. The resulting mzXML files were processed by DIA-Umpire (v2.0). Both HYE124 and HYE110 samples were processed using same parameters listed in Supplementary Table 5. In brief, for detection of precursor ion signal, the following parameters were used: 30 p.p.m mass tolerance, charge state range from 1+ to 5+ for MS1 precursor ions, 2+ to 4+ for MS2 unfragmented precursor ions. For detection of fragment ions, 40 p.p.m mass tolerance was used. The maximum retention time range was set to 1.5 minutes. The minimum intensity threshold for each DIA acquisition scheme (i.e. all data acquired on the same instrument and using the same window setting) was set manually (Supplementary Table 5) and the automatic background detection was not used.
The generated pseudo MS/MS spectra were searched using X! Tandem41, Comet35 and MSGF+42 search engines using the following parameters - allow tryptic peptides only, up to two missed cleavages, and methionine oxidation as variable modification and cysteine carbamidomethylation as static modification. Note that X! Tandem by default adds the following variable modifications: -17.0265 Da (-NH3) or -18.0106 Da (-H2O) on N-terminal Q or E, -17.0265 Da (-NH3) on N-terminal cysteine, and N-terminal acetylation (42.0106 Da). The precursor-ion mass tolerance and the fragment-ion mass tolerance were set to 30 p.p.m. and 40 p.p.m., respectively. We used the same FASTA file used as for searching the DDA data. The fasta file contained corresponding reversed sequences, which were considered as decoys for target-decoy analysis. The output files from the search engines were further analyzed by PeptideProphet43 and combined by iProphet44.
False discovery rate (FDR) of peptide ion identifications was estimated using target-decoy approach based on maximum iProphet probabilities for each peptide ion (peptide sequence, charge state, modification and modification site) individually for each SWATH-MS run. If the maximum iProphet probability of a peptide ion passed the desired FDR threshold, then all detections of same peptide ion across all files within the same data acquisition scheme were accepted. Protein inference was done by ProteinProphet43 independently for each SWATH-MS acquisition using iProphet results. A 1% protein FDR global (“master”) protein list for each individual SWATH-MS run was generated using the target-decoy approach45 based on maximum peptide ion iProphet probability. The protein list for each individual SWATH-MS run was then determined by mapping its locally identified peptides (at 1% peptide ion FDR) to the master protein list for the corresponding data acquisitions scheme.
All peptide ions identified within 1% FDR were used to generate an internal spectral library for DIA-Umpire’s targeted re-extraction in each SWATH-MS run to reduce the number of missing quantifications across the dataset. For quantification analysis, protein-level quantification was performed using the default peptide and fragment selection procedure (Top6pep/Top6fra, Freq > 0.5), as described in the DIA-Umpire manuscript. In the first iteration of HYE124 sample quantification, all detected fragment ions were included for fragment selection procedure implemented in DIA-Umpire v1.4. In the second iteration of HYE124 result, fragment ions below 350 m/z were excluded from the fragment selection procedure and the quantification was performed by DIA-Umpire v2.0. For the HYE110 quantification analysis, DIA-Umpire v2.0 was used with the addition of 350 m/z fragment filtering.
Software changes after first iteration
Both OpenSWATH and Spectronaut modified the respective background subtraction algorithms. Skyline adapted a different workflow by interrogating centroid data, which notably reduced the noise input. SWATH 2.0 disabled the cross-annotation and reporting of single-hit proteins DIA-Umpire excluded fragment ions below 350 m/z for quantification, and switched to a different raw data converter for centroiding, improving quantification precision.
Benchmark analysis with LFQbench
To provide a fair comparison of the quantification performance of the SWATH software tools tested, the developers of the respective software tools jointly established the data integration and evaluation criteria. First, the result exports from different software tools were processed by the FSWE module of the LFQbench package to generate homogenous peptide and protein quantification report files (function FSWE.generateReports). In this study, we established an FDR threshold of 1% for all software tool reports (report files from each software tool were whether previously filtered - Spectronaut and DIA-Umpire, or filtered by FSWE – OpenSWATH, SWATH 2.0, and Skyline). In the case of SWATH 2.0, the iteration 2 has been performed by filtering results by FDR in FSWE, and thus the original file is the same for both iterations. In addition to the built-in protein quantification reported from DIA-Umpire and SWATH 2.0, provided peptide quantification data from each tool were used to quantify proteins using the TOP346,47 quantification model implemented in LFQbench and agreed among software developers in this work. TOP3 is a popular approach to estimate absolute protein quantities based on the average intensity of the three most intense peptides detected46.
The quantitative readings of different software packages were transformed to a reference range of values by linear scaling (function FSWE.scaleIntensities) (Supplementary Figure 25). To determine scaling factors, we used peptide quantification readings of SWATH 2.0 software as reference. We adapted the precursor reports produced by each tool by summarizing the peptide intensity as the sum of all precursors identified with the same peptide sequence and modifications. The scaling factors were applied to transform both corresponding peptide and protein quantification reports.
The software reports of each dataset (sample, instrument, swath windows setup) were processed separately. The homogenized quantification reports were collected in separate subfolders for each dataset in a file system structure as specified for the root folder for the subsequent LFQbench analysis. Using the core module of LFQbench, collected peptide and quantification reports of the four datasets were analyzed (function LFQbench.batchProcessRootFolder) and the result sets for all datasets and software tools were stored to a file for the subsequent creation of figures and tables provided in this study.
We repeated the analysis for single hit proteins (proteins identified by only one peptide) by using the parameter “singleHits = True” at the FSWE.generateReports function.
For the reproducibility of this analysis, all LFQbench analysis steps described in this section and used LFQbench parameters as well as the definition of the analyzed datasets were scripted in R-files (see Supplementary Material at ProteomeXchange: scripts).
Metrics
LFQbench reports a set of metrics values, including: identification rate (number of identified proteins for benchmark species), technical variance (the median CV for the background species), global accuracy (defined as the median deviation of log-ratios to the expected value), global precision of quantification (the standard deviation of log-ratios), and global species overlap (defined as the area under the ROC curve between a species pair). Additionally for this work, we have included the averaged standard deviations and averaged deviations from the expected value of data tertiles as corresponding local metrics (Supplementary Table 1), and tertile box plots (Figure 2 and Supplementary Figures 7-9, and 11-13). To determine statistical significance between results provided by different iterations of the software tools, we performed a one-sided Wilcoxon rank sum tests based on the absolute deviations from the expected log2 values for each protein or peptide.
Peptide and Protein overlap analysis
Peptide and protein identifications were read from the LFQbench-compatible reports generated by FSWE module. For compatibility, all peptide modifications are converted to UniMod.
Peak retention time and intensity match analysis
One of the injections (lgillet_I150211_008) of the TripleTOF 6600 – 64 windows dataset was selected to compare intensity and peak retention time values reported by each software tool. For determining if a software tool reports a different peak compared to other tools (Figure 4 upper panel), the standard deviations of the reported retention times of each peak (identified as peptide + precursor charge state) were calculated, considering only peaks reported for at least three software tools. If the standard deviation of a group of peaks was higher than 0.2 minutes, the reported peak (of one software tool) that deviated most from the average retention time was considered an outlier. To avoid ambiguous cases, in which more than one reported peak is deviated from the average, we removed from the standard deviation calculation the most deviated outlier, and checked if the new standard deviation was below the selected threshold (0.2 minutes). Intensity peaks were paired by using the intensity value reported by each tool (Figure 4 lower panel).
Analysis Reproducibility and Code Availability
A set of scripts that run LFQbench (LFQbench is available in: https://github.com/IFIproteomics/LFQbench), and arrange the final figures of this work are provided at the ProteomeXchange. Given the set of software tool reports (folder provided in ProteomeXchange in a zipped file), only minimal changes (i.e. file paths, selecting some variable values,…) are necessary to reproduce all the analyses of this work.
The script process_hye_samples.R runs LFQbench analyses for all datasets studied in this manuscript (four datasets of HYE124 – including 2 iterations each – and four datasets of HYE110), and produces the Supplementary Figures 17 and 25. After the execution of process_hye_samples.R, the script generate_figures.R reproduces (or produces the necessary data) for most of the figures and tables of the manuscript (Figure 2, Table 1, Supplementary Tables 1-3, 6, and 9, Supplementary Figures 5, 7-14, 17, 18, and 22). For analyses, which require to cross data from multiple datasets or software tools the following scripts are provided. The script Int.Correlations.TechReplicates.and.Datasets.R analyses intensity correlations of technical replicates and datasets (Supplementary Table 4, and Supplementary Figures 4 and 6). The script pair.RTs.and.Intensities.R displays the intensity and retention time correlations among the different software tools (Figure 4 and Supplementary Figures 15 and 21). For reproducing peptide and protein overlap Venn diagrams, the reader can run the scripts peptideOverlap.R (Supplementary Figure 20), peptideOverlapTertiles.R (Supplementary Figure 22), and proteinOverlap.R (Figure 3). The comparison of the signal to noise ratios obtained with the different swath isolation modes (32 fixed windows fixed, 32 variable windows, 64 fixed windows, and 64 variable windows) is run by the script SignalNoiseRatios.R (Supplementary Figure 3). The script significance.tests.R produces the significance tests for the Supplementary Table 1. The R markdown file ionlibrary_statistics.Rmd generates the DDA ion library statistics shown in Supplementary Table 8, and the comparison between the DDA ion library and the fragments used by DIA-Umpire (Supplementary Figure 16) is performed by match_fragments_DIAumpire_DDAlibrary.R. Finally, the script ExperimentSimulation.R performs several simulations of LFQ experiments and performs the corresponding LFQanalysis (Supplementary Figure 2).
LFQbench software
LFQbench is an open source R library for the automated evaluation of label-free quantitation based on the interpretation of the quantitative analysis results of hybrid proteome sample set data16.
Input data format
To deal with differences in result reporting formats among different data analysis solutions, we defined a simple data input format. For evaluation with LFQbench, the input data has to be converted to a delimiter separated (e.g. tabulator separated data as .tsv files) having column names in the first row. First column must contain identification names of quantified proteins or peptides. One of the other columns must be named “species” and must contain the species names as plain text e.g. HUMAN, YEAST, ECOLI, PIG, etc. of the quantified protein or peptide. All other columns should contain quantitation readings in different experiment runs in the order of samples and in equal numbers of replicate experiments for each sample (e.g. A1, A2, …, An, B1, B2, …, Bn).
File format conversion
The FSWE (Format SoftWare Exports) module homogenizes the results exports from different software tools, and applies a peptide-protein quantitation model. This module accepts plain text format reports in both long and wide formats, and it can be easily adapted for reading any kind of quantitation report provided in plain text format by all software tools. For each file format, the following parameters must be configured: value name for the quantitative value (quantitative.var), protein name (protein.var) (protein names must include a species tag), injection filename (filename.var), sequence including modifications (sequence.mod.var), precursor charge state (charge.var), string to report missing values (nastrings), the input format (input_format, options: “long”, “wide”). Additionally, Q-value column (qvalue.var) and a threshold (q_filter_threshold) may be reported for filtering by Q-values. LFQbench provides predefined settings for a set of software tools namely DIA-Umpire, OpenSWATH, SWATH 2.0 (PeakView), Skyline and Spectronaut including parameter schemas for the built-in protein quantitation in DIA-Umpire and SWATH 2.0. The interface function FSWE.addSoftwareConfiguration allows an easy definition of further parameter schemas if needed. The species tags, experiments, samples and injection names must be specified before converting software reports. The interface function FSWE.generateReports produces two output files for each software tool report: a peptide report and a protein report. The peptide report sums the quantitative values (quantitative.var) of the different precursor charge states (charge.var) reported for each peptide (sequence.mod.var). It converts reported modifications to the UniMod format, then it removes duplicated precursor extractions (based on sequence.mod.var and charge.var), it filters the data by a Q-value threshold (q_filter_threshold), and it removes precursors labeled as decoy (decoy.tag), and peptides shared between species. The protein report estimates a quantitation value for each protein group (protein.var) by using a TOP3 approach: the three most intense peptide quantitative values of each individual run are averaged (a minimum of two peptides is required). Produced peptide and protein reports can be directly used in the main LFQbench module. FSWE filters results at the protein level, only proteins having quantification values in at least two technical replicates in at least one of the samples are considered for further analysis. If a quantification value is absent in one or two technical replicates, LFQbench calculates the average of the reported values. If a quantification value is missing in all three replicates this leads to an invalid quantification ratio.
Intensity scaling
Software tools may report quantitation values in different ways. To enable a direct comparison between different software tools, peptide and protein reports can be scaled to a reference using the interface function FSWE.scaleIntensities. The function scales quantitation values of each input file in the specified folder by using a linear regression through the origin of the data within the 98th percentile of the peptide quantitation values of each software tool to the peptide quantitation values of the specified reference software.
LFQbench analysis
For the main analysis, LFQbench reads quantitative values from a valid input file, process them in multiple steps and produces a result set object which summarizes the input data and contains statistics and evaluation metrics based on the evaluated data. A first process checks input data validity (see Input data format section). The second step removes from the dataset quantitative amounts below a user-defined threshold. Next, missing value and identification statistics are calculated. At the next stage, peptides or protein amounts are optionally converted to relative values by transforming the original quantitative values to parts per million of the total amount in individual experiment runs. For the evaluation of technical reproducibility, LFQbench calculates dispersion of quantitative values as coefficients of variation for each identified peptide or protein among technical replicates of each sample. After assessing the technical variance, quantitative values in replicate runs are used to calculate sample average amounts for each peptide or protein. To generate a basis for the evaluation of the relative quantitation performance, logarithmic (log2) ratios of sample average amounts are calculated for each identification and each sample pair in the present dataset (e.g. log2(A/B)). LFQbench estimates the validity range of log-ratios as a maximum difference of a user controlled factor (default 5) times the standard deviation from the average log-ratio value for each species. Outlier log-ratios that are out of validity range are dropped and remaining log-ratios are shifted by the median log-ratio of the predefined background species to center the data (Supplementary Table 9).
Finally, global and local metrics for the evaluation of precision and accuracy of quantification, and species separation ability are calculated and stored in the result set. User can explore calculated result sets for the processed data and evaluation metrics directly in an R environment or visualize results and export identification and quantification metrics using LFQbench functions:
-
-
LFQbench.getMetrics
-
-
LFQbench.showMetrics
-
-
LFQbench.showDistributionDensityPlot
-
-
LFQbench.showLogRatioBoxPlot
-
-
LFQbench.showScatterAndBoxPlot
-
-
LFQbench.showScatterAndDensityPlot
-
-
LFQbench.showScatterPlot.
Batch analysis
For a combined evaluation of multiple input files, the main LFQbench analysis can be run in batch mode using the interface function LFQbench.batchProcessRootFolder. In batch mode, LFQbench discovers the input files from a subfolder of a data root folder structure, processes them and produces corresponding result sets. The calculated result sets are automatically visualized, and statistics as well as identification and quantification metrics are exported to files.
Supplementary Material
Editorial summary.
LFQbench, a software tool to assess the quality of label-free quantitative proteomics analyses, enables developers to benchmark and improve analytic methods
Acknowledgements
We want to thank Ruben Spohrer for his excellent sample preparation, and Lyle Burton, Adam Lau, and Gordana Ivosev for their support with SWATH 2.0. P.N. and S.T. are supported by grants from BMBF (Express2Present 0316179C), the DFG (ST599/1-1) and Mainz University (Research Center for Immunotherapy (FZI)). H.L.R. is supported by SNF grant P2EZP3_162268. Y.P-R. is supported by the BBSRC ‘PROCESS’ grant [BB/K01997X/1]. A.I.N is supported by U.S. National Institutes of Health grant no. 5R01GM94231. R.A. was supported by ERC AdG #233226 (Proteomics v3.0) and ERC -2014-AdG # 670821 (Proteomicxs 4D), by the PhosphonetX project of SystemsX.ch and by the Swiss National Science Foundation grant #3100A_166435.
Footnotes
Author contributions
P.N. and S.T. designed and supervised the study, U.D. and L.C.G. prepared the samples and performed the MS measurements, L.C.G., G.R., and H.L.R. executed and supervised the OpenSWATH analyses, P.N. and S.A.T. executed and supervised the SWATH 2.0 analyses, P.N. and B.M. executed and supervised the Skyline analyses, P.N., O.M.B., and L.R. executed and supervised the Spectronaut analyses, C.C.T. and A.N. executed and supervised the DIA-Umpire analyses, J.K., P.N., and Y.P-R. developed LFQbench, P.N., S.T., J.K., B.M., O.M.B. performed the benchmark analyses, L.C.G., O.M.B., B.M., H.L.R., S.A.T., C.C.T., L.R., G.R., A.I.N., and R.A. provided critical input into the project, P.N., J.K., and S.T. wrote the manuscript.
Competing financial interests
The authors declare the following competing financial interests: S.A.T. is employed by SCIEX, O.M.B. and L.R. are employed by Biognosys AG.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository30 with the dataset identifier PXD002952.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Mallick P, Kuster B. Proteomics: a pragmatic perspective. Nat Biotechnol. 2010;28:695–709. doi: 10.1038/nbt.1658. [DOI] [PubMed] [Google Scholar]
- 3.Distler U, Kuharev J, Tenzer S. Biomedical applications of ion mobility-enhanced data-independent acquisition-based label-free quantitative proteomics. Expert Rev Proteomics. 2014;11:675–684. doi: 10.1586/14789450.2014.971114. [DOI] [PubMed] [Google Scholar]
- 4.Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geromanos SJ, Hughes C, Ciavarini S, Vissers JPC, Langridge JI. Using ion purity scores for enhancing quantitative accuracy and precision in complex proteomics samples. Anal Bioanal Chem. 2012;404:1127–1139. doi: 10.1007/s00216-012-6197-y. [DOI] [PubMed] [Google Scholar]
- 6.Geiger T, Cox J, Mann M. Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol Cell Proteomics. 2010;9:2252–2261. doi: 10.1074/mcp.M110.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 8.Li G-Z, et al. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 9.Michalski A, Cox J, Mann M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J Proteome Res. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 10.Gatto L, et al. Testing and Validation of Computational Methods for Mass Spectrometry. J Proteome Res. 2015 doi: 10.1021/acs.jproteome.5b00852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dufresne Craig, H D, I AR, A K, B M, P B, R K, R P, S B, S S, C CM. ABRF Research Group Development and Characterization of a Proteomics Normalization Standard Consisting of 1,000 Stable Isotope Labeled Peptides. Journal of Biomolecular Techniques : JBT. 2014;25:S1. [Google Scholar]
- 12.Yates JR, et al. Toward objective evaluation of proteomic algorithms. Nat Methods. 2012;9:455–456. doi: 10.1038/nmeth.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leprevost FDV, Barbosa VC, Francisco EL, Perez-Riverol Y, Carvalho PC. On best practices in the development of bioinformatics software. Front Genet. 2014;5:199. doi: 10.3389/fgene.2014.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pak H, et al. Clustering and filtering tandem mass spectra acquired in data-independent mode. J Am Soc Mass Spectrom. 2013;24:1862–1871. doi: 10.1007/s13361-013-0720-z. [DOI] [PubMed] [Google Scholar]
- 15.The difficulty of a fair comparison. Nat Meth. 2015;12:273–273. doi: 10.1038/nmeth.3359. [DOI] [PubMed] [Google Scholar]
- 16.Kuharev J, Navarro P, Distler U, Jahn O, Tenzer S. In-depth evaluation of software tools for data-independent acquisition based label-free quantification. Proteomics. 2015;15:3140–3151. doi: 10.1002/pmic.201400396. [DOI] [PubMed] [Google Scholar]
- 17.Sajic T, Liu Y, Aebersold R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteomics Clin Appl. 2015;9:307–321. doi: 10.1002/prca.201400117. [DOI] [PubMed] [Google Scholar]
- 18.Röst HL, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotechnol. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 19.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruderer R, et al. Extending the Limits of Quantitative Proteome Profiling with Data-Independent Acquisition and Application to Acetaminophen-Treated Three-Dimensional Liver Microtissues. Mol Cell Proteomics. 2015;14:1400–1410. doi: 10.1074/mcp.M114.044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiter L, et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 22.Tsou C-C, et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat Methods. 2015;12:258–64. doi: 10.1038/nmeth.3255. 7 p following 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox J, et al. MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navarro P, et al. General Statistical Framework for Quantitative Proteomics by Stable Isotope Labeling. J Proteome Res. 2014 doi: 10.1021/pr4006958. [DOI] [PubMed] [Google Scholar]
- 25.Bell AW, et al. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat Methods. 2009;6:423–430. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schubert OT, et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat Protocols. 2015;10:426–441. doi: 10.1038/nprot.2015.015. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberger G, et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci Data. 2014;1:140031. doi: 10.1038/sdata.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shteynberg D, Nesvizhskii AI, Moritz RL, Deutsch EW. Combining results of multiple search engines in proteomics. Mol Cell Proteomics. 2013;12:2383–2393. doi: 10.1074/mcp.R113.027797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Z-F, Lin S, Molden RC, Garcia BA. Evaluation of proteomic search engines for the analysis of histone modifications. J Proteome Res. 2014;13:4470–4478. doi: 10.1021/pr5008015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vizcaíno JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Research. 2013;41:D1063–9. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Distler U, et al. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat Methods. 2014;11:167–170. doi: 10.1038/nmeth.2767. [DOI] [PubMed] [Google Scholar]
- 32.Fonslow BR, et al. Digestion and depletion of abundant proteins improves proteomic coverage. Nat Methods. 2013;10:54–56. doi: 10.1038/nmeth.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 34.Escher C, et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12:1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eng JK, Jahan TA, Hoopmann MR. Comet: An open-source MS/MS sequence database search tool. Proteomics. 2012;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 36.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. ELECTROPHORESIS. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Reiter L, et al. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics. 2009;8:2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lam H, et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 39.Deutsch EW, et al. TraML--a standard format for exchange of selected reaction monitoring transition lists. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.R111.015040. R111.015040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunszt P, et al. iPortal: the swiss grid proteomics portal: Requirements and new features based on experience and usability considerations. Concurrency Computat: Pract Exper. 2014;27:433–445. [Google Scholar]
- 41.F D, Beavis RC. A Method for Assessing the Statistical Significance of Mass Spectrometry-Based Protein Identifications Using General Scoring Schemes. Anal Chem. 2003 doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 42.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5 SP:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nesvizhskii AI, Keller A, Kolker E. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry - Analytical Chemistry (ACS Publications) Analytical …. 2003 doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 44.Shteynberg D, et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.007690. M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 46.Silva JC, Gorenstein MV, Li G-Z, Vissers JPC, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Ning K, Fermin D, Nesvizhskii AI. Comparative analysis of different label-free mass spectrometry based protein abundance estimates and their correlation with RNA-Seq gene expression data. J Proteome Res. 2012;11:2261–2271. doi: 10.1021/pr201052x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.