Abstract
Mutation of the AMP deaminase 1 (AMPD1) gene, the predominate AMPD gene expressed in skeletal muscle, is one of the most common inherited defects in the Caucasian population; 2–3% of individuals in this ethnic group are homozygous for defects in the AMPD1 gene. Several studies of human subjects have reported variable results with some studies suggesting this gene defect may cause symptoms of a metabolic myopathy and/or easy fatigability while others indicate individuals with this inherited defect are completely asymptomatic. Because of confounding problems in assessing muscle symptoms and performance in human subjects with different genetic backgrounds and different environmental experiences such as prior exercise conditioning and diet, a strain of inbred mice with selective disruption of the AMPD1 was developed to study the consequences of muscle AMPD deficiency in isolation. Studies reported here demonstrate that these animals are a good metabolic phenocopy of human AMPD1 deficiency but they exhibit no abnormalities in muscle performance in three different exercise protocols.
Keywords: AMP deaminase, Nucleotide metabolism, Muscle
1. Introduction
The AMP deaminase 1 (AMPD1) gene, one of the three members of the AMPD multigene family, encodes the predominant AMPD (adenosine monophosphate (AMP) deaminase: EC 3.5.4.6) isoform expressed in skeletal muscle [1]. AMPD1 deficiency is one of the most common inherited defects in the Caucasian population with a number of studies documenting that 2–3% of individuals in this ethnic group are homozygous for defects in the AMPD1 gene [2]. It is not unreasonable to assume that deficiency of this enzyme activity which is essential for purine nucleotide interconversion and energy metabolism [1] might be associated with muscle symptoms and/or impaired muscle performance. Since the original report of this enzyme deficiency by Fishbein et al. [3] in a group of patients with symptoms consistent with a metabolic myopathy, there have been over 25 studies of individuals with this enzyme defect [1]. This body of literature encompasses conflicting reports that describe a range of findings from asymptomatic individuals, patients with symptoms consistent with those of a metabolic myopathy, individuals with impaired muscle performance on exercise testing as well as individuals with normal exercise performance. This variability in findings in human subjects might be explained by a host of factors including underlying genetic differences in the human population, differences in exercise conditioning, differences in diet, differences in pain threshold, and differences in motivation to perform in exercise tests—all of which might contribute to differences in symptoms and/or exercise performance.
Given the high prevalence of mutations in the AMPD1 gene leading to deficiency in this enzyme activity in skeletal muscle and the conundrum it creates due to the frequent diagnosis of AMPD deficiency in clinical settings, we developed a murine model in which disruption of the AMPD1 gene is the sole genetic difference in an inbred strain of mice. Using a model system such as this it is possible to eliminate differences in exercise activity prior to physiological testing, differences in diet, and presumably other environmental and/or psychological factors that might affect exercise performance. In this murine model in which the AMPD1 gene has been disrupted, we find that these animals exhibit all the metabolic defects reported in humans with inherited defects in the AMPD1 gene but exhibit no changes in exercise performance in three different protocols for assessing muscle function.
2. Material and methods
2.1. Isolation of mouse AMPD1 cDNA and gene
Mouse AMPD1 cDNA clones were isolated from DNA fragments amplified by reverse transcription polymerase chain amplification (RT-PCR) using RNA isolated from P19 mouse embryonal carcinoma cells derived from 129/sv mice. PCR was carried out using primers designed from the sequence of human AMPD1 cDNA. Mouse AMPD1 genomic clones were isolated from PCR-amplified DNA fragments of genomic DNA of P19 cells. The 3′ fragment of the AMPD1 gene was obtained using an adaptor cassette and cassette primer following the manufacturer's protocol (LA PCR in vitro Cloning Kit, Takara).
2.2. Generation of AMPD1 mutant animals
The targeting vector was linearized with NotI and electroporated into D3 embryonic stem cells. Transfected cells were selected by exposure to neomycin for 7 days, after which isolated colonies were screened by PCR. Homologous recombination was confirmed by Southern hybridization on both the long- and short-arm sides. After undergoing a karyotype test, cells were injected into blastocysts derived from C57BL/6 mice to produce chimeric mice. Chimeric pups were identified by their agouti coat color, after which chimeric males were bred to C57BL/6J females and germ-line transmission was determined. Mice with the targeted allele were backcrossed to C57BL/6 mice more than 10 times before the analysis. All of the animals had free access to food (CE-2, CLEAR) and water, and were housed in a controlled SPF environment with a 12-hour light–dark cycle and constant temperature.
2.3. Genotyping, DNA, and RNA analysis
Total DNA was extracted from ES cells or mouse tails. PCR assays for genotype screening were performed using the following primers: for the wild-type AMPD1 gene; 5′-ACAGGCCCTTGTGCAATTAGA-3′ (AM33-) and 5′-TTTCGCACAGAGGACCTTCC-3′ (AM12 +), for the knock-out AMPD1 gene; 5′-ACAGGCCCTTGTGCAATTAGA-3′ (AM33-) and 5′-CCGATTCGCAGCGCATCGCC-3′ (AM11 +). Southern hybridization was performed using a 5′ or 3′ AMPD1 probe with 5′-GTAATACGACTCACTATCGGGC–3′ and 5′-AATTAACCCTCACTAAAGGG–3′. Total RNA extraction and northern hybridization were performed using a standard protocol. A cDNA probe was produced by PCR using the primer set 5′-GGCATGAATACATTTCTGTTTC-3′ and 5′-CTGAGCGACATTTGTCCTC-3′.
For real-time RT-PCR, total RNA was extracted from tissues using a standard protocol. Total RNA (1 μg) was reverse-transcribed in a 20-μl reaction mixture containing random primers and Superscript III enzyme (Invitrogen, Carlsbad, CA). Quantitative real-time PCR was performed with an ABI Prism 7000 Sequence Detection System using an SYBR Green PCR Master Mix Reagent Kit (Applied Biosystems, Foster City, CA).
2.4. AMPD activity assay
Tissues were homogenized with an extraction buffer (100 mM K-phosphate, pH 6.5, 180 mM KCl, 1 mM DTT). The supernatant was dialyzed against 50 mM imidazole/HCl (pH 6.5), 150 mM KCl, and 1 mM DTT for 17 h at 4 °C. Fifty microliter samples were mixed with a 150-μl reaction buffer (25 mM imidazole/HCl (pH 6.5), 150 mM KCl, 10 mM AMP) and incubated at 37 °C for 30, 60, 120, or 180 min. The amount of IMP produced was measured using HPLC with a Capcell Pak C18 column, and a mobile phase including phosphoric acid and diethylethanolamine (Sigma-Aldrich, St. Louis, USA). AMPD activity was assayed by measuring the production of IMP. One unit of enzyme activity was defined as the amount that catalyzed formation of 1 μmol of IMP.
2.5. Protocol for exercise testing
A treadmill movement test was performed according to a previously described protocol [4]. Mice at 12 weeks of age were first acclimated to the treadmill by placing them on an unmoving treadmill with a 20° incline, thereafter the speed of the treadmill was increased from 5 to 7, 10, 12, and 15 m/min for 5 min intervals on 3 successive days before testing.
For the endurance exercise, the treadmill was set at a 20° incline and an initial speed of 5 m/min, with the speed increased every 150 s to 7, 10, 12 or 15 m/min. Thereafter, mice were run for a total 85 min, a point at which they could not maintain sufficient speed to avoid persistent electric shock. The number of electric stimulations was recorded as a measure of fatigability. For sprint exercise testing, the treadmill was set at a 20° incline and an initial speed of 15 m/min for 30 s. For femoral artery ligation ischemia of the hindlimb, male mice were anesthetized, then after a skin incision, the entire femoral artery was dissected free. Arteries in both hindlimbs were ligated with 4–0 silk. Twenty minutes were allowed for recovery from anesthetization, then the mice were subjected to 30 s of the sprint exercise as described above.
At the conclusion of the respective exercise tests, gastrocnemius specimens were snap frozen in liquid nitrogen and stored at − 80 °C. Muscle samples were also collected from animals not subjected to the respective exercise protocols and used as “resting” controls.
2.6. Nucleotide, lactate and adenosine levels
For nucleotide levels tissues were homogenized in 0.4 mol/l perchloric acid. After centrifugation, the clear supernatant was neutralized with 5 M K2CO3 to pH 6.6–8.0, then 10 μl of neutralized supernatant was applied to a Capcell Pak C18 column (Shiseido, Tokyo, Japan) for HPLC (Lachrom Elite, Hitachi, Japan) analysis [5].
Tissue lactate levels were measured by the enzymatic method using lactate oxidase. Tissue samples were homogenized in 0.4 mol/l perchloric acid and the clear supernatant was neutralized with 5 M K2CO3, then the neutralized supernatant was used for the measurement.
Tissue adenosine levels were measured with fluorescence-HPLC, using a method modified by Katayama et al. [6]. Tissue samples were homogenized in 0.4 mol/l perchloric acid and the clear supernatant was neutralized with 5 M K2CO3, then the neutralized supernatant was used for reacting with 2-chloroacetaldehyde, vidarabine, and acetate buffer, using the same protocol noted above. Finally, 10 μl was injected into the HPLC device and the assay was performed using a mobile phase of 0.05 M citric acid, 0.1 M disodium hydrogen phosphate (pH 4.2), and methanol (90 + 10), with a column temperature of 23 °C and flow-rate of 0.8 ml/min.
2.7. Western blot analysis
Muscle samples were sonicated and homogenized in lysis buffer (1 × PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 100 μg/ml phenylmethylsulfonyl fluoride, 45 μg/ml aprotinin, 100 mM sodium orthovanadate). The homogenate supernatant was subjected to protein determination with a BCA Protein Assay Kit (Pierce, IL, USA) and then applied onto SDS-PAGE gels for electrophoresis. After electrophoresis, the gels were transferred to polyvinylidene difluoride membranes, then incubated with the primary antibody, followed by the secondary antibody. Signals were detected with an ECL kit (Amersham, Piscataway, USA).
2.8. Statistical analysis
Results from knockout mice were compared with those of their wild-type littermates using the Mann–Whitney U test, with P < 0.05 considered to indicate statistical significance.
3. Results
3.1. AMPD1 knock-out mice
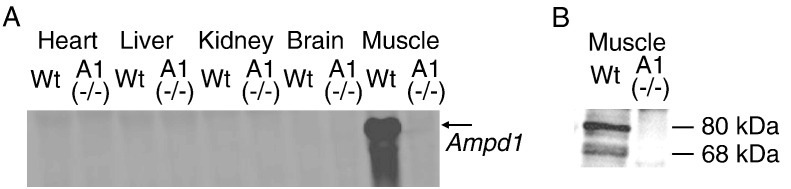
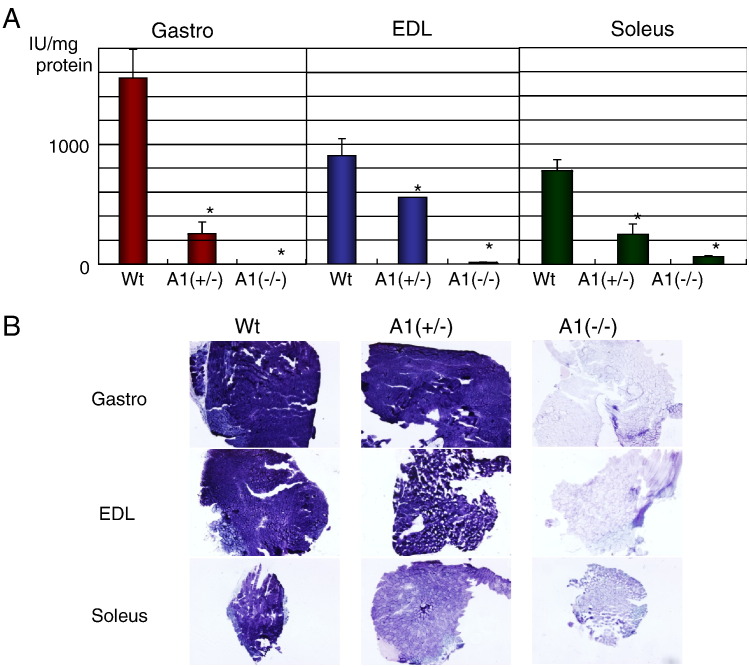
AMPD1 knockout mice were generated using a standard gene targeting method (Supplemental Fig. S1A). Disruption of the AMPD1 gene was confirmed by PCR and Southern blotting (Supplemental Fig. S1B). Northern and western blots confirmed that the AMPD1 knockout homozygote [A1(−/−)] mice did not show any expression of AMPD1 RNA or protein (Figs. 1A and B). Furthermore, AMPD activity was decreased in all muscle types of AMPD1 knockout heterozygote [A1(+/−)] mice (Fig. 2A), and almost undetectable in the gastrocnemius and extensor digitorum longus (EDL) of A1(−/−) mice. In the soleus muscle of A1(−/−) mice, approximately 10% of AMPD activity remained when compared to the wild type, due to the expression of the AMPD3 gene in this muscle type [7]. AMPD1 activity as assessed by histochemical staining (Fig. 2C), a common test used in analysis of human muscle biopsies, also showed reduction or loss of AMPD enzyme activity in muscle samples from the A1(+/−) and A1(−/−) animals, respectively.
Fig. 1.
Expression of AMPD1 gene in AMPD1 knockout mice. A: Expression of AMPD1 mRNA in various tissues including skeletal muscle (gastrocnemius) studied by northern blot analysis (Wt: wild type, A1(−/−): AMPD1 null), B: Expression of AMPD1 protein in skeletal muscle (gastrocnemius) by western blot analysis (Wt: wild type, A1(−/−): AMPD1 null).
Fig. 2.
AMPD activity in skeletal muscles. A: AMPD activity in gastrocnemius (Gastro), EDL, and soleus muscle tissues. Values are shown as the mean ± SE (n = 3 for wild type, n = 5 for AMPD1 heterozygote, n = 5 for AMPD1 null). *P < 0.05. (Wt: wild type, A1(+/−): AMPD1 heterozygote, A1(−/−): AMPD1 null), B: AMPD histoenzymatic stain in gastrocnemius (Gastro), EDL, and soleus muscle tissues. (Wt: wild type, A1(+/−): AMPD1 heterozygote, A1(−/−): AMPD1 null).
3.2. Phenotypic observations of AMPD1 knockout mice
No detectable phenotypic differences were observed between A1(−/−) and wild-type mice in a controlled SPF environment with a 12-hour light–dark cycle, constant temperature (25˚C), and free access to food (CE-2, CLEAR) and water. A1(−/−) mice were born with the expected Mendelian ratio, litter sizes were normal and male and female knockout mice were fertile. A1(−/−) mice exhibited no significant changes in regard to body weight, rates of growth, body composition, food intake, and lifespan as compared with wild-type mice. Hematoxylin and eosin (H&E) stained sections of the liver, kidney, lung, brain, heart, spleen, and stomach showed no significant changes (data not shown). Gastrocnemius, EDL, and soleus muscle samples were analyzed with H&E, periodic acid-Schiff (PAS), oil red O, succinate dehydrogenase (SDH), cytochrome oxidase, acid phosphatase, Gomori's Trichrome, Sudan black B, and NADH staining (data not shown), and there were no detectable differences between A1(−/−) and wild-type animals indicating there was no change in fiber-type change in the A1(−/−) mice.
3.3. Assessment of muscle performance
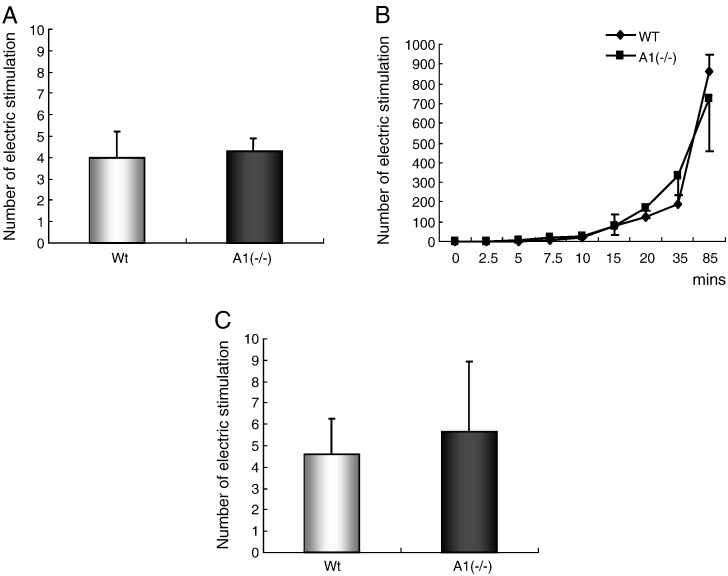
Muscle function was assessed in 12-week-old male mice using both an endurance (85 min) and sprint (30 s) exercise protocol. No significant differences were observed in fatigability after sprint (Fig. 3A) and endurance (Fig. 3B) exercise between the wild-type and A1(−/−) mice. In addition, exercise performance was assessed under ischemic conditions, i.e. following arterial ligation, and no difference in exercise performance was noted between wild-type and A1(−/−) animals (Fig. 3C).
Fig. 3.
Exercise test in the endurance or sprint condition. A: Numbers of electric stimulations after sprint exercise for 30 s (n = 4). B: Numbers of electric stimulations during endurance exercise for 2.5, 5, 7.5, 10, 15, 20, 35, and 85 min (n = 9 for wild type, n = 8 for AMPD1 null, but n = 3 for 15 min for both wild type and AMPD1 null). C: Numbers of electric stimulations after sprint exercise for 30 s with femoral artery ligation ischemia (n = 3 for wild type, n = 5 for AMPD1 null). Values are shown as the mean ± SE. *P < 0.05. (Wt: wild type, A1(−): AMPD1 null).
Nucleotide levels (IMP, AMP, ADP, ATP) were determined in the gastrocnemius muscle before and after performing sprint and endurance exercises (Table 1). IMP in the A1(−/−) group was not detected before or after exercise, indicating that the degradation pathway of AMP to IMP was blocked in the muscle tissues of those mice. After 85 min endurance exercise, the increase in AMP and decrease in ATP levels were similar in both the wild-type and A1(−/−) mice. In the sprint protocol, the level of IMP was significantly increased in the wild-type mice after 30 s exercise, whereas AMP, ADP and ATP levels were not significantly different between the wild-type and A1(−/−) mice in this condition.
Table 1.
The nucleotide levels in the gastrocnemius muscles before and after endurance (85 min) and sprint (30 s) exercise.
| Mean ± SE (nmol/mg wet weight) | ||||||
|---|---|---|---|---|---|---|
| n | ATP | ADP | AMP | IMP | ||
| Pre | Wt | 4 | 10.0 ± 0.2 | 2.79 ± 0.13 | 0.36 ± 0.01 | 0.26 ± 0.04 |
| A1(−/−) | 3 | 9.9 ± 0.3 | 2.78 ± 0.13 | 0.40 ± 0.03 | ND | |
| Endurance ex | Wt | 3 | 9.2 ± 0.4⁎ | 2.20 ± 0.10 | 0.62 ± 0.26⁎ | 0.15 ± 0.08 |
| A1(−/−) | 3 | 9.0 ± 0.3⁎ | 2.30 ± 0.22 | 1.00 ± 0.45⁎ | ND | |
| Sprint ex | Wt | 3 | 10.8 ± 0.4 | 2.82 ± 0.04 | 0.36 ± 0.03 | 0.41 ± 0.02# |
| A1(−/−) | 3 | 12.5 ± 0.2 | 2.98 ± 0.08 | 0.43 ± 0.04 | ND | |
Pre: pre-exercise, Sprint ex: sprint exercise, Endurance ex: endurance exercise.
n: number of animals.
ND: not detected.
P < 0.05 between Pre and Endurance ex.
P < 0.05 between Pre and Sprint ex.
We also measured nucleotide levels in the gastrocnemius muscle before and after sprint exercise in mice with femoral artery ligation ischemia (Table 2). During this short burst of exercise (30 s), no apparent difference of performance was observed between the wild-type and A1(−/−) mice (Fig. 3C). Prior to exercise, ATP levels were decreased in both groups due to acute ischemia, while IMP was increased in the wild-type but not A1(−/−) mice. After 30 s of sprint exercise, the ATP level in the wild-type was further decreased and that of IMP was further increased. Furthermore, ATP, ADP, and AMP levels were higher in the A1(−/−) gastrocnemius muscle than in that of the wild type mice in this condition.
Table 2.
The nucleotide levels in the gastrocnemius muscles under ischemia before and after short sprint exercise (30 s).
| mean ± SE (nmol/mg wet weight) | ||||||
|---|---|---|---|---|---|---|
| n | ATP | ADP | AMP | IMP | ||
| Pre | Wt | 4 | 10.0 ± 0.2 | 2.79 ± 0.13 | 0.36 ± 0.01 | 0.26 ± 0.04 |
| A1(−/−) | 3 | 9.9 ± 0.3 | 2.78 ± 0.13 | 0.40 ± 0.03 | ND | |
| Post-ischemia | Wt | 3 | 7.9 ± 0.7⁎ | 2.98 ± 0.26 | 0.45 ± 0.13 | 1.30 ± 0.34⁎ |
| A1(−/−) | 3 | 8.1 ± 1.0⁎ | 3.20 ± 0.10 | 0.57 ± 0.03⁎ | ND | |
| Post-ischemia | Wt | 3 | 4.8 ± 0.2⁎ | 2.50 ± 0.16 | 0.41 ± 0.06 | 4.50 ± 0.48⁎ |
| + Ex | A1(−/−) | 3 | 8.3 ± 0.5# | 4.20 ± 0.41# | 0.91 ± 0.12# | ND |
Pre: pre-exercise and ischemia, Post- ischemia + Ex: Post-ischemia and exercise.
n: number of animals.
ND: not detected.
P < 0.05 between Pre and and Post-ischemia, or Post-ischemia and Post-ischemia + Ex.
P < 0.05 between genotypes.
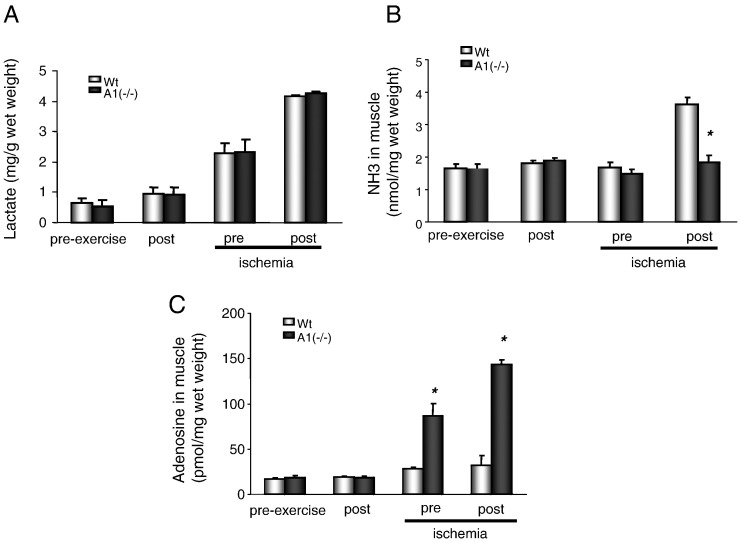
Lactate production was measured after sprint exercise under non-ischemic or ischemic conditions (Fig. 4A). A1(−/−) mice exhibited no differences as compared to the wild-type mice with regard to lactate production. In contrast, NH3 accumulation in the gastrocnemius muscle was abolished after exercise in the AMPD1 knockout animals (Fig. 4B). In addition, adenosine production was increased significantly in the A1(−/−) gastrocnemius muscle following exercise under ischemic conditions (Fig. 4C).
Fig. 4.
Assessment of metabolites in muscles of wild-type (Wt) and A1(−/−) mice after treadmill exercise. A: Lactate in muscles before and after exercise under ischemia (n = 3 for wild type, n = 5 for AMPD1 null). B: NH3 in muscles before and after exercise under ischemia (n = 3 for wild type, n = 6 for AMPD1 null). C: adenosine production in muscles before and after exercise under ischemia (n = 3 for wild type, n = 5 for AMPD1 null).
4. Discussion
The results of this study demonstrate that disruption of the AMPD1 gene in this murine model [A1(−/−)] mimics essentially all of the metabolic features AMPD1 deficiency in human subjects-AMPD enzyme activity is markedly reduced in skeletal muscle but not entirely eliminated due to expression of other members of this gene family, NH3 production is markedly reduced upon ischemic challenge, IMP is virtually undetectable even after strenuous exercise, the pool of adenine nucleotides although redistributed among the ATP/ADP/AMP components is maintained in the face of strenuous exercise and ischemia, and adenosine is produced in increased amounts. All of these metabolic signatures have been observed in muscle samples from human subjects with mutations in the AMPD1 gene [1]. Thus, we conclude that this murine model is a good metabolic phenocopy for human AMPD1 deficiency.
Disruption of the AMPD1 gene did not adversely affect the general health of these mice in comparison to mice with a normal AMPD1 gene with regard to fetal development, postnatal growth and development, appetite, and activity in their cages. Also, muscle fiber type was not different in the A1(−/−) mice compared with the wild-type mice.
Different exercise protocols were employed to assess muscle performance and metabolic changes in three different physiological situations, i.e. endurance exercise, sprint exercise, and exercise performed during ischemia. These conditions were selected to mimic situations which have been used in various studies of human subjects with AMPD1 deficiency. In all three exercise protocols there was no demonstrable difference in exercise performance between mice with disruption of the AMPD1 gene and mice with normal AMPD1.
We conclude from this study that isolated deficiency of AMPD1 does not lead to muscle dysfunction under the various conditions employed in this study. This does not however preclude that AMPD1 deficiency may contribute to muscle dysfunction in association when other genetic or environmental differences are present. The results of this study suggest that in patients with muscle symptoms and AMPD1 deficiency it may be prudent to search for other causes of symptoms and/or muscle dysfunction.
While this study does not support a role for AMPD1 deficiency in isolation as a cause of skeletal muscle dysfunction, it does not rule out effects of loss of AMPD1 activity in skeletal muscle on other metabolic pathways such as insulin mediated pathways of carbohydrate and lipid metabolism [8].
The following is the supplementary data related to this article.
Disruption of AMPD1 gene. A: Mouse AMPD1 gene and gene targeting strategy. Long- and short-arm genomic fragments of the mouse AMPD1 gene were reconstructed into a pSK vector. The long and short-arm fragments, divided by the insertion of neomycin resistant gene driven by PGK promoter, contained exons 3 to 8 and introns. As a result of homologous recombination, exon 6 of AMPD1 was inserted using a neomycin resistant gene. Upstream of the long arm, the diphtheria toxin A-chain gene was inserted to function as a negative selectable marker for random integration. B and C: PCR screening and Southern blot analysis for genotyping of AMPD1 knockout mice.
Acknowledgments
We express our thanks to the members of the Department of Bioscience and Genetics for their technical support. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and grants from the Japan Science and Technology Corporation, the Ministry of Health, Labour and Welfare of Japan, the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation (NIBIO), and the National Natural Science Foundation of China (Grant No 81070673).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Norman B., Sabina R.L. Myoadenylate deaminase deficiency. In: Valle D., Beaudet A.L., Vogelstein B., Kinzler K.W., Antonarakis S.E., Ballabio A., editors. The Online Metabolic & Molecular Bases of Inherited Disease, New York, McGraw-Hill. 2010. [Google Scholar]
- 2.Morisaki T., Gross M., Morisaki H. Molecular basis of AMP deaminase deficiency in skeletal muscle. Proc. Natl. Acad. Sci. U. S. A. 1992;89:6457–6461. doi: 10.1073/pnas.89.14.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fishbein W.N., Armbrustmacher V.W., Griffin J.L. Myo-adenylate deaminase deficiency: a new disease of muscle. Science. 1978;200:545–548. doi: 10.1126/science.644316. [DOI] [PubMed] [Google Scholar]
- 4.Lightfoot J.T., Turner M.J., Debate K.A. Interstrain variation in murine aerobic capacity. Med. Sci. Sports Exerc. 2001;33:2053–2057. doi: 10.1097/00005768-200112000-00012. [DOI] [PubMed] [Google Scholar]
- 5.Norman B., Glenmark B., Jansson E. Muscle amp deaminase deficiency in 2% of a healthy population. Muscle Nerve. 1995;18:239–241. doi: 10.1002/mus.880180216. [DOI] [PubMed] [Google Scholar]
- 6.Katayama M., Matsuda Y., Shimokawa K. Simultaneous determination of six adenyl purines in human plasma by high-performance liquid chromatography with fluorescence derivatization. J. Chromatogr. B Biomed. Sci. Appl. 2001;760:159–163. doi: 10.1016/s0378-4347(01)00265-1. [DOI] [PubMed] [Google Scholar]
- 7.Wang X., Morisaki H., Sermsuvitayawong K. Cloning and expression of cDNA encoding hearttype isoform of AMP deaminase. Gene. 1997;188:285–290. doi: 10.1016/s0378-1119(96)00818-9. [DOI] [PubMed] [Google Scholar]
- 8.Goodarzi M.O., Taylor K.D., Guo X. Variation in the gene for muscle-specific amp deaminase is associated with insulin clearance, a highly heritable trait. Diabetes. 2005;54:1222–1227. doi: 10.2337/diabetes.54.4.1222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Disruption of AMPD1 gene. A: Mouse AMPD1 gene and gene targeting strategy. Long- and short-arm genomic fragments of the mouse AMPD1 gene were reconstructed into a pSK vector. The long and short-arm fragments, divided by the insertion of neomycin resistant gene driven by PGK promoter, contained exons 3 to 8 and introns. As a result of homologous recombination, exon 6 of AMPD1 was inserted using a neomycin resistant gene. Upstream of the long arm, the diphtheria toxin A-chain gene was inserted to function as a negative selectable marker for random integration. B and C: PCR screening and Southern blot analysis for genotyping of AMPD1 knockout mice.