Abstract
Genome wide association studies (GWAS) have shown that SNPs in non-coding regions are associated with inherited susceptibility to cancer. The effect of one single SNP, however, is weak. To identify potential co-factors of SNPs, we investigated the underlying mechanism by which SNPs affect lung cancer susceptibility. We found that rs2853677 is located within the Snail1 binding site in a TERT enhancer. This enhancer increases TERT transcription when juxtaposed to the TERT promoter. The binding of Snail1 to the enhancer disrupts enhancer-promoter colocalization and silences TERT transcription. The high risk variant of rs2853677 disrupts the Snail1 binding site and derepresses TERT expression in response to Snail1 upregulation, thus increasing lung adenocarcinoma susceptibility. Our data suggest that Snail1 may be a co-factor of rs2853677 for predicting lung adenocarcinoma susceptibility and prognosis.
Keywords: rs2853677, Snail1, TERT, enhancer
INTRODUCTION
Cancer is frequently characterized by genetic alterations accompanied by epigenetic changes. These changes result in the aberrant activation and/or silencing of tumor related genes and confer premalignant epithelial cells with the capacity for unrestrained proliferation, resistance to cell death, evasion of immune destruction, and progression to frank malignancy [1–3]. Genetic alterations include somatic mutations, gene amplification, gene deletions, and SNPs. Whereas somatic mutations contribute to sporadically arising cancers, SNPs, which are found throughout the genome and can be inherited, play important roles in both sporadic and familial forms of cancer. A better understanding of the SNPs underlying the development of cancer is strongly needed to elucidate the etiology of the disease and to identify high-risk individuals for targeted screening and/or prevention.
GWAS have identified more than 150 loci associated with increased susceptibility to cancer, according to the Catalog of Published Genome-wide Association Studies [4]. Some SNPs are located in the protein-coding region and affect cancer susceptibility by changing protein function. Most SNPs, however, are located outside of known protein-coding sequences [5]. When present in the promoter region, SNPs can create or delete transcription factor binding sites and have appreciable effects on both gene transcription and protein yield [6, 7]. When present in the 3′ UTR region, SNPs can affect the binding affinity of miRNA [8]. However, how SNPs that are located in introns or inter-gene regions alter cancer susceptibility is largely uncharacterized.
Lung cancer is the leading cause of cancer-related death in most countries. Lung cancer associated SNPs were analyzed by GWAS in patients of various ethnic backgrounds. A Chinese group identified two intronic SNPs (rs2736100 at 5p15.33 and rs4488809 at 3q28) and two intergenic SNPs (rs753955 at 13q12.12 and rs12296850 at 12q23.1) with MAFs > 0.25 that are associated with lung cancer in a Chinese population [9]. Another two SNPs (rs2853677 at 5p15.33 and rs2741354 at 8q21.1) have been reported to be associated with lung cancer in Japanese and European populations, respectively [10, 11]. However, how these SNPs affect lung cancer susceptibility is completely unknown. Here, we investigated these 6 previously reported risk loci and confirmed that rs2853677 in the second intron of the telomerase reverse transcriptase (TERT) gene is associated with a high risk of lung adenocarcinoma in the Han Chinese population. A DNA fragment encompassing rs2853677 functions as an enhancer, which increases TERT transcription when colocalizing with TERT promoter. Snail1 binds to the enhancer, reconfigures the chromatin structure within the TERT gene, and represses TERT transcription. The high risk allele of rs2853677 disrupts the Snail1 binding site, causing derepression of TERT transcription in response to Snail1 upregulation. Our data implicate that rs2853677 may be a potential biomarker for prognosis in Snail1 associated cancer.
RESULTS
rs2853677 is associated with an increased risk of lung adenocarcinoma
We successfully genotyped 6 SNPs using SBE assay. All SNPs conformed to HWE in the controls (Table 1). After adjustment for gender and age, none of the SNPs reached statistical significance (Table 2).
Table 1. HWE tests in controls.
| SNP ID | HWE P value in controls |
|---|---|
| rs753955 | 0.62 |
| rs2853677 | 0.63 |
| rs2736100 | 0.58 |
| rs4488809 | 1 |
| rs2741354 | 0.71 |
| rs12296850 | 0.056 |
Table 2. Associations between 6 SNPs with lung cancer risk.
| SNP | Genotype | Control, n (%) | Case, n (%) | OR | 95% CI | P |
|---|---|---|---|---|---|---|
| rs753955 | T/T | 159 (47.2) | 158 (40.4) | Reference | Referent | |
| C/T | 142 (42.1) | 174 (44.5) | 1.18 | 0.74–1.88 | 0.493 | |
| C/C | 36 (10.7) | 59 (15.1) | 1.74 | 0.87–3.47 | 0.116 | |
| Dominant | 1.29 | 0.83–2.00 | 0.26 | |||
| Recessive | 1.6 | 0.84–3.07 | 0.15 | |||
| rs2853677 | T/T | 143 (42.4) | 137 (35) | Reference | Reference | |
| C/T | 157 (46.6) | 194 (49.6) | 1.56 | 0.97–2.51 | 0.66 | |
| C/C | 37 (11) | 60 (15.3) | 1.86 | 0.92–3.78 | 0.085 | |
| Dominant | 1.62 | 1.03–2.54 | 0.035 | |||
| Recessive | 1.45 | 0.76–2.80 | 0.26 | |||
| rs2736100 | T/T | 117 (34.7) | 109 (27.9) | Reference | Reference | |
| G/T | 159 (47.2) | 201 (51.4) | 1.27 | 0.77–2.09 | 0.359 | |
| G/G | 61 (18.1) | 81 (20.7) | 1.29 | 0.69–2.42 | 0.421 | |
| Dominant | 1.27 | 0.79–2.05 | 0.32 | |||
| Recessive | 1.12 | 0.65–1.93 | 0.69 | |||
| rs4488809 | T/T | 100 (29.7) | 115 (29.4) | Reference | Reference | |
| C/T | 168 (49.9) | 179 (45.8) | 0.98 | 0.58–1.63 | 0.927 | |
| C/C | 69 (20.5) | 97 (24.8) | 0.97 | 0.52–1.82 | 0.934 | |
| Dominant | 0.98 | 0.60–1.59 | 0.93 | |||
| Recessive | 0.99 | 0.58–1.68 | 0.97 | |||
| rs2741354 | G/G | 159 (47.2) | 175 (44.8) | Reference | Reference | |
| G/A | 143 (42.4) | 187 (47.8) | 1.46 | 0.92–2.31 | 0.104 | |
| A/A | 35 (10.4) | 29 (7.4) | 0.83 | 0.37–1.86 | 0.644 | |
| Dominant | 1.33 | 0.86–2.06 | 0.2 | |||
| Recessive | 0.69 | 0.32–1.50 | 0.35 | |||
| rs12296850 | A/A | 171 (50.7) | 210 (53.7) | Reference | Reference | |
| G/A | 148 (43.9) | 159 (40.7) | 1.03 | 0.66–1.62 | 0.894 | |
| G/G | 18 (5.3) | 22 (5.6) | 0.91 | 0.34–2.42 | 0.843 | |
| Dominant | 1.02 | 0.66–1.57 | 0.94 | |||
| Recessive | 0.89 | 0.34–2.33 | 0.82 |
NOTE: Statistically significant (P < 0.0083) associations are in bold.
Logistic regression was adjusted for gender and age.
Following subgroup analysis stratified by tumor histology, rs2853677 was associated with adenocarcinoma in a dominant model [ORCC/CT VS. TT = 2.33 (1.33–4.06), P = 0.0024] (Table 3). It was not associated with either squamous cell carcinoma or small cell lung cancer (SCLC). The other SNPs were not associated with any subtype of lung cancer (Table 3).
Table 3. Associations between SNPs with differential subtypes of lung cancer.
| SNP | Genotype | Squamous | SCLC | Adenocarcinoma | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | ||
| rs753955 | T/T | Reference | Reference | Reference | Reference | Reference | Reference | |||
| C/T | 1.05 | 0.50–2.19 | 0.903 | 0.81 | 0.38–1.72 | 0.578 | 1.03 | 0.60–1.77 | 0.922 | |
| C/C | 2.38 | 0.85–6.68 | 0.098 | 2.7 | 0.99–7.33 | 0.052 | 1.16 | 0.48–2.78 | 0.741 | |
| Dominant | 1.27 | 0.64–2.55 | 0.49 | 1.1 | 0.55–2.19 | 0.78 | 1.05 | 0.63–1.76 | 0.85 | |
| Recessive | 2.33 | 0.90–6.01 | 0.082 | 2.99 | 1.17–7.62 | 0.026 | 1.14 | 0.50–2.63 | 0.75 | |
| rs2853677 | T/T | Reference | Reference | Reference | Reference | Reference | Reference | |||
| C/T | 1.03 | 0.50–2.09 | 0.945 | 1.07 | 0.52–2.19 | 0.859 | 2.27 | 1.26–4.06 | 0.006 | |
| C/C | 1.15 | 0.36–3.68 | 0.811 | 0.89 | 0.26–3.02 | 0.858 | 2.55 | 1.12–5.78 | 0.025 | |
| Dominant | 1.05 | 0.53–2.07 | 0.89 | 1.04 | 0.52–2.07 | 0.92 | 2.33 | 1.33–4.06 | 0.0024 | |
| Recessive | 1.14 | 0.38–3.41 | 0.82 | 0.86 | 0.27–2.75 | 0.8 | 1.58 | 1.75–3.29 | 0.23 | |
| rs2736100 | T/T | Reference | Reference | Reference | Reference | Reference | Reference | |||
| G/T | 0.85 | 0.40–1.82 | 0.672 | 0.75 | 0.34–1.64 | 0.475 | 1.65 | 0.89–3.05 | 0.111 | |
| G/G | 0.77 | 0.28–2.07 | 0.602 | 0.84 | 0.32–2.19 | 0.715 | 1.74 | 0.83–3.63 | 0.142 | |
| Dominant | 0.83 | 0.40–1.70 | 0.61 | 0.78 | 0.37–1.61 | 0.5 | 1.67 | 0.94–2.99 | 0.08 | |
| Recessive | 0.85 | 0.36–2.04 | 0.72 | 0.99 | 0.42–2.31 | 0.98 | 1.26 | 0.68–2.33 | 0.47 | |
| rs4488809 | T/T | Reference | Reference | Reference | Reference | Reference | Reference | |||
| C/T | 1.41 | 0.61–3.25 | 0.426 | 0.67 | 0.30–1.48 | 0.319 | 1 | 0.55–1.81 | 0.99 | |
| C/C | 1.18 | 0.45–3.13 | 0.733 | 0.81 | 0.31–2.07 | 0.656 | 0.89 | 0.43–1.84 | 0.747 | |
| Dominant | 1.33 | 0.60–2.94 | 0.48 | 0.71 | 0.34–1.48 | 0.37 | 0.96 | 0.55–1.69 | 0.89 | |
| Recessive | 0.94 | 0.43–2.05 | 0.87 | 1.03 | 0.45–2.34 | 0.94 | 0.89 | 0.48–1.66 | 0.71 | |
| rs2741354 | G/G | Reference | Reference | Reference | Reference | Reference | Reference | |||
| G/A | 1.71 | 0.84–3.50 | 0.142 | 1.45 | 0.70–2.98 | 0.317 | 1.72 | 1.00–2.95 | 0.049 | |
| A/A | 1.56 | 0.48–5.06 | 0.458 | 1.3 | 0.37–4.49 | 0.681 | 0.79 | 0.28–2.23 | 0.663 | |
| Dominant | 1.68 | 0.85–3.32 | 0.13 | 1.42 | 0.71–2.82 | 0.32 | 1.53 | 0.91–2.57 | 0.1 | |
| Recessive | 1.2 | 0.39–3.68 | 0.75 | 1.1 | 0.33–3.61 | 0.88 | 0.61 | 0.23–1.66 | 0.33 | |
| rs12296850 | A/A | Reference | Reference | Reference | Reference | Reference | Reference | |||
| G/A | 1.36 | 0.67–2.74 | 0.391 | 1.42 | 0.70–2.87 | 0.33 | 0.67 | 0.39–1.15 | 0.148 | |
| G/G | 1.29 | 0.34–5.00 | 0.708 | 0.69 | 0.11–4.35 | 0.693 | 0.76 | 0.24–2.41 | 0.644 | |
| Dominant | 1.35 | 0.69–2.65 | 0.38 | 1.34 | 0.67–2.67 | 0.41 | 0.68 | 0.41–1.15 | 0.15 | |
| Recessive | 1.12 | 0.30–4.17 | 0.86 | 0.58 | 0.09–3.52 | 0.53 | 0.89 | 0.29–2.78 | 0.85 | |
NOTE: Statistically significant (P < 0.0028) associations are in bold.
The region encompassing rs2853677 functions as an enhancer of TERT
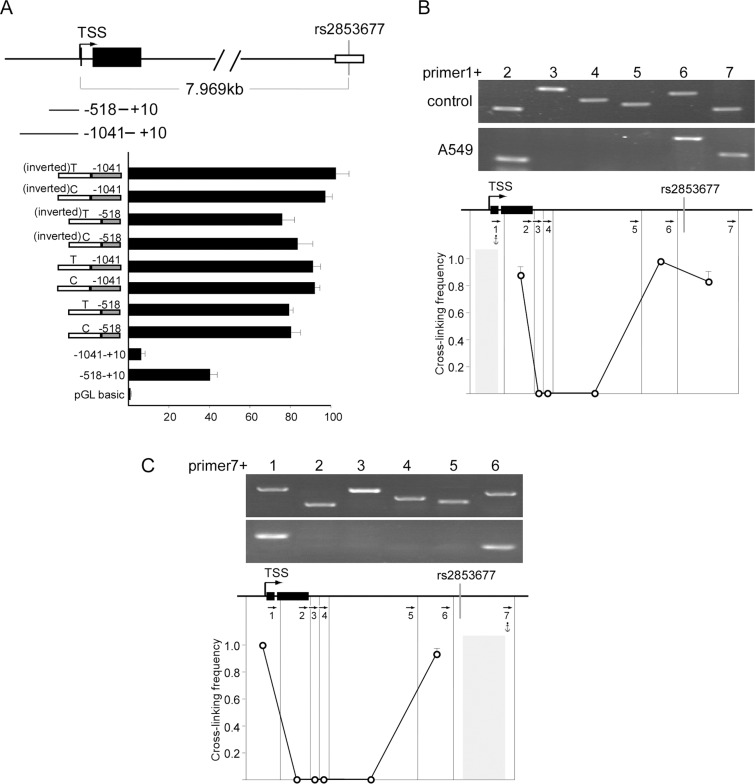
Rs2853677 is located in the second intron at + 7969 base pairs (bp) from the transcription start site of the TERT gene. The TERT gene encodes the catalytic subunit of telomerase, the ribonucleoprotein complex that maintains telomere length. To test whether rs2853677 resides in a cis-regulatory element of TERT, a 600 bp fragment encompassing the rs2853677 was amplified from human A549 cellular DNA that carries the T/C genotype of rs2853677. PCR products with the T or C allele were inserted into a construct containing a luciferase reporter gene, upstream of the TERT promoter in the original or inverted orientation. Luciferase reporter constructs were transfected into HEK293 cells, and the − 518 to + 10 fragment exhibited basal promoter activity. Extension of the fragment to − 1041 repressed promoter activity, suggesting that a silencer exists within − 1041 to − 518 (Figure 1A). The DNA fragment encompassing rs2853677 increased expression of the reporter gene 2-fold when inserted directly upstream of the basic promoter (− 518–+ 10) and 10-fold when inserted directly upstream of − 1041–+10, in the forward or inverted orientation (Figure 1A). Both the T and the C allele exhibited the same effect on TERT promoter activity. These data suggest that rs2853677 resides in a transcriptional enhancer of TERT, whose function may be to abolish silencer activity.
Figure 1. Rs2853677 is located inside of an enhancer.
(A) Luciferase reporter studies with serial extensions of the TERT promoter containing DNA fragment encompassing rs2853677 with T or C allele. The DNA fragment carrying T or C allele was amplified from A549 cells and placed directly upstream of the TERT promoter in forward or inverted orientation. The constructs were transiently transfected along with pRL-CMV Renilla luciferase reporter into HEK293 cells. luciferase activity was measured after 24 hrs' transfection. Mean ± SD indicates three independent transfections. (B) 3C was used to calculate cross linking frequency between chromatin segments to assess proximity in A549 cells. Vertical lines represent Pst I restriction sites. Arrows indicate PCR primer sites and direction. Anchor symbol marks anchoring primer. Cross linking frequency between the TERT promoter and different segments is shown. Top panels show representative PCR products. Mean ± SD of 3 independent chromatin preparations is shown. (C) 3C assay was performed with the DNA fragment harboring rs2853677 as an anchor fragment. Top panels show representative PCR products. Mean ± SD indicate 3 independent chromatin preparations.
We then explored whether the intronic enhancer encompassing rs2853677 is functional in vivo. Long range communication requires physical interaction between cis-regulatory elements with their target promoter [18, 19]. We used chromosome conformation capture (3C) to examine the physical proximity between the intronic enhancer and the TERT promoter. In brief, cross-linked chromatin was digested with PstI, diluted and re-ligated, and long range association frequencies were assessed with PCR. We used Image Quant software to quantitate the PCR signals. The highest cross-linking frequency was set to 1.0 to normalize the variation between separate chromatin preparations. Very strong associations were detected between the TERT promoter and the intronic enhancer when the TERT promoter fragment was used as a PCR anchor in A549 cells (Figure 1B). Consistently, the same strong interaction between the intronic enhancer and the TERT promoter was detected when using the intronic enhancer as a PCR anchor (Figure 1C). Our data indicate that rs2853677 is located in a functional enhancer of TERT. Thus, the polymorphism of rs2853677 may affect the transcription of TERT by altering the binding affinity of a transcription factor.
rs2853677-C disrupts a binding site for Snail1 and abolishes the suppressive role of Snail1 in TERT transcription
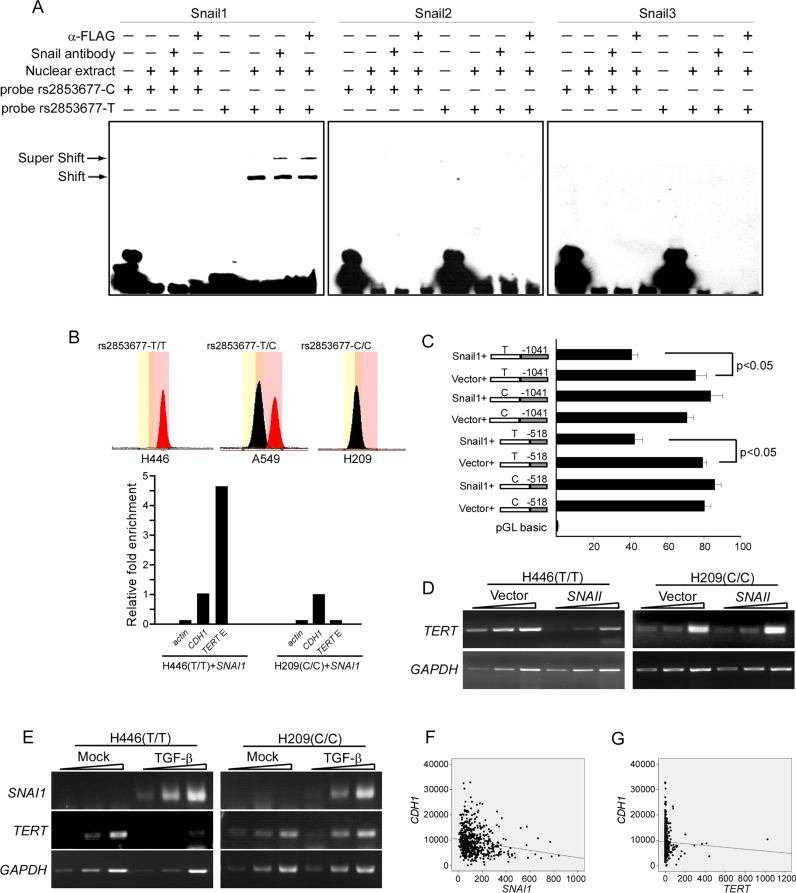
Bioinformatic analysis revealed that CG[C/T]CTG is a potential binding site of the Snail family of proteins. We first evaluated the interaction between this consensus sequence and Snail proteins (Snail1, Snail2 and Snail3) and its modulation by rs2853677 in vitro using an EMSA assay. The 25 bp oligonucleotides encompassing the rs2853677-T or -C allele were labeled with biotin at their 3′ termini and incubated with nuclear extract extracted from Snail1-FLAG, Snail2-FLAG or Snail3-FLAG overexpressing H446 cells. Supershift assays were performed to confirm the association of the tagged Snail proteins with the respective oligonucleotide probes. A shifted band was observed when the oligos containing the T allele were incubated with nuclear extract containing Snail1-FLAG, but not Snail2-FLAG or Snail3-FLAG. The shifted band was supershifted when FLAG antibody or Snail1 antibody was added (Figure 2A). The C allele oligos, however, were not shifted by any nuclear extract (Figure 2A). This result indicates that Snail1 binds to the oligos carrying the T allele, but not the C allele. Neither Snail2 or Snail3 binds to either oligo. We then examined the effect of polymorphism rs2853677 on the in vivo occupancy of Snail1 to the consensus binding site using ChIP analysis. We analyzed the genotype of rs2853677 in 5 human lung cancer cell lines, including A549, H1229, H69, H209 and H446. H446 and H209, which carry the T/T and C/C genotypes, respectively, were selected for further analysis. Snail1 conjugated with FLAG at its C-terminus was transfected into these two cell lines. ChIP was performed using FLAG antibody. Enrichment of Snail1 in the CDH1 promoter was set to 1.0. Occupancy of Snail1 with the TERT enhancer was detected in H446 cells, but not in H209 cells (Figure 2B). These data indicate that the rs2853677-C allele disrupts the Snail1 binding site.
Figure 2. Rs2853677 affects TERT transcriptional regulation by Snail1.
(A) EMSA shows mobility super shift (S.S.) of DNA fragment containing rs2853677. SNAI1-FLAG or SNAI2-FLAG or SNAI3-FLAG were overexpressed in H446 cells. Oligos carrying T or C allele were labeled with biotin in its 3′ terminus and incubated with nuclear extract that was purified from H446 expressing Snail1/2/3-FLAG. The antibody against Snail1/2/3 or FLAG was added to confirm the binding of Snail proteins to the oligos. Oligos harboring T allele but not the C allele can be bound by Snail1. (B) ChIP shows the occupancy of Snail1 in the TERT intronic enhancer in vivo. SNAI1-FLAG was overexpressed in H446 or H209 cells. ChIP was performed with FLAG antibody. Enrichment fold was evaluated by real-time PCR. Upper panels showed genotype of rs2853677 in H446 and H209 cells. Lower panels showed relative fold enrichment of Snail1 in the TERT intronic enhancer. CDH1 promoter was used as a positive control and its enrichment was set to 1.0. Actin was used as a negative control. Association of Snail1-Flag with the TERT intronic enhancer was shown. (C) Luciferase reporter study shows effect of Snail1 cotransfection on the TERT intronic enhancer activity. Snail1 or empty vector was cotransfected with the enhancer-TERT promoter constructs into HEK293 cells. Luciferase activity was measured after 24 hrs' transfection. Mean ± SD indicate three independent transfections. (D) Semiquantitative RT-PCR was performed to measure TERT transcription in control or Snail1 expressing cells. 1 ul, 3 ul, 9 ul of reverse transcribed cDNA was analyzed by PCR. (E) Semiquantitative RT-PCR was performed to measure TERT and SNAI1 transcription in control or TGF-β treated cells. 1 ul, 3u l, 9 ul of reverse transcribed cDNA was analyzed by PCR. (F) Expression of Snail1 and E-cadherin was assessed for correlation in lung adenocarcinoma using TCGA databases. SNAI1 negatively correlated with CDH1 (r = − 0.169, p = 4.545 × 10−5). (G) Expression of TERT and E-cadherin was assessed for correlation in lung adenocarcinoma using TCGA databases. TERT did not correlate with CDH1 (r = − 0.041, p = 0.321).
We next tested whether this differential binding could modulate TERT gene expression in response to Snail1 upregulation. Luciferase assays of HEK293 cells showed that cotransfection of SNAI1 with the luciferase reporter partially repressed enhancer activity when the enhancer harbored the T allele but had no effect on the enhancer carrying the C allele (Figure 2C), indicating that Snail1 inhibits enhancer activity upon association with the binding site. Accordingly, RT-PCR with one-, two-, or three-fold template DNA concentrations showed that transient expression of Snail1 repressed TERT transcription in H446 cells, but not in H209 cells (Figure 2D). Furthermore, TERT transcription was inhibited in H446 cells after 24 hr treatment with 10 μg/ml TGF-β, but not in TGF-β treated H209 cells, although both exhibited enhanced Snail1 expression upon TGF-β treatment (Figure 2E). These data indicate that Snail1 can bind to the enhancer and repress TERT transcription by inhibiting enhancer activity. The C allele of rs2853677 disrupts the association of Snail1 and prevents TERT downregulation in response to Snail1 upregulation.
The well-known function of Snail1 is to promote the epithelial-mesenchymal transition (EMT), a process that is critical for cancer malignancy and metastasis. We accessed The Cancer Genome Atlas (TCGA) public database to analyze any correlation among Snail1, TERT and E-cadherin (an important cell-cell adherent junction protein whose downregulation is a golden mark of the EMT) in lung adenocarcinoma. Expression of E-cadherin correlated negatively with Snail1 (r = − 0.169, p = 4.545 × 10−5), but not with TERT (r = − 0.041, p = 0.321) (Figure 2F). Accordingly, rs2853677 is not associated with lung adenocarcinoma metastasis [ORCC/CT VS. TT = 0.090 (0.45–1.83), P = 0.78] (Table 4). These results suggest that the rs2853677-C allele has no effect on the EMT of lung adenocarcinoma.
Table 4. Association between rs2853677 and lung adenocarcinoma metastasis.
| Model | Genotype | OR (95% CI) | P |
|---|---|---|---|
| Codominant | T/T | Reference | 0.6 |
| C/T | 0.80 (0.38–1.69) | ||
| C/C | 1.29 (0.47–3.50) | ||
| Dominant | T/T | Reference | 0.78 |
| C/T-C/C | 0.90 (0.45–1.83) | ||
| Recessive | T/T-C/T | Reference | 0.4 |
| C/C | 1.46 (0.60–3.60) |
NOTE: Logistic regression was adjusted for gender and age.
Snail1 binds to the TERT enhancer and disrupts long range communication between the enhancer and the promoter
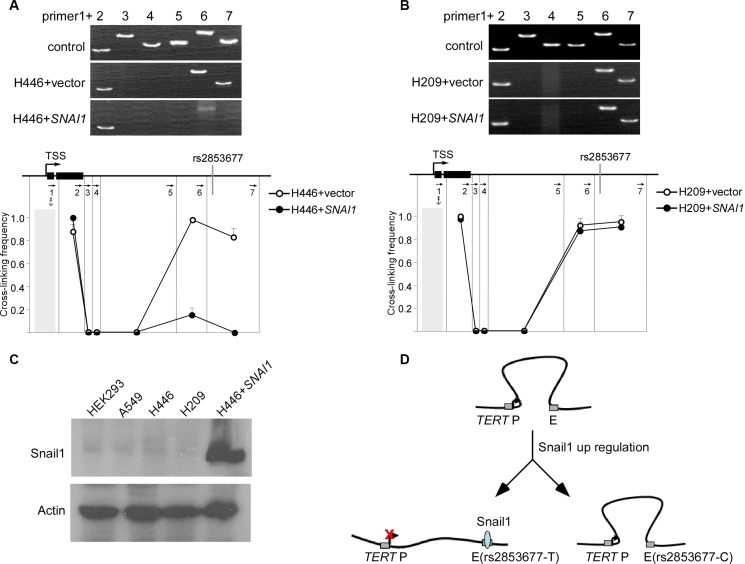
We next investigated the mechanism by which Snail1 regulates the activity of the intronic enhancer. We have previously shown that the intronic enhancer physically interacts with the TERT promoter by looping out the intervening sequences. We examined whether Snail1 could change this chromatin structure using 3C. Transient expression of Snail1 caused delocalization of the intronic enhancer from the TERT promoter in H446 cells (Figure 3A), but not in H209 cells (Figure 3B). Notably, the TERT gene in both H446 and H209 cells exhibited physical interaction between the promoter and the intronic enhancer (Figure 3A and 3B), and the intronic enhancer carrying either the C or the T allele exhibited the same effect on TERT promoter activity in a luciferase assay in HEK293 cells. This leads us to postulate that Snail1 may not be expressed in these cells. We then examined the expression of Snail1 in HEK293, H446, and H209 cells. As expected, these three cell lines and A549 do not express Snail1 (Figure 3C). These data support a model whereby TERT transcription requires long-range physical interaction between the enhancer and the TERT promoter. Snail1 associates with the enhancer, disrupting enhancer-promoter physical interaction, downregulating TERT transcription. The C allele of rs2853677 disrupts the binding site of Snail1, thereby abolishing the suppressive effect of Snail1 on TERT transcription (Figure 3D).
Figure 3. Snail1 disrupts long range communication between the intronic enhancer and the TERT promoter.
(A) Snail1 was overexpressed in H446 cells that harbor T/T genotype of rs2853677. 3C was performed to test the effect of Snail1 overexpression on proximity of the TERT enhancer and the promoter in H446 cells. Upper panels show representative PCR products. The lower panel shows the relative cross-linking frequencies between the TERT promoter and other DNA fragments. Mean ± SD indicate 3 independent chromatin preparations. (B) Snail1 was overexpressed in H209 cells that harbor C/C genotype of rs2853677. 3C was performed to test the effect of Snail1 overexpression on proximity of the TERT enhancer and the promoter in H209 cells. (C) Western blot showing the expression of Snail1 in variant cells. H446 expressing Snail1 was a positive control. (D) Schematic showing formation of a transcriptionally active complex containing the TERT promoter (TERT P) and the enhancer (E) Upon binding of Snail1 (shaded circles) to enhancer (T allele), the interaction is disrupted and TERT is downregulated.
DISCUSSION
Genetic factors contribute to cancer susceptibility. Here, we confirmed that rs2853677, which is located in the second intron of TERT, is associated with a high risk of lung adenocarcinoma in the Han Chinese population. The high risk variant disrupts the Snail1 binding site and prevents repression of TERT in response to the upregulation of Snail1, thus resulting in a high level expression of TERT in the presence of Snail1.
TERT expression level determines telomere length, and overexpression of TERT immortalizes normal cells [20–22]. Several lines of evidence have shown that immortalized human cells are susceptible to transformation via the introduction of an oncogene [23–25]. There is also evidence showed that cotransfection of TERT with H-ras can transform immortalized cells [26]. Therefore, high levels of TERT expression in oncogene (for example, Snail1) expressing cells promote tumorigenesis. TERT promoter mutations at sites − 57, − 256, or − 228 are associated with melanoma, non-small cell lung cancer and bladder cancer, respectively [7, 27, 28]. Due to its importance, transcriptional regulation of TERT has been studied, and multiple cis-regulatory elements have been found within or upstream of the TERT promoter [29, 30]. In this report, we identified a new cis-regulatory element residing within the second intron. This cis-regulatory element attenuates silencer activity when juxtaposed to the TERT promoter by looping out the intervening DNA and increasing TERT transcription. An oncogenic transcription factor, Snail1, binds to the enhancer and disrupts this physical interaction, repressing TERT transcription.
Snail1 is a member of the Snail superfamily of zinc-finger transcription factors, which also includes Snail2 (Slug) and Snail3 (Smuc). Although the three members share a conserved DNA binding domain, and their regulated genes overlap, only Snail1 can bind to the sequence harboring rs2853677. Elevated Snail1 expression is detected in many types of cancer and correlates with tumor malignancy [31, 32]. Although Snail1 is generally considered to be an oncogene, our results clearly indicate that in addition to repressing the expression of E-cadherin, the enhanced expression of Snail1 also downregulates TERT transcription, which inhibits tumorigenesis. Therefore, Snail1 executes dual functions in tumorigenesis. The C allele of rs2853677, however, disrupts the Snail1 binding site and derepresses the expression of TERT by Snail1, blocking the inhibitory effect of Snail1 on tumorigenesis and thus increasing lung cancer susceptibility. Although the well-known function of Snail1 is to promote the EMT, rs2853677 does not seem to affect the EMT.
Unexpectedly, rs2853677 is associated only with lung adenocarcinoma and is not related to SCLC, although SCLC expresses similar levels of Snail1 compared with lung adenocarcinoma (Supplementary Figure S1) and SCLC expresses more TERT than lung adenocarcinoma [33, 34]. This may be due to other transcription factors or signaling pathways that are activated in SCLC and may block the effect of Snail1 on TERT transcriptional regulation. It is known that nearly all patients with SCLC are missing RB1 and have more frequent mutations in TP53 than patients with lung adenocarcinoma [35]. The p53 protein is a potent inhibitor of the TERT promoter [36]. Mutation of p53 can relieve repression of the TERT promoter and therefore lead to elevated TERT expression in SCLC. In addition, the PI3-kinase (PI3K) pathway, another positive TERT transcriptional regulator, is also activated in SCLC [37, 38]. The mechanism by which these signaling pathways activate the TERT promoter is not clear. It is possible that Snail1 induced TERT transcriptional inhibition is not feasible in the presence of these signaling pathways.
In summary, we identified Snail1 as a co-factor of rs2853677 for predicting susceptibility to and the prognosis of lung adenocarcinoma. Given that Snail1 expression is enhanced in many types of cancers, the association of rs2853677 with other cancer types needs to be studied.
MATERIALS AND METHODS
Cell lines
HEK293, A549, H1229, H69, H209, and H446 cell lines were from ATCC, and were cultured in RPMI1640 containing 10% FBS, 100 U of penicillin/ml, and 100 μg of streptomycin/ml.
Study population
This case control study consisted of 391 lung cancer cases and 337 controls. The 391 patients with histologically confirmed lung cancer were recruited from Tianjin Medical University Cancer Institute & Hospital in China. The 337 controls were randomly selected from volunteers enrolled for physical examination at Tianjin General Hospital. All the controls did not have a history of cancer. Informed consent was obtained from all participants. This study was approved by Ethics Committee of Tianjin Medical University. The detail of the subject characteristics are listed in Table 5.
Table 5. Characteristics of cases and controls.
| Controls, n (%) | Cases, n (%) | P | |
|---|---|---|---|
| Total subjects | 337 (100%) | 391 (100%) | |
| Gender | 0.969 | ||
| Male | 228 (67.7%) | 264 (67.5%) | |
| Female | 109 (32.3%) | 127 (32.5%) | |
| Age | |||
| Mean ± SD | 38.8 ± 10.7 | 58.63 ± 8.8 | 0.000 |
| Tumor type | |||
| Squamous | 107 (27.4%) | ||
| SCLC | 64 (16.4%) | ||
| Adenocarcinoma | 183 (46.8%) | ||
| Other | 37 (9.5%) |
SNP selection and genotyping
6 previously reported risk loci located in the intron or inter-gene region were selected [9–14]. The information of six SNPs was listed in Table 6. The genotyping of 6 SNPs were examined in one PCR reaction. 2 μl of the PCR product was cleaned up with 1 μl of ExoSAP-IT (Amersham Biosciences, USA). Multiplex SBE reactions were performed in a total volume of 5 μL comprising 2 μL of SNaPshot ready reaction mix (Applied Biosystems, USA), 1.5 μL of cleaned PCR products and 1 μL of SBE primer mix. The SBE products were purified again using ExoSAP-IT. Genotyping for SNPs was performed by ABI 3130 Genetic Analyzer (Applied Biosystems, USA) according to the supplier's instructions. Primers were listed in Table 7.
Table 6. Information of six SNPs.
| SNPs | Allele | Locus | Location |
|---|---|---|---|
| rs753955 | C/T | 13q12.12 | intergenic |
| rs2853677 | C/T | 5p15.33 | intron of TERT |
| rs2736100 | G/T | 5p15.33 | intron of TERT |
| rs4488809 | C/T | 3q28 | intron of TP63 |
| rs2741354 | G/A | 8q21.1 | intergenic |
| rs12296850 | G/A | 12q23.1 | intergenic |
Table 7. Primers and probes used in experiment.
| primers used for genotyping of SNPs | |
| rs753955-F | 5′-AATATAGGTGGGCCCTGTCC-3′ |
| rs753955-R | 5′-GGGAAAGACAATGCTGTGGT-3′ |
| rs2853677-F | 5′-CCAATCCAGTCTGACAGTCG-3′ |
| rs2853677-R | 5′-GAAACAAGGGAACGAGGACA-3′ |
| rs2736100-F | 5′-GTGCTGTTTTCCCTGCTGAC-3′ |
| rs2736100-R | 5′-GGGAACAAAGGAGGAAAAGC-3′ |
| rs4488809-F | 5′-ATGCAAGCATCTGCTCTTGA-3′ |
| rs4488809-R | 5′-TGTGCATTCCTGTGTTTCCT-3′ |
| rs2741354-F | 5′-GGCCAACACAAGGACTGACT-3′ |
| rs2741354-R | 5′-ATTTCGCTGCAGCTTCTTTC-3′ |
| rs12296850-F | 5′-AGGATTCATGGGATCAGTGG-3′ |
| rs12296850-R | 5′-GTAGGTCCCACAGGGAGTGA-3′ |
| rs753955-SBE | 5′- (GACT)1ATCATGTGAAGGCTTGAA-3′ |
| rs2853677-SBE | 5′- (GACT)3TTTGTCACTAGAGACCCG-3′ |
| rs2736100-SBE | 5′- (GACT)5TCCGTGTTGAGTGTTTCT-3′ |
| rs4488809-SBE | 5′- (GACT)7TGCTCTTGAGGCAGTAAA-3′ |
| rs2741354-SBE | 5′- (GACT)9GGTATCACCCTAAACCAAG-3′ |
| rs12296850-SBE | 5′- (GACT)10CACATATAAGTAAAAGGGCTTAC-3′ |
| primers used for cloning | |
| SNAI1-CDS-F-Nflag | 5′-CGGGATCCGCCACCATGGACTACAAGGACGACG ATGACAAGCCGCGCTCTTTCCTCGTCAGGA-3′ |
| SNAI1-CDS-R | 5′-CGGGATCCTCAGCGGGGACATCCTGAGCAGCCG GACTCTTGGT-3′ |
| primers used for luciferase assay | |
| TERT-A | 5′-AGGAAGCTTCGCTGCCTGAAACTCGC-3 |
| TERT-S500 | 5′-TCACTCGAGGCATTCGTGGTGCCCG-3′ |
| TERT-S1000 | 5′-GTCCTCGAGTGTCCTGCGGTTGTGCC-3′ |
| SNP-F | 5′-CGGGGTACCATGTGAGGCATTGTTAGGTGCAT-3′ |
| SNP-R | 5′-CGGGAGCTCCTAAGTCAGCAGGGAAAACAGCA-3′ |
| primers used for EMSA | |
| Snail-EMSA-F1-biotin | 5′-ACTAGAGACCCGCCTGGTGCACTCTG-biotin |
| Snail-EMSA-R1-biotin | 5′-CAGAGTGCACCAGGCGGGTCTCTAGT-biotin |
| Snail-EMSA-F2-biotin | 5′-ACTAGAGACCCGTCTGGTGCACTCTG-biotin |
| Snail-EMSA-R2-biotin | 5′-CAGAGTGCACCAGACGGGTCTCTAGT-biotin |
| primers used for ChIP | |
| ChIP-S | 5′-ACTCTTGGACTCAAGGGATC-3′ |
| ChIP-A | 5′-ACACTCGGCAGGAAACG-3′ |
| E-Cad-S | 5′-CAACTCCAGGCTAGAGGGTCACCGC-3′ |
| E-Cad-A | 5′-AACTGACTTCCGCAAGCTCACAGG-3′ |
| β-actin-S | 5′-GCCAACCGCGAGAAGATGACCCAGA-3′ |
| β-actin-A | 5′-GAGTCCATCACGATGCCAGTAG-3′ |
| primers used for RT-PCR | |
| TERT-Q-F | 5′-CCGATTGTGAACATGGACTACG-3′ |
| TERT-Q-R | 5′-CACGCTGAACAGTGCCTTC-3′ |
| SNAI1-Q-F | 5′-ACTGCAACAAGGAATACCTCAG-3′ |
| SNAI1-Q-R | 5′-GCACTGGTACTTCTTGACATCTG-3′ |
| GAPDH-F | 5′-GTCAACGGATTTGGTCGTATT-3′ |
| GAPDH-R | 5′-AGTCTTCTGGGTGGCAGTGAT-3′ |
| primers used for 3C | |
| 3C-1 | 5′-CCAGCCCCTCCCCTTCCTTT-3′ |
| 3C-2 | 5′-GCTCCAGGCACAACGAACGC-3′ |
| 3C-3 | 5′-GGAAGATGAGCGTGCGGGACT-3′ |
| 3C-4 | 5′-CGGTGGCTCACGCCGGTAAT-3′ |
| 3C-5 | 5′-CCCCAGGTGTCCTTGGCGTTTG-3′ |
| 3C-6 | 5′-GTTTGGTAGCATTTATGTGAGGCATTG-3′ |
| 3C-7 | 5′-TGTGCGGTGTCTGGATGG-3′ |
| control 1-S | 5′-GGGGAACCAGCGACATGC-3′ |
| control 1-A | 5′-CCCTGAACACCCACAAACACT-3′ |
| control 2-S | 5′-GCTTGCTCCTGAATGTTTGCT-3′ |
| control 2-A | 5′-TGGTGAGAAACAAGGGAACGAG-3′ |
| control 3-A | 5′-CTCCATACATCCAGCTCACC-3′ |
| control 3-S | 5′-TGGGAACCAGGACAAAGG-3′ |
Data analysis
Differences of gender and age between cases and controls were compared by χ2 tests and student t tests respectively. Hardy-Weinberg Equilibrium (HWE) was assessed in the control samples by applying an exact test and then genotype frequencies were compared using χ2 tests. The association between lung cancer and each SNP was examined using logistic regression under Codominant, dominant and recessive models after adjusting for sex and age. The significance level was set at 0.018. These analyses were carried out using SNPStats [15] and SPSS 16.0.
Cloning and transduction
Human SNAI1 was amplified from H1155 cDNA. SNAI2 or SNAI3 was amplified from Beas-2B cDNA. FLAG was conjugated to the C-terminus. SNAI1-FLAG or SNAI2-FLAG or SNAI3-FLAG was ligated into the lentiviral shuttle pCCL.PPT.hPGK.IRES.GFP/pre. The plasmid was used to produce lentivirus in 293T cells with the packaging plasmids pMD2.BSBG, pMDLg/pRRE and pRSV-REV.
Luciferase assay
A 528 bp and 1051 bp DNA fragment upstream of TERT TSS were amplified from A549 cells DNA using primers listed in Table 7. The DNA fragments were inserted into the pGLbasic. Two analogous 600 bp DNA fragments encompassing the rs2853677 site were amplified from A549 cells and the T or the C-allele were subsequently inserted in forward or inverted orientation into upstream of the TERT promoter. Plasmids were cotransfected into HEK293 cells using Lipofectamine 2000 (Invitrogen) with pRL-CMV Renilla luciferase reporter, which was used for normalization (Promega).
Chromatin immunoprecipitation
SNAI1-FLAG was overexpressed in H446 and H209 cells. ChIP was performed using M2-argarsose as described previously [16]. Fold enrichment was analyzed by PCR assays. β-actin promoter was treated as a negative control and CDH1 promoter was treated as a positive control. Primers used in this study are listed in Table 7.
EMSA
Nuclear proteins from H446 expressing Snail1 or Snail2 or Snail3 cells were extracted using a NucBuster protein extraction kit (Novagen) according to the manufacturer's instructions. Double-stranded oligonucleotides corresponding to the potential Snail1 binding sites 5′-ACTAGAGACCCG[C/T]CTGGTGCACTCTG was end-labeled with biotin in its 3′ terminus. Binding assays were performed in 10 μl of reaction mixture with 2 μg of nuclear protein extracts and 1 nM labeled probes at room temperature for 30 min in binding buffer (10 mM Tris-Cl, 55 mM KCl, 2.5 mM MgCl2, 0.25 mM EDTA, 1 mM DTT, 0.05% NP-40, 5% Glycerol and 1 μg poly dI-dC). In supershift assay, Snail1 or Snail2 or Snail3 antibodys or FLAG antibody was added. Reactions were analyzed by electrophoresis on a 6.0% non-denaturing polyacrylamide gel at 100 V for 1 h. After transfer, the membrane was immediately cross-linked and bands detected by chemiluminescence (Pierce, USA).
Chromosome conformation capture
3C assays were performed as described previously [17]. Briefly, 106 cells were cross-linked, lysed, and nuclei were digested with Pst I. After ligation and subsequent DNA purification, the cross-linking frequencies between the anchor and test fragments were estimated by PCR reactions using primers listed in Table 7. To create a standard for normalization of relative PCR efficiencies, three PCR products together containing all the sequence from the TERT gene TSS to10 Kb downstream were amplified, digested with Pst I, and ligated at high concentrations to generate equimolar mixtures of all possible ligation products. The cross-linking and ligation efficiencies between different samples were normalized by setting the highest cross-linking frequency to 1.0.
SUPPLEMENTARY MATERIALS FIGURE
Acknowledgments
We thank Dr. Weidong Li at Tianjin Medical University for his critical review.
Abbreviations
The abbreviations used are
- SNP
single nucleotide polymorphism
- GWAS
genome-wide association studies
- TERT
telomerase reverse transcriptase
- HWE
Hardy-Weinberg Equilibrium
- SBE
single base extension
- 3C
chromosome comformation capture
- ChIP
chromatin immunoprecipitation
- EMT
epithelial-mesenchymal transition
- RT-PCR
reverse transcription-polymerase chain reaction
- SCLC
small cell lung cancer
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
GRANT SUPPORT
This work was supported by the National Natural Science Foundation of China (91519331, 31371295, 81572271, 81372307, 81572882), Ministry of Science and Technology of China (2014CB910104), the Tianjin Municipal Science and Technology Commission (15JCZDJC34800), the specialized Fund for the Doctoral Program of Higher Education (20121202110001).
REFERENCES
- 1.Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 2.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011;130:59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- 5.Sur I, Tuupanen S, Whitington T, Aaltonen LA, Taipale J. Lessons from functional analysis of genome-wide association studies. Cancer Res. 2013;73:4180–4184. doi: 10.1158/0008-5472.CAN-13-0789. [DOI] [PubMed] [Google Scholar]
- 6.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, Onel K, Yip L, Hwang SJ, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 7.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 8.Ryan BM, Robles AI, McClary AC, Haznadar M, Bowman ED, Pine SR, Brown D, Khan M, Shiraishi K, Kohno T, Okayama H, Modali R, Yokota J, et al. Identification of a functional SNP in the 3′ UTR of CXCR2 that is associated with reduced risk of lung cancer. Cancer Res. 2015;75:566–575. doi: 10.1158/0008-5472.CAN-14-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, Li Z, Deng Q, Wang J, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet. 2011;43:792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 10.Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, Ohnami S, Shimada Y, Ashikawa K, Saito A, Watanabe S, Tsuta K, Kamatani N, et al. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet. 2012;44:900–903. doi: 10.1038/ng.2353. [DOI] [PubMed] [Google Scholar]
- 11.Brenner DR, Brennan P, Boffetta P, Amos CI, Spitz MR, Chen C, Goodman G, Heinrich J, Bickeböller H, Rosenberger A, Risch A, Muley T, McLaughlin JR, et al. Hierarchical modeling identifies novel lung cancer susceptibility variants in inflammation pathways among 10,140 cases and 11,012 controls. Hum Genet. 2013;132:579–589. doi: 10.1007/s00439-013-1270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miki D, Kubo M, Takahashi A, Yoon KA, Kim J, Lee GK, Zo JI, Lee JS, Hosono N, Morizono T, Tsunoda T, Kamatani N, Chayama K, et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet. 2010;42:893–896. doi: 10.1038/ng.667. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Z, Li C, Yang L, Zhang X, Zhao X, Song X, Li X, Wang J, Qian J, Yang Y, Jin L, Chen H, Lu D. Significant association of 5p15. 33 (TERT-CLPTM1L genes) with lung cancer in Chinese Han population. Exp Lung Res. 2013;39:91–98. doi: 10.3109/01902148.2012.762436. [DOI] [PubMed] [Google Scholar]
- 14.Dong J, Jin G, Wu C, Guo H, Zhou B, Lv J, Lu D, Shi Y, Shu Y, Xu L, Chu M, Wang C, Zhang R, et al. Genome-wide association study identifies a novel susceptibility locus at 12q23.1 for lung squamous cell carcinoma in Han Chinese. PLoS Genet. 2013;9:e1003190. doi: 10.1371/journal.pgen.1003190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3′ boundary sequence spanning 46 kilobases. Mol Cell Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Xu Z, Du W, Zhang Z, Wei Y, Wang H, Zhu Z, Qin L, Wang L, Niu Q, Zhao X, Girard L, Gong Y, et al. Aiolos Promotes Anchorage Independence by Silencing p66Shc Transcription in Cancer Cells. Cancer Cell. 2014;25:575–589. doi: 10.1016/j.ccr.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 19.Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 20.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 21.Vaziri H, Squire JA, Pandita TK, Bradley G, Kuba RM, Zhang H, Gulyas S, Hill RP, Nolan GP, Benchimol S. Analysis of genomic integrity and p53-dependent G1 checkpoint in telomerase-induced extended-life-span human fibroblasts. Mol Cell Biol. 1999;19:2373–2379. doi: 10.1128/mcb.19.3.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harley CB. Telomerase is not an oncogene. Oncogene. 2002;21:494–502. doi: 10.1038/sj.onc.1205076. [DOI] [PubMed] [Google Scholar]
- 23.Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 24.Yu J, Boyapati A, Rundell K. Critical role for SV40 small-t antigen in human cell transformation. Virology. 2001;290:192–198. doi: 10.1006/viro.2001.1204. [DOI] [PubMed] [Google Scholar]
- 25.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, Popescu NC, Hahn WC, Weinberg RA. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart SA, Hahn WC, O'Connor BF, Banner EN, Lundberg AS, Modha P, Mizuno H, Brooks MW, Fleming M, Zimonjic DB, Popescu NC, Weinberg RA. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc Natl Acad Sci USA. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu D, Dwyer J, Li H, Duan W, Liu JP. Ets2 maintains hTERT gene expression and breast cancer cell proliferation by interacting with c-Myc. J Biol Chem. 2008;283:23567–23580. doi: 10.1074/jbc.M800790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Wu S, Wang H, Bi X, Yang Z, Du Y, He L, Cai Z, Wang J, Fan Z. The C228T mutation of TERT promoter frequently occurs in bladder cancer stem cells and contributes to tumorigenesis of bladder cancer. Oncotarget. 2015;6:19542–19551. doi: 10.18632/oncotarget.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horikawa I, Chiang YJ, Patterson T, Feigenbaum L, Leem SH, Michishita E, Larionov V, Hodes RJ, Barrett JC. Differential cis-regulation of human versus mouse TERT gene expression in vivo: Identification of a human-specific repressive element. Proc Natl Acad Sci USA. 2005;102:18437–18442. doi: 10.1073/pnas.0508964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran DA, Wong TC, Schep AN, Drewell RA. Characterization of an ultra-conserved putative cis-regulatory module at the mammalian telomerase reverse transcriptase gene. DNA Cell Biol. 2010;29:499–508. doi: 10.1089/dna.2009.0994. [DOI] [PubMed] [Google Scholar]
- 31.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 32.Kaufhold S, Bonavida B. Central role of Snail1 in the regulation of EMT and resistance in cancer: a target for therapeutic intervention. J Exp Clin Cancer Res. 2014;33:62. doi: 10.1186/s13046-014-0062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lantuejoul S, Soria JC, Moro-Sibilot D, Morat L, Veyrenc S, Lorimier P, Brichon PY, Sabatier L, Brambilla C, Brambilla E. Differential expression of telomerase reverse transcriptase (hTERT) in lung tumours. Br J Cancer. 2004;90:1222–1229. doi: 10.1038/sj.bjc.6601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaffaroni N, De Polo D, Villa R, Della Porta C, Collini P, Fabbri A, Pilotti S, Daidone MG. Differential expression of telomerase activity in neuroendocrine lung tumours: correlation with gene product immunophenotyping. J Pathol. 2003;201:127–133. doi: 10.1002/path.1401. [DOI] [PubMed] [Google Scholar]
- 35.Wistuba II, Gazdar AF, Minna JD. Molecular genetics of small cell lung carcinoma. Semin Oncol. 2001;28:3–13. [PubMed] [Google Scholar]
- 36.Yao Y, Bellon M, Shelton S, Nicot C. Tumor Suppressors p53, p63TAα, p63TAy, p73α, and p73β Use Distinct Pathways to Repress Telomerase Expression. Journal of Biol Chem. 2012;287:20737–20747. doi: 10.1074/jbc.M111.319236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellon M, Nicot C. Central role of PI3K in transcriptional activation of hTERT in HTLV-I-infected cells. Blood. 2008;112:2946–2955. doi: 10.1182/blood-2008-01-134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voortman J, Lee JH, Killian JK, Suuriniemi M, Wang Y, Lucchi M, Smith WI, Jr, Meltzer P, Wang Y, Ciaccone G. Array comparative genomic hybridization-based characterization of genetic alterations in pulmonary neuroendocrine tumors. Proc Natl Acad Sci USA. 2010;107:13040–13045. doi: 10.1073/pnas.1008132107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osborne JK, Larsen JE, Shields MD, Gonzales JX, Shames DS, Sato M, Kulkarni A, Wistuba II, Girard L, Minna JD, Cobb MH. NeuroD1 regulates survival and migration of neuroendocrine lung carcinomas via signaling molecules TrkB and NCAM. Proc. Natl. Acad. Sci. USA. 2013;110:6524–6529. doi: 10.1073/pnas.1303932110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.