Abstract
HnRNPA2B1 encodes an RNA binding protein associated with neurodegeneration. However, its function in the nervous system is unclear. Transcriptome-wide cross-linking and immunoprecipitation in mouse spinal cord discover UAGG motifs enriched within ~2,500 hnRNP A2/B1 binding sites and an unexpected role for hnRNP A2/B1 in alternative polyadenylation. HnRNP A2/B1 loss results in alternative splicing (AS), including skipping of an exon in amyotrophic lateral sclerosis (ALS)-associated D-amino acid oxidase (DAO) that reduces D-serine metabolism. ALS-associated hnRNP A2/B1 D290V mutant patient fibroblasts and motor neurons differentiated from induced pluripotent stem cells (iPSC-MNs) demonstrate abnormal splicing changes, likely due to increased nuclear-insoluble hnRNP A2/B1. Mutant iPSC-MNs display decreased survival in long-term culture, and exhibit hnRNP A2/B1 localization to cytoplasmic granules as well as exacerbated changes in gene expression and splicing upon cellular stress. Our findings provide a cellular resource and reveal RNA networks relevant to neurodegeneration, regulated by normal and mutant hnRNP A2/B1.
Introduction
Altered levels or mutations in RNA binding proteins (RBPs) are associated with neurological diseases, including spinal muscular atrophy, fragile X syndrome, amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD) (Mohagheghi et al., 2016) and Alzheimer’s disease (AD) (Belzil et al., 2013; Nussbacher et al., 2015; Polymenidou et al., 2012). ALS is a fatal neurodegenerative disorder characterized by progressive loss of upper and lower motor neurons. The causes of ALS remain largely unknown; 90% of cases are sporadic and 10% have a hereditary component (Pasinelli and Brown, 2006). Mutations in RBPs, including TAR DNA Binding Protein (TDP-43), Fused in Sarcoma (FUS) and TAF15 (Couthouis et al., 2011; Kabashi et al., 2008; Van Deerlin et al., 2008; Vance et al., 2009) cause familial ALS. Both familial and sporadic cases of ALS show TDP-43 or FUS positive aggregates or amyloid-like fibrils, even when neither gene is mutated (Mackenzie et al., 2007; Neumann et al., 2006).
FUS/TLS and TDP-43 are members of the heterogeneous nuclear ribonucleoprotein particle protein (hnRNP) family of RBPs, which includes hnRNP A2/B1, thought to be sequestered by CGG repeats in the FMR1 transcript. In a Drosophila fragile X-associated tremor/ataxia syndrome (FXTAS) model, hnRNP A2/B1 over-expression rescued the neurodegenerative phenotype (Sofola et al., 2007). In AD, hnRNP A2/B1 is depleted in patient brains, and this loss is mediated by the death of cholinergic neurons (Berson et al., 2012). Multisystem proteinopathy (MSP) is an autosomal dominant disorder characterized by muscle weakness and skeletal abnormalities. Mutations in the gene encoding Valosin Containing Protein (VCP) have been shown to cause MSP with motor neuron degeneration (Johnson et al., 2010; Watts et al., 2004) and whole exome sequencing of a multiplex MSP family recently revealed a p.D290V mutation in the HNRNPA2B1 gene (Kim et al., 2013).
HnRNP A2/B1 has two isoforms, A2 and B1 (341 and 353 amino acids, respectively), both transcribed from the HNRNPA2B1 gene. The putative functions of hnRNP A2/B1 include pre-mRNA splicing (Clower et al., 2010; Hutchison et al., 2002), mRNA trafficking (Shan et al., 2003), transcript stability (Fahling et al., 2006), and translational control (Kosturko et al., 2006). Like TDP-43, hnRNP A2/B1 contains two RNA recognition motifs (RRMs) and has a glycine-rich domain (GRD) near the C-terminal end. HnRNP A2/B1 is primarily localized to the nucleus; however, nuclear-cytoplasmic trafficking does occur and is controlled in part by a nuclear localization signal in the GRD (Siomi et al., 1997). The pathogenic D290V mutation lies in the GRD of hnRNP A2/B1, akin to TDP-43, where most ALS-causing mutations occur in its GRD (Lagier-Tourenne et al., 2010). While we and others have characterized the function of hnRNP A2/B1 in human cell-lines (Huelga et al., 2012), the role that hnRNP A2/B1 plays in vivo in CNS tissues is largely unknown. Furthermore, the effects of pathogenic mutations in hnRNP A2/B1 on RNA metabolism are not understood.
Results
Discovery of hnRNP A2/B1 RNA binding sites in mouse spinal cord and human iPSC-MNs
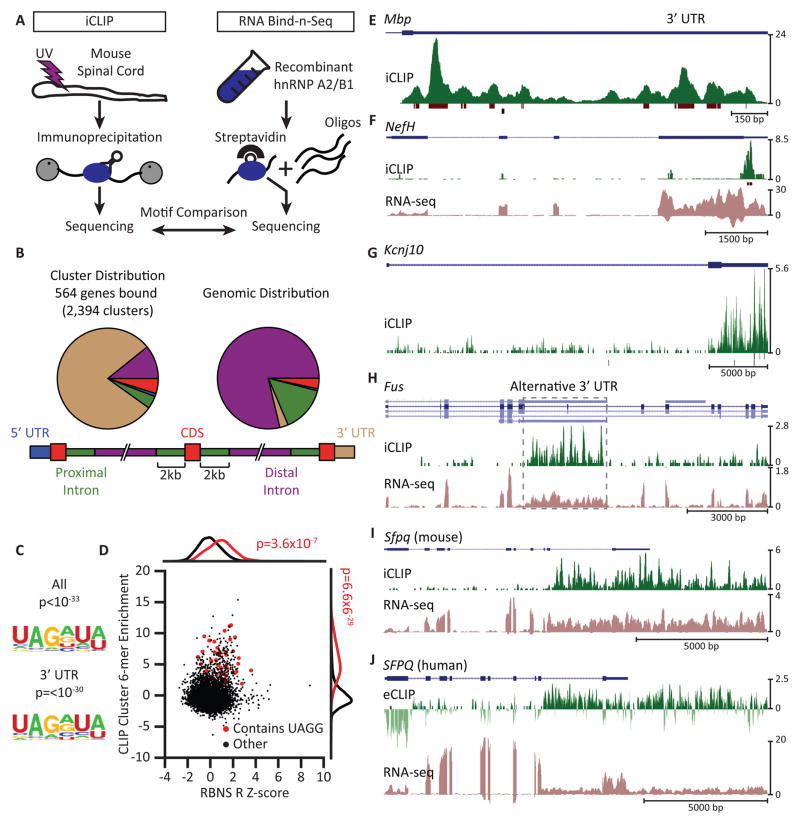
We constructed transcriptome-wide maps of hnRNP A2/B1 binding sites in 8-week old mouse spinal cord by immunoprecipitating UV-crosslinked protein-RNA complexes (Figure 1A (left); Figures S1A and S1B). Individual-nucleotide CLIP (iCLIP) procedures identified 2,394 clusters of hnRNP A2/B1 binding sites in 564 genes (Figure 1B; Data S1A). Gene ontology (GO) analyses of mRNA targets revealed significantly enriched (p < 10−5) categories, such as protein binding, myelination, axon and neural projection. Most binding sites were within 3′ untranslated regions (3′UTRs) of protein-coding genes (Figure 1B). Motif analysis revealed a significantly enriched UAG[A/G] sequence within these clusters (Figure 1C) that resembled the previously identified binding sequence of hnRNP A2/B1 (Huelga et al., 2012). We confirmed this motif using RNA Bind-N-Seq (RBNS) (Lambert et al., 2014). We found that hexamers containing the UAGG core sequence were shifted significantly towards higher hnRNP A2/B1 RBNS enrichment and also showed strong enrichment in hnRNP A2/B1 CLIP binding sites (Figure 1D; Figure S1C). We conclude that hnRNP A2/B1 binds to UAGG motifs within RNAs without requiring co-factor associations.
Figure 1. HnRNP A2/B1 recognizes UAGG and predominantly binds 3′ untranslated regions (3′UTRs).
(A) Experimental approaches. Individual nucleotide crosslinking followed by immunoprecipitation (iCLIP) in mouse spinal cord was used to identify hnRNP A2/B1 protein-RNA binding sites in vivo (left). RNA binding followed by sequencing (RNA bind-n-seq, RBNS) using the recombinant RNA binding domain of hnRNP A2/B1 was used to identify high-affinity RNA motifs in vitro (right). Motifs enriched by both methods were compared.
(B) hnRNP A2/B1 iCLIP-derived clusters are enriched in 3′UTRs of protein coding genes (left) when compared to the expected distribution of gene regions (5′ and 3′UTRs, exons, and exon-proximal and distal portions of introns; right). Proximal intron regions are defined as extending up to 2 kb from an exon-intron junction (bottom).
(C) The UAGG motif is significantly enriched in clusters from all regions (top) and those restricted to 3′UTRs (bottom). p-values were determined by the HOMER algorithm.
(D) Hexamers from RNA sequences found by RBNS and iCLIP that contain UAGG are significantly enriched (red dots) compared to those that did not contain UAGG (black dots). p-values were calculated by a Kolmogorov-Smirnov test on the distributions represented by UAGG (red) and other k-mers (black).
(E–J) Genome browser views of CLIP reads (green, plotted in the positive direction of the y axis) and clusters (maroon) mapped to selected genes characteristic of the spinal cord (blue). y axes are scaled to the read numbers indicated to the right of each plot. In (H), alternative transcript isoforms are indicated in light blue. Reads from strand-specific RNA-seq analyses are plotted in mauve in the negative direction of the y axis in (F), (H) and (I). (E) – (I) show iCLIP analyses from mouse spinal cord; (J) shows eCLIP analysis from human iPSC-derived motor neurons, represented as log2 ratio of antibody-enriched immunoprecipitate (IP) over size-matched input. The dark green track is eCLIP reads enriched in IP. The light green track is eCLIP reads enriched in input. The vertical axes denote RPM.
We observed abundant 3′UTR binding in the myelin basic protein (Mbp) gene, a known hnRNP A2/B1 RNA substrate (Ainger et al., 1997) (Figure 1E), the neurofilament heavy chain gene (Nefh) (Figure 1F) and the astroglial inward rectifying potassium channel gene Kcnj10 (Figure 1G). Thus, hnRNP A2/B1 binds transcripts characteristic of oligodendrocytes (Mbp), neurons (Nefh) and astroglia (Kcnj10). We also evaluated if hnRNP A2/B1 protein interacts with RNA encoded by ALS-associated genes. We found seven genes, of which five exhibited hnRNP A2/B1 binding within 3′UTRs, including Hnrnpa2b1 itself, the glial glutamate transporter Slc1a2, and Ubqln2 (Figure S1D). Fus contained hnRNP A2/B1 binding sites within an intronic region that may be processed as an alternative 3′UTR (Figure 1H). HnRNP A2/B1 binding was also found within long, unannotated 3′UTRs, as illustrated by the murine splicing factor proline/glutamine-rich (Sfpq) gene (Figure 1I) and its human ortholog (Figure 1J). The region bound in the SFPQ gene illustrates an intronic region that may be retained to lengthen the 3′UTR. We conclude that hnRNP A2/B1 binds to 3′UTRs within CNS transcripts, including ALS-associated genes.
To characterize the RNA binding properties of hnRNP A2/B1 in human cells, we differentiated human iPSCs to motor neurons (iPSC-MNs; Figure S6A; Data S1B) and employed enhanced CLIP (eCLIP; (Van Nostrand et al., 2016); Figure S1E). Cluster identification in iPSC-MNs revealed 866 clusters in 527 genes enriched above input (Figure S1E; Data S1C). Compared to mouse spinal cord, the regional distribution of hnRNP A2/B1 binding did not concentrate as heavily in 3′UTRs but was still highly enriched above expectation (Figure S1F). The UAGG motif, previously identified by iCLIP and RBNS, was significantly enriched near the center of eCLIP clusters (Figure S1G). Despite differences in heterogeneity and maturity of cell-types, the 35 target transcripts conserved between adult mouse spinal cord and human iPSC-MNs (Figure S1H) included a surprisingly large number of RBPs (yellow) and motor proteins (green) (Figure S1I). Many genes had similar binding patterns across transcripts of homologous genes, such as PNISR and LENG8 (Figures S1J and S1K). We conclude that hnRNP A2/B1 binds similar regions in mouse spinal cord and human iPSC-MNs and harbors a conserved propensity to bind transcripts encoding RBPs.
HnRNP A2/B1 depletion in vivo results in limited changes in gene expression
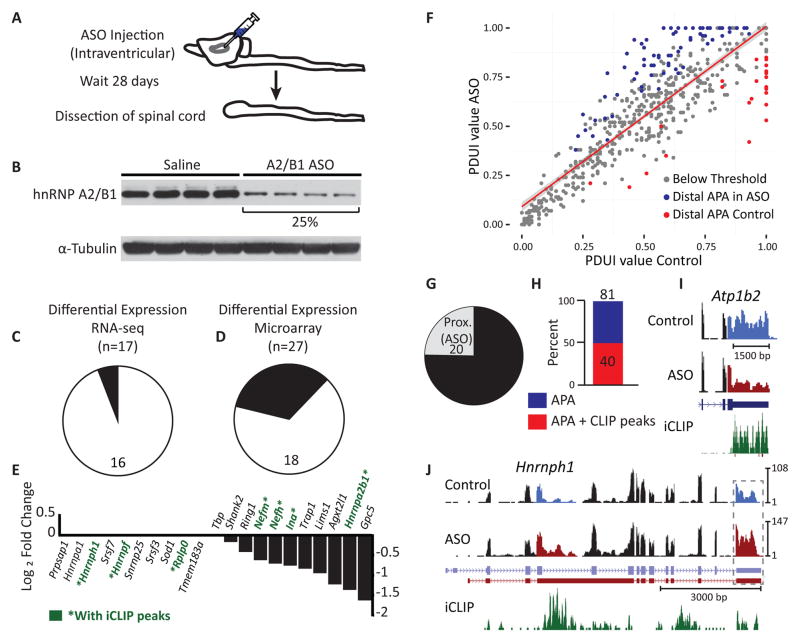
To investigate molecular pathways controlled by hnRNP A2/B1, we targeted its transcript with antisense oligonucleotides (ASOs) in mouse spinal cord (Figure 2A). We achieved ~75% protein depletion after 28 days (Figure 2B). Surprisingly, we detected only a small number of gene expression changes either by RNA-seq (Figure 2C; Data S1D) or microarray analysis (Figure 2D; Data S1E). Specifically, we identified only ten significantly down-regulated genes, including Hnrnpa2b1 itself. Interestingly, of the similar numbers of upregulated genes, we detected reproducible increases in mRNA levels of splicing factors Hnrnpa1, Hnrnph1, and Srsf7. These were validated by quantitative RT-PCR (Figures 2E, S2A–S2C). We also observed increased protein levels of hnRNP A1 and SRSF7 (Figure S2D), consistent with our previous report of cross-regulation (Huelga et al., 2012). We did not find altered protein levels for hnRNP H1, M or U (Figure S2D). We conclude that loss of hnRNP A2/B1 does not lead to widespread changes in transcript levels in the nervous system, in contrast to other ALS-associated RBPs such as TDP-43, FUS/TLS and TAF15 evaluated in mouse brain (Kapeli et al., 2016; Lagier-Tourenne et al., 2012; Polymenidou et al., 2011).
Figure 2. HnRNP A2/B1 depletion results in alternative polyadenylation changes.
(A) Experimental approach. An antisense oligonucleotide (ASO) targeting the Hnrnpa2b1 transcript, or a vehicle control (saline) solution, was injected into the lateral ventricles of mice (n = 4 mice per treatment) and RNA was isolated from spinal cords 28 days post-injection.
(B) Western blotting shows that ASO treatment leads to a ~75% reduction in hnRNP A2/B1 protein levels compared to saline controls. Tubulin was the loading control.
(C–D) Statistically significant changes in transcript levels were identified for 17 and 27 genes when analyzed by RNA-seq (C) and microarray (D), respectively. Down- and upregulated genes are indicated in black and white, respectively. Significance was defined using a false-discovery rate threshold of 0.05 for Benjamini-Hochberg corrected values for multiple hypothesis testing.
(E) Quantitative RT-PCR validation of the results from (C) and (D) confirmed increased transcript levels for nine of ten genes, including Srsf7, Hnrnpa1, Hnrnph and Hnrnpf (left), and decreased transcript levels for all ten genes, including Hnrbpa2b1 itself (right), upon ASO-mediated depletion of hnRNP A2/B1. Tbp was the reference gene.
(F) DaPars analysis of RNA-seq data from the mouse spinal cord samples revealed that depletion of hnRNP A2/B1 leads to changes in poly(A) site (PAS) utilization. The scatterplot of distal PAS (dPAS) usage indexes (PDUIs) in control and hnRNP A2/B1 depletion samples is shown. Significantly (FDR < 0.05, |ΔPDUI| ≥ 0.2, and |dPDUI| ≥ 0.2) shortened and lengthened transcripts are colored in red and blue, respectively. Gray dots indicate transcripts that did not pass the significance threshold.
(G) Upon hnRNP A2/B1 depletion, 3′UTR shortening and lengthening was observed for 20 and 61 transcripts, respectively.
(H) Of the 81 transcripts displaying differential PAS utilization, 40 contained significant hnRNP A2/B1 occupancy in their 3′UTRs, as identified by iCLIP.
(I–J) Examples of genes with differential PAS utilization that are bound by hnRNP A2/B1 in their 3′UTRs. Regions of alternative usage are highlighted in blue (control) and red (ASO treated) in the corresponding RNA-seq track. Vertical axes are RPM.
HnRNP A2/B1 depletion in vivo and in vitro has minimal effect on mature microRNA levels
A previous study reported a role for hnRNP A2/B1 in microRNA biogenesis (Alarcon et al., 2015). We performed small RNA sequencing in mouse spinal cords and human iPSC-MNs treated with hnRNP A2/B1-targeting and control ASOs (Figure 2B; Figures 6E and 6F) and detected ~500 distinct mature microRNAs. Upon hnRNP A2/B1 depletion in human iPSC-MNs, none were changed (adjusted p < 0.05). In mouse spinal cord, only a single microRNA (miR-146a) was differentially expressed (upregulated by two-fold) (Figure S2E; Data S1F and S1G). We conclude that hnRNP A2/B1 does not play a major role in regulating the steady-state levels of microRNAs in the CNS.
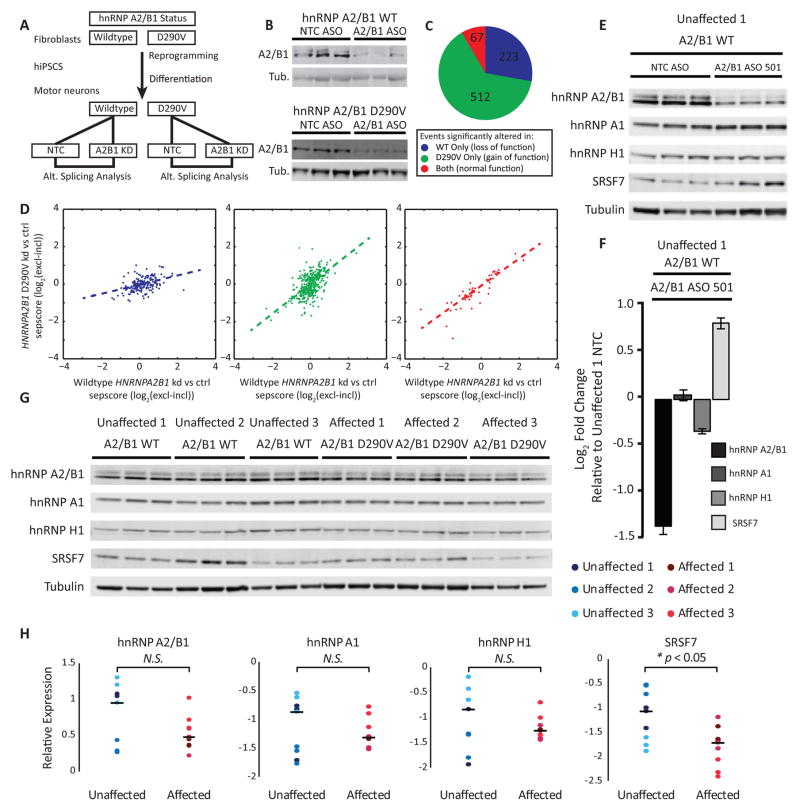
Figure 6. iPSC-MNs from patients with mutations in HNRNPA2B1 exhibit splicing defects.
(A) Schematic of experimental design. Fibroblasts expressing wildtype and hnRNP A2/B1 D290V were reprogrammed to iPSCs. iPSCs were differentiated to motor neurons. Motor neurons were treated with ASO against HNRNPA2B1 or non-targeting control (NTC). ASO treated and NTC treated RNA from both individuals was subjected to splicing sensitive microarray analysis.
(B) Depletion of hnRNP A2/B1 protein by ASO in motor neurons confirmed by western blotting. Tubulin (Tub.) was the loading control.
(C) Classification of 802 alternative cassette splicing changes into loss of function, gain of function, or normal function based on whether the event was detected in one genotype only, or both.
(D) Scatterplots comparing the sepscore (ASO vs. NTC) for WT HNRNPA2B1 vs. HNRNPA2B1 p.D290V. Dotted line is the least squares linear regression. The colors denote events detected in the WT sample only (blue), the mutant sample (green), or both samples (red). See the Supplemental Experimental Procedures for a definition of sepscore.
(E) Western blotting for hnRNP A2/B1, hnRNP A1, hnRNP H1 and SRSF7 after ASO-mediated hnRNP A2/B1 depletion, or non-targeting control (NTC) ASO, in iPSC-MNs from an unaffected individual. Tubulin was the loading control. Each lane represents a technical replicate of ASO treatment.
(F) Densitometry quantitation of the blot in (E) shows efficient depletion of hnRNP A2/B1, a small decrease in levels of hnRNP H1 and a modest increase in levels of SRSF7. Averages are plotted with error bars representing SEM. Three replicates per condition.
(G) Western blots of hnRNP A2/B1, hnRNP A1, hnRNP H1 and SRSF7 in iPSC-MNs from three unaffected and three affected individuals. Tubulin served as a loading control. Each lane contains lysate from a separate tissue culture well.
(H) Densitometry quantitation of the blot in (G). Dots represent band intensities normalized to tubulin loading control, color-coded according to the individual. Lines represent the median levels across all nine samples for each indicated protein. A two sample, two tailed, homoscedastic t-test was performed with n = 9 to generate the relevant p-values.
HnRNP A2/B1 regulates alternative 3′ cleavage choice in vivo
As hnRNP A2/B1 binds in 3′UTRs, we next evaluated whether hnRNP A2/B1 affects alternative polyadenylation. To identify alternative 3′ cleavage sites, we used the DaPars algorithm (Xia et al., 2014) to analyze the RNA-seq data from ASO-treated spinal cords. We identified 81 alternative polyA site (PAS) shifts (Figure 2F; Data S1H), of which 61 (~80%) resulted in distal polyA site usage to generate a longer 3′UTR upon depletion of hnRNP A2/B1, with the remainder exhibiting 3′UTR shortening (proximal polyA usage; Figure 2G). Strikingly, half (40) of the 81 APA changes contained hnRNPA2/B1 binding sites within the 3′UTR (Figure 2H). To illustrate, we identified a 3′UTR shortening in the ATPase, Na+/K+ Transporting, Beta 2 polypeptide (Atp1b2) gene (Figure 2I), 3′UTR and a lengthening in Hnrnph1 (Figure 2J). Quantitative RT-PCR validated PAS shifts within these genes (Figure S2F). We conclude that hnRNP A2/B1 binding within 3′UTRs is associated with hnRNP A2/B1-dependent alternative polyadenylation choice.
Hundreds of alternative splicing events depend on hnRNP A2/B1 protein level in vivo
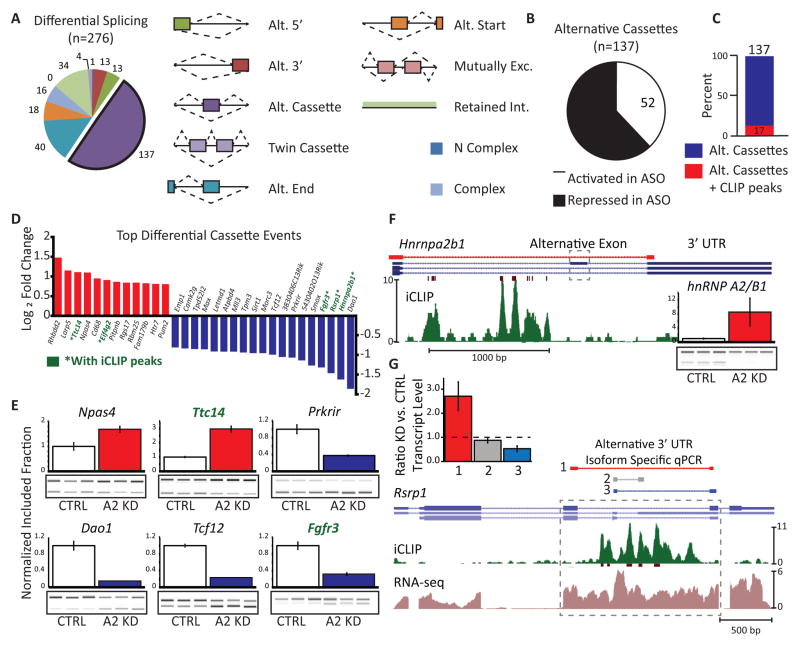
We identified 276 AS changes from our splicing-sensitive microarrays (Figure 3A, Data S1I). The largest differences were observed in alternative cassette exons, which were more frequently skipped (repressed) upon depletion of hnRNP A2/B1 than included (activated) (Figure 3B). Interestingly, while only 12% (17) of cassette events harbored hnRNP A2/B1 binding sites in flanking intronic regions (Figure 3C), five of the top 20 cassette events had hnRNP A2/B1 binding proximal to the exon (genes marked in green in Figure 3D). RT-PCR validation of AS events confirmed the microarray data (Figure 3E; Figure S3A). Intriguingly, we identified an alternative cassette exon within the 3′UTR of the Hnrnpa2b1 gene itself (Figure 3F). iCLIP data indicated that hnRNP A2/B1 binds upstream of the exon to enhance its use, resulting in an isoform that is expected to be subjected to nonsense-mediated decay (NMD) (McGlincy et al., 2010). This mode of autoregulation is consistent with our and others’ reports of splicing factors controlling their own levels (Huelga et al., 2012; McGlincy et al., 2010). We also observed AS events within 3′UTRs of other genes. HnRNP A2/B1 binding was observed in a region that alternatively encodes either an intron or a 3′UTR for a shorter isoform in the Arginine/serine-rich protein 1 (Rsrp1) gene. Depletion of hnRNP A2/B1 result in removal of the intron that generates a longer isoform (Figure 3G). We also found that decreased levels of hnRNP A2/B1 affect the AS of cassette exons within other ALS-associated genes, namely D-amino acid oxidase (Dao), Ataxin-2 (Atxn2) and inositol triphosphate receptor, type 2 (Itpr2) (Figure S3B). We conclude that loss of hnRNP A2/B1 affects hundreds of AS events in vivo, including an exon within its own 3′UTR.
Figure 3. HnRNP A2/B1 depletion in mouse spinal cord affects alternative splicing.
(A) Pie chart shows the classification of 276 significantly changing (q value < 0.05, sepscore > 0.5) splicing events identified by splicing-sensitive microarray. The legend illustrates the types of splicing events represented on the array.
(B) 137 alternative cassette events are altered upon loss of hnRNP A2/B1. 85 events were repressed upon knockdown and 52 were activated.
(C) Of the 137 transcripts displaying differential inclusion of cassette exons, 17 contained significant hnRNP A2/B1 occupancy, as identified by iCLIP.
(D) Ranked list of the 30 alternative cassette events with the largest absolute fold change. Activated cassettes are in red. Repressed cassettes are in blue. Gene names shown in green have significant hnRNP A2/B1 occupancy, as identified by iCLIP.
(E) RT-PCR validation of six alternative cassette events. Samples from control mice (white bars) are normalized to 1. Samples from ASO treated mice are shown in red (activated) and blue (repressed). Of the four samples from each treatment group used to generate the bar graphs, three are shown in the gel images that were used to generate the bar graphs. Error bars are SEM computed with four replicates per condition.
(F) The alternative exon (dashed box) regulated by hnRNP A2/B1 within its own 3′UTR has nearby iCLIP clusters. Color coding of the browser tracks is as in Figures 1E–J. The red browser track shows the position of PCR primers used for validation. Inset: RT-PCR validation of the event. Bars are as in Figure 3E.
(G) An example of alternative 3′UTR usage (gray box) regulated by hnRNP A2/B1, in the Rsrp1 gene with nearby iCLIP clusters. Color coding of the browser tracks is as in Figures 1E–J. The red, gray and green lines above the gene models indicate the position of primers used for qPCR validation. Colors correspond to the bar graph in the inset. Inset: qPCR validation of alternative 3′UTR usage. Tbp was used as the reference gene. Error bars are SEM computed with three replicates per condition.
HnRNP A2/B1 interacts with RNA components of the spliceosome
To evaluate if hnRNP A2/B1 interacts with components of the splicing machinery, we used northern blotting to measure snRNA levels in hnRNP A2/B1 immunoprecipitates of mouse spinal cord. We confirmed specific enrichment of U2, U4, U5, and U6 but not U1 snRNA (Figure S3C). Total snRNA levels were unchanged upon of hnRNP A2/B1 depletion (Figure S3D). We conclude that hnRNP A2/B1 interacts with snRNAs but loss of hnRNP A2/B1 does not affect snRNA levels.
HnRNP A2/B1-dependent splicing of the D-amino acid oxidase transcript results in reduced expression and enzymatic activity
The AS event with the largest magnitude change in our splicing microarray analysis is a 118-nucleotide cassette exon (exon 9) in the Dao gene (Figure 4A). This event is prominent because of previous studies implicating Dao in ALS (Kosuge et al., 2009; Sasabe et al., 2007). Dao codes for an enzyme important in the metabolism of D-serine and is largely expressed in the brainstem and spinal cord. D-serine is an agonist of the NMDA receptor and increased D-serine levels have been reported to contribute to excitotoxicity in ALS (Sasabe et al., 2007). Depletion of hnRNP A2/B1 protein did not change overall Dao mRNA expression (Figure 4B), but substantially decreased the abundance of the longer transcript relative to the shorter transcript isoform (Figure 4C). This alteration is predicted to cause a reading frame shift and early termination of the protein, but the location of the premature termination codon is not predicted to lead to NMD. In agreement with the mRNA data, we observed a 30% reduction in protein levels of the long isoform while the short isoform was not detected by western blotting (Figure 4D). The short isoform is predicted to lack two alpha helices and three beta sheets (Figure 4E).
Figure 4. hnRNP A2/B1-dependent AS in the D-amino acid oxidase (DAO) gene results in reduced DAO protein expression and enzymatic activity.
(A) Gene models of the long and short mRNA isoforms produced by AS of the hnRNP A2/B1-dependent cassette exon (in white) in the Dao pre-mRNA. Unaffected exons are shown as black boxes. Introns and UTRs are shown as thin boxes and lines, respectively. The lengths of the predicted protein products are indicated. Amino acid (AA).
(B) The transcript levels of Dao mRNA in control and hnRNP A2/B1 depleted mouse spinal cord (n = 4 mice per group) determined by qPCR. Tbp was used as reference. Error bars are SEM computed with four replicates. The mean transcript level for the control mice is normalized to 1.
(C) Gel-like image of AS of the Dao cassette exon in mice, determined by RT-PCR, shows exon exclusion upon hnRNP A2/B1 depletion.
(D) Western blotting shows a reduction in DAO protein levels to ~70% of non-target control levels in mice treated with hnRNP A2/B1 targeting ASO. Actin was the loading control.
(E) X-ray crystallography structure of human DAO (left; Protein Database ID 2E48, Kawazoe et al. (2006)) and the predicted structure of the short isoform (right). The arrows denote two alpha helices and three beta sheets that are absent in the predicted shorter isoform.
(F) Schematic of constructs used to express DAO long and short isoforms in stable cell lines. Western blotting shows reduced protein levels in clones expressing the short isoform. Tubulin was the loading control.
(G) No difference in transcript level of Dao mRNA from the clones shown in (F) determined by qPCR. Tbp was used as a reference transcript. Error bars are SEM computed with four replicates per condition.
(H) DAO activity in the clones shown in (F). Bars represent the ratio of substrate to non-substrate dependent activity. Error bars are SEM computed with four replicates per condition.
(I) Western blotting of DAO protein translated in rabbit reticulocyte lysate reveals similar levels in cells expressing either isoform.
(J) DAO activity in the lysates shown in (J). The wedge below the bars denotes serial two-fold dilutions of the lysates.
(K) Depletion of hnRNP A2/B1 in the human U-251 glioblastoma cell line. Three different targeting shRNAs and one non-targeting control shRNA plasmids were used in triplicates. Splicing-sensitive RT-PCR (top) for DAO shows that under conditions where the short DAO isoform cannot be detected in the NTC-shRNA treated samples, the short DAO isoform was consistently detected in samples treated with the two most effective shRNA constructs. hnRNP A2/B1 depletion was verified by western blotting (bottom). Tubulin was the loading control.
We stably expressed both isoforms in human 293 FRT cell lines (Figure 4F) and found that while both were similarly expressed at the mRNA level (Figure 4G), the protein level of the short isoform was significantly reduced (Figure 4F), suggesting that it is subjected to reduced translation or protein stability. We treated cell lines stably expressing each isoform with the proteasome inhibitor MG-132 and performed immunoblotting over 24 hours. We found that while the short isoform accumulated, the levels of the long isoform remained stable (Figures S4A and S4B). We measured DAO enzymatic activity in a cell-based assay (Figure S4C) and found significant DAO activity in cells expressing the long isoform, while cells expressing the short isoform had no DAO activity above the background level (Figure 4H). We measured the activity of both isoforms produced in cell-free extracts (Figure 4I) and found that the short isoform has an 85% reduction in enzymatic activity (Figure 4J). Taken together, we conclude that hnRNP A2/B1 depletion leads to production of a shorter isoform of Dao, which is subject to proteasomal degradation and exhibits ~six-fold reduction in enzymatic activity.
To determine if AS of Dao is evolutionarily conserved, we inspected the orthologous 118bp exon in human DAO, which has high amino acid conservation to the murine exon but is not annotated as being alternatively spliced. We were unable to detect expression of DAO mRNA human iPSC-MNs and iPSC-Astrocytes (data not shown). As expression of DAO is thought to be region- and cell-type specific (Horiike et al., 1994), we consulted the Brain RNA-seq database (Zhang et al., 2014). Little to no expression of DAO in any of the samples in this database was reported (Figures S4D and S4E). In a panel of human CNS RNA samples, we detected DAO mRNA expression only in astrocytes from human cerebellum and spinal cord (Figure S4F, top). Sanger sequencing of the bands confirmed a longer isoform as human mRNA containing exons 8, 9, and 10 and a shorter isoform lacking exon 9 (Figure S4F, bottom). We conclude that DAO mRNA is expressed primarily in human cerebellum and spinal cord astrocytes and that its AS is conserved in mouse spinal cord. Importantly, depletion of hnRNP A2/B1 in a human glioblastoma multiforme cell line (U-251) revealed an increase in the shorter isoform, consistent with exon 9 skipping (Figure 4K). This result confirms conservation of hnRNP A2/B1-dependent splicing of DAO exon 9 in mice and humans. Thus, an unanticipated hnRNP A2/B1-dependent AS of an exon in Dao dramatically reduces D-serine metabolism, implicating lower levels of hnRNP A2/B1 with DAO-mediated pathogenicity observed in disease.
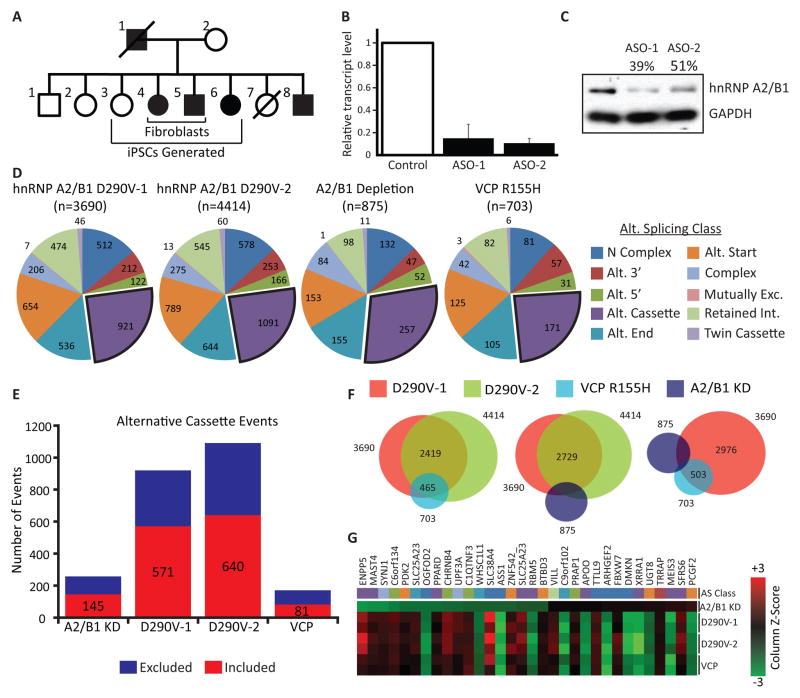
HnRNP A2/B1 D290V mutation results in splicing defects in human fibroblasts
As mutations within the hnRNP A2/B1 gene are associated with MSP and ALS (Kim et al., 2013), we next examined if the D290V mutation affects AS. RNA from fibroblasts with the disease-associated hnRNP A2/B1 variant (two affected individuals) and from unaffected control fibroblasts were used in splicing array analysis (Figure 5A; Data S1J). To evaluate if splicing changes could be attributed to a mutant-dependent loss of normal splicing, we also depleted hnRNP A2/B1 in fibroblasts from unaffected control individuals (Figures 5B and 5C). For splicing-sensitive array analysis, hnRNP A2/B1 mutant and depletion samples were compared to two unrelated, unaffected control samples. For hnRNP A2/B1 D290V mutants, ~4,000 splicing events were altered relative to controls (Figure 5D). This is in contrast to hnRNP A2/B1-depleted fibroblasts, where only 875 events were significantly different (Figure 5D). Interestingly, in fibroblasts harboring a mutation in the only other known gene related to MSP, the VCP R155H mutation, 703 AS events were altered (Figure 5D). We compared the overlap of significantly changed alternative cassette events between the different groups of samples (Figures 5E and 5F). For the VCP R155H mutant fibroblasts, we found that 465 (129 are alternative cassette events) out of 703 (171 are alternative cassettes) AS events were shared between both of the hnRNP A2/B1 mutant samples. This indicates a very strong and statistically significant (p < 10−25) overlap between the splicing signatures of these two disease-causing variants (Figure 5F). However, each of the hnRNP A2/B1 D290V samples had about 1,000 alternative cassette events that were not in common with the VCP mutant samples, indicating potentially divergent molecular processes (Figure 5F). In contrast, hnRNP A2/B1 knockdown samples had only a small, statistically insignificant overlap in cassette events with hnRNP A2/B1 D290V or VCP R155H mutant samples (p = 0.21 and p = 0.11, respectively). To further investigate the difference between hnRNP A2/B1 depletion and the D290V mutation, we analyzed the 32 splicing events that were significantly different in all four comparisons tested and found that the direction of splicing changes in the three mutant samples was usually anti-correlated with the knockdown sample (Figure 5G). We conclude that in fibroblasts, disease-causing mutations in hnRNP A2/B1 result in thousands of splicing changes, the majority of which were not observed when hnRNP A2/B1 was depleted. Furthermore, we find a remarkable (66%) overlap in significant splicing changes in patient samples with mutations in the only two genes known to cause MSP (hnRNP A2/B1 and VCP).
Figure 5. Fibroblasts from patients with mutations in HNRNPA2B1 and VCP exhibit widespread splicing defects.
(A) A multiplex family affected by MSP, where affected (filled in black) individuals harbor p.D290V mutations in the HNRNPA2B1 gene. Fibroblasts and iPSCs used in experiments in Figures 5–7 were obtained from the indicated individuals.
(B) Depletion of HNRNPA2B1 mRNA in fibroblasts from an unaffected individual by treatment with ASOs, as measured by qPCR using GAPDH as reference. Error bars are SEM computed with three replicates per condition. The control transcript level is normalized to 1. The error for the control samples was added to the error for the other conditions by error propagation.
(C) Western blot analysis showing successful depletion of hnRNP A2/B1 protein. GAPDH was the loading control.
(D) Splicing-sensitive microarrays detected splicing changes in fibroblasts obtained from individuals with the indicated genotype. hnRNP A2/B1 depletion is from samples in Figures 5B and 5C. Significant splicing changes are determined by comparing to two different unrelated control samples. Color coding and splicing event categories are as in Figure 3A.
(E) Quantification of alternative cassette events detected in the various samples. Colors indicate the direction of the splicing change. Excluded refers to excluded in the depleted or affected sample. Included refers to included in the depleted or affected sample.
(F) Venn diagrams illustrating overlapping splicing events between the indicated groups. Overlapping events refers to identical splicing events flagged as significant in two or more samples, regardless of the directionality of the changes.
(G) Heat map of 32 splicing changes that were flagged as significant in all four groups. Red changes are activated compared to controls. Green changes are repressed. Color mapping is performed according to the column-wise Z-score.
Altered hnRNP A2/B1-dependent alternative splicing in human iPSC-MNs
To evaluate if RNA processing was altered in hnRNP A2/B1 D290V iPSC-MNs, we generated iPSCs from affected and unaffected fibroblasts (Figures 5A and S5A). Normal ploidy and pluripotency was confirmed by array CGH (Figure S5A) and immunofluorescence (Figure S5B), respectively. PSCs were differentiated to motor neurons using a modified dual-SMAD inhibition protocol (Burkhardt et al., 2013; Chambers et al., 2009) (Figure S6A) for up to 53 days, at which point cells abundantly expressed pan-neuronal markers, such as MAP2 and phosphorylated Neurofilament (SMI31). We also observed robust expression of the motor neuron-specific markers ISLET1, HB9 and cholinergic marker CHT1 (Figure S6B). Multi-electrode array analysis showed high frequency action potentials and synchronous firing (Figures S6C–S6G), confirming mature and active motor neuron networks.
To identify AS events affected by depletion of hnRNP A2/B1 in normal iPSC-MNs and to study if these events are similarly affected in the D290V mutant lines, iPSC-MNs differentiated from one non-affected and one D290V mutant iPSC line were treated with non-targeting control (NTC) and hnRNP A2/B1 targeting ASOs (Figure 6A). HnRNP A2/B1 protein was successfully reduced in normal and D290V mutant human iPSC-MNs (Figure 6B) and mRNA subjected to splicing-sensitive microarrays (Data S1K–S1M; Supplemental Experimental Procedures). We found 67 differentially altered AS events that were indicative of loss of normal hnRNP A2/B1 function, 512 events that were mutant-dependent gain-of-function, and 223 mutant-dependent loss-of-function events (Figures 6C and 6D). Notably, nearly two-thirds of all splicing events grouped into the mutant-dependent gain-of-function category, corroborating our findings in patient fibroblasts (Figure 5).
We hypothesized that hnRNP A2/B1 D290V mutant protein may be aggregating in the nucleus, thus causing widespread splicing changes. To test this, we performed cellular fractionation of PSC-MNs from unaffected and MSP patients. We observed increased levels of nuclear, insoluble hnRNP A2/B1 in samples from a VCP R155H patient and one of two hnRNP A2/B1 D290V patients (Figure S7A), suggesting that these mutant proteins are indeed more aggregation-prone than wildtype protein and may start to aggregate in the nucleus; this aggregation may lead to dysfunction affecting other RBPs. We next measured SRSF7, hnRNP A1 and hnRNP H1 protein levels, previously shown to be subjected to cross-regulation by hnRNP A2/B1 (Huelga et al., 2012). In iPSC-MNs from one control individual, hnRNP A2/B1 depletion led to no change in hnRNP A1 or hnRNP H1 (Figures 6E and 6F). We observed a significant increase in SRSF7, consistent with our result observed in mouse spinal cord (Figure S2D). We next compared the protein levels between three unaffected and three affected individuals (p > 0.05) and found that SRSF7 levels were significantly lower in affected samples compared to unaffected samples (p < 0.05) (Figures 6G and 6H). Our findings indicate divergent regulation of SRSF7 at the protein level by mutant and wild-type hnRNP A2/B1. We conclude that AS is abnormal in MSP iPSC-MNs, potentially caused in part by increased nuclear insolubility of mutant hnRNP A2/B1.
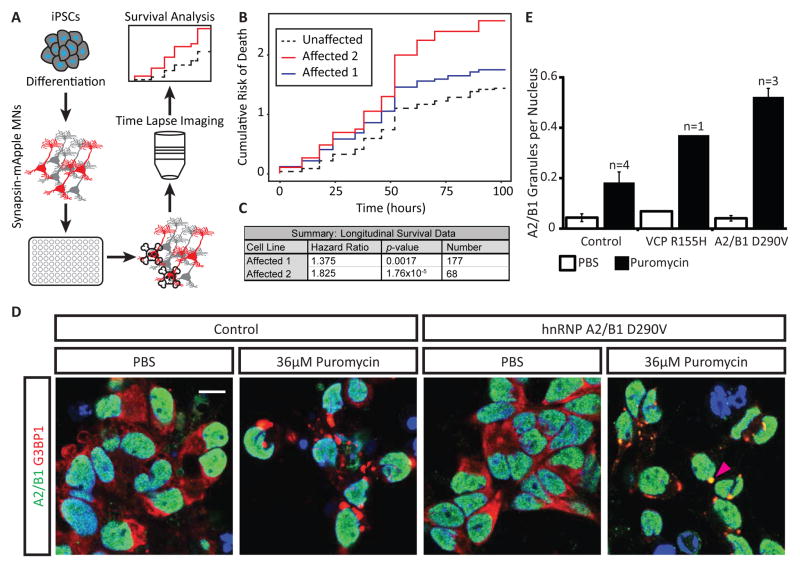
hnRNP A2/B1-mutant neurons display increased risk of death during long-term culture
We hypothesized that increased nuclear insolubility of hnRNP A2/B1 and abnormal RNA processing in hnRNP A2/B1 D290V MNs might cause decreased survival over time. To monitor the progressive death of motor neurons – a defining characteristic of ALS – we tracked iPSC-MNs, transfected with a mApple reporter under the control of the synapsin promoter (marking neurons), from affected and unaffected individuals (Figure 7A), according to methods previously described (Skibinski and Finkbeiner, 2013). We found a significantly higher risk of death in iPSC-MNs from two affected individuals compared to two unrelated controls (Figures 7B and 7C). We conclude that our in vitro iPSC-MN model of hnRNP A2/B1 D290V proteinopathy exhibits progressive death.
Figure 7. hnRNP A2/B1 D290V motor neurons exhibit increased risk of death and accumulate excess hnRNP A2/B1 in stress granules.
(A) Schematic of experimental design for long term imaging and survival analysis. iPSCs from affected and unaffected individuals were differentiated into iPSC-MNs, transfected with Synapsin-mApple and plated in 96-well plates. Fluorescent cells were tracked over time using time lapse imaging. Survival of motor neurons was evaluated using Kaplan-Meier analysis.
(B) Cumulative risk of death curves derived from Kaplan-Meier survival curves for iPSC-MNs differentiated from two affected (hnRNP A2/B1 D290V) and two unaffected individuals. Data was recorded over four days. Cells from both unaffected individuals are combined into a single line for analysis purposes.
(C) Summary of Cox proportional hazard analysis of survival data depicted in (B). Affected cells have a significantly higher hazard ratio compared to unaffected cells. A hazard ratio greater than 1.0 indicates increased risk of death as compared to the control lines from healthy individuals. See supplemental experimental procedures for a discussion of hazard ratio and p-value calculations.
(D) Immunofluorescence staining of hnRNP A2/B1 and the stress granule marker G3BP1 in iPSC-MNs from an unaffected and an affected individual. iPSC-MNs from both individuals accumulate foci positive for G3BP1 when treated with puromycin, but not vehicle (PBS). iPSC-MNs derived from the affected individual also accumulate hnRNP A2/B1 in the foci. White arrow: G3BP1 positive stress granule. Magenta arrow: G3BP1 and hnRNP A2/B1 positive stress granule. Scale bar = 10 μm.
(E) Quantification of images in (A). hnRNP A2/B1 positive granules per nucleus were counted. “n” refers to the number of individuals analyzed with each genotype. Error bars are SEM computed with the indicated sample size.
hnRNP A2/B1-mutant iPSC-MNs exhibit excess hnRNP A2/B1 in stress granules
Another hallmark of ALS and other related diseases is the accumulation of protein aggregates in the cytoplasm of affected cells (Mackenzie et al., 2007; Neumann et al., 2006; Van Deerlin et al., 2008). HnRNP A2/B1-positive aggregates were previously reported for patient muscle biopsy samples from individuals with the hnRNP A2/B1 D290V mutation (Kim et al., 2013). However, hnRNP A2/B1 aggregation has not been observed in human neurons with endogenous expression of mutant RBPs. In unstressed PSC-MNs, we do not have biochemical evidence for increased insoluble hnRNP A2/B1 in the cytoplasm (Figure 7D; Figures S7A–D). Indeed, hnRNP A2/B1 was localized to the nucleus and cytoplasmic aggregates were not observed in either affected individuals or controls (Figures 7D–E and S7B–D). We challenged PSC-MNs with puromycin for 24 hours to induce the formation of stress granules (Kedersha et al., 2000; Liu-Yesucevitz et al., 2010). PSC-MNs developed dense cytoplasmic aggregates positive for the stress granule markers G3BP1 (white arrows in Figure 7D), TIA1, PABP, and TDP-43 (Figure S7B). In iPSC-MNs from patients with D290V mutations, these aggregates were positive for hnRNP A2/B1 (magenta arrows in Figure 7D; Figures S7B and S7C). In contrast, control samples rarely accumulated any cytoplasmic hnRNP A2/B1 even during puromycin treatment (Figures 7D and 7E). Interestingly, iPSC-MNs from a VCP R155H patient had increased hnRNP A2/B1-containing granules upon puromycin induction (Figure S7C). We conclude that iPSC-MNs from hnRNP A2/B1 D290V patients exhibit reproducible disease-relevant cellular attributes such as increased propensity for stress granule accumulation.
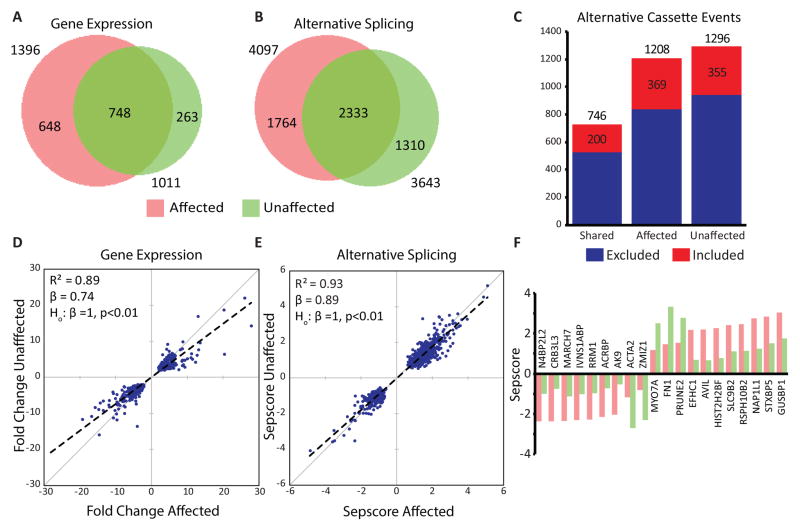
MSP/ALS iPSC-MNs exhibit exacerbated transcriptome changes in response to stress
To determine if iPSC-MNs derived from MSP patients would exhibit unusual AS and expression changes in response to stress, we exposed PSC-MNs to puromycin treatment and measured the changes using splicing sensitive microarrays (Figure S8A; Data S1N and S1O). We used cells from three hnRNP A2/B1 D290V individuals, one VCP R155H individual, and three unaffected individuals. The majority of gene expression and AS changes were shared among all seven samples (Figures 8A and 8B). A large fraction of gene expression and AS changes was observed only in affected samples (pink crescent in Figures 8A and 8B). A smaller fraction was observed only in control cells (green crescent in Figures 8A and 8B). Analysis of alternative cassette events showed a clear bias towards exon skipping upon stress in all sample types (Figure 8C). Hierarchical clustering showed a clear delineation of stressed and unstressed samples (Figure S8B). As expected, we found significant upregulation of the Hsp70-like gene HSPA14, the chaperone DNAJC12 and early response genes JUN and FOS in all samples (Data S1N). We also observed significant AS changes in the transcription factor HSF2, the chaperones DNAJC12, DNAJB2, DNAJC120, DNAJC13, and DNAJC21 in all samples tested (Data S1O).
Figure 8. iPSC-MNs from affected patients display an exaggerated response to stress.
(A–B) Venn diagrams showing the overlap of stress-induced gene expression (A) and AS (B) changes between iPSC-MNs from four affected (pink) and three unaffected (green) individuals. For both analyses, a plurality of events is shared.
(C) For each group in (B), the type of alternative cassette exon events (excluded vs. included) were counted. Most stress-induced changes are exon-skipping in all groups.
(D–E) Scatterplots generated by calculating the median magnitude change from affected (x axis) and unaffected (y axis) samples for each shared event shown in (A–B). Dashed lines represent the fit obtained by linear regression, with the coefficient of determination (R2) and slope (beta) indicated. A Student’s t-test shows that the slopes are significantly different from one (p < 0.01) for both analyses, indicating that the magnitude of the stress-induced events are more pronounced in affected than in unaffected samples. See Supplemental Experimental Procedures for a definition of sepscore.
(F) The 20 splicing changes with the largest difference in Sepscore between affected and unaffected samples are plotted along with the genes they are found in. While the direction (exclusion vs. inclusion) is the same regardless of disease status, for 15 of the 20 events the magnitudes of the stress-induced splicing changes were larger in affected samples than in unaffected samples.
To identify stress-responsive, mutant-specific AS and expression changes, we performed hierarchical clustering using pairwise fold-changes (748 genes) and AS differences (2333 events) in all samples. We found two distinct clusters, one containing affected samples, and another containing the unaffected controls (Figures S8C and S8D). Regression analysis showed a highly significant slope value less than one when comparing median fold-changes or AS differences from affected to unaffected samples (Figure 8D and 8E). This result indicates a trend towards increased magnitude expression and splicing changes in affected samples compared to controls upon stress. Indeed, the median change was larger in magnitude for affected samples than unaffected samples for 15 of the top 20 AS events (Figure 8F). Therefore, we find that stress induces both shared and disease-specific changes in gene expression and AS, with the iPSC-MNs derived from patients with MSP exhibiting an exacerbated stress response compared to controls.
Discussion
Our study provides a systematic transcriptome-wide analysis of hnRNP A2/B1 binding and function in the nervous system. Strikingly, the binding pattern of hnRNP A2/B1 in in vivo adult nervous system is largely distinct from the binding modes observed for FUS/TLS, TDP-43 and TAF15, which bind mostly in introns. We find that for hnRNP A2/B1, the majority of binding sites are in 3′UTRs. While FUS and TDP-43 each bind thousands of transcripts (Lagier-Tourenne et al., 2012; Polymenidou et al., 2011), hnRNP A2/B1 binds a much more modest number. These qualitative differences indicate distinct roles in RNA processing. Previous data has shown that FUS/TLS associates with PolII complexes and may be deposited co-transcriptionally (Schwartz et al., 2012). Similarly, TDP-43 may be involved in transcriptional elongation and PolII pausing (Lalmansingh et al., 2011). Our finding of hnRNP A2/B1 in 3′UTR regions and not “coating” introns indicates that hnRNP A2/B1 is not deposited until the nascent mRNA is almost completely transcribed. These findings are consistent with our observation that very few genes (<30) exhibit significant expression changes after depletion of hnRNP A2/B1.
When we compared AS changes induced by hnRNP A2/B1 depletion in vivo to hnRNP A2/B1 binding sites, we found that while the most significantly ranked AS changes did harbor proximal binding sites, the vast majority of splicing changes did not. This is also in contrast to other ALS proteins (FUS/TLS, and TDP-43), where binding sites are frequently observed proximal to AS events and depletion is associated with widespread gene expression changes (Lagier-Tourenne et al., 2012; Polymenidou et al., 2011). Aside from a strong intronic binding site downstream of an auto-regulated alternative exon in the 3′UTR of HNRNPA2B1, our data supports our conclusion that the majority of the AS changes observed after depletion of hnRNP A2/B1 are not likely due to direct pre-mRNA binding of hnRNP A2/B1. Significant hnRNP A2/B1 association to the U2, U5 and U6 snRNAs suggests that hnRNP A2/B1 may play a role in spliceosome assembly or snRNA recycling and thereby affects AS without direct interaction with the affected transcripts. Another possible mechanism is indirect action through regulation of other splicing factors, such as SRSF7.
Aside from AS regulation we unexpectedly identified alternative polyadenylation events affected by loss of hnRNP A2/B1 in vivo. Importantly, half exhibited hnRNP A2/B1 binding within the 3′UTRs. Two notable transcripts – RSRP1 and HNRNPH1 – encoding splicing factors showed large differences in 3′UTR selection preference near hnRNP A2/B1 binding sites after depletion and could also be responsible for a large number of secondary splicing changes in other genes.
The most significant and robust splicing change observed after depletion of hnRNP A2/B1 in the mouse spinal cord is an exon within the gene encoding D-amino-acid-oxidase. Although low in abundance, D-amino acids are important receptor co-agonists in the CNS with disease implications (Paul and de Belleroche, 2014). D-serine is a known agonist for the NMDA receptor and has been implicated in schizophrenia and ALS (Burnet et al., 2008; Mitchell et al., 2010; Paul et al., 2014; Yang et al., 2013). Previous studies have shown that D-serine metabolism is affected in the SOD1 transgenic mouse model of ALS and mutant mice that do not express DAO protein exhibit muscle weakness, motor neuron loss, and other ALS-like symptoms (Sasabe et al., 2012; Thompson et al., 2012). Recently, a large study of ALS patients found polymorphisms in DAO to be associated with decreased survival (Cirulli et al., 2015). In our study, we observed exon skipping of a 118 nt exon in the Dao transcript after depletion of hnRNP A2/B1. The resulting protein isoform is highly unstable and compromised in its enzymatic activity. We also demonstrated that AS of DAO exon 9 is evolutionarily conserved in human and mouse. Determining if D-serine metabolism is perturbed by mutations in hnRNP A2/B1 may require new transgenic mouse models or the analysis of post-mortem human tissue harboring HNRNPA2B1 mutations. We opine that this connection between AS and regulation of D-amino acid metabolism is relevant to neurodegeneration. Reduced levels of hnRNP A2/B1 have been associated with AD and this depletion was specifically linked to loss of cholinergic neurons (Berson et al., 2012). Our unexpected findings link the levels of an RBP to a physiologically important enzyme.
Our data provides compelling hypotheses regarding how mutations in hnRNP A2/B1 may lead to neurodegeneration. iPSC-MNs derived from patients with MSP mutations showed reduced survival in long-term culture, recapitulating a defining characteristic of ALS, namely progressive neuronal death. We also found a substantial number of differential AS events in hnRNP A2/B1 D290V mutant cells that were not present upon hnRNP A2/B1 depletion. This indicates that at least by AS, the effects of hnRNP A2/B1 D290V are not equivalent to simple loss-of-function. Instead, our observation is reminiscent of a recent study showing that a toxic FUS mutation causes motor neuron degeneration in mice not by loss-of-function, but by a combination of loss- and gain-of-function (Scekic-Zahirovic et al., 2016). One possible mechanism underlying the abnormal AS changes we observe is suggested by the presence of increased nuclear, insoluble hnRNP A2/B1 protein in unstressed mutant motor neurons. This aberrant assembly of protein-RNA granules likely sequesters hnRNP A2/B1 protein, as well as other RBPs, from their RNA substrates, leading to both loss- and gain-of-function effects in RNA processing. As hnRNP A2/B1 also affects the levels of other RBPs, it is reasonable to expect that the normal control of AS is impacted.
We expected that the propensity of hnRNP A2/B1 to form aggregates would also be reflected in an increase in insoluble hnRNP A2/B1 in the cytoplasm of unstressed motor neurons. However, chemical stress was required to induce hnRNP A2/B1 to move to cytoplasmic aggregates, called stress granules (Kim et al., 2013; Molliex et al., 2015; Xiang et al., 2015). Similar phenomena have been observed for other ALS-related RBPs including TDP-43, FUS/TLS, and EWSR1. It is hypothesized that cytoplasmic aggregation represents a loss-of-function and may be the cause of neurodegeneration in ALS (Iguchi et al., 2013; Sun et al., 2015; Wu et al., 2012; Yang et al., 2014). In light of our data, we put forward an alternative interpretation. The RBPs mentioned above are also normally localized to the nucleus. Although aggregation is typically described in the cytoplasm, nuclear aggregates of TDP-43 have been reported (Burkhardt et al., 2013; Udan-Johns et al., 2014). Thus, cytoplasmic aggregation may represent an end-stage symptom of dysfunction that begins in the nucleus.
Cells with hnRNP A2/B1 D290V mutations and VCP R155H mutations also responded to stress with numerous changes in gene expression and AS. Some of these changes were unique to affected cells; however, most were also detected in controls. Surprisingly, when we examined these common changes we found that affected cells systematically responded with changes in the same genes but at greater magnitude than controls. We believe that hnRNP A2/B1 D290V mutant motor neurons respond to stress in largely the same way as controls, but their response is exacerbated. This may indicate that drugs that dampen the stress response or reduce the assembly of stress granules may be useful in treating ALS or other neurodegenerative diseases.
Experimental Procedures
RNA Bind-n-Seq (RBNS)
RBNS was performed with N-terminally GST-tagged HNRNPA2B1 amino acids 1–197 (containing both RRMs) essentially as previously described (Lambert et al., 2014), except that binding was at 4 °C. Motif enrichment (R) values we re calculated for 6-mers as the motif frequency in the RBP-selected pool over the frequency in the input RNA library. R values were considered significant at Z-score ≥ 2.
CLIP analysis
iCLIP using flash-frozen, ground and UV-irradiated spinal cords of 8-week old, female, C57/BL6J mice was performed as described (Huppertz et al., 2014) and CLIP-seq libraries sequenced in single-end 50 bp mode. CLIP-seq peaks were identified as previously described (Zisoulis et al., 2010). eCLIP using UV-irradiated day 28-day iPSC-MN was performed as described (Van Nostrand et al., 2016) and libraries sequenced in paired-end 50 bp. eCLIP peaks were identified using CLIPPER (Lovci et al., 2013). Peaks were called significant if the number of reads in the IP sample was greater than the number of reads in the size-matched input sample and the peaks had a Bonferroni-corrected Fisher exact test p-value < 0.05.
DAO activity assays
For cell-based assays, stable cell lines expressing the short and long isoforms of DAO were generated using the FLP-In-293 system (Thermo Fisher Scientific). D-serine was added to the media (50 mM final concentration) and H2O2 released by DAO-catalyzed oxidation of D-serine measured with the Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Thermo Fisher Scientific) using a fluorescence plate reader (Infinite 200 PRO; Tecan). For cell-free assays, 3xFLAG-tagged versions of the long and short isoforms of mouse Dao were expressed in rabbit reticulocyte lysates (TNT Quick Coupled Reticulocyte Lysate kit; Promega) and assayed in the presence of 50 mM D-serine and 500 nM FAD, as described above.
Stress granule assays
iPSC-MNs were generated from healthy and affected individuals. Cells were treated with 36 μM puromycin or vehicle for 24 hours starting 28 days after differentiation. iPSC-MNs were fixed and processed for immunofluorescence. Four images were taken for each condition from each cell line used. Images were blinded and then nuclei and G3BP1 and hnRNP A2/B1 foci were counted manually. For microarray experiments, iPSC-MNs were harvested with Trizol following puromycin treatment. RNA was prepared and hybridized to Affymetrix HTA 2.0 microarrays. Gene expression and alternative splicing changes were determined as described elsewhere (Huelga et al., 2012).
Long term imaging experiments
iPSC lines from two ALS patients and two unaffected individuals were differentiated into motor neurons according to (Burkhardt et al., 2013; Chambers et al., 2009) with modifications from (Du et al., 2015). MNs were dissociated and plated in 96-well plates, then lipofected with a construct containing mApple under the control of the Synapsin promoter. iPSC-MNs were imaged starting 2 days after transfection, once every 6 hours. Individual neurons were tracked and analyzed for survival as described (Skibinski and Finkbeiner, 2013).
Data Availability
Small RNA-seq, RNA-seq, iCLIP, eCLIP and microarray data are available at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE86464.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Yeo lab, especially Stefan Aigner for critical reading of the manuscript. S.C.H. and G.P. were funded by National Science Foundation Graduate Research Fellowships. F.J.M. was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number T32GM008666. This work was supported by grants from the National Institute of Health (HG004659, NS075449, and U54HG007005 to G.W.Y), the California Institute of Regenerative Medicine (RB3-05009 and RB4-06045 to G.W.Y.) and ALS Association (VC8K27 to G.W.Y). The neuronal survival studies were supported by ALS Association and Taube/Koret Center for Neurodegenerative Disease Research awarded to S.F. This work was partially supported by NIH grant U54HG007005 to C.B.B. We would like to thank Ionis Pharmaceuticals for sharing reagents. G.W.Y. is an Alfred P. Sloan Research Fellow.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, G.W.Y; Methodology, F.J.M; Investigation, F.J.M., K.K., P.F., S.J.C., K.L., C.G-B., L.F., H.W., J.K.N., R.L., B.S., S.M.B. and F.R.; Validation, F.J.M, H.W., L.J., A.J. and B.S.; Formal Analysis, F.J.M., G.A.P., E.L.V.N., R.B., S.C.H., P.F., J.P.D., Resources, J.L-A., F.B., M.A., C.B.B., H.J.K., S.F., J.P.T. and G.W.Y.; Writing – Original Draft, F.J.M. and G.W.Y.; Writing – Review & Editing, F.J.M. and G.W.Y.; Funding Acquisition C.B.B. and G.W.Y.; Visualization, F.J.M. and G.W.Y.; Supervision, G.W.Y.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, Carson JH. Transport and Localization Elements in Myelin Basic Protein mRNA. The Journal of Cell Biology. 1997;138:1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, Gendron TF, Petrucelli L. RNA-mediated toxicity in neurodegenerative disease. Mol Cell Neurosci. 2013;56:406–419. doi: 10.1016/j.mcn.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson A, Barbash S, Shaltiel G, Goll Y, Hanin G, Greenberg DS, Ketzef M, Becker AJ, Friedman A, Soreq H. Cholinergic-associated loss of hnRNP-A/B in Alzheimer’s disease impairs cortical splicing and cognitive function in mice. EMBO Mol Med. 2012;4:730–742. doi: 10.1002/emmm.201100995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt MF, Martinez FJ, Wright S, Ramos C, Volfson D, Mason M, Garnes J, Dang V, Lievers J, Shoukat-Mumtaz U, et al. A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. Mol Cell Neurosci. 2013;56:355–364. doi: 10.1016/j.mcn.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnet PW, Eastwood SL, Bristow GC, Godlewska BR, Sikka P, Walker M, Harrison PJ. D-amino acid oxidase activity and expression are increased in schizophrenia. Mol Psychiatry. 2008;13:658–660. doi: 10.1038/mp.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger BJ, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A. 2010;107:1894–1899. doi: 10.1073/pnas.0914845107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A. 2011;108:20881–20890. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZW, Chen H, Liu H, Lu J, Qian K, Huang CL, Zhong X, Fan F, Zhang SC. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nature communications. 2015;6:6626. doi: 10.1038/ncomms7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahling M, Mrowka R, Steege A, Martinka P, Persson PB, Thiele BJ. Heterogeneous nuclear ribonucleoprotein-A2/B1 modulate collagen prolyl 4-hydroxylase, alpha (I) mRNA stability. J Biol Chem. 2006;281:9279–9286. doi: 10.1074/jbc.M510925200. [DOI] [PubMed] [Google Scholar]
- Horiike K, Tojo H, Arai R, Nozaki M, Maeda T. D-amino-acid oxidase is confined to the lower brain stem and cerebellum in rat brain: regional differentiation of astrocytes. Brain Res. 1994;652:297–303. doi: 10.1016/0006-8993(94)90240-2. [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell reports. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D’Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, Konig J, Ule J. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison S, LeBel C, Blanchette M, Chabot B. Distinct sets of adjacent heterogeneous nuclear ribonucleoprotein (hnRNP) A1/A2 binding sites control 5′ splice site selection in the hnRNP A1 mRNA precursor. J Biol Chem. 2002;277:29745–29752. doi: 10.1074/jbc.M203633200. [DOI] [PubMed] [Google Scholar]
- Iguchi Y, Katsuno M, Niwa J, Takagi S, Ishigaki S, Ikenaka K, Kawai K, Watanabe H, Yamanaka K, Takahashi R, et al. Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain. 2013;136:1371–1382. doi: 10.1093/brain/awt029. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kapeli K, Pratt GA, Vu AQ, Hutt KR, Martinez FJ, Sundararaman B, Batra R, Freese P, Lambert NJ, Huelga SC, et al. Distinct and shared functions of ALS-associated proteins TDP-43, FUS and TAF15 revealed by multisystem analyses. Nature communications. 2016;7:12143. doi: 10.1038/ncomms12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosturko LD, Maggipinto MJ, Korza G, Lee JW, Carson JH, Barbarese E. Heterogeneous nuclear ribonucleoprotein (hnRNP) E1 binds to hnRNP A2 and inhibits translation of A2 response element mRNAs. Mol Biol Cell. 2006;17:3521–3533. doi: 10.1091/mbc.E05-10-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuge Y, Sekikawa-Nishida K, Negi H, Ishige K, Ito Y. Characterization of chronic glutamate-mediated motor neuron toxicity in organotypic spinal cord culture prepared from ALS model mice. Neurosci Lett. 2009;454:165–169. doi: 10.1016/j.neulet.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalmansingh AS, Urekar CJ, Reddi PP. TDP-43 is a transcriptional repressor: the testis-specific mouse acrv1 gene is a TDP-43 target in vivo. J Biol Chem. 2011;286:10970–10982. doi: 10.1074/jbc.M110.166587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, Kwong LK, Forman MS, Ravits J, Stewart H, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- McGlincy NJ, Tan LY, Paul N, Zavolan M, Lilley KS, Smith CW. Expression proteomics of UPF1 knockdown in HeLa cells reveals autoregulation of hnRNP A2/B1 mediated by alternative splicing resulting in nonsense-mediated mRNA decay. BMC Genomics. 2010;11:565. doi: 10.1186/1471-2164-11-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J, Paul P, Chen HJ, Morris A, Payling M, Falchi M, Habgood J, Panoutsou S, Winkler S, Tisato V, et al. Familial amyotrophic lateral sclerosis is associated with a mutation in D-amino acid oxidase. Proc Natl Acad Sci USA. 2010;107:7556–7561. doi: 10.1073/pnas.0914128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohagheghi F, Prudencio M, Stuani C, Cook C, Jansen-West K, Dickson DW, Petrucelli L, Buratti E. TDP-43 functions within a network of hnRNP proteins to inhibit the production of a truncated human SORT1 receptor. Hum Mol Genet. 2016;25:534–545. doi: 10.1093/hmg/ddv491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nussbacher JK, Batra R, Lagier-Tourenne C, Yeo GW. RNA-binding proteins in neurodegeneration: Seq and you shall receive. Trends Neurosci. 2015;38:226–236. doi: 10.1016/j.tins.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Paul P, de Belleroche J. The role of D-serine and glycine as co-agonists of NMDA receptors in motor neuron degeneration and amyotrophic lateral sclerosis (ALS) Front Synaptic Neurosci. 2014;6:10. doi: 10.3389/fnsyn.2014.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P, Murphy T, Oseni Z, Sivalokanathan S, de Belleroche JS. Pathogenic effects of amyotrophic lateral sclerosis-linked mutation in D-amino acid oxidase are mediated by D-serine. Neurobiol Aging. 2014;35:876–885. doi: 10.1016/j.neurobiolaging.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Bennett CF, Cleveland DW, Yeo GW. Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res. 2012;1462:3–15. doi: 10.1016/j.brainres.2012.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Mazur C, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe J, Chiba T, Yamada M, Okamoto K, Nishimoto I, Matsuoka M, Aiso S. D-serine is a key determinant of glutamate toxicity in amyotrophic lateral sclerosis. EMBO J. 2007;26:4149–4159. doi: 10.1038/sj.emboj.7601840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe J, Miyoshi Y, Suzuki M, Mita M, Konno R, Matsuoka M, Hamase K, Aiso S. D-amino acid oxidase controls motoneuron degeneration through D-serine. Proc Natl Acad Sci USA. 2012;109:627–632. doi: 10.1073/pnas.1114639109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scekic-Zahirovic J, Sendscheid O, El Oussini H, Jambeau M, Sun Y, Mersmann S, Wagner M, Dieterle S, Sinniger J, Dirrig-Grosch S, et al. Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J. 2016;35:1077–1097. doi: 10.15252/embj.201592559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, Cech TR. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 2012;26:2690–2695. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Munro TP, Barbarese E, Carson JH, Smith R. A molecular mechanism for mRNA trafficking in neuronal dendrites. J Neurosci. 2003;23:8859–8866. doi: 10.1523/JNEUROSCI.23-26-08859.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Eder PS, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski G, Finkbeiner S. Longitudinal measures of proteostasis in live neurons: features that determine fate in models of neurodegenerative disease. FEBS Lett. 2013;587:1139–1146. doi: 10.1016/j.febslet.2013.02.043. [DOI] [PubMed] [Google Scholar]
- Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Ling SC, Qiu J, Albuquerque CP, Zhou Y, Tokunaga S, Li H, Qiu H, Bui A, Yeo GW, et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nature communications. 2015;6:6171. doi: 10.1038/ncomms7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M, Marecki JC, Marinesco S, Labrie V, Roder JC, Barger SW, Crow JP. Paradoxical roles of serine racemase and D-serine in the G93A mSOD1 mouse model of amyotrophic lateral sclerosis. J Neurochem. 2012;120:598–610. doi: 10.1111/j.1471-4159.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan-Johns M, Bengoechea R, Bell S, Shao J, Diamond MI, True HL, Weihl CC, Baloh RH. Prion-like nuclear aggregation of TDP-43 during heat shock is regulated by HSP40/70 chaperones. Hum Mol Genet. 2014;23:157–170. doi: 10.1093/hmg/ddt408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nature methods. 2016;13:508–514. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Wu LS, Cheng WC, Shen CK. Targeted depletion of TDP-43 expression in the spinal cord motor neurons leads to the development of amyotrophic lateral sclerosis-like phenotypes in mice. J Biol Chem. 2012;287:27335–27344. doi: 10.1074/jbc.M112.359000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nature communications. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, McKnight SL. The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang H, Qiao T, Yang B, Aliaga L, Qiu L, Tan W, Salameh J, McKenna-Yasek DM, Smith T, et al. Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2014;111:E1121–1129. doi: 10.1073/pnas.1322641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HC, Liu CM, Liu YL, Chen CW, Chang CC, Fann CS, Chiou JJ, Yang UC, Chen CH, Faraone SV, et al. The DAO gene is associated with schizophrenia and interacts with other genes in the Taiwan Han Chinese population. PLoS One. 2013;8:e60099. doi: 10.1371/journal.pone.0060099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Small RNA-seq, RNA-seq, iCLIP, eCLIP and microarray data are available at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE86464.